Tauopathies: Deciphering Disease Mechanisms to Develop Effective Therapies
Abstract
1. MAPT and Tauopathy Spectrum Disorders
2. Molecular Mechanisms of Tau Pathology
2.1. Protein Post-Translational Modifications
2.2. Tau Misfolding and Phase Transition
2.3. Tau Spreading and the Prion-Like Model
2.4. Protein Degradation Failure
2.5. Neuronal Stress Responses to Tau and Stress Granules Formation
2.6. Disruption of Mitochondrial Function and ER Unfolded Protein Stress Response
2.7. Disruption of the RNA Splicing Machinery
2.8. The Role of Neural Inflammation in Neurodegeneration
3. Tau-Directed Therapeutics
3.1. Modulators of MAPT Expression
3.2. Inhibitors of Tau PTMs
3.3. Tau Aggregation Inhibitors
3.4. Modulators of Tau Clearance by Autophagy and the Proteasome
3.5. Anti-Tau Immunotherapy
3.6. Tau-Targeted Therapeutics with Bifunctional Degraders
4. Concluding Remarks
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s disease |
| ASO | Anti-sense oligonucleotide |
| BBB | Blood-brain barrier |
| CBD | Corticobasal degeneration |
| CNS | Central nervous system |
| CSF | Cerebral spinal fluid |
| ER | Endoplasmic reticulum |
| FTD | Frontotemporal dementia |
| GPCR | G-coupled protein receptors |
| GSK-3 | Glycogen synthase kinase 3 |
| HSPGs | Heparan sulfate proteoglycans |
| iPSC | Induced pluripotent stem cells |
| MAPT | Microtubule-associated protein Tau |
| MW | Molecular weight |
| NFT | Neurofibrillary tangle |
| OGA | O-GlcNAcase |
| P-Tau | Phosphorylated Tau |
| PDE | Phosphodiesterase |
| PET | Positron emission tomography |
| PHF | Paired helical filament |
| PK | Pharmacokinetics |
| PP2A | Protein phosphatase 2 |
| PROTAC | Proteolysis targeting chimera |
| PSP | Progressive supranuclear palsy |
| PTM | Post-translational modification |
| SG | Stress granule |
| TTBK1 | Tau tubulin kinase 1 |
| UPR | Unfolded protein response |
| UPS | Ubiquitin proteasome system |
References
- Cornutiu, G. The Epidemiological Scale of Alzheimer’s Disease. J. Clin. Med. Res. 2015, 7, 657–666. [Google Scholar] [CrossRef]
- Association, A.S. Alzheimer’s Disease Facts and Figures. Alzheimers Dement. 2019, 15, 321–387. [Google Scholar]
- Holtzman, D.M.; Morris, J.C.; Goate, A.M. Alzheimer’s disease: The challenge of the second century. Sci. Transl. Med. 2011, 3, 77sr71. [Google Scholar] [CrossRef]
- Gigant, B.; Landrieu, I.; Fauquant, C.; Barbier, P.; Huvent, I.; Wieruszeski, J.M.; Knossow, M.; Lippens, G. Mechanism of Tau-promoted microtubule assembly as probed by NMR spectroscopy. J. Am. Chem. Soc. 2014, 136, 12615–12623. [Google Scholar] [CrossRef] [PubMed]
- Trinczek, B.; Ebneth, A.; Mandelkow, E.M.; Mandelkow, E. Tau regulates the attachment/detachment but not the speed of motors in microtubule-dependent transport of single vesicles and organelles. J. Cell Sci. 1999, 112 Pt 14, 2355–2367. [Google Scholar]
- Preuss, U.; Biernat, J.; Mandelkow, E.M.; Mandelkow, E. The ‘jaws’ model of tau-microtubule interaction examined in CHO cells. J. Cell Sci. 1997, 110 Pt 6, 789–800. [Google Scholar]
- Dorostkar, M.M.; Zou, C.; Blazquez-Llorca, L.; Herms, J. Analyzing dendritic spine pathology in Alzheimer’s disease: Problems and opportunities. Acta Neuropathol. 2015, 130, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Marciniak, E.; Leboucher, A.; Caron, E.; Ahmed, T.; Tailleux, A.; Dumont, J.; Issad, T.; Gerhardt, E.; Pagesy, P.; Vileno, M.; et al. Tau deletion promotes brain insulin resistance. J. Exp. Med. 2017, 214, 2257–2269. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M.; Spillantini, M.G.; Jakes, R.; Rutherford, D.; Crowther, R.A. Multiple isoforms of human microtubule-associated protein tau: Sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron 1989, 3, 519–526. [Google Scholar] [CrossRef]
- Trabzuni, D.; Wray, S.; Vandrovcova, J.; Ramasamy, A.; Walker, R.; Smith, C.; Luk, C.; Gibbs, J.R.; Dillman, A.; Hernandez, D.G.; et al. MAPT expression and splicing is differentially regulated by brain region: Relation to genotype and implication for tauopathies. Hum. Mol. Genet. 2012, 21, 4094–4103. [Google Scholar] [CrossRef]
- Kosik, K.S.; Orecchio, L.D.; Bakalis, S.; Neve, R.L. Developmentally regulated expression of specific tau sequences. Neuron 1989, 2, 1389–1397. [Google Scholar] [CrossRef]
- Park, S.A.; Ahn, S.I.; Gallo, J.M. Tau mis-splicing in the pathogenesis of neurodegenerative disorders. BMB Rep. 2016, 49, 405–413. [Google Scholar] [CrossRef]
- Dunker, A.K.; Silman, I.; Uversky, V.N.; Sussman, J.L. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 2008, 18, 756–764. [Google Scholar] [CrossRef] [PubMed]
- Kovacech, B.; Skrabana, R.; Novak, M. Transition of tau protein from disordered to misordered in Alzheimer’s disease. Neurodegener. Dis. 2010, 7, 24–27. [Google Scholar] [CrossRef] [PubMed]
- Lee, V.M.; Goedert, M.; Trojanowski, J.Q. Neurodegenerative tauopathies. Annu. Rev. Neurosci. 2001, 24, 1121–1159. [Google Scholar] [CrossRef] [PubMed]
- Josephs, K.A.; Hodges, J.R.; Snowden, J.S.; Mackenzie, I.R.; Neumann, M.; Mann, D.M.; Dickson, D.W. Neuropathological background of phenotypical variability in frontotemporal dementia. Acta Neuropathol. 2011, 122, 137–153. [Google Scholar] [CrossRef] [PubMed]
- Crary, J.F.; Trojanowski, J.Q.; Schneider, J.A.; Abisambra, J.F.; Abner, E.L.; Alafuzoff, I.; Arnold, S.E.; Attems, J.; Beach, T.G.; Bigio, E.H.; et al. Primary age-related tauopathy (PART): A common pathology associated with human aging. Acta Neuropathol. 2014, 128, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Boxer, A.L.; Yu, J.T.; Golbe, L.I.; Litvan, I.; Lang, A.E.; Hoglinger, G.U. Advances in progressive supranuclear palsy: New diagnostic criteria, biomarkers, and therapeutic approaches. Lancet Neurol. 2017, 16, 552–563. [Google Scholar] [CrossRef]
- Irwin, D.J.; Brettschneider, J.; McMillan, C.T.; Cooper, F.; Olm, C.; Arnold, S.E.; Van Deerlin, V.M.; Seeley, W.W.; Miller, B.L.; Lee, E.B.; et al. Deep clinical and neuropathological phenotyping of Pick disease. Ann. Neurol. 2016, 79, 272–287. [Google Scholar] [CrossRef]
- Rankin, K.P.; Mayo, M.C.; Seeley, W.W.; Lee, S.; Rabinovici, G.; Gorno-Tempini, M.L.; Boxer, A.L.; Weiner, M.W.; Trojanowski, J.Q.; DeArmond, S.J.; et al. Behavioral variant frontotemporal dementia with corticobasal degeneration pathology: Phenotypic comparison to bvFTD with Pick’s disease. J. Mol. Neurosci. 2011, 45, 594–608. [Google Scholar] [CrossRef][Green Version]
- Dickson, D.W.; Yen, S.H.; Suzuki, K.I.; Davies, P.; Garcia, J.H.; Hirano, A. Ballooned neurons in select neurodegenerative diseases contain phosphorylated neurofilament epitopes. Acta Neuropathol. 1986, 71, 216–223. [Google Scholar] [CrossRef] [PubMed]
- Braak, H.; Braak, E. Argyrophilic grain disease: Frequency of occurrence in different age categories and neuropathological diagnostic criteria. J. Neural Transm. 1998, 105, 801–819. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, M.J.; Litvan, I.; Lang, A.E.; Bak, T.H.; Bhatia, K.P.; Borroni, B.; Boxer, A.L.; Dickson, D.W.; Grossman, M.; Hallett, M.; et al. Criteria for the diagnosis of corticobasal degeneration. Neurology 2013, 80, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Day, G.S.; Lim, T.S.; Hassenstab, J.; Goate, A.M.; Grant, E.A.; Roe, C.M.; Cairns, N.J.; Morris, J.C. Differentiating cognitive impairment due to corticobasal degeneration and Alzheimer disease. Neurology 2017, 88, 1273–1281. [Google Scholar] [CrossRef] [PubMed]
- Hoglinger, G.U.; Respondek, G.; Stamelou, M.; Kurz, C.; Josephs, K.A.; Lang, A.E.; Mollenhauer, B.; Muller, U.; Nilsson, C.; Whitwell, J.L.; et al. Clinical diagnosis of progressive supranuclear palsy: The movement disorder society criteria. Mov. Disord. 2017, 32, 853–864. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, E.G.; Mandelli, M.L.; Miller, Z.A.; Santos-Santos, M.A.; Wilson, S.M.; Agosta, F.; Grinberg, L.T.; Huang, E.J.; Trojanowski, J.Q.; Meyer, M.; et al. Typical and atypical pathology in primary progressive aphasia variants. Ann. Neurol. 2017, 81, 430–443. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, G.G.; Robinson, J.L.; Xie, S.X.; Lee, E.B.; Grossman, M.; Wolk, D.A.; Irwin, D.J.; Weintraub, D.; Kim, C.F.; Schuck, T.; et al. Evaluating the Patterns of Aging-Related Tau Astrogliopathy Unravels Novel Insights into Brain Aging and Neurodegenerative Diseases. J. Neuropathol. Exp. Neurol. 2017, 76, 270–288. [Google Scholar] [CrossRef]
- Rodriguez, R.D.; Suemoto, C.K.; Molina, M.; Nascimento, C.F.; Leite, R.E.; de Lucena Ferretti-Rebustini, R.E.; Farfel, J.M.; Heinsen, H.; Nitrini, R.; Ueda, K.; et al. Argyrophilic Grain Disease: Demographics, Clinical, and Neuropathological Features from a Large Autopsy Study. J. Neuropathol. Exp. Neurol. 2016, 75, 628–635. [Google Scholar] [CrossRef]
- Arena, J.D.; Smith, D.H.; Lee, E.B.; Gibbons, G.S.; Irwin, D.J.; Robinson, J.L.; Lee, V.M.; Trojanowski, J.Q.; Stewart, W.; Johnson, V.E. Tau immunophenotypes in chronic traumatic encephalopathy recapitulate those of ageing and Alzheimer’s disease. Brain 2020, 143, 1572–1587. [Google Scholar] [CrossRef]
- Hutton, M. Molecular genetics of chromosome 17 tauopathies. Ann. N. Y. Acad. Sci. 2000, 920, 63–73. [Google Scholar] [CrossRef]
- Wang, Y.; Mandelkow, E. Tau in physiology and pathology. Nat. Rev. Neurosci. 2016, 17, 5–21. [Google Scholar] [CrossRef] [PubMed]
- Furman, J.L.; Vaquer-Alicea, J.; White, C.L., 3rd; Cairns, N.J.; Nelson, P.T.; Diamond, M.I. Widespread tau seeding activity at early Braak stages. Acta Neuropathol. 2017, 133, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Jucker, M.; Walker, L.C. Propagation and spread of pathogenic protein assemblies in neurodegenerative diseases. Nat. Neurosci. 2018, 21, 1341–1349. [Google Scholar] [CrossRef] [PubMed]
- Kuret, J.; Congdon, E.E.; Li, G.; Yin, H.; Yu, X.; Zhong, Q. Evaluating triggers and enhancers of tau fibrillization. Microsc. Res. Tech. 2005, 67, 141–155. [Google Scholar] [CrossRef] [PubMed]
- Mandelkow, E.M.; Mandelkow, E. Biochemistry and cell biology of tau protein in neurofibrillary degeneration. Cold Spring Harb. Perspect. Med. 2012, 2, a006247. [Google Scholar] [CrossRef]
- von Bergen, M.; Barghorn, S.; Biernat, J.; Mandelkow, E.M.; Mandelkow, E. Tau aggregation is driven by a transition from random coil to beta sheet structure. Biochim. Biophys. Acta 2005, 1739, 158–166. [Google Scholar] [CrossRef]
- Sillen, A.; Leroy, A.; Wieruszeski, J.M.; Loyens, A.; Beauvillain, J.C.; Buee, L.; Landrieu, I.; Lippens, G. Regions of tau implicated in the paired helical fragment core as defined by NMR. Chembiochem 2005, 6, 1849–1856. [Google Scholar] [CrossRef]
- Kovacs, G.G.; Lutz, M.I.; Ricken, G.; Strobel, T.; Hoftberger, R.; Preusser, M.; Regelsberger, G.; Honigschnabl, S.; Reiner, A.; Fischer, P.; et al. Dura mater is a potential source of Abeta seeds. Acta Neuropathol. 2016, 131, 911–923. [Google Scholar] [CrossRef]
- Spillantini, M.G.; Goedert, M. Tau pathology and neurodegeneration. Lancet Neurol. 2013, 12, 609–622. [Google Scholar] [CrossRef]
- Mez, J.; Daneshvar, D.H.; Kiernan, P.T.; Abdolmohammadi, B.; Alvarez, V.E.; Huber, B.R.; Alosco, M.L.; Solomon, T.M.; Nowinski, C.J.; McHale, L.; et al. Clinicopathological Evaluation of Chronic Traumatic Encephalopathy in Players of American Football. JAMA 2017, 318, 360–370. [Google Scholar] [CrossRef]
- Jadhav, S.; Avila, J.; Scholl, M.; Kovacs, G.G.; Kovari, E.; Skrabana, R.; Evans, L.D.; Kontsekova, E.; Malawska, B.; de Silva, R.; et al. A walk through tau therapeutic strategies. Acta Neuropathol. Commun. 2019, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Khanna, M.R.; Kovalevich, J.; Lee, V.M.; Trojanowski, J.Q.; Brunden, K.R. Therapeutic strategies for the treatment of tauopathies: Hopes and challenges. Alzheimers Dement. 2016, 12, 1051–1065. [Google Scholar] [CrossRef]
- Feinstein, S.C.; Wilson, L. Inability of tau to properly regulate neuronal microtubule dynamics: A loss-of-function mechanism by which tau might mediate neuronal cell death. Biochim. Biophys. Acta 2005, 1739, 268–279. [Google Scholar] [CrossRef] [PubMed]
- Trojanowski, J.Q.; Lee, V.M. Pathological tau: A loss of normal function or a gain in toxicity? Nat. Neurosci. 2005, 8, 1136–1137. [Google Scholar] [CrossRef]
