The Role of Histone Acetylation-/Methylation-Mediated Apoptotic Gene Regulation in Hepatocellular Carcinoma
Abstract
1. Introduction
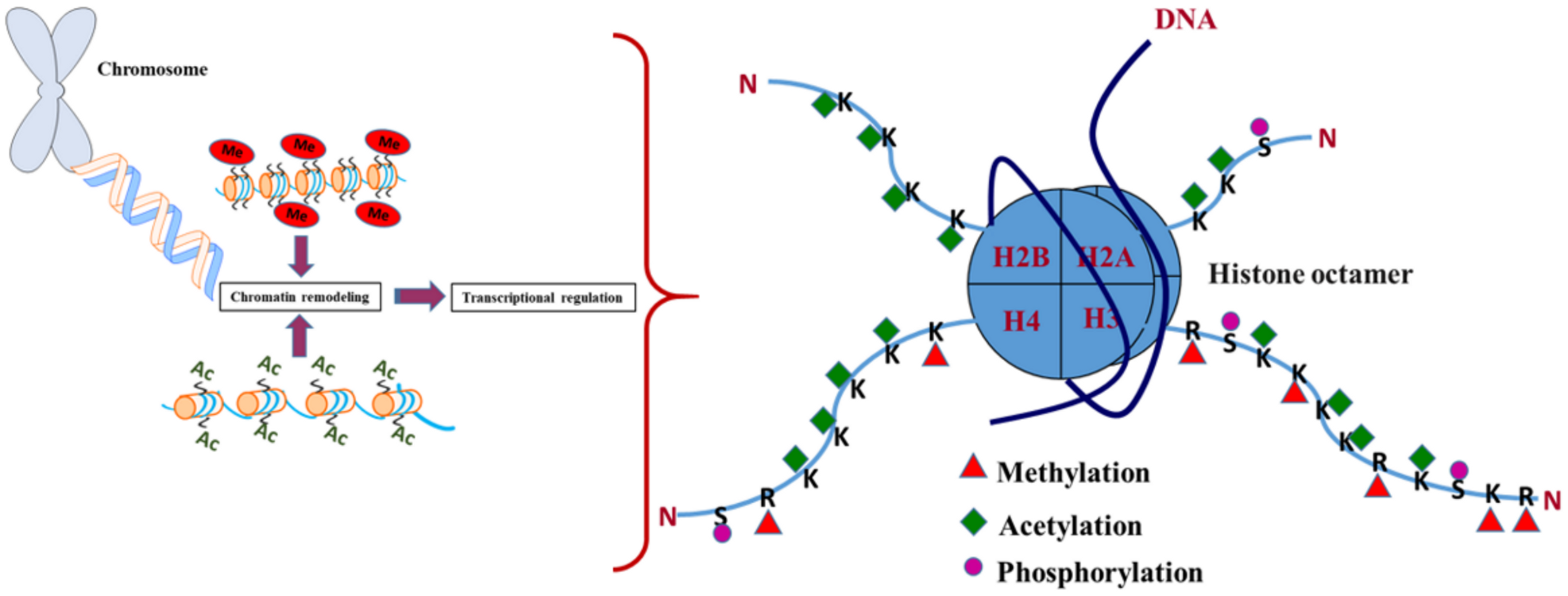
2. Histone Modifications and Its Biological Importance
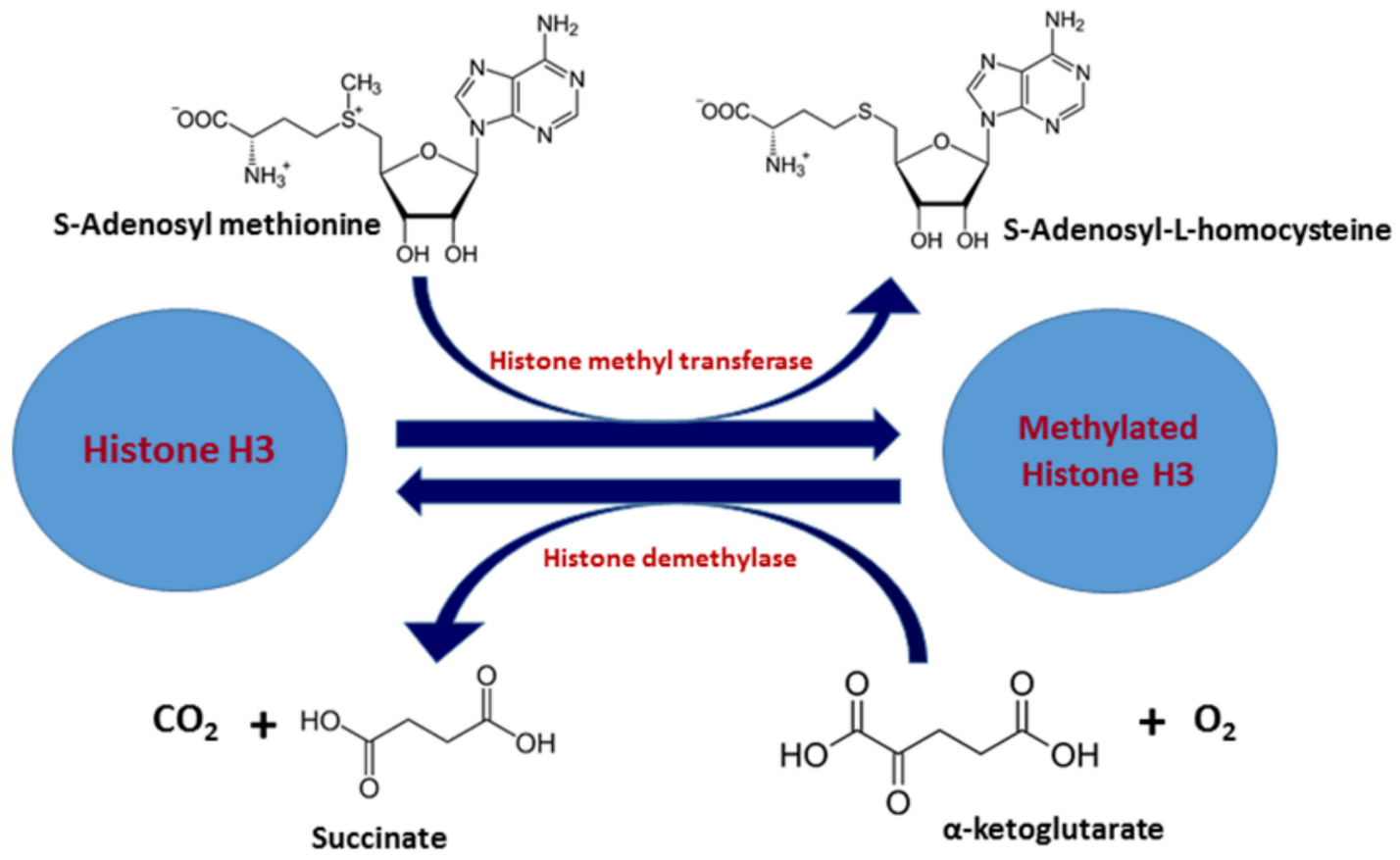
2.1. Histone Methylation: Epigenetic Mechanism and Factors Involved
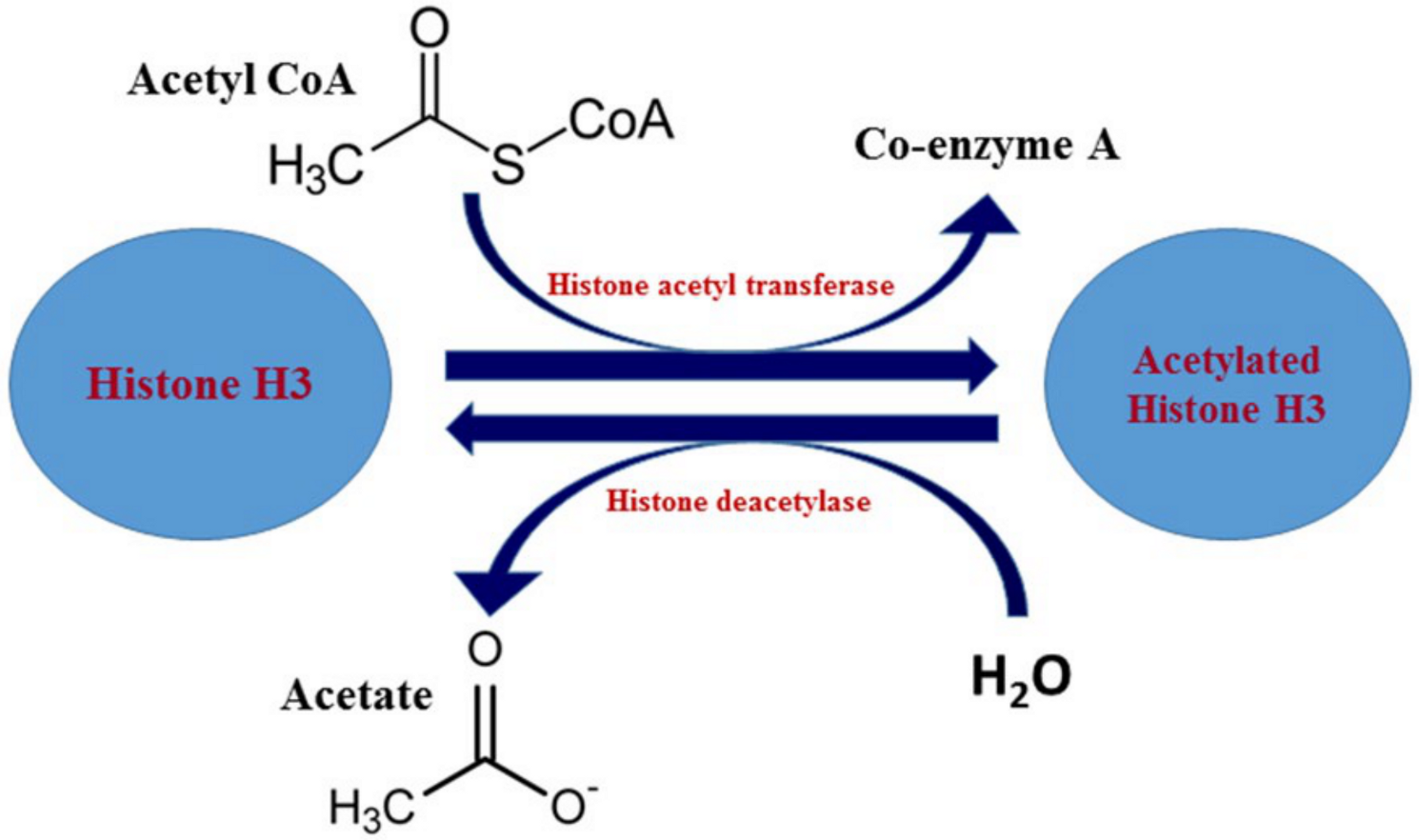
2.2. Histone Acetylation: Epigenetic Mechanism and Factors Involved
| Classification | Family/Type | Enzymes | References | ||
|---|---|---|---|---|---|
| HMTs | PRMTs | TYPE I | PRMT 1, PRMT 3, PRMT 4/CRM1, PRMT-6, PRMT-8 | [42,43,44,55,56] | |