- Cripps, D.; Thomas, S.N.; Jeng, Y.; Yang, F.; Davies, P.; Yang, A.J. Alzheimer disease-specific conformation of hyperphosphorylated paired helical filament-Tau is polyubiquitinated through Lys-48, Lys-11, and Lys-6 ubiquitin conjugation. J. Biol. Chem. 2006, 281, 10825–10838. [Google Scholar] [CrossRef]
- Ramachandran, G.; Udgaonkar, J.B. Mechanistic studies unravel the complexity inherent in tau aggregation leading to Alzheimer’s disease and the tauopathies. Biochemistry 2013, 52, 4107–4126. [Google Scholar] [CrossRef] [PubMed]
- Tepper, K.; Biernat, J.; Kumar, S.; Wegmann, S.; Timm, T.; Hubschmann, S.; Redecke, L.; Mandelkow, E.M.; Muller, D.J.; Mandelkow, E. Oligomer formation of tau protein hyperphosphorylated in cells. J. Biol. Chem. 2014, 289, 34389–34407. [Google Scholar] [CrossRef]
- Martin, L.; Latypova, X.; Terro, F. Post-translational modifications of tau protein: Implications for Alzheimer’s disease. Neurochem. Int. 2011, 58, 458–471. [Google Scholar] [CrossRef]
- Reynolds, C.H.; Garwood, C.J.; Wray, S.; Price, C.; Kellie, S.; Perera, T.; Zvelebil, M.; Yang, A.; Sheppard, P.W.; Varndell, I.M.; et al. Phosphorylation regulates tau interactions with Src homology 3 domains of phosphatidylinositol 3-kinase, phospholipase Cgamma1, Grb2, and Src family kinases. J. Biol. Chem. 2008, 283, 18177–18186. [Google Scholar] [CrossRef]
- Cohen, T.J.; Guo, J.L.; Hurtado, D.E.; Kwong, L.K.; Mills, I.P.; Trojanowski, J.Q.; Lee, V.M. The acetylation of tau inhibits its function and promotes pathological tau aggregation. Nat. Commun. 2011, 2, 252. [Google Scholar] [CrossRef]
- Kamah, A.; Huvent, I.; Cantrelle, F.X.; Qi, H.; Lippens, G.; Landrieu, I.; Smet-Nocca, C. Nuclear magnetic resonance analysis of the acetylation pattern of the neuronal Tau protein. Biochemistry 2014, 53, 3020–3032. [Google Scholar] [CrossRef] [PubMed]
- Min, S.W.; Cho, S.H.; Zhou, Y.; Schroeder, S.; Haroutunian, V.; Seeley, W.W.; Huang, E.J.; Shen, Y.; Masliah, E.; Mukherjee, C.; et al. Acetylation of tau inhibits its degradation and contributes to tauopathy. Neuron 2010, 67, 953–966. [Google Scholar] [CrossRef] [PubMed]
- Kellogg, E.H.; Hejab, N.M.A.; Poepsel, S.; Downing, K.H.; DiMaio, F.; Nogales, E. Near-atomic model of microtubule-tau interactions. Science 2018, 360, 1242–1246. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S.; Wegmann, S.; Cho, H.; DeVos, S.L.; Commins, C.; Roe, A.D.; Nicholls, S.B.; Carlson, G.A.; Pitstick, R.; Nobuhara, C.K.; et al. Neuronal uptake and propagation of a rare phosphorylated high-molecular-weight tau derived from Alzheimer’s disease brain. Nat. Commun. 2015, 6, 8490. [Google Scholar] [CrossRef]
- Biernat, J.; Mandelkow, E.M. The development of cell processes induced by tau protein requires phosphorylation of serine 262 and 356 in the repeat domain and is inhibited by phosphorylation in the proline-rich domains. Mol. Biol. Cell 1999, 10, 727–740. [Google Scholar] [CrossRef]
- Mandell, J.W.; Banker, G.A. A spatial gradient of tau protein phosphorylation in nascent axons. J. Neurosci. 1996, 16, 5727–5740. [Google Scholar] [CrossRef]
- Augustinack, J.C.; Schneider, A.; Mandelkow, E.M.; Hyman, B.T. Specific tau phosphorylation sites correlate with severity of neuronal cytopathology in Alzheimer’s disease. Acta Neuropathol. 2002, 103, 26–35. [Google Scholar] [CrossRef]
- Neddens, J.; Temmel, M.; Flunkert, S.; Kerschbaumer, B.; Hoeller, C.; Loeffler, T.; Niederkofler, V.; Daum, G.; Attems, J.; Hutter-Paier, B. Phosphorylation of different tau sites during progression of Alzheimer’s disease. Acta Neuropathol. Commun. 2018, 6, 52. [Google Scholar] [CrossRef]
- Pascual, G.; Wadia, J.S.; Zhu, X.; Keogh, E.; Kukrer, B.; van Ameijde, J.; Inganas, H.; Siregar, B.; Perdok, G.; Diefenbach, O.; et al. Immunological memory to hyperphosphorylated tau in asymptomatic individuals. Acta Neuropathol. 2017, 133, 767–783. [Google Scholar] [CrossRef]
- Guillozet-Bongaarts, A.L.; Cahill, M.E.; Cryns, V.L.; Reynolds, M.R.; Berry, R.W.; Binder, L.I. Pseudophosphorylation of tau at serine 422 inhibits caspase cleavage: In vitro evidence and implications for tangle formation in vivo. J. Neurochem. 2006, 97, 1005–1014. [Google Scholar] [CrossRef]
- Vana, L.; Kanaan, N.M.; Ugwu, I.C.; Wuu, J.; Mufson, E.J.; Binder, L.I. Progression of tau pathology in cholinergic Basal forebrain neurons in mild cognitive impairment and Alzheimer’s disease. Am. J. Pathol. 2011, 179, 2533–2550. [Google Scholar] [CrossRef]
- Dillon, G.M.; Henderson, J.L.; Bao, C.; Joyce, J.A.; Calhoun, M.; Amaral, B.; King, K.W.; Bajrami, B.; Rabah, D. Acute inhibition of the CNS-specific kinase TTBK1 significantly lowers tau phosphorylation at several disease relevant sites. PLoS ONE 2020, 15, e0228771. [Google Scholar] [CrossRef]
- Sato, S.; Cerny, R.L.; Buescher, J.L.; Ikezu, T. Tau-tubulin kinase 1 (TTBK1), a neuron-specific tau kinase candidate, is involved in tau phosphorylation and aggregation. J. Neurochem. 2006, 98, 1573–1584. [Google Scholar] [CrossRef] [PubMed]
- Bao, C.; Bajrami, B.; Marcotte, D.J.; Chodaparambil, J.V.; Kerns, H.M.; Henderson, J.; Wei, R.; Gao, B.; Dillon, G.M. Mechanisms of Regulation and Diverse Activities of Tau-Tubulin Kinase (TTBK) Isoforms. Cell. Mol. Neurobiol. 2020. [Google Scholar] [CrossRef]
- Alafuzoff, I.; Arzberger, T.; Al-Sarraj, S.; Bodi, I.; Bogdanovic, N.; Braak, H.; Bugiani, O.; Del-Tredici, K.; Ferrer, I.; Gelpi, E.; et al. Staging of neurofibrillary pathology in Alzheimer’s disease: A study of the BrainNet Europe Consortium. Brain Pathol. 2008, 18, 484–496. [Google Scholar] [CrossRef] [PubMed]
- Mondragon-Rodriguez, S.; Perry, G.; Luna-Munoz, J.; Acevedo-Aquino, M.C.; Williams, S. Phosphorylation of tau protein at sites Ser(396–404) is one of the earliest events in Alzheimer’s disease and Down syndrome. Neuropathol. Appl. Neurobiol. 2014, 40, 121–135. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Prokop, S.; Gorion, K.M.; Kim, J.D.; Sorrentino, Z.A.; Bell, B.M.; Manaois, A.N.; Chakrabarty, P.; Davies, P.; Giasson, B.I. Tau Ser208 phosphorylation promotes aggregation and reveals neuropathologic diversity in Alzheimer’s disease and other tauopathies. Acta Neuropathol. Commun. 2020, 8, 88. [Google Scholar] [CrossRef] [PubMed]
- Barthelemy, N.R.; Mallipeddi, N.; Moiseyev, P.; Sato, C.; Bateman, R.J. Tau Phosphorylation Rates Measured by Mass Spectrometry Differ in the Intracellular Brain vs. Extracellular Cerebrospinal Fluid Compartments and Are Differentially Affected by Alzheimer’s Disease. Front. Aging Neurosci. 2019, 11, 121. [Google Scholar] [CrossRef] [PubMed]
- Dujardin, S.; Commins, C.; Lathuiliere, A.; Beerepoot, P.; Fernandes, A.R.; Kamath, T.V.; De Los Santos, M.B.; Klickstein, N.; Corjuc, D.L.; Corjuc, B.T.; et al. Tau molecular diversity contributes to clinical heterogeneity in Alzheimer’s disease. Nat. Med. 2020, 26, 1256–1263. [Google Scholar] [CrossRef]
- Johnson, G.V.; Stoothoff, W.H. Tau phosphorylation in neuronal cell function and dysfunction. J. Cell Sci. 2004, 117, 5721–5729. [Google Scholar] [CrossRef]
- Noble, W.; Hanger, D.P.; Miller, C.C.; Lovestone, S. The importance of tau phosphorylation for neurodegenerative diseases. Front. Neurol. 2013, 4, 83. [Google Scholar] [CrossRef] [PubMed]
- Nozal, V.; Martinez, A. Tau Tubulin Kinase 1 (TTBK1), a new player in the fight against neurodegenerative diseases. Eur. J. Med. Chem. 2019, 161, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Shi, J.; Yin, X.; Iqbal, K.; Grundke-Iqbal, I.; Gong, C.X.; Liu, F. PP2A regulates tau phosphorylation directly and also indirectly via activating GSK-3beta. J. Alzheimers Dis. 2010, 19, 1221–1229. [Google Scholar] [CrossRef] [PubMed]
- Gotz, J.; Gladbach, A.; Pennanen, L.; van Eersel, J.; Schild, A.; David, D.; Ittner, L.M. Animal models reveal role for tau phosphorylation in human disease. Biochim. Biophys. Acta 2010, 1802, 860–871. [Google Scholar] [CrossRef]
- Abreha, M.H.; Dammer, E.B.; Ping, L.; Zhang, T.; Duong, D.M.; Gearing, M.; Lah, J.J.; Levey, A.I.; Seyfried, N.T. Quantitative Analysis of the Brain Ubiquitylome in Alzheimer’s Disease. Proteomics 2018, 18, e1800108. [Google Scholar] [CrossRef]
- Nakayama, Y.; Sakamoto, S.; Tsuji, K.; Ayaki, T.; Tokunaga, F.; Ito, H. Identification of linear polyubiquitin chain immunoreactivity in tau pathology of Alzheimer’s disease. Neurosci. Lett. 2019, 703, 53–57. [Google Scholar] [CrossRef]
- Cohen, T.J.; Friedmann, D.; Hwang, A.W.; Marmorstein, R.; Lee, V.M. The microtubule-associated tau protein has intrinsic acetyltransferase activity. Nat. Struct. Mol. Biol. 2013, 20, 756–762. [Google Scholar] [CrossRef]
- Cook, C.; Carlomagno, Y.; Gendron, T.F.; Dunmore, J.; Scheffel, K.; Stetler, C.; Davis, M.; Dickson, D.; Jarpe, M.; DeTure, M.; et al. Acetylation of the KXGS motifs in tau is a critical determinant in modulation of tau aggregation and clearance. Hum. Mol. Genet. 2014, 23, 104–116. [Google Scholar] [CrossRef]
- Min, S.W.; Chen, X.; Tracy, T.E.; Li, Y.; Zhou, Y.; Wang, C.; Shirakawa, K.; Minami, S.S.; Defensor, E.; Mok, S.A.; et al. Critical role of acetylation in tau-mediated neurodegeneration and cognitive deficits. Nat. Med. 2015, 21, 1154–1162. [Google Scholar] [CrossRef]
- Irwin, D.J.; Cohen, T.J.; Grossman, M.; Arnold, S.E.; McCarty-Wood, E.; Van Deerlin, V.M.; Lee, V.M.; Trojanowski, J.Q. Acetylated tau neuropathology in sporadic and hereditary tauopathies. Am. J. Pathol. 2013, 183, 344–351. [Google Scholar] [CrossRef]
- Rane, J.S.; Kumari, A.; Panda, D. An acetylation mimicking mutation, K274Q, in tau imparts neurotoxicity by enhancing tau aggregation and inhibiting tubulin polymerization. Biochem. J. 2019, 476, 1401–1417. [Google Scholar] [CrossRef] [PubMed]
- Tracy, T.E.; Sohn, P.D.; Minami, S.S.; Wang, C.; Min, S.W.; Li, Y.; Zhou, Y.; Le, D.; Lo, I.; Ponnusamy, R.; et al. Acetylated Tau Obstructs KIBRA-Mediated Signaling in Synaptic Plasticity and Promotes Tauopathy-Related Memory Loss. Neuron 2016, 90, 245–260. [Google Scholar] [CrossRef] [PubMed]
- Binder, L.I.; Guillozet-Bongaarts, A.L.; Garcia-Sierra, F.; Berry, R.W. Tau, tangles, and Alzheimer’s disease. Biochim. Biophys. Acta 2005, 1739, 216–223. [Google Scholar] [CrossRef]
- Carmel, G.; Mager, E.M.; Binder, L.I.; Kuret, J. The structural basis of monoclonal antibody Alz50′s selectivity for Alzheimer’s disease pathology. J. Biol. Chem. 1996, 271, 32789–32795. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Sierra, F.; Mondragon-Rodriguez, S.; Basurto-Islas, G. Truncation of tau protein and its pathological significance in Alzheimer’s disease. J. Alzheimers Dis. 2008, 14, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.P.; Corbett, N.J.; Kellett, K.A.B.; Hooper, N.M. Tau Proteolysis in the Pathogenesis of Tauopathies: Neurotoxic Fragments and Novel Biomarkers. J. Alzheimers Dis. 2018, 63, 13–33. [Google Scholar] [CrossRef] [PubMed]
- Gamblin, T.C.; Chen, F.; Zambrano, A.; Abraha, A.; Lagalwar, S.; Guillozet, A.L.; Lu, M.; Fu, Y.; Garcia-Sierra, F.; LaPointe, N.; et al. Caspase cleavage of tau: Linking amyloid and neurofibrillary tangles in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2003, 100, 10032–10037. [Google Scholar] [CrossRef] [PubMed]
- Novak, M.; Kabat, J.; Wischik, C.M. Molecular characterization of the minimal protease resistant tau unit of the Alzheimer’s disease paired helical filament. EMBO J. 1993, 12, 365–370. [Google Scholar] [CrossRef]
- Wischik, C.M.; Novak, M.; Thogersen, H.C.; Edwards, P.C.; Runswick, M.J.; Jakes, R.; Walker, J.E.; Milstein, C.; Roth, M.; Klug, A. Isolation of a fragment of tau derived from the core of the paired helical filament of Alzheimer disease. Proc. Natl. Acad. Sci. USA 1988, 85, 4506–4510. [Google Scholar] [CrossRef]
- Kudo, L.C.; Parfenova, L.; Ren, G.; Vi, N.; Hui, M.; Ma, Z.; Lau, K.; Gray, M.; Bardag-Gorce, F.; Wiedau-Pazos, M.; et al. Puromycin-sensitive aminopeptidase (PSA/NPEPPS) impedes development of neuropathology in hPSA/TAU(P301L) double-transgenic mice. Hum. Mol. Genet. 2011, 20, 1820–1833. [Google Scholar] [CrossRef]
- Fitzpatrick, A.W.P.; Falcon, B.; He, S.; Murzin, A.G.; Murshudov, G.; Garringer, H.J.; Crowther, R.A.; Ghetti, B.; Goedert, M.; Scheres, S.H.W. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 2017, 547, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, S.K.; Sanders, D.W.; Thomas, T.L.; Ruchinskas, A.J.; Vaquer-Alicea, J.; Sharma, A.M.; Miller, T.M.; Diamond, M.I. Tau Prion Strains Dictate Patterns of Cell Pathology, Progression Rate, and Regional Vulnerability in Vivo. Neuron 2016, 92, 796–812. [Google Scholar] [CrossRef] [PubMed]
- Falcon, B.; Zivanov, J.; Zhang, W.; Murzin, A.G.; Garringer, H.J.; Vidal, R.; Crowther, R.A.; Newell, K.L.; Ghetti, B.; Goedert, M.; et al. Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules. Nature 2019, 568, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M.; Falcon, B.; Zhang, W.; Ghetti, B.; Scheres, S.H.W. Distinct Conformers of Assembled Tau in Alzheimer’s and Pick’s Diseases. Cold Spring Harb. Symp. Quant. Biol. 2018, 83, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Jarrett, J.T.; Lansbury, P.T., Jr. Seeding “one-dimensional crystallization” of amyloid: A pathogenic mechanism in Alzheimer’s disease and scrapie? Cell 1993, 73, 1055–1058. [Google Scholar] [CrossRef]
- Koo, E.H.; Lansbury, P.T., Jr.; Kelly, J.W. Amyloid diseases: Abnormal protein aggregation in neurodegeneration. Proc. Natl. Acad. Sci. USA 1999, 96, 9989–9990. [Google Scholar] [CrossRef]
- Majewski, J.; Jones, E.M.; Vander Zanden, C.M.; Biernat, J.; Mandelkow, E.; Chi, E.Y. Lipid membrane templated misfolding and self-assembly of intrinsically disordered tau protein. Sci. Rep. 2020, 10, 13324. [Google Scholar] [CrossRef]
- Brangwynne, C.P. Phase transitions and size scaling of membrane-less organelles. J. Cell Biol. 2013, 203, 875–881. [Google Scholar] [CrossRef]
- Hyman, A.A.; Weber, C.A.; Julicher, F. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 2014, 30, 39–58. [Google Scholar] [CrossRef]
- Kanaan, N.M.; Hamel, C.; Grabinski, T.; Combs, B. Liquid-liquid phase separation induces pathogenic tau conformations in vitro. Nat. Commun. 2020, 11, 2809. [Google Scholar] [CrossRef]
- Ambadipudi, S.; Biernat, J.; Riedel, D.; Mandelkow, E.; Zweckstetter, M. Liquid-liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein Tau. Nat. Commun. 2017, 8, 275. [Google Scholar] [CrossRef] [PubMed]
- Wegmann, S.; Eftekharzadeh, B.; Tepper, K.; Zoltowska, K.M.; Bennett, R.E.; Dujardin, S.; Laskowski, P.R.; MacKenzie, D.; Kamath, T.; Commins, C.; et al. Tau protein liquid-liquid phase separation can initiate tau aggregation. EMBO J. 2018, 37. [Google Scholar] [CrossRef]
- Zhang, X.; Lin, Y.; Eschmann, N.A.; Zhou, H.; Rauch, J.N.; Hernandez, I.; Guzman, E.; Kosik, K.S.; Han, S. RNA stores tau reversibly in complex coacervates. PLoS Biol. 2017, 15, e2002183. [Google Scholar] [CrossRef] [PubMed]
- Boyko, S.; Qi, X.; Chen, T.H.; Surewicz, K.; Surewicz, W.K. Liquid-liquid phase separation of tau protein: The crucial role of electrostatic interactions. J. Biol. Chem. 2019, 294, 11054–11059. [Google Scholar] [CrossRef] [PubMed]
- Cruz, A.; Verma, M.; Wolozin, B. The Pathophysiology of Tau and Stress Granules in Disease. Adv. Exp. Med. Biol. 2019, 1184, 359–372. [Google Scholar] [CrossRef]
- Ukmar-Godec, T.; Wegmann, S.; Zweckstetter, M. Biomolecular condensation of the microtubule-associated protein tau. Semin. Cell Dev. Biol. 2020, 99, 202–214. [Google Scholar] [CrossRef] [PubMed]
- Mitrea, D.M.; Kriwacki, R.W. Phase separation in biology; functional organization of a higher order. Cell Commun. Signal. 2016, 14, 1. [Google Scholar] [CrossRef]
- Kontaxi, C.; Piccardo, P.; Gill, A.C. Lysine-Directed Post-translational Modifications of Tau Protein in Alzheimer’s Disease and Related Tauopathies. Front. Mol. Biosci. 2017, 4, 56. [Google Scholar] [CrossRef]
- Park, S.; Lee, J.H.; Jeon, J.H.; Lee, M.J. Degradation or aggregation: The ramifications of post-translational modifications on tau. BMB Rep. 2018, 51, 265–273. [Google Scholar] [CrossRef]
- Ferreon, J.C.; Jain, A.; Choi, K.J.; Tsoi, P.S.; MacKenzie, K.R.; Jung, S.Y.; Ferreon, A.C. Acetylation Disfavors Tau Phase Separation. Int. J. Mol. Sci. 2018, 19, 1360. [Google Scholar] [CrossRef]
- Lee, M.J.; Lee, J.H.; Rubinsztein, D.C. Tau degradation: The ubiquitin-proteasome system versus the autophagy-lysosome system. Prog. Neurobiol. 2013, 105, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Nguyen, M.D.; Dobbin, M.M.; Fischer, A.; Sananbenesi, F.; Rodgers, J.T.; Delalle, I.; Baur, J.A.; Sui, G.; Armour, S.M.; et al. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer’s disease and amyotrophic lateral sclerosis. EMBO J. 2007, 26, 3169–3179. [Google Scholar] [CrossRef] [PubMed]
- Braak, H.; Braak, E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991, 82, 239–259. [Google Scholar] [CrossRef] [PubMed]
- Hyman, B.T.; Van Hoesen, G.W.; Damasio, A.R.; Barnes, C.L. Alzheimer’s disease: Cell-specific pathology isolates the hippocampal formation. Science 1984, 225, 1168–1170. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, S.K.; Del Tredici, K.; Thomas, T.L.; Braak, H.; Diamond, M.I. Tau seeding activity begins in the transentorhinal/entorhinal regions and anticipates phospho-tau pathology in Alzheimer’s disease and PART. Acta Neuropathol. 2018, 136, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Sanders, D.W.; Kaufman, S.K.; Holmes, B.B.; Diamond, M.I. Prions and Protein Assemblies that Convey Biological Information in Health and Disease. Neuron 2016, 89, 433–448. [Google Scholar] [CrossRef] [PubMed]
- Clavaguera, F.; Akatsu, H.; Fraser, G.; Crowther, R.A.; Frank, S.; Hench, J.; Probst, A.; Winkler, D.T.; Reichwald, J.; Staufenbiel, M.; et al. Brain homogenates from human tauopathies induce tau inclusions in mouse brain. Proc. Natl. Acad. Sci. USA 2013, 110, 9535–9540. [Google Scholar] [CrossRef]
- Clavaguera, F.; Bolmont, T.; Crowther, R.A.; Abramowski, D.; Frank, S.; Probst, A.; Fraser, G.; Stalder, A.K.; Beibel, M.; Staufenbiel, M.; et al. Transmission and spreading of tauopathy in transgenic mouse brain. Nat. Cell Biol. 2009, 11, 909–913. [Google Scholar] [CrossRef]
- de Calignon, A.; Polydoro, M.; Suarez-Calvet, M.; William, C.; Adamowicz, D.H.; Kopeikina, K.J.; Pitstick, R.; Sahara, N.; Ashe, K.H.; Carlson, G.A.; et al. Propagation of tau pathology in a model of early Alzheimer’s disease. Neuron 2012, 73, 685–697. [Google Scholar] [CrossRef]
- Frost, B.; Jacks, R.L.; Diamond, M.I. Propagation of tau misfolding from the outside to the inside of a cell. J. Biol. Chem. 2009, 284, 12845–12852. [Google Scholar] [CrossRef]
- Guo, J.L.; Narasimhan, S.; Changolkar, L.; He, Z.; Stieber, A.; Zhang, B.; Gathagan, R.J.; Iba, M.; McBride, J.D.; Trojanowski, J.Q.; et al. Unique pathological tau conformers from Alzheimer’s brains transmit tau pathology in nontransgenic mice. J. Exp. Med. 2016, 213, 2635–2654. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Cirrito, J.R.; Stewart, F.R.; Jiang, H.; Finn, M.B.; Holmes, B.B.; Binder, L.I.; Mandelkow, E.M.; Diamond, M.I.; Lee, V.M.; et al. In vivo microdialysis reveals age-dependent decrease of brain interstitial fluid tau levels in P301S human tau transgenic mice. J. Neurosci. 2011, 31, 13110–13117. [Google Scholar] [CrossRef] [PubMed]
- Arriagada, P.V.; Growdon, J.H.; Hedley-Whyte, E.T.; Hyman, B.T. Neurofibrillary tangles but not senile plaques parallel duration and severity of Alzheimer’s disease. Neurology 1992, 42, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Brunello, C.A.; Merezhko, M.; Uronen, R.L.; Huttunen, H.J. Mechanisms of secretion and spreading of pathological tau protein. Cell. Mol. Life Sci. 2020, 77, 1721–1744. [Google Scholar] [CrossRef]
- Pooler, A.M.; Phillips, E.C.; Lau, D.H.; Noble, W.; Hanger, D.P. Physiological release of endogenous tau is stimulated by neuronal activity. EMBO Rep. 2013, 14, 389–394. [Google Scholar] [CrossRef]
- Yamada, K.; Holth, J.K.; Liao, F.; Stewart, F.R.; Mahan, T.E.; Jiang, H.; Cirrito, J.R.; Patel, T.K.; Hochgrafe, K.; Mandelkow, E.M.; et al. Neuronal activity regulates extracellular tau in vivo. J. Exp. Med. 2014, 211, 387–393. [Google Scholar] [CrossRef]
- Wang, Y.; Balaji, V.; Kaniyappan, S.; Kruger, L.; Irsen, S.; Tepper, K.; Chandupatla, R.; Maetzler, W.; Schneider, A.; Mandelkow, E.; et al. The release and trans-synaptic transmission of Tau via exosomes. Mol. Neurodegener. 2017, 12, 5. [Google Scholar] [CrossRef]
- Yamada, K. Extracellular Tau and Its Potential Role in the Propagation of Tau Pathology. Front. Neurosci. 2017, 11, 667. [Google Scholar] [CrossRef]
- Chen, J.J.; Nathaniel, D.L.; Raghavan, P.; Nelson, M.; Tian, R.; Tse, E.; Hong, J.Y.; See, S.K.; Mok, S.A.; Hein, M.Y.; et al. Compromised function of the ESCRT pathway promotes endolysosomal escape of tau seeds and propagation of tau aggregation. J. Biol. Chem. 2019, 294, 18952–18966. [Google Scholar] [CrossRef]
- Holmes, B.B.; DeVos, S.L.; Kfoury, N.; Li, M.; Jacks, R.; Yanamandra, K.; Ouidja, M.O.; Brodsky, F.M.; Marasa, J.; Bagchi, D.P.; et al. Heparan sulfate proteoglycans mediate internalization and propagation of specific proteopathic seeds. Proc. Natl. Acad. Sci. USA 2013, 110, E3138–E3147. [Google Scholar] [CrossRef]
- Michel, C.H.; Kumar, S.; Pinotsi, D.; Tunnacliffe, A.; St George-Hyslop, P.; Mandelkow, E.; Mandelkow, E.M.; Kaminski, C.F.; Kaminski Schierle, G.S. Extracellular monomeric tau protein is sufficient to initiate the spread of tau protein pathology. J. Biol. Chem. 2014, 289, 956–967. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Esko, J.D. Demystifying heparan sulfate-protein interactions. Annu. Rev. Biochem. 2014, 83, 129–157. [Google Scholar] [CrossRef] [PubMed]
- Rauch, J.N.; Chen, J.J.; Sorum, A.W.; Miller, G.M.; Sharf, T.; See, S.K.; Hsieh-Wilson, L.C.; Kampmann, M.; Kosik, K.S. Tau Internalization is Regulated by 6-O Sulfation on Heparan Sulfate Proteoglycans (HSPGs). Sci. Rep. 2018, 8, 6382. [Google Scholar] [CrossRef] [PubMed]
- Stopschinski, B.E.; Holmes, B.B.; Miller, G.M.; Manon, V.A.; Vaquer-Alicea, J.; Prueitt, W.L.; Hsieh-Wilson, L.C.; Diamond, M.I. Specific glycosaminoglycan chain length and sulfation patterns are required for cell uptake of tau versus alpha-synuclein and beta-amyloid aggregates. J. Biol. Chem. 2018, 293, 10826–10840. [Google Scholar] [CrossRef]
- Kanekiyo, T.; Zhang, J.; Liu, Q.; Liu, C.C.; Zhang, L.; Bu, G. Heparan sulphate proteoglycan and the low-density lipoprotein receptor-related protein 1 constitute major pathways for neuronal amyloid-beta uptake. J. Neurosci. 2011, 31, 1644–1651. [Google Scholar] [CrossRef]
- Rauch, J.N.; Luna, G.; Guzman, E.; Audouard, M.; Challis, C.; Sibih, Y.E.; Leshuk, C.; Hernandez, I.; Wegmann, S.; Hyman, B.T.; et al. LRP1 is a master regulator of tau uptake and spread. Nature 2020, 580, 381–385. [Google Scholar] [CrossRef]
- Nakajima, C.; Kulik, A.; Frotscher, M.; Herz, J.; Schafer, M.; Bock, H.H.; May, P. Low density lipoprotein receptor-related protein 1 (LRP1) modulates N-methyl-D-aspartate (NMDA) receptor-dependent intracellular signaling and NMDA-induced regulation of postsynaptic protein complexes. J. Biol. Chem. 2013, 288, 21909–21923. [Google Scholar] [CrossRef]
- Wegmann, S.; Bennett, R.E.; Delorme, L.; Robbins, A.B.; Hu, M.; McKenzie, D.; Kirk, M.J.; Schiantarelli, J.; Tunio, N.; Amaral, A.C.; et al. Experimental evidence for the age dependence of tau protein spread in the brain. Sci. Adv. 2019, 5, eaaw6404. [Google Scholar] [CrossRef]
- Lasagna-Reeves, C.A.; Castillo-Carranza, D.L.; Sengupta, U.; Clos, A.L.; Jackson, G.R.; Kayed, R. Tau oligomers impair memory and induce synaptic and mitochondrial dysfunction in wild-type mice. Mol. Neurodegener. 2011, 6, 39. [Google Scholar] [CrossRef]
- Puangmalai, N.; Bhatt, N.; Montalbano, M.; Sengupta, U.; Gaikwad, S.; Ventura, F.; McAllen, S.; Ellsworth, A.; Garcia, S.; Kayed, R. Internalization mechanisms of brain-derived tau oligomers from patients with Alzheimer’s disease, progressive supranuclear palsy and dementia with Lewy bodies. Cell Death Dis. 2020, 11, 314. [Google Scholar] [CrossRef]
- Scheres, S.H.; Zhang, W.; Falcon, B.; Goedert, M. Cryo-EM structures of tau filaments. Curr. Opin. Struct. Biol. 2020, 64, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Hayashi, M.; Nakano, H.; Shimazaki, M.; Sugimori, K.; Koshino, Y. Correlation between astrocyte apoptosis and Alzheimer changes in gray matter lesions in Alzheimer’s disease. J. Alzheimers Dis. 2004, 6, 623–632; discussion 673–681. [Google Scholar] [CrossRef] [PubMed]
- Su, J.H.; Nichol, K.E.; Sitch, T.; Sheu, P.; Chubb, C.; Miller, B.L.; Tomaselli, K.J.; Kim, R.C.; Cotman, C.W. DNA damage and activated caspase-3 expression in neurons and astrocytes: Evidence for apoptosis in frontotemporal dementia. Exp. Neurol. 2000, 163, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Narasimhan, S.; Guo, J.L.; Changolkar, L.; Stieber, A.; McBride, J.D.; Silva, L.V.; He, Z.; Zhang, B.; Gathagan, R.J.; Trojanowski, J.Q.; et al. Pathological Tau Strains from Human Brains Recapitulate the Diversity of Tauopathies in Nontransgenic Mouse Brain. J. Neurosci. 2017, 37, 11406–11423. [Google Scholar] [CrossRef]
- Narasimhan, S.; Changolkar, L.; Riddle, D.M.; Kats, A.; Stieber, A.; Weitzman, S.A.; Zhang, B.; Li, Z.; Roberson, E.D.; Trojanowski, J.Q.; et al. Human tau pathology transmits glial tau aggregates in the absence of neuronal tau. J. Exp. Med. 2020, 217. [Google Scholar] [CrossRef]
- Goodwin, M.S.; Croft, C.L.; Futch, H.S.; Ryu, D.; Ceballos-Diaz, C.; Liu, X.; Paterno, G.; Mejia, C.; Deng, D.; Menezes, K.; et al. Utilizing minimally purified secreted rAAV for rapid and cost-effective manipulation of gene expression in the CNS. Mol. Neurodegener. 2020, 15, 15. [Google Scholar] [CrossRef]
- DeVos, S.L.; Miller, R.L.; Schoch, K.M.; Holmes, B.B.; Kebodeaux, C.S.; Wegener, A.J.; Chen, G.; Shen, T.; Tran, H.; Nichols, B.; et al. Tau reduction prevents neuronal loss and reverses pathological tau deposition and seeding in mice with tauopathy. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef]
- Wobst, H.J.; Sharma, A.; Diamond, M.I.; Wanker, E.E.; Bieschke, J. The green tea polyphenol (-)-epigallocatechin gallate prevents the aggregation of tau protein into toxic oligomers at substoichiometric ratios. FEBS Lett. 2015, 589, 77–83. [Google Scholar] [CrossRef]
- Yanamandra, K.; Kfoury, N.; Jiang, H.; Mahan, T.E.; Ma, S.; Maloney, S.E.; Wozniak, D.F.; Diamond, M.I.; Holtzman, D.M. Anti-tau antibodies that block tau aggregate seeding in vitro markedly decrease pathology and improve cognition in vivo. Neuron 2013, 80, 402–414. [Google Scholar] [CrossRef]
- Ciechanover, A. Proteolysis: From the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell Biol. 2005, 6, 79–87. [Google Scholar] [CrossRef]
- Wong, E.; Cuervo, A.M. Integration of clearance mechanisms: The proteasome and autophagy. Cold Spring Harb. Perspect. Biol. 2010, 2, a006734. [Google Scholar] [CrossRef] [PubMed]
- Bain, H.D.C.; Davidson, Y.S.; Robinson, A.C.; Ryan, S.; Rollinson, S.; Richardson, A.; Jones, M.; Snowden, J.S.; Pickering-Brown, S.; Mann, D.M.A. The role of lysosomes and autophagosomes in frontotemporal lobar degeneration. Neuropathol. Appl. Neurobiol. 2019, 45, 244–261. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K.; Akiyama, H.; Arai, T.; Kondo, H.; Haga, C.; Iritani, S.; Tsuchiya, K. Alz-50/Gallyas-positive lysosome-like intraneuronal granules in Alzheimer’s disease and control brains. Neurosci. Lett. 1998, 258, 113–116. [Google Scholar] [CrossRef]
- Lim, F.; Hernandez, F.; Lucas, J.J.; Gomez-Ramos, P.; Moran, M.A.; Avila, J. FTDP-17 mutations in tau transgenic mice provoke lysosomal abnormalities and Tau filaments in forebrain. Mol. Cell. Neurosci. 2001, 18, 702–714. [Google Scholar] [CrossRef]
- Menzies, F.M.; Fleming, A.; Rubinsztein, D.C. Compromised autophagy and neurodegenerative diseases. Nat. Rev. Neurosci. 2015, 16, 345–357. [Google Scholar] [CrossRef]
- Nixon, R.A.; Yang, D.S. Autophagy failure in Alzheimer’s disease—Locating the primary defect. Neurobiol. Dis. 2011, 43, 38–45. [Google Scholar] [CrossRef]
- Rubinsztein, D.C.; DiFiglia, M.; Heintz, N.; Nixon, R.A.; Qin, Z.H.; Ravikumar, B.; Stefanis, L.; Tolkovsky, A. Autophagy and its possible roles in nervous system diseases, damage and repair. Autophagy 2005, 1, 11–22. [Google Scholar] [CrossRef]
- Morimoto, R.I.; Cuervo, A.M. Proteostasis and the aging proteome in health and disease. J. Gerontol. A Biol. Sci. Med. Sci. 2014, 69 (Suppl. 1), S33–S38. [Google Scholar] [CrossRef]
- Wang, Y.; Mandelkow, E. Degradation of tau protein by autophagy and proteasomal pathways. Biochem. Soc. Trans. 2012, 40, 644–652. [Google Scholar] [CrossRef]
- Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009, 78, 477–513. [Google Scholar] [CrossRef]
- Pickart, C.M.; VanDemark, A.P. Opening doors into the proteasome. Nat. Struct. Biol. 2000, 7, 999–1001. [Google Scholar] [CrossRef] [PubMed]
- Mori, H.; Kondo, J.; Ihara, Y. Ubiquitin is a component of paired helical filaments in Alzheimer’s disease. Science 1987, 235, 1641–1644. [Google Scholar] [CrossRef] [PubMed]
- Perry, G.; Friedman, R.; Shaw, G.; Chau, V. Ubiquitin is detected in neurofibrillary tangles and senile plaque neurites of Alzheimer disease brains. Proc. Natl. Acad. Sci. USA 1987, 84, 3033–3036. [Google Scholar] [CrossRef] [PubMed]
- Morishima-Kawashima, M.; Hasegawa, M.; Takio, K.; Suzuki, M.; Titani, K.; Ihara, Y. Ubiquitin is conjugated with amino-terminally processed tau in paired helical filaments. Neuron 1993, 10, 1151–1160. [Google Scholar] [CrossRef]
- Paine, S.; Bedford, L.; Thorpe, J.R.; Mayer, R.J.; Cavey, J.R.; Bajaj, N.; Sheppard, P.W.; Lowe, J.; Layfield, R. Immunoreactivity to Lys63-linked polyubiquitin is a feature of neurodegeneration. Neurosci. Lett. 2009, 460, 205–208. [Google Scholar] [CrossRef]
- Babu, J.R.; Geetha, T.; Wooten, M.W. Sequestosome 1/p62 shuttles polyubiquitinated tau for proteasomal degradation. J. Neurochem. 2005, 94, 192–203. [Google Scholar] [CrossRef]
- Petrucelli, L.; Dickson, D.; Kehoe, K.; Taylor, J.; Snyder, H.; Grover, A.; De Lucia, M.; McGowan, E.; Lewis, J.; Prihar, G.; et al. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum. Mol. Genet. 2004, 13, 703–714. [Google Scholar] [CrossRef]
- Shimura, H.; Schwartz, D.; Gygi, S.P.; Kosik, K.S. CHIP-Hsc70 complex ubiquitinates phosphorylated tau and enhances cell survival. J. Biol. Chem. 2004, 279, 4869–4876. [Google Scholar] [CrossRef]
- Pickart, C.M. Ubiquitin enters the new millennium. Mol. Cell 2001, 8, 499–504. [Google Scholar] [CrossRef]
- Thrower, J.S.; Hoffman, L.; Rechsteiner, M.; Pickart, C.M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000, 19, 94–102. [Google Scholar] [CrossRef]
- Dickey, C.A.; Yue, M.; Lin, W.L.; Dickson, D.W.; Dunmore, J.H.; Lee, W.C.; Zehr, C.; West, G.; Cao, S.; Clark, A.M.; et al. Deletion of the ubiquitin ligase CHIP leads to the accumulation, but not the aggregation, of both endogenous phospho- and caspase-3-cleaved tau species. J. Neurosci. 2006, 26, 6985–6996. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.M.; Wong, E.S.; Kirkpatrick, D.S.; Pletnikova, O.; Ko, H.S.; Tay, S.P.; Ho, M.W.; Troncoso, J.; Gygi, S.P.; Lee, M.K.; et al. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum. Mol. Genet. 2008, 17, 431–439. [Google Scholar] [CrossRef] [PubMed]
- David, D.C.; Layfield, R.; Serpell, L.; Narain, Y.; Goedert, M.; Spillantini, M.G. Proteasomal degradation of tau protein. J. Neurochem. 2002, 83, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Bennett, E.J.; Bence, N.F.; Jayakumar, R.; Kopito, R.R. Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Mol. Cell 2005, 17, 351–365. [Google Scholar] [CrossRef]
- Keck, S.; Nitsch, R.; Grune, T.; Ullrich, O. Proteasome inhibition by paired helical filament-tau in brains of patients with Alzheimer’s disease. J. Neurochem. 2003, 85, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Nachman, E.; Wentink, A.S.; Madiona, K.; Bousset, L.; Katsinelos, T.; Allinson, K.; Kampinga, H.; McEwan, W.A.; Jahn, T.R.; Melki, R.; et al. Disassembly of Tau fibrils by the human Hsp70 disaggregation machinery generates small seeding-competent species. J. Biol. Chem. 2020, 295, 9676–9690. [Google Scholar] [CrossRef]
- Oddo, S.; Billings, L.; Kesslak, J.P.; Cribbs, D.H.; LaFerla, F.M. Abeta immunotherapy leads to clearance of early, but not late, hyperphosphorylated tau aggregates via the proteasome. Neuron 2004, 43, 321–332. [Google Scholar] [CrossRef]
- Rubinsztein, D.C. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 2006, 443, 780–786. [Google Scholar] [CrossRef]
- Myeku, N.; Clelland, C.L.; Emrani, S.; Kukushkin, N.V.; Yu, W.H.; Goldberg, A.L.; Duff, K.E. Tau-driven 26S proteasome impairment and cognitive dysfunction can be prevented early in disease by activating cAMP-PKA signaling. Nat. Med. 2016, 22, 46–53. [Google Scholar] [CrossRef]
- Dickey, C.A.; Kamal, A.; Lundgren, K.; Klosak, N.; Bailey, R.M.; Dunmore, J.; Ash, P.; Shoraka, S.; Zlatkovic, J.; Eckman, C.B.; et al. The high-affinity HSP90-CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. J. Clin. Investig. 2007, 117, 648–658. [Google Scholar] [CrossRef]
- Dolan, P.J.; Johnson, G.V. A caspase cleaved form of tau is preferentially degraded through the autophagy pathway. J. Biol. Chem. 2010, 285, 21978–21987. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.R.; Bondada, V.; Keller, J.N.; Thorpe, J.; Geddes, J.W. Proteasome or calpain inhibition does not alter cellular tau levels in neuroblastoma cells or primary neurons. J. Alzheimers Dis. 2005, 7, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Delobel, P.; Leroy, O.; Hamdane, M.; Sambo, A.V.; Delacourte, A.; Buee, L. Proteasome inhibition and Tau proteolysis: An unexpected regulation. FEBS Lett. 2005, 579, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Feuillette, S.; Blard, O.; Lecourtois, M.; Frebourg, T.; Campion, D.; Dumanchin, C. Tau is not normally degraded by the proteasome. J. Neurosci. Res. 2005, 80, 400–405. [Google Scholar] [CrossRef]
- Wang, Y.; Martinez-Vicente, M.; Kruger, U.; Kaushik, S.; Wong, E.; Mandelkow, E.M.; Cuervo, A.M.; Mandelkow, E. Tau fragmentation, aggregation and clearance: The dual role of lysosomal processing. Hum. Mol. Genet. 2009, 18, 4153–4170. [Google Scholar] [CrossRef]
- Myeku, N.; Duff, K.E. Targeting the 26S Proteasome To Protect Against Proteotoxic Diseases. Trends Mol. Med. 2018, 24, 18–29. [Google Scholar] [CrossRef]
- Keller, J.N.; Hanni, K.B.; Markesbery, W.R. Impaired proteasome function in Alzheimer’s disease. J. Neurochem. 2000, 75, 436–439. [Google Scholar] [CrossRef]
- Iwata, A.; Riley, B.E.; Johnston, J.A.; Kopito, R.R. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J. Biol. Chem. 2005, 280, 40282–40292. [Google Scholar] [CrossRef]
- Ravikumar, B.; Duden, R.; Rubinsztein, D.C. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 2002, 11, 1107–1117. [Google Scholar] [CrossRef]
- De Duve, C.; Wattiaux, R. Functions of lysosomes. Annu. Rev. Physiol. 1966, 28, 435–492. [Google Scholar] [CrossRef]
- Mizushima, N.; Levine, B.; Cuervo, A.M.; Klionsky, D.J. Autophagy fights disease through cellular self-digestion. Nature 2008, 451, 1069–1075. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Klionsky, D.J. An overview of the molecular mechanism of autophagy. Curr. Top. Microbiol. Immunol. 2009, 335, 1–32. [Google Scholar] [CrossRef]
- Berger, Z.; Ravikumar, B.; Menzies, F.M.; Oroz, L.G.; Underwood, B.R.; Pangalos, M.N.; Schmitt, I.; Wullner, U.; Evert, B.O.; O’Kane, C.J.; et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum. Mol. Genet. 2006, 15, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Ravikumar, B.; Vacher, C.; Berger, Z.; Davies, J.E.; Luo, S.; Oroz, L.G.; Scaravilli, F.; Easton, D.F.; Duden, R.; O’Kane, C.J.; et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004, 36, 585–595. [Google Scholar] [CrossRef]
- Mandrioli, J.; D’Amico, R.; Zucchi, E.; Gessani, A.; Fini, N.; Fasano, A.; Caponnetto, C.; Chio, A.; Dalla Bella, E.; Lunetta, C.; et al. Rapamycin treatment for amyotrophic lateral sclerosis: Protocol for a phase II randomized, double-blind, placebo-controlled, multicenter, clinical trial (RAP-ALS trial). Medicine 2018, 97, e11119. [Google Scholar] [CrossRef]
- Wang, Y.; Kruger, U.; Mandelkow, E.; Mandelkow, E.M. Generation of tau aggregates and clearance by autophagy in an inducible cell model of tauopathy. Neurodegener. Dis. 2010, 7, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.; Zhou, J.; Lynch, G. Lysosomal protease inhibitors induce meganeurites and tangle-like structures in entorhinohippocampal regions vulnerable to Alzheimer’s disease. Exp. Neurol. 1999, 158, 312–327. [Google Scholar] [CrossRef]
- Lamark, T.; Kirkin, V.; Dikic, I.; Johansen, T. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle 2009, 8, 1986–1990. [Google Scholar] [CrossRef]
- Wong, E.; Cuervo, A.M. Autophagy gone awry in neurodegenerative diseases. Nat. Neurosci. 2010, 13, 805–811. [Google Scholar] [CrossRef]
- Wong, E.S.; Tan, J.M.; Soong, W.E.; Hussein, K.; Nukina, N.; Dawson, V.L.; Dawson, T.M.; Cuervo, A.M.; Lim, K.L. Autophagy-mediated clearance of aggresomes is not a universal phenomenon. Hum. Mol. Genet. 2008, 17, 2570–2582. [Google Scholar] [CrossRef]
- Silva, M.C.; Cheng, C.; Mair, W.; Almeida, S.; Fong, H.; Biswas, M.H.U.; Zhang, Z.; Huang, Y.; Temple, S.; Coppola, G.; et al. Human iPSC-Derived Neuronal Model of Tau-A152T Frontotemporal Dementia Reveals Tau-Mediated Mechanisms of Neuronal Vulnerability. Stem Cell Rep. 2016, 7, 325–340. [Google Scholar] [CrossRef] [PubMed]
- Schaeffer, V.; Lavenir, I.; Ozcelik, S.; Tolnay, M.; Winkler, D.T.; Goedert, M. Stimulation of autophagy reduces neurodegeneration in a mouse model of human tauopathy. Brain 2012, 135, 2169–2177. [Google Scholar] [CrossRef]
- Hamano, T.; Gendron, T.F.; Causevic, E.; Yen, S.H.; Lin, W.L.; Isidoro, C.; Deture, M.; Ko, L.W. Autophagic-lysosomal perturbation enhances tau aggregation in transfectants with induced wild-type tau expression. Eur. J. Neurosci. 2008, 27, 1119–1130. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Ravikumar, B.; Rubinsztein, D.C. Autophagic clearance of aggregate-prone proteins associated with neurodegeneration. Methods Enzymol. 2009, 453, 83–110. [Google Scholar] [CrossRef]
- Caccamo, A.; Majumder, S.; Richardson, A.; Strong, R.; Oddo, S. Molecular interplay between mammalian target of rapamycin (mTOR), amyloid-beta, and Tau: Effects on cognitive impairments. J. Biol. Chem. 2010, 285, 13107–13120. [Google Scholar] [CrossRef]
- Lin, A.L.; Jahrling, J.B.; Zhang, W.; DeRosa, N.; Bakshi, V.; Romero, P.; Galvan, V.; Richardson, A. Rapamycin rescues vascular, metabolic and learning deficits in apolipoprotein E4 transgenic mice with pre-symptomatic Alzheimer’s disease. J. Cereb. Blood Flow Metab. 2017, 37, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Ozcelik, S.; Fraser, G.; Castets, P.; Schaeffer, V.; Skachokova, Z.; Breu, K.; Clavaguera, F.; Sinnreich, M.; Kappos, L.; Goedert, M.; et al. Rapamycin attenuates the progression of tau pathology in P301S tau transgenic mice. PLoS ONE 2013, 8, e62459. [Google Scholar] [CrossRef] [PubMed]
- Siman, R.; Cocca, R.; Dong, Y. The mTOR Inhibitor Rapamycin Mitigates Perforant Pathway Neurodegeneration and Synapse Loss in a Mouse Model of Early-Stage Alzheimer-Type Tauopathy. PLoS ONE 2015, 10, e0142340. [Google Scholar] [CrossRef] [PubMed]
- Spilman, P.; Podlutskaya, N.; Hart, M.J.; Debnath, J.; Gorostiza, O.; Bredesen, D.; Richardson, A.; Strong, R.; Galvan, V. Inhibition of mTOR by rapamycin abolishes cognitive deficits and reduces amyloid-beta levels in a mouse model of Alzheimer’s disease. PLoS ONE 2010, 5, e9979. [Google Scholar] [CrossRef]
- Congdon, E.E.; Wu, J.W.; Myeku, N.; Figueroa, Y.H.; Herman, M.; Marinec, P.S.; Gestwicki, J.E.; Dickey, C.A.; Yu, W.H.; Duff, K.E. Methylthioninium chloride (methylene blue) induces autophagy and attenuates tauopathy in vitro and in vivo. Autophagy 2012, 8, 609–622. [Google Scholar] [CrossRef]
- Kim, S.; Choi, K.J.; Cho, S.J.; Yun, S.M.; Jeon, J.P.; Koh, Y.H.; Song, J.; Johnson, G.V.; Jo, C. Fisetin stimulates autophagic degradation of phosphorylated tau via the activation of TFEB and Nrf2 transcription factors. Sci. Rep. 2016, 6, 24933. [Google Scholar] [CrossRef] [PubMed]
- Kruger, U.; Wang, Y.; Kumar, S.; Mandelkow, E.M. Autophagic degradation of tau in primary neurons and its enhancement by trehalose. Neurobiol. Aging 2012, 33, 2291–2305. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.C.; Nandi, G.A.; Tentarelli, S.; Gurrell, I.K.; Jamier, T.; Lucente, D.; Dickerson, B.C.; Brown, D.G.; Brandon, N.J.; Haggarty, S.J. Prolonged tau clearance and stress vulnerability rescue by pharmacological activation of autophagy in tauopathy neurons. Nat. Commun. 2020, 11, 3258. [Google Scholar] [CrossRef] [PubMed]
- Medina, M. An Overview on the Clinical Development of Tau-Based Therapeutics. Int. J. Mol. Sci. 2018, 19, 1160. [Google Scholar] [CrossRef]
- Rubinsztein, D.C.; Bento, C.F.; Deretic, V. Therapeutic targeting of autophagy in neurodegenerative and infectious diseases. J. Exp. Med. 2015, 212, 979–990. [Google Scholar] [CrossRef]
- Schaeffer, V.; Goedert, M. Stimulation of autophagy is neuroprotective in a mouse model of human tauopathy. Autophagy 2012, 8, 1686–1687. [Google Scholar] [CrossRef]
- Lopez, A.; Lee, S.E.; Wojta, K.; Ramos, E.M.; Klein, E.; Chen, J.; Boxer, A.L.; Gorno-Tempini, M.L.; Geschwind, D.H.; Schlotawa, L.; et al. A152T tau allele causes neurodegeneration that can be ameliorated in a zebrafish model by autophagy induction. Brain 2017, 140, 1128–1146. [Google Scholar] [CrossRef]
- Baughman, H.E.R.; Clouser, A.F.; Klevit, R.E.; Nath, A. HspB1 and Hsc70 chaperones engage distinct tau species and have different inhibitory effects on amyloid formation. J. Biol. Chem. 2018, 293, 2687–2700. [Google Scholar] [CrossRef]
- Baughman, H.E.R.; Pham, T.T.; Adams, C.S.; Nath, A.; Klevit, R.E. Release of a disordered domain enhances HspB1 chaperone activity toward tau. Proc. Natl. Acad. Sci. USA 2020, 117, 2923–2929. [Google Scholar] [CrossRef] [PubMed]
- Weickert, S.; Wawrzyniuk, M.; John, L.H.; Rudiger, S.G.D.; Drescher, M. The mechanism of Hsp90-induced oligomerizaton of Tau. Sci. Adv. 2020, 6, eaax6999. [Google Scholar] [CrossRef]
- Yu, A.; Fox, S.G.; Cavallini, A.; Kerridge, C.; O’Neill, M.J.; Wolak, J.; Bose, S.; Morimoto, R.I. Tau protein aggregates inhibit the protein-folding and vesicular trafficking arms of the cellular proteostasis network. J. Biol. Chem. 2019, 294, 7917–7930. [Google Scholar] [CrossRef] [PubMed]
- Calamini, B.; Silva, M.C.; Madoux, F.; Hutt, D.M.; Khanna, S.; Chalfant, M.A.; Saldanha, S.A.; Hodder, P.; Tait, B.D.; Garza, D.; et al. Small-molecule proteostasis regulators for protein conformational diseases. Nat. Chem. Biol. 2011, 8, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P.; Kedersha, N. Stress granules: The Tao of RNA triage. Trends Biochem. Sci. 2008, 33, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Ramaswami, M.; Taylor, J.P.; Parker, R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell 2013, 154, 727–736. [Google Scholar] [CrossRef]
- Wolozin, B. Regulated protein aggregation: Stress granules and neurodegeneration. Mol. Neurodegener. 2012, 7, 56. [Google Scholar] [CrossRef]
- Bentmann, E.; Haass, C.; Dormann, D. Stress granules in neurodegeneration—Lessons learnt from TAR DNA binding protein of 43 kDa and fused in sarcoma. FEBS J. 2013, 280, 4348–4370. [Google Scholar] [CrossRef]
- Hoover, B.R.; Reed, M.N.; Su, J.; Penrod, R.D.; Kotilinek, L.A.; Grant, M.K.; Pitstick, R.; Carlson, G.A.; Lanier, L.M.; Yuan, L.L.; et al. Tau mislocalization to dendritic spines mediates synaptic dysfunction independently of neurodegeneration. Neuron 2010, 68, 1067–1081. [Google Scholar] [CrossRef]
- Zempel, H.; Mandelkow, E. Lost after translation: Missorting of Tau protein and consequences for Alzheimer disease. Trends Neurosci. 2014, 37, 721–732. [Google Scholar] [CrossRef]
- Wolozin, B.; Apicco, D. RNA binding proteins and the genesis of neurodegenerative diseases. Adv. Exp. Med. Biol. 2015, 822, 11–15. [Google Scholar] [CrossRef]
- Vanderweyde, T.; Apicco, D.J.; Youmans-Kidder, K.; Ash, P.E.A.; Cook, C.; Lummertz da Rocha, E.; Jansen-West, K.; Frame, A.A.; Citro, A.; Leszyk, J.D.; et al. Interaction of tau with the RNA-Binding Protein TIA1 Regulates tau Pathophysiology and Toxicity. Cell Rep. 2016, 15, 1455–1466. [Google Scholar] [CrossRef]
- Thomas, M.G.; Loschi, M.; Desbats, M.A.; Boccaccio, G.L. RNA granules: The good, the bad and the ugly. Cell. Signal. 2011, 23, 324–334. [Google Scholar] [CrossRef] [PubMed]
- Vanderweyde, T.; Yu, H.; Varnum, M.; Liu-Yesucevitz, L.; Citro, A.; Ikezu, T.; Duff, K.; Wolozin, B. Contrasting pathology of the stress granule proteins TIA-1 and G3BP in tauopathies. J. Neurosci. 2012, 32, 8270–8283. [Google Scholar] [CrossRef] [PubMed]
- Brunello, C.A.; Yan, X.; Huttunen, H.J. Internalized Tau sensitizes cells to stress by promoting formation and stability of stress granules. Sci. Rep. 2016, 6, 30498. [Google Scholar] [CrossRef] [PubMed]
- Apicco, D.J.; Ash, P.E.A.; Maziuk, B.; LeBlang, C.; Medalla, M.; Al Abdullatif, A.; Ferragud, A.; Botelho, E.; Ballance, H.I.; Dhawan, U.; et al. Reducing the RNA binding protein TIA1 protects against tau-mediated neurodegeneration in vivo. Nat. Neurosci. 2018, 21, 72–80. [Google Scholar] [CrossRef]
- Jiang, L.; Ash, P.E.A.; Maziuk, B.F.; Ballance, H.I.; Boudeau, S.; Abdullatif, A.A.; Orlando, M.; Petrucelli, L.; Ikezu, T.; Wolozin, B. TIA1 regulates the generation and response to toxic tau oligomers. Acta Neuropathol. 2019, 137, 259–277. [Google Scholar] [CrossRef]
- Frandemiche, M.L.; De Seranno, S.; Rush, T.; Borel, E.; Elie, A.; Arnal, I.; Lante, F.; Buisson, A. Activity-dependent tau protein translocation to excitatory synapse is disrupted by exposure to amyloid-beta oligomers. J. Neurosci. 2014, 34, 6084–6097. [Google Scholar] [CrossRef]
- Lin, M.T.; Beal, M.F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 2006, 443, 787–795. [Google Scholar] [CrossRef]
- Wang, W.; Zhao, F.; Ma, X.; Perry, G.; Zhu, X. Mitochondria dysfunction in the pathogenesis of Alzheimer’s disease: Recent advances. Mol. Neurodegener. 2020, 15, 30. [Google Scholar] [CrossRef]
- Swerdlow, R.H. Mitochondria and Mitochondrial Cascades in Alzheimer’s Disease. J. Alzheimers Dis. 2018, 62, 1403–1416. [Google Scholar] [CrossRef]
- Hirai, K.; Aliev, G.; Nunomura, A.; Fujioka, H.; Russell, R.L.; Atwood, C.S.; Johnson, A.B.; Kress, Y.; Vinters, H.V.; Tabaton, M.; et al. Mitochondrial abnormalities in Alzheimer’s disease. J. Neurosci. 2001, 21, 3017–3023. [Google Scholar] [CrossRef]
- Pickett, E.K.; Rose, J.; McCrory, C.; McKenzie, C.A.; King, D.; Smith, C.; Gillingwater, T.H.; Henstridge, C.M.; Spires-Jones, T.L. Region-specific depletion of synaptic mitochondria in the brains of patients with Alzheimer’s disease. Acta Neuropathol. 2018, 136, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Su, B.; Lee, H.G.; Li, X.; Perry, G.; Smith, M.A.; Zhu, X. Impaired balance of mitochondrial fission and fusion in Alzheimer’s disease. J. Neurosci. 2009, 29, 9090–9103. [Google Scholar] [CrossRef]
- Dixit, R.; Ross, J.L.; Goldman, Y.E.; Holzbaur, E.L. Differential regulation of dynein and kinesin motor proteins by tau. Science 2008, 319, 1086–1089. [Google Scholar] [CrossRef] [PubMed]
- Ebneth, A.; Godemann, R.; Stamer, K.; Illenberger, S.; Trinczek, B.; Mandelkow, E. Overexpression of tau protein inhibits kinesin-dependent trafficking of vesicles, mitochondria, and endoplasmic reticulum: Implications for Alzheimer’s disease. J. Cell Biol. 1998, 143, 777–794. [Google Scholar] [CrossRef]
- Gilley, J.; Seereeram, A.; Ando, K.; Mosely, S.; Andrews, S.; Kerschensteiner, M.; Misgeld, T.; Brion, J.P.; Anderton, B.; Hanger, D.P.; et al. Age-dependent axonal transport and locomotor changes and tau hypophosphorylation in a “P301L” tau knockin mouse. Neurobiol. Aging 2012, 33, 621.e1–621.e15. [Google Scholar] [CrossRef]
- Kopeikina, K.J.; Carlson, G.A.; Pitstick, R.; Ludvigson, A.E.; Peters, A.; Luebke, J.I.; Koffie, R.M.; Frosch, M.P.; Hyman, B.T.; Spires-Jones, T.L. Tau accumulation causes mitochondrial distribution deficits in neurons in a mouse model of tauopathy and in human Alzheimer’s disease brain. Am. J. Pathol. 2011, 179, 2071–2082. [Google Scholar] [CrossRef]
- Wee, M.; Chegini, F.; Power, J.H.T.; Majd, S. Tau Positive Neurons Show Marked Mitochondrial Loss and Nuclear Degradation in Alzheimer’s Disease. Curr. Alzheimer Res. 2018, 15, 928–937. [Google Scholar] [CrossRef]
- Vossel, K.A.; Xu, J.C.; Fomenko, V.; Miyamoto, T.; Suberbielle, E.; Knox, J.A.; Ho, K.; Kim, D.H.; Yu, G.Q.; Mucke, L. Tau reduction prevents Abeta-induced axonal transport deficits by blocking activation of GSK3beta. J. Cell Biol. 2015, 209, 419–433. [Google Scholar] [CrossRef]
- Lovell, M.A.; Xiong, S.; Xie, C.; Davies, P.; Markesbery, W.R. Induction of hyperphosphorylated tau in primary rat cortical neuron cultures mediated by oxidative stress and glycogen synthase kinase-3. J. Alzheimers Dis. 2004, 6, 659–671; discussion 673–681. [Google Scholar] [CrossRef]
- Thornton, C.; Bright, N.J.; Sastre, M.; Muckett, P.J.; Carling, D. AMP-activated protein kinase (AMPK) is a tau kinase, activated in response to amyloid beta-peptide exposure. Biochem. J. 2011, 434, 503–512. [Google Scholar] [CrossRef]
- Moreira, P.I.; Siedlak, S.L.; Wang, X.; Santos, M.S.; Oliveira, C.R.; Tabaton, M.; Nunomura, A.; Szweda, L.I.; Aliev, G.; Smith, M.A.; et al. Increased autophagic degradation of mitochondria in Alzheimer disease. Autophagy 2007, 3, 614–615. [Google Scholar] [CrossRef] [PubMed]
- Martin-Maestro, P.; Gargini, R.; Perry, G.; Avila, J.; Garcia-Escudero, V. PARK2 enhancement is able to compensate mitophagy alterations found in sporadic Alzheimer’s disease. Hum. Mol. Genet. 2016, 25, 792–806. [Google Scholar] [CrossRef] [PubMed]
- Brooks, W.M.; Lynch, P.J.; Ingle, C.C.; Hatton, A.; Emson, P.C.; Faull, R.L.; Starkey, M.P. Gene expression profiles of metabolic enzyme transcripts in Alzheimer’s disease. Brain Res. 2007, 1127, 127–135. [Google Scholar] [CrossRef]
- Liang, W.S.; Dunckley, T.; Beach, T.G.; Grover, A.; Mastroeni, D.; Ramsey, K.; Caselli, R.J.; Kukull, W.A.; McKeel, D.; Morris, J.C.; et al. Altered neuronal gene expression in brain regions differentially affected by Alzheimer’s disease: A reference data set. Physiol. Genomics 2008, 33, 240–256. [Google Scholar] [CrossRef]
- Sorrentino, V.; Romani, M.; Mouchiroud, L.; Beck, J.S.; Zhang, H.; D’Amico, D.; Moullan, N.; Potenza, F.; Schmid, A.W.; Rietsch, S.; et al. Enhancing mitochondrial proteostasis reduces amyloid-beta proteotoxicity. Nature 2017, 552, 187–193. [Google Scholar] [CrossRef]
- Gerakis, Y.; Hetz, C. Emerging roles of ER stress in the etiology and pathogenesis of Alzheimer’s disease. FEBS J. 2018, 285, 995–1011. [Google Scholar] [CrossRef] [PubMed]
- Hetz, C.; Mollereau, B. Disturbance of endoplasmic reticulum proteostasis in neurodegenerative diseases. Nat. Rev. Neurosci. 2014, 15, 233–249. [Google Scholar] [CrossRef] [PubMed]
- Rutkowski, D.T.; Hegde, R.S. Regulation of basal cellular physiology by the homeostatic unfolded protein response. J. Cell Biol. 2010, 189, 783–794. [Google Scholar] [CrossRef]
- Roussel, B.D.; Kruppa, A.J.; Miranda, E.; Crowther, D.C.; Lomas, D.A.; Marciniak, S.J. Endoplasmic reticulum dysfunction in neurological disease. Lancet Neurol. 2013, 12, 105–118. [Google Scholar] [CrossRef]
- Wang, M.; Kaufman, R.J. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature 2016, 529, 326–335. [Google Scholar] [CrossRef]
- Harding, H.P.; Zhang, Y.; Ron, D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 1999, 397, 271–274. [Google Scholar] [CrossRef]
- Bocai, N.I.; Marcora, M.S.; Belfiori-Carrasco, L.F.; Morelli, L.; Castano, E.M. Endoplasmic Reticulum Stress in Tauopathies: Contrasting Human Brain Pathology with Cellular and Animal Models. J. Alzheimers Dis. 2019, 68, 439–458. [Google Scholar] [CrossRef]
- Ho, Y.S.; Yang, X.; Lau, J.C.; Hung, C.H.; Wuwongse, S.; Zhang, Q.; Wang, J.; Baum, L.; So, K.F.; Chang, R.C. Endoplasmic reticulum stress induces tau pathology and forms a vicious cycle: Implication in Alzheimer’s disease pathogenesis. J. Alzheimers Dis. 2012, 28, 839–854. [Google Scholar] [CrossRef]
- Resende, R.; Ferreiro, E.; Pereira, C.; Oliveira, C.R. ER stress is involved in Abeta-induced GSK-3beta activation and tau phosphorylation. J. Neurosci. Res. 2008, 86, 2091–2099. [Google Scholar] [CrossRef]
- Bordi, M.; Berg, M.J.; Mohan, P.S.; Peterhoff, C.M.; Alldred, M.J.; Che, S.; Ginsberg, S.D.; Nixon, R.A. Autophagy flux in CA1 neurons of Alzheimer hippocampus: Increased induction overburdens failing lysosomes to propel neuritic dystrophy. Autophagy 2016, 12, 2467–2483. [Google Scholar] [CrossRef]
- Bugallo, R.; Marlin, E.; Baltanas, A.; Toledo, E.; Ferrero, R.; Vinueza-Gavilanes, R.; Larrea, L.; Arrasate, M.; Aragon, T. Fine tuning of the unfolded protein response by ISRIB improves neuronal survival in a model of amyotrophic lateral sclerosis. Cell Death Dis. 2020, 11, 397. [Google Scholar] [CrossRef]
- Apicco, D.J.; Zhang, C.; Maziuk, B.; Jiang, L.; Ballance, H.I.; Boudeau, S.; Ung, C.; Li, H.; Wolozin, B. Dysregulation of RNA Splicing in Tauopathies. Cell Rep. 2019, 29, 4377–4388.e4. [Google Scholar] [CrossRef]
- Koike, M.; Tsukada, S.; Tsuzuki, K.; Kijima, H.; Ozawa, S. Regulation of kinetic properties of GluR2 AMPA receptor channels by alternative splicing. J. Neurosci. 2000, 20, 2166–2174. [Google Scholar] [CrossRef]
- Pei, W.; Huang, Z.; Wang, C.; Han, Y.; Park, J.S.; Niu, L. Flip and flop: A molecular determinant for AMPA receptor channel opening. Biochemistry 2009, 48, 3767–3777. [Google Scholar] [CrossRef]
- Yirmiya, R.; Goshen, I. Immune modulation of learning, memory, neural plasticity and neurogenesis. Brain Behav. Immun. 2011, 25, 181–213. [Google Scholar] [CrossRef]
- Perry, V.H.; Cunningham, C.; Holmes, C. Systemic infections and inflammation affect chronic neurodegeneration. Nat. Rev. Immunol. 2007, 7, 161–167. [Google Scholar] [CrossRef]
- Wendeln, A.C.; Degenhardt, K.; Kaurani, L.; Gertig, M.; Ulas, T.; Jain, G.; Wagner, J.; Hasler, L.M.; Wild, K.; Skodras, A.; et al. Innate immune memory in the brain shapes neurological disease hallmarks. Nature 2018, 556, 332–338. [Google Scholar] [CrossRef]
- Allen, M.; Wang, X.; Burgess, J.D.; Watzlawik, J.; Serie, D.J.; Younkin, C.S.; Nguyen, T.; Malphrus, K.G.; Lincoln, S.; Carrasquillo, M.M.; et al. Conserved brain myelination networks are altered in Alzheimer’s and other neurodegenerative diseases. Alzheimers Dement. 2018, 14, 352–366. [Google Scholar] [CrossRef]
- Cribbs, D.H.; Berchtold, N.C.; Perreau, V.; Coleman, P.D.; Rogers, J.; Tenner, A.J.; Cotman, C.W. Extensive innate immune gene activation accompanies brain aging, increasing vulnerability to cognitive decline and neurodegeneration: A microarray study. J. Neuroinflamm. 2012, 9, 179. [Google Scholar] [CrossRef]
- Mostafavi, S.; Gaiteri, C.; Sullivan, S.E.; White, C.C.; Tasaki, S.; Xu, J.; Taga, M.; Klein, H.U.; Patrick, E.; Komashko, V.; et al. A molecular network of the aging human brain provides insights into the pathology and cognitive decline of Alzheimer’s disease. Nat. Neurosci. 2018, 21, 811–819. [Google Scholar] [CrossRef]
- Seyfried, N.T.; Dammer, E.B.; Swarup, V.; Nandakumar, D.; Duong, D.M.; Yin, L.; Deng, Q.; Nguyen, T.; Hales, C.M.; Wingo, T.; et al. A Multi-network Approach Identifies Protein-Specific Co-expression in Asymptomatic and Symptomatic Alzheimer’s Disease. Cell Syst. 2017, 4, 60–72.e4. [Google Scholar] [CrossRef]
- Heneka, M.T.; Golenbock, D.T.; Latz, E. Innate immunity in Alzheimer’s disease. Nat. Immunol. 2015, 16, 229–236. [Google Scholar] [CrossRef]
- Keren-Shaul, H.; Spinrad, A.; Weiner, A.; Matcovitch-Natan, O.; Dvir-Szternfeld, R.; Ulland, T.K.; David, E.; Baruch, K.; Lara-Astaiso, D.; Toth, B.; et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 2017, 169, 1276–1290.e17. [Google Scholar] [CrossRef] [PubMed]
- Leyns, C.E.G.; Holtzman, D.M. Glial contributions to neurodegeneration in tauopathies. Mol. Neurodegener. 2017, 12, 50. [Google Scholar] [CrossRef]
- Kunkle, B.W.; Grenier-Boley, B.; Sims, R.; Bis, J.C.; Damotte, V.; Naj, A.C.; Boland, A.; Vronskaya, M.; van der Lee, S.J.; Amlie-Wolf, A.; et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat. Genet. 2019, 51, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Hueston, C.M.; O’Leary, J.D.; Hoban, A.E.; Kozareva, D.A.; Pawley, L.C.; O’Leary, O.F.; Cryan, J.F.; Nolan, Y.M. Chronic interleukin-1beta in the dorsal hippocampus impairs behavioural pattern separation. Brain Behav. Immun. 2018, 74, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Liddelow, S.A.; Guttenplan, K.A.; Clarke, L.E.; Bennett, F.C.; Bohlen, C.J.; Schirmer, L.; Bennett, M.L.; Munch, A.E.; Chung, W.S.; Peterson, T.C.; et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 2017, 541, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Krasemann, S.; Madore, C.; Cialic, R.; Baufeld, C.; Calcagno, N.; El Fatimy, R.; Beckers, L.; O’Loughlin, E.; Xu, Y.; Fanek, Z.; et al. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity 2017, 47, 566–581.e9. [Google Scholar] [CrossRef] [PubMed]
- Gratuze, M.; Leyns, C.E.; Sauerbeck, A.D.; St-Pierre, M.K.; Xiong, M.; Kim, N.; Serrano, J.R.; Tremblay, M.E.; Kummer, T.T.; Colonna, M.; et al. Impact of TREM2R47H variant on tau pathology-induced gliosis and neurodegeneration. J. Clin. Investig. 2020, 130, 4954–4968. [Google Scholar] [CrossRef]
- Gratuze, M.; Leyns, C.E.G.; Holtzman, D.M. New insights into the role of TREM2 in Alzheimer’s disease. Mol. Neurodegener. 2018, 13, 66. [Google Scholar] [CrossRef]
- Cruchaga, C.; Kauwe, J.S.; Harari, O.; Jin, S.C.; Cai, Y.; Karch, C.M.; Benitez, B.A.; Jeng, A.T.; Skorupa, T.; Carrell, D.; et al. GWAS of cerebrospinal fluid tau levels identifies risk variants for Alzheimer’s disease. Neuron 2013, 78, 256–268. [Google Scholar] [CrossRef]
- Lill, C.M.; Rengmark, A.; Pihlstrom, L.; Fogh, I.; Shatunov, A.; Sleiman, P.M.; Wang, L.S.; Liu, T.; Lassen, C.F.; Meissner, E.; et al. The role of TREM2 R47H as a risk factor for Alzheimer’s disease, frontotemporal lobar degeneration, amyotrophic lateral sclerosis, and Parkinson’s disease. Alzheimers Dement. 2015, 11, 1407–1416. [Google Scholar] [CrossRef]
- Suarez-Calvet, M.; Kleinberger, G.; Araque Caballero, M.A.; Brendel, M.; Rominger, A.; Alcolea, D.; Fortea, J.; Lleo, A.; Blesa, R.; Gispert, J.D.; et al. sTREM2 cerebrospinal fluid levels are a potential biomarker for microglia activity in early-stage Alzheimer’s disease and associate with neuronal injury markers. EMBO Mol. Med. 2016, 8, 466–476. [Google Scholar] [CrossRef]
- Buerger, K.; Ewers, M.; Pirttila, T.; Zinkowski, R.; Alafuzoff, I.; Teipel, S.J.; DeBernardis, J.; Kerkman, D.; McCulloch, C.; Soininen, H.; et al. CSF phosphorylated tau protein correlates with neocortical neurofibrillary pathology in Alzheimer’s disease. Brain 2006, 129, 3035–3041. [Google Scholar] [CrossRef]
- Gjoneska, E.; Pfenning, A.R.; Mathys, H.; Quon, G.; Kundaje, A.; Tsai, L.H.; Kellis, M. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nature 2015, 518, 365–369. [Google Scholar] [CrossRef]
- Wang, H.; Li, Y.; Ryder, J.W.; Hole, J.T.; Ebert, P.J.; Airey, D.C.; Qian, H.R.; Logsdon, B.; Fisher, A.; Ahmed, Z.; et al. Genome-wide RNAseq study of the molecular mechanisms underlying microglia activation in response to pathological tau perturbation in the rTg4510 tau transgenic animal model. Mol. Neurodegener. 2018, 13, 65. [Google Scholar] [CrossRef] [PubMed]
- Sayed, F.A.; Telpoukhovskaia, M.; Kodama, L.; Li, Y.; Zhou, Y.; Le, D.; Hauduc, A.; Ludwig, C.; Gao, F.; Clelland, C.; et al. Differential effects of partial and complete loss of TREM2 on microglial injury response and tauopathy. Proc. Natl. Acad. Sci. USA 2018, 115, 10172–10177. [Google Scholar] [CrossRef] [PubMed]
- Bemiller, S.M.; McCray, T.J.; Allan, K.; Formica, S.V.; Xu, G.; Wilson, G.; Kokiko-Cochran, O.N.; Crish, S.D.; Lasagna-Reeves, C.A.; Ransohoff, R.M.; et al. TREM2 deficiency exacerbates tau pathology through dysregulated kinase signaling in a mouse model of tauopathy. Mol. Neurodegener. 2017, 12, 74. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J.; Feldman, H.H.; Scheltens, P. The “rights” of precision drug development for Alzheimer’s disease. Alzheimers Res. Ther. 2019, 11, 76. [Google Scholar] [CrossRef]
- Tan, C.C.; Yu, J.T.; Wang, H.F.; Tan, M.S.; Meng, X.F.; Wang, C.; Jiang, T.; Zhu, X.C.; Tan, L. Efficacy and safety of donepezil, galantamine, rivastigmine, and memantine for the treatment of Alzheimer’s disease: A systematic review and meta-analysis. J. Alzheimers Dis. 2014, 41, 615–631. [Google Scholar] [CrossRef]
- Congdon, E.E.; Sigurdsson, E.M. Tau-targeting therapies for Alzheimer disease. Nat. Rev. Neurol. 2018, 14, 399–415. [Google Scholar] [CrossRef]
- Gotz, J.; Xia, D.; Leinenga, G.; Chew, Y.L.; Nicholas, H. What Renders TAU Toxic. Front. Neurol. 2013, 4, 72. [Google Scholar] [CrossRef]
- Banks, W.A. Developing drugs that can cross the blood-brain barrier: Applications to Alzheimer’s disease. BMC Neurosci. 2008, 9 (Suppl. 3), S2. [Google Scholar] [CrossRef]
- Golde, T.E.; DeKosky, S.T.; Galasko, D. Alzheimer’s disease: The right drug, the right time. Science 2018, 362, 1250–1251. [Google Scholar] [CrossRef]
- Karran, E.; Hardy, J. A critique of the drug discovery and phase 3 clinical programs targeting the amyloid hypothesis for Alzheimer disease. Ann. Neurol. 2014, 76, 185–205. [Google Scholar] [CrossRef]
- VandeVrede, L.; Boxer, A.L.; Polydoro, M. Targeting tau: Clinical trials and novel therapeutic approaches. Neurosci. Lett. 2020, 731, 134919. [Google Scholar] [CrossRef] [PubMed]
- Tabrizi, S.J.; Leavitt, B.R.; Landwehrmeyer, G.B.; Wild, E.J.; Saft, C.; Barker, R.A.; Blair, N.F.; Craufurd, D.; Priller, J.; Rickards, H.; et al. Targeting Huntingtin Expression in Patients with Huntington’s Disease. N. Engl. J. Med. 2019, 380, 2307–2316. [Google Scholar] [CrossRef] [PubMed]
- Devi, G.; Scheltens, P. Heterogeneity of Alzheimer’s disease: Consequence for drug trials? Alzheimers Res. Ther. 2018, 10, 122. [Google Scholar] [CrossRef] [PubMed]
- Lei, P.; Ayton, S.; Moon, S.; Zhang, Q.; Volitakis, I.; Finkelstein, D.I.; Bush, A.I. Motor and cognitive deficits in aged tau knockout mice in two background strains. Mol. Neurodegener. 2014, 9, 29. [Google Scholar] [CrossRef] [PubMed]
- Zhukareva, V.; Vogelsberg-Ragaglia, V.; Van Deerlin, V.M.; Bruce, J.; Shuck, T.; Grossman, M.; Clark, C.M.; Arnold, S.E.; Masliah, E.; Galasko, D.; et al. Loss of brain tau defines novel sporadic and familial tauopathies with frontotemporal dementia. Ann. Neurol. 2001, 49, 165–175. [Google Scholar] [CrossRef]
- Mignon, L.; Kordasiewicz, H.; Lane, R.; Smith, A.; Miller, T.; Narayanan, P.; Swayze, E.; Norris, D.; Fitzsimmons, B.; Bennett, F. Design of the First-in-Human Study of IONIS-MAPTRx, a Tau-lowering Antisense Oligonucleotide, in Patients with Alzheimer Disease (S2.006). Neurology 2018, 90, S2.006. [Google Scholar]
- Xu, H.; Rosler, T.W.; Carlsson, T.; de Andrade, A.; Fiala, O.; Hollerhage, M.; Oertel, W.H.; Goedert, M.; Aigner, A.; Hoglinger, G.U. Tau silencing by siRNA in the P301S mouse model of tauopathy. Curr. Gene Ther. 2014, 14, 343–351. [Google Scholar] [CrossRef]
- Schoch, K.M.; DeVos, S.L.; Miller, R.L.; Chun, S.J.; Norrbom, M.; Wozniak, D.F.; Dawson, H.N.; Bennett, C.F.; Rigo, F.; Miller, T.M. Increased 4R-Tau Induces Pathological Changes in a Human-Tau Mouse Model. Neuron 2016, 90, 941–947. [Google Scholar] [CrossRef]
- Dolan, P.J.; Johnson, G.V. The role of tau kinases in Alzheimer’s disease. Curr. Opin. Drug Discov. Dev. 2010, 13, 595–603. [Google Scholar]
- Wang, J.Z.; Grundke-Iqbal, I.; Iqbal, K. Kinases and phosphatases and tau sites involved in Alzheimer neurofibrillary degeneration. Eur. J. Neurosci. 2007, 25, 59–68. [Google Scholar] [CrossRef]
- Noble, W.; Planel, E.; Zehr, C.; Olm, V.; Meyerson, J.; Suleman, F.; Gaynor, K.; Wang, L.; LaFrancois, J.; Feinstein, B.; et al. Inhibition of glycogen synthase kinase-3 by lithium correlates with reduced tauopathy and degeneration in vivo. Proc. Natl. Acad. Sci. USA 2005, 102, 6990–6995. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.P.; Xie, J.W.; Wang, C.Y.; Wang, T.; Wang, X.; Wang, S.L.; Teng, W.P.; Wang, Z.Y. Valproate reduces tau phosphorylation via cyclin-dependent kinase 5 and glycogen synthase kinase 3 signaling pathways. Brain Res. Bull. 2011, 85, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Sereno, L.; Coma, M.; Rodriguez, M.; Sanchez-Ferrer, P.; Sanchez, M.B.; Gich, I.; Agullo, J.M.; Perez, M.; Avila, J.; Guardia-Laguarta, C.; et al. A novel GSK-3beta inhibitor reduces Alzheimer’s pathology and rescues neuronal loss in vivo. Neurobiol. Dis. 2009, 35, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Lovestone, S.; Boada, M.; Dubois, B.; Hull, M.; Rinne, J.O.; Huppertz, H.J.; Calero, M.; Andres, M.V.; Gomez-Carrillo, B.; Leon, T.; et al. A phase II trial of tideglusib in Alzheimer’s disease. J. Alzheimers Dis. 2015, 45, 75–88. [Google Scholar] [CrossRef]
- Chin, J.; Palop, J.J.; Puolivali, J.; Massaro, C.; Bien-Ly, N.; Gerstein, H.; Scearce-Levie, K.; Masliah, E.; Mucke, L. Fyn kinase induces synaptic and cognitive impairments in a transgenic mouse model of Alzheimer’s disease. J. Neurosci. 2005, 25, 9694–9703. [Google Scholar] [CrossRef]
- Kaufman, A.C.; Salazar, S.V.; Haas, L.T.; Yang, J.; Kostylev, M.A.; Jeng, A.T.; Robinson, S.A.; Gunther, E.C.; van Dyck, C.H.; Nygaard, H.B.; et al. Fyn inhibition rescues established memory and synapse loss in Alzheimer mice. Ann. Neurol. 2015, 77, 953–971. [Google Scholar] [CrossRef]
- Hebron, M.L.; Javidnia, M.; Moussa, C.E. Tau clearance improves astrocytic function and brain glutamate-glutamine cycle. J. Neurol. Sci. 2018, 391, 90–99. [Google Scholar] [CrossRef]
- Hebron, M.L.; Lonskaya, I.; Moussa, C.E. Nilotinib reverses loss of dopamine neurons and improves motor behavior via autophagic degradation of alpha-synuclein in Parkinson’s disease models. Hum. Mol. Genet. 2013, 22, 3315–3328. [Google Scholar] [CrossRef]
- Pagan, F.; Hebron, M.; Valadez, E.H.; Torres-Yaghi, Y.; Huang, X.; Mills, R.R.; Wilmarth, B.M.; Howard, H.; Dunn, C.; Carlson, A.; et al. Nilotinib Effects in Parkinson’s disease and Dementia with Lewy bodies. J. Parkinsons Dis. 2016, 6, 503–517. [Google Scholar] [CrossRef]
- McKenzie-Nickson, S.; Chan, J.; Perez, K.; Hung, L.W.; Cheng, L.; Sedjahtera, A.; Gunawan, L.; Adlard, P.A.; Hayne, D.J.; McInnes, L.E.; et al. Modulating Protein Phosphatase 2A Rescues Disease Phenotype in Neurodegenerative Tauopathies. ACS Chem. Neurosci. 2018, 9, 2731–2740. [Google Scholar] [CrossRef]
- Chen, X.; Li, Y.; Wang, C.; Tang, Y.; Mok, S.A.; Tsai, R.M.; Rojas, J.C.; Karydas, A.; Miller, B.L.; Boxer, A.L.; et al. Promoting tau secretion and propagation by hyperactive p300/CBP via autophagy-lysosomal pathway in tauopathy. Mol. Neurodegener. 2020, 15, 2. [Google Scholar] [CrossRef] [PubMed]
- Ryan, P.; Xu, M.; Davey, A.K.; Danon, J.J.; Mellick, G.D.; Kassiou, M.; Rudrawar, S. O-GlcNAc Modification Protects against Protein Misfolding and Aggregation in Neurodegenerative Disease. ACS Chem. Neurosci. 2019, 10, 2209–2221. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Shi, J.; Tanimukai, H.; Gu, J.; Gu, J.; Grundke-Iqbal, I.; Iqbal, K.; Gong, C.X. Reduced O-GlcNAcylation links lower brain glucose metabolism and tau pathology in Alzheimer’s disease. Brain 2009, 132, 1820–1832. [Google Scholar] [CrossRef] [PubMed]
- Hastings, N.B.; Wang, X.; Song, L.; Butts, B.D.; Grotz, D.; Hargreaves, R.; Fred Hess, J.; Hong, K.K.; Huang, C.R.; Hyde, L.; et al. Inhibition of O-GlcNAcase leads to elevation of O-GlcNAc tau and reduction of tauopathy and cerebrospinal fluid tau in rTg4510 mice. Mol. Neurodegener. 2017, 12, 39. [Google Scholar] [CrossRef]
- Selnick, H.G.; Hess, J.F.; Tang, C.; Liu, K.; Schachter, J.B.; Ballard, J.E.; Marcus, J.; Klein, D.J.; Wang, X.; Pearson, M.; et al. Discovery of MK-8719, a Potent O-GlcNAcase Inhibitor as a Potential Treatment for Tauopathies. J. Med. Chem. 2019, 62, 10062–10097. [Google Scholar] [CrossRef]
- Brunden, K.R.; Ballatore, C.; Crowe, A.; Smith, A.B., 3rd; Lee, V.M.; Trojanowski, J.Q. Tau-directed drug discovery for Alzheimer’s disease and related tauopathies: A focus on tau assembly inhibitors. Exp. Neurol. 2010, 223, 304–310. [Google Scholar] [CrossRef]
- Panza, F.; Solfrizzi, V.; Seripa, D.; Imbimbo, B.P.; Lozupone, M.; Santamato, A.; Zecca, C.; Barulli, M.R.; Bellomo, A.; Pilotto, A.; et al. Tau-Centric Targets and Drugs in Clinical Development for the Treatment of Alzheimer’s Disease. Biomed. Res. Int. 2016, 2016, 3245935. [Google Scholar] [CrossRef]
- Bulic, B.; Pickhardt, M.; Schmidt, B.; Mandelkow, E.M.; Waldmann, H.; Mandelkow, E. Development of tau aggregation inhibitors for Alzheimer’s disease. Angew. Chem. Int. Ed. 2009, 48, 1740–1752. [Google Scholar] [CrossRef]
- Panza, F.; Seripa, D.; Solfrizzi, V.; Imbimbo, B.P.; Santamato, A.; Lozupone, M.; Capozzo, R.; Prete, C.; Pilotto, A.; Greco, A.; et al. Tau aggregation inhibitors: The future of Alzheimer’s pharmacotherapy? Expert Opin. Pharmacother. 2016, 17, 457–461. [Google Scholar] [CrossRef][Green Version]
- Hochgrafe, K.; Sydow, A.; Matenia, D.; Cadinu, D.; Konen, S.; Petrova, O.; Pickhardt, M.; Goll, P.; Morellini, F.; Mandelkow, E.; et al. Preventive methylene blue treatment preserves cognition in mice expressing full-length pro-aggregant human Tau. Acta Neuropathol. Commun. 2015, 3, 25. [Google Scholar] [CrossRef]
- Wischik, C.M.; Staff, R.T.; Wischik, D.J.; Bentham, P.; Murray, A.D.; Storey, J.M.; Kook, K.A.; Harrington, C.R. Tau aggregation inhibitor therapy: An exploratory phase 2 study in mild or moderate Alzheimer’s disease. J. Alzheimers Dis. 2015, 44, 705–720. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, S.; Feldman, H.H.; Schneider, L.S.; Wilcock, G.K.; Frisoni, G.B.; Hardlund, J.H.; Moebius, H.J.; Bentham, P.; Kook, K.A.; Wischik, D.J.; et al. Efficacy and safety of tau-aggregation inhibitor therapy in patients with mild or moderate Alzheimer’s disease: A randomised, controlled, double-blind, parallel-arm, phase 3 trial. Lancet 2016, 388, 2873–2884. [Google Scholar] [CrossRef]
- Chirita, C.; Necula, M.; Kuret, J. Ligand-dependent inhibition and reversal of tau filament formation. Biochemistry 2004, 43, 2879–2887. [Google Scholar] [CrossRef] [PubMed]
- Congdon, E.E.; Necula, M.; Blackstone, R.D.; Kuret, J. Potency of a tau fibrillization inhibitor is influenced by its aggregation state. Arch. Biochem. Biophys. 2007, 465, 127–135. [Google Scholar] [CrossRef][Green Version]
- Feng, B.Y.; Toyama, B.H.; Wille, H.; Colby, D.W.; Collins, S.R.; May, B.C.; Prusiner, S.B.; Weissman, J.; Shoichet, B.K. Small-molecule aggregates inhibit amyloid polymerization. Nat. Chem. Biol. 2008, 4, 197–199. [Google Scholar] [CrossRef]
- Pickhardt, M.; von Bergen, M.; Gazova, Z.; Hascher, A.; Biernat, J.; Mandelkow, E.M.; Mandelkow, E. Screening for inhibitors of tau polymerization. Curr. Alzheimer Res. 2005, 2, 219–226. [Google Scholar] [CrossRef]
- Khlistunova, I.; Biernat, J.; Wang, Y.; Pickhardt, M.; von Bergen, M.; Gazova, Z.; Mandelkow, E.; Mandelkow, E.M. Inducible expression of Tau repeat domain in cell models of tauopathy: Aggregation is toxic to cells but can be reversed by inhibitor drugs. J. Biol. Chem. 2006, 281, 1205–1214. [Google Scholar] [CrossRef]
- Larbig, G.; Pickhardt, M.; Lloyd, D.G.; Schmidt, B.; Mandelkow, E. Screening for inhibitors of tau protein aggregation into Alzheimer paired helical filaments: A ligand based approach results in successful scaffold hopping. Curr. Alzheimer Res. 2007, 4, 315–323. [Google Scholar] [CrossRef][Green Version]
- Pickhardt, M.; Larbig, G.; Khlistunova, I.; Coksezen, A.; Meyer, B.; Mandelkow, E.M.; Schmidt, B.; Mandelkow, E. Phenylthiazolyl-hydrazide and its derivatives are potent inhibitors of tau aggregation and toxicity in vitro and in cells. Biochemistry 2007, 46, 10016–10023. [Google Scholar] [CrossRef]
- Brunden, K.R.; Trojanowski, J.Q.; Lee, V.M. Evidence that non-fibrillar tau causes pathology linked to neurodegeneration and behavioral impairments. J. Alzheimers Dis. 2008, 14, 393–399. [Google Scholar] [CrossRef]
- Caccamo, A.; Magri, A.; Medina, D.X.; Wisely, E.V.; Lopez-Aranda, M.F.; Silva, A.J.; Oddo, S. mTOR regulates tau phosphorylation and degradation: Implications for Alzheimer’s disease and other tauopathies. Aging Cell 2013, 12, 370–380. [Google Scholar] [CrossRef] [PubMed]
- Majumder, S.; Richardson, A.; Strong, R.; Oddo, S. Inducing autophagy by rapamycin before, but not after, the formation of plaques and tangles ameliorates cognitive deficits. PLoS ONE 2011, 6, e25416. [Google Scholar] [CrossRef] [PubMed]
- Chresta, C.M.; Davies, B.R.; Hickson, I.; Harding, T.; Cosulich, S.; Critchlow, S.E.; Vincent, J.P.; Ellston, R.; Jones, D.; Sini, P.; et al. AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res. 2010, 70, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Nixon, R.A. Autophagy, amyloidogenesis and Alzheimer disease. J. Cell Sci. 2007, 120, 4081–4091. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.J.; Chandswangbhuvana, D.; Margol, L.; Glabe, C.G. Loss of endosomal/lysosomal membrane impermeability is an early event in amyloid Abeta1-42 pathogenesis. J. Neurosci. Res. 1998, 52, 691–698. [Google Scholar] [CrossRef]
- Cortes, C.J.; La Spada, A.R. TFEB dysregulation as a driver of autophagy dysfunction in neurodegenerative disease: Molecular mechanisms, cellular processes, and emerging therapeutic opportunities. Neurobiol. Dis. 2019, 122, 83–93. [Google Scholar] [CrossRef]
- Luo, W.; Dou, F.; Rodina, A.; Chip, S.; Kim, J.; Zhao, Q.; Moulick, K.; Aguirre, J.; Wu, N.; Greengard, P.; et al. Roles of heat-shock protein 90 in maintaining and facilitating the neurodegenerative phenotype in tauopathies. Proc. Natl. Acad. Sci. USA 2007, 104, 9511–9516. [Google Scholar] [CrossRef]
- Guo, X.; Wang, X.; Wang, Z.; Banerjee, S.; Yang, J.; Huang, L.; Dixon, J.E. Site-specific proteasome phosphorylation controls cell proliferation and tumorigenesis. Nat. Cell Biol. 2016, 18, 202–212. [Google Scholar] [CrossRef]
- Ranek, M.J.; Terpstra, E.J.; Li, J.; Kass, D.A.; Wang, X. Protein kinase g positively regulates proteasome-mediated degradation of misfolded proteins. Circulation 2013, 128, 365–376. [Google Scholar] [CrossRef]
- Lee, B.H.; Lu, Y.; Prado, M.A.; Shi, Y.; Tian, G.; Sun, S.; Elsasser, S.; Gygi, S.P.; King, R.W.; Finley, D. USP14 deubiquitinates proteasome-bound substrates that are ubiquitinated at multiple sites. Nature 2016, 532, 398–401. [Google Scholar] [CrossRef]
- Boselli, M.; Lee, B.H.; Robert, J.; Prado, M.A.; Min, S.W.; Cheng, C.; Silva, M.C.; Seong, C.; Elsasser, S.; Hatle, K.M.; et al. An inhibitor of the proteasomal deubiquitinating enzyme USP14 induces tau elimination in cultured neurons. J. Biol. Chem. 2017, 292, 19209–19225. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.H.; Lee, M.J.; Park, S.; Oh, D.C.; Elsasser, S.; Chen, P.C.; Gartner, C.; Dimova, N.; Hanna, J.; Gygi, S.P.; et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 2010, 467, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Osta, A.; Cuadrado-Tejedor, M.; Garcia-Barroso, C.; Oyarzabal, J.; Franco, R. Phosphodiesterases as therapeutic targets for Alzheimer’s disease. ACS Chem. Neurosci. 2012, 3, 832–844. [Google Scholar] [CrossRef] [PubMed]
- Bittar, A.; Bhatt, N.; Kayed, R. Advances and considerations in AD tau-targeted immunotherapy. Neurobiol. Dis. 2020, 134, 104707. [Google Scholar] [CrossRef]
- Sandusky-Beltran, L.A.; Sigurdsson, E.M. Tau immunotherapies: Lessons learned, current status and future considerations. Neuropharmacology 2020, 175, 108104. [Google Scholar] [CrossRef]
- Yu, Y.J.; Watts, R.J. Developing therapeutic antibodies for neurodegenerative disease. Neurotherapeutics 2013, 10, 459–472. [Google Scholar] [CrossRef]
- Hickman, D.T.; Lopez-Deber, M.P.; Ndao, D.M.; Silva, A.B.; Nand, D.; Pihlgren, M.; Giriens, V.; Madani, R.; St-Pierre, A.; Karastaneva, H.; et al. Sequence-independent control of peptide conformation in liposomal vaccines for targeting protein misfolding diseases. J. Biol. Chem. 2011, 286, 13966–13976. [Google Scholar] [CrossRef]
- Theunis, C.; Crespo-Biel, N.; Gafner, V.; Pihlgren, M.; Lopez-Deber, M.P.; Reis, P.; Hickman, D.T.; Adolfsson, O.; Chuard, N.; Ndao, D.M.; et al. Efficacy and safety of a liposome-based vaccine against protein Tau, assessed in tau.P301L mice that model tauopathy. PLoS ONE 2013, 8, e72301. [Google Scholar] [CrossRef]
- Paholikova, K.; Salingova, B.; Opattova, A.; Skrabana, R.; Majerova, P.; Zilka, N.; Kovacech, B.; Zilkova, M.; Barath, P.; Novak, M. N-terminal truncation of microtubule associated protein tau dysregulates its cellular localization. J. Alzheimers Dis. 2015, 43, 915–926. [Google Scholar] [CrossRef]
- Lee, S.H.; Le Pichon, C.E.; Adolfsson, O.; Gafner, V.; Pihlgren, M.; Lin, H.; Solanoy, H.; Brendza, R.; Ngu, H.; Foreman, O.; et al. Antibody-Mediated Targeting of Tau in Vivo Does Not Require Effector Function and Microglial Engagement. Cell Rep. 2016, 16, 1690–1700. [Google Scholar] [CrossRef]
- Sopko, R.; Golonzhka, O.; Arndt, J.; Quan, C.; Czerkowicz, J.; Cameron, A.; Smith, B.; Murugesan, Y.; Gibbons, G.; Kim, S.J.; et al. Characterization of tau binding by gosuranemab. Neurobiol. Dis. 2020, 146, 105120. [Google Scholar] [CrossRef] [PubMed]
- Braak, H.; Del Tredici, K. The pathological process underlying Alzheimer’s disease in individuals under thirty. Acta Neuropathol. 2011, 121, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Kfoury, N.; Holmes, B.B.; Jiang, H.; Holtzman, D.M.; Diamond, M.I. Trans-cellular propagation of Tau aggregation by fibrillar species. J. Biol. Chem. 2012, 287, 19440–19451. [Google Scholar] [CrossRef]
- Gu, J.; Congdon, E.E.; Sigurdsson, E.M. Two novel Tau antibodies targeting the 396/404 region are primarily taken up by neurons and reduce Tau protein pathology. J. Biol. Chem. 2013, 288, 33081–33095. [Google Scholar] [CrossRef]
- Pedersen, J.T.; Sigurdsson, E.M. Tau immunotherapy for Alzheimer’s disease. Trends Mol. Med. 2015, 21, 394–402. [Google Scholar] [CrossRef] [PubMed]
- Novak, P.; Schmidt, R.; Kontsekova, E.; Zilka, N.; Kovacech, B.; Skrabana, R.; Vince-Kazmerova, Z.; Katina, S.; Fialova, L.; Prcina, M.; et al. Safety and immunogenicity of the tau vaccine AADvac1 in patients with Alzheimer’s disease: A randomised, double-blind, placebo-controlled, phase 1 trial. Lancet Neurol. 2017, 16, 123–134. [Google Scholar] [CrossRef]
- Sakamoto, K.M.; Kim, K.B.; Kumagai, A.; Mercurio, F.; Crews, C.M.; Deshaies, R.J. Protacs: Chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. USA 2001, 98, 8554–8559. [Google Scholar] [CrossRef]
- Cromm, P.M.; Samarasinghe, K.T.G.; Hines, J.; Crews, C.M. Addressing Kinase-Independent Functions of Fak via PROTAC-Mediated Degradation. J. Am. Chem. Soc. 2018. [Google Scholar] [CrossRef]
- Gechijian, L.N.; Buckley, D.L.; Lawlor, M.A.; Reyes, J.M.; Paulk, J.; Ott, C.J.; Winter, G.E.; Erb, M.A.; Scott, T.G.; Xu, M.; et al. Functional TRIM24 degrader via conjugation of ineffectual bromodomain and VHL ligands. Nat. Chem. Biol. 2018, 14, 405–412. [Google Scholar] [CrossRef]
- Chamberlain, P.P.; Lopez-Girona, A.; Miller, K.; Carmel, G.; Pagarigan, B.; Chie-Leon, B.; Rychak, E.; Corral, L.G.; Ren, Y.J.; Wang, M.; et al. Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs. Nat. Struct. Mol. Biol. 2014, 21, 803–809. [Google Scholar] [CrossRef]
- Collins, I.; Wang, H.; Caldwell, J.J.; Chopra, R. Chemical approaches to targeted protein degradation through modulation of the ubiquitin-proteasome pathway. Biochem. J. 2017, 474, 1127–1147. [Google Scholar] [CrossRef] [PubMed]
- Fischer, E.S.; Bohm, K.; Lydeard, J.R.; Yang, H.; Stadler, M.B.; Cavadini, S.; Nagel, J.; Serluca, F.; Acker, V.; Lingaraju, G.M.; et al. Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature 2014, 512, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Kronke, J.; Udeshi, N.D.; Narla, A.; Grauman, P.; Hurst, S.N.; McConkey, M.; Svinkina, T.; Heckl, D.; Comer, E.; Li, X.; et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 2014, 343, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Middleton, R.E.; Sun, H.; Naniong, M.; Ott, C.J.; Mitsiades, C.S.; Wong, K.K.; Bradner, J.E.; Kaelin, W.G., Jr. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science 2014, 343, 305–309. [Google Scholar] [CrossRef]
- Metzger, M.B.; Pruneda, J.N.; Klevit, R.E.; Weissman, A.M. RING-type E3 ligases: Master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta 2014, 1843, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Bondeson, D.P.; Mares, A.; Smith, I.E.; Ko, E.; Campos, S.; Miah, A.H.; Mulholland, K.E.; Routly, N.; Buckley, D.L.; Gustafson, J.L.; et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat. Chem. Biol. 2015, 11, 611–617. [Google Scholar] [CrossRef]
- Churcher, I. Protac-Induced Protein Degradation in Drug Discovery: Breaking the Rules or Just Making New Ones? J. Med. Chem. 2018, 61, 444–452. [Google Scholar] [CrossRef]
- Lu, M.; Liu, T.; Jiao, Q.; Ji, J.; Tao, M.; Liu, Y.; You, Q.; Jiang, Z. Discovery of a Keap1-dependent peptide PROTAC to knockdown Tau by ubiquitination-proteasome degradation pathway. Eur. J. Med. Chem. 2018, 146, 251–259. [Google Scholar] [CrossRef]
- Silva, M.C.; Ferguson, F.M.; Cai, Q.; Donovan, K.A.; Nandi, G.; Patnaik, D.; Zhang, T.; Huang, H.T.; Lucente, D.E.; Dickerson, B.C.; et al. Targeted degradation of aberrant tau in frontotemporal dementia patient-derived neuronal cell models. Elife 2019, 8. [Google Scholar] [CrossRef]
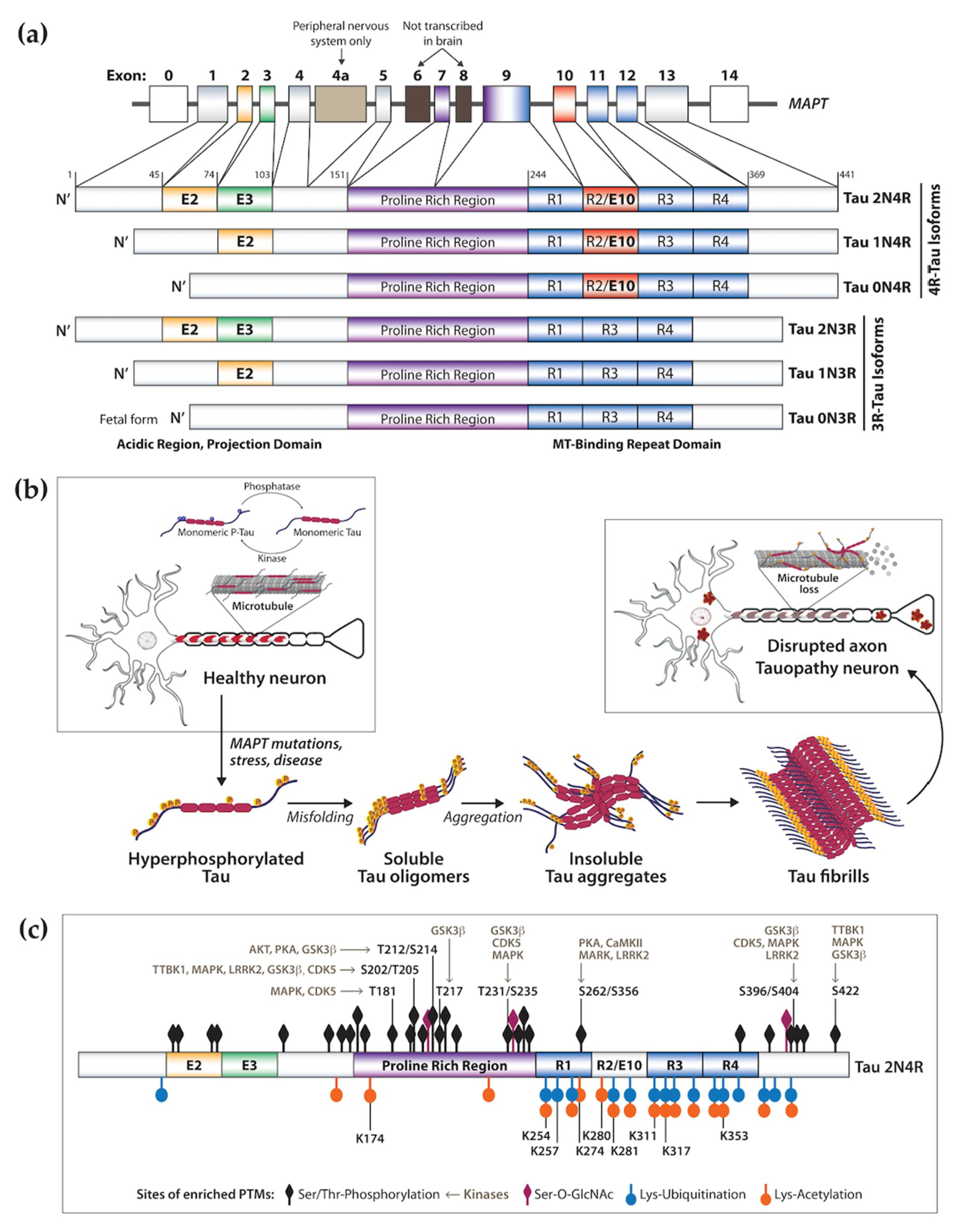
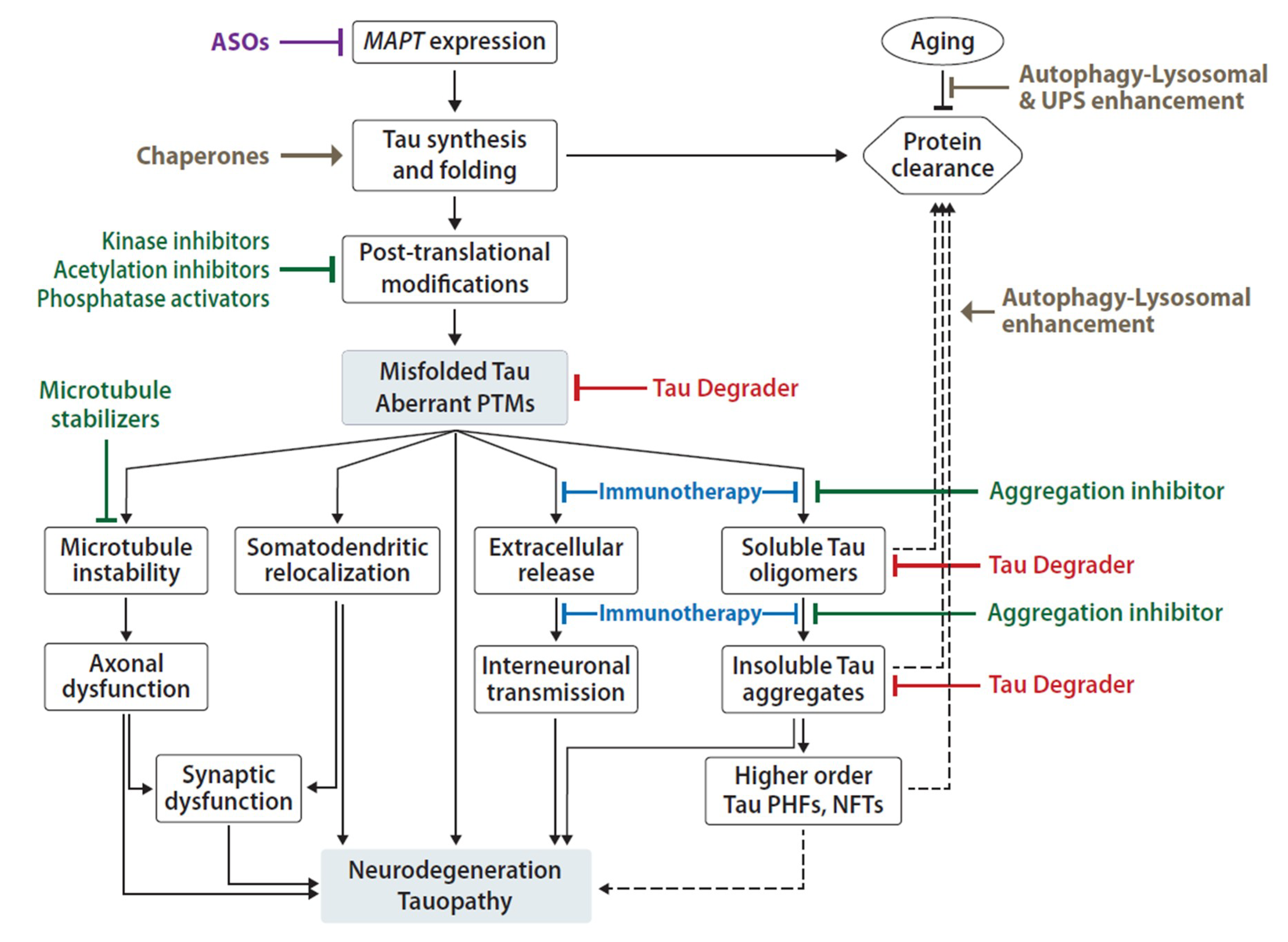
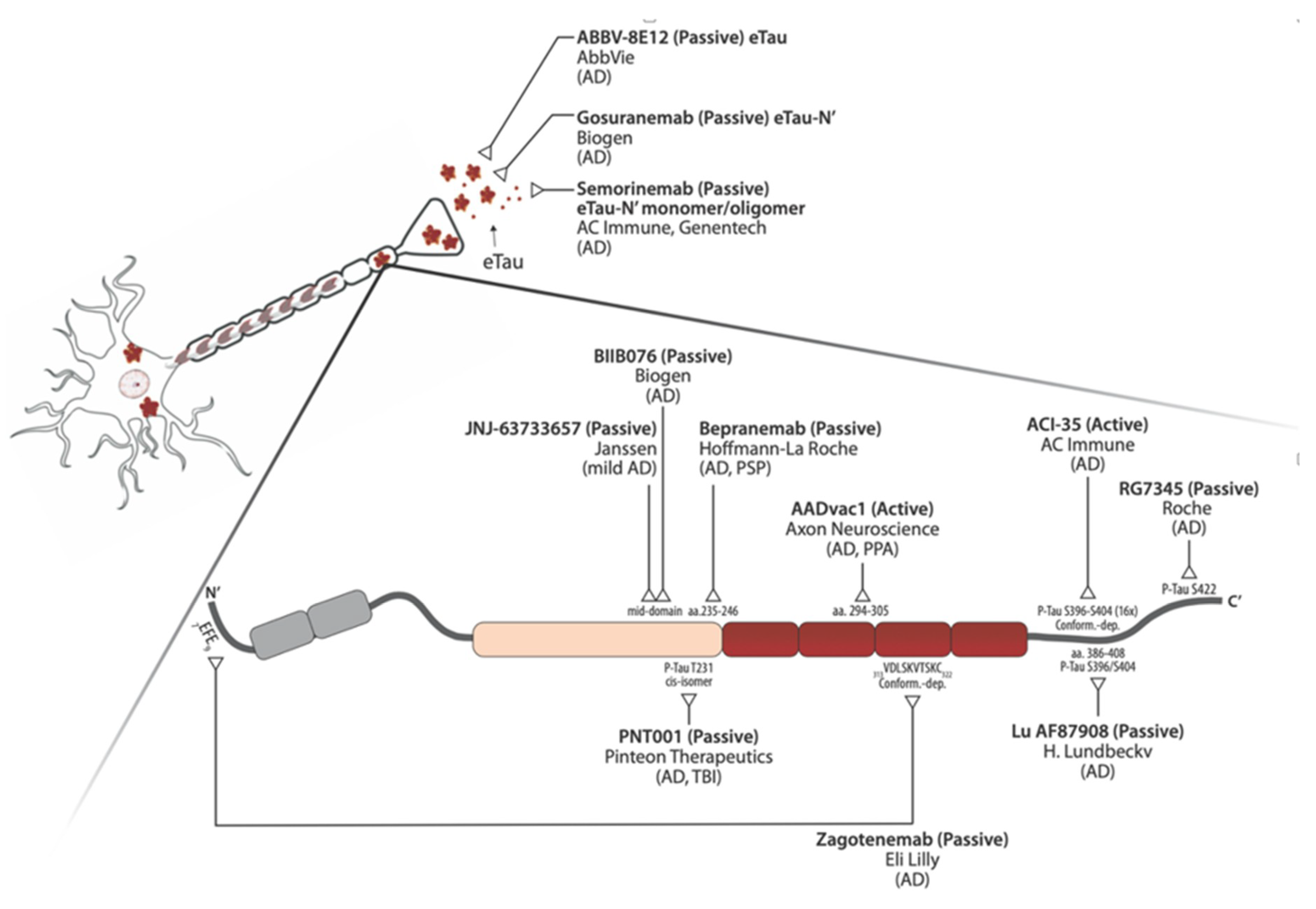
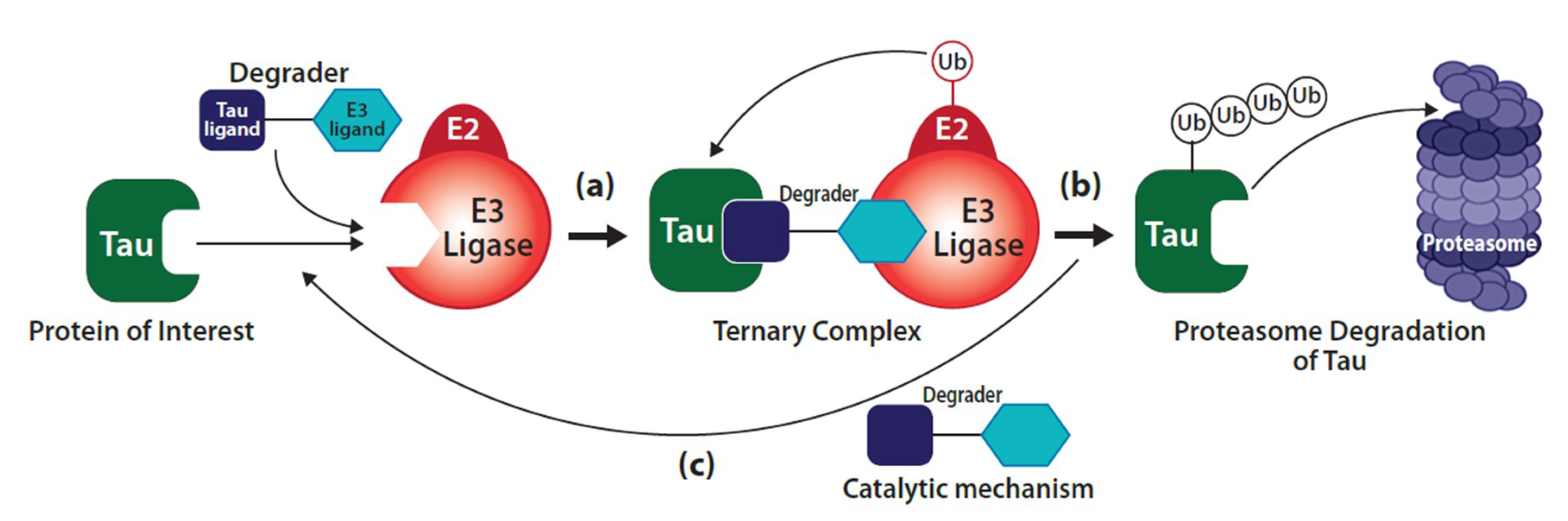
| Clinical | Symptomology | Tau | Neuronal Pathology | Glia Pathology | Affected Brain Regions | |
|---|---|---|---|---|---|---|
| Primary Tauopathy | Pick’s disease (PiD) | Behavioral change, social disinhibition, eating disorder, absent/late parkinsonism. | 3R | Round cytoplasmic inclusions (Pick bodies), rare NFTs. | Ramified astrocytes. | Dentate gyrus of the hippocampus, frontal and temporal neocortical layers II, IV. Frontal, insular and anterior temporal cortices. |
| Behavioral variant of FTD (bvFTD) | Behavioral disinhibition, apathy, empathy loss, compulsiveness, executive and cognitive dysfunction. | 3R > 4R | Cytoplasmic NFTs, short dystrophic neurites. | Orbitofrontal, dorsolateral prefrontal, medial prefrontal cortices. Subcortical brain nuclei. Temporal-parietal lobes. | ||
| Progressive supranuclear palsy (PSP) | Apathy, anxiety, sleep disturbance. Spectrum from pure motor to pure cognitive presentations. | 4R | NFTs, neuropile threads. | Tufted astrocytes, somatodendritic coiled bodies. | Subthalamic nucleus, basal ganglia, brainstem. Posterior mesencephalic cortex. | |
| Corticobasal syndrome (CBS) | Asymmetric motor symptoms, apraxia, sensory impairment. Spectrum from pure motor to pure cognitive presentation. | 4R | NFTs, neuropile threads, ballooned neurons, pleomorphic inclusions (pre-tangles). | Annular clusters of short fuzzy cell processes, astrocytic Tau plaques, argyrophilic inclusions. | Frontoparietal cortex, striatum, substantia nigra. | |
| Argyrophilic grain disease (AGD) | Personality change, emotional imbalance, memory failure. | 4R | Argyrophilic grains, dendritic straight filaments and smooth tubules. | Thorn-shaped astrocytes, coiled bodies. | Medial temporal lobe, entorhinal cortex, hippocampus, amygdala. | |
| Aging-related Tau astrogliopathy (ARTAG) | Cognitive decline. | 4R | - | Thorn-shaped and granular-fuzzy astrocytes. | Medial temporal lobe, lobar (frontal, parietal, occipital, lateral temporal), subcortical, brainstem. | |
| Globular glial tauopathy (GGT) | Behavior change, cognitive decline, motor neuron disease (Parkinsonism). | 4R | - | Globular inclusions in astrocytes and oligodendrocytes. | White matter, limbic and isocortical regions. Hippocampus. | |
| Primary progressive aphasia (PPA) | Language deterioration, loss of semantic memory. | 3R, 4R | NFTs, amyloid plaques | Globular astrocytic inclusions. | Anterior and temporal lobes, parietal lobe. Frontoinsular cortex | |
| Primary age-related tauopathy (PART) | Cognitive impairment. | 3R, 4R | NFTs, neuropile threads | Medial temporal lobe. | Medial temporal lobe. | |
| Tangle-only dementia (TOD) | Late-onset dementia. | 3R, 4R | Intracellular PHFs, NFTs and neuropil threads. | Hippocampus. | ||
| Secondary Tauopathy | Alzheimer’s disease (AD) | Memory loss, cognitive dysfunction, social behavior changes. | 3R, 4R | NFTs, neuropile threads, neuritic plaques. | Entorhinal cortex, hippocampus, cerebral cortex. | |
| Chronic traumatic encephalop-athy (CTE) | Memory loss, confusion, personality/behavior changes. Motor decline. | 3R, 4R | P-Tau aggregates around small vessels, TDP-43 cytoplasmic inclusions. | P-Tau aggregates around small vessels. | Cortical sulci, isocortex layers II–III, hippocampus, subcortical nuclei. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, M.C.; Haggarty, S.J. Tauopathies: Deciphering Disease Mechanisms to Develop Effective Therapies. Int. J. Mol. Sci. 2020, 21, 8948. https://doi.org/10.3390/ijms21238948
Silva MC, Haggarty SJ. Tauopathies: Deciphering Disease Mechanisms to Develop Effective Therapies. International Journal of Molecular Sciences. 2020; 21(23):8948. https://doi.org/10.3390/ijms21238948
Chicago/Turabian StyleSilva, M. Catarina, and Stephen J. Haggarty. 2020. "Tauopathies: Deciphering Disease Mechanisms to Develop Effective Therapies" International Journal of Molecular Sciences 21, no. 23: 8948. https://doi.org/10.3390/ijms21238948
APA StyleSilva, M. C., & Haggarty, S. J. (2020). Tauopathies: Deciphering Disease Mechanisms to Develop Effective Therapies. International Journal of Molecular Sciences, 21(23), 8948. https://doi.org/10.3390/ijms21238948