| TYPE II | PRMT 5, PRMT 9/FBXO11 | [57] | |||
| TYPE III | PRMT7 | [57] | |||
| KMTs | SET | SET I | EZH I, H3K27 | [46,47] | |
| SET 2 | NSD1-3, SETD2, SMYD2 | [58,59,60,61] | |||
| SUV39 | SUV39H1, SUV39H2, G9a GLP, ESET//SETDB1 CLLL8/SETDB2 | [62,63,64,65,66] | |||
| RIZ | RIZ 1, BLIMP1/PRDM1 PFM1/CRS2 | [67] | |||
| Nongroup | SET7/9, SET8, SUV4-20H1 SUV4-20H2 | [61,68] | |||
| Seven-β-strand (7BS) | Dot1/DOT1L | [49] | |||
| HDMs | KDM1 | KDM1A, KDM1B | [36] | ||
| JMJC | KDM2-7/8 | [51,52] | |||
| HATs | GNAT | KAT2A, KAT2B | [69,70] | ||
| MYST | KAT7, KAT8, KAT5, KAT6A KAT6B | [71,72] | |||
| p300/CBP | KAT3B | [73] | |||
| Transcription coactivators | KAT4, KAT12 | [74] | |||
| Steroid receptor | KAT13A, KAT13B KAT13C, KAT13D | [74] | |||
| Cytoplasmic | HAT1, HAT4 | [75] | |||
| HDACs | CLASS 1 | HDAC1, HDAC2 HDAC3, HDAC8 | [76,77,78,79] | ||
| CLASS II a | HDAC4, HDAC5 HDAC7, HDAC9 | [80,81,82,83] | |||
| CLASS II b | HDAC6, HDAC10 | [84] | |||
| CLASS III | Sirtuins (SIRT 1-7) | [85] | |||
| CLASS IV | HDAC 11 | [86] | |||
3. Regulatory Mechanism of Apoptosis by Histone Acetylation/Methylation
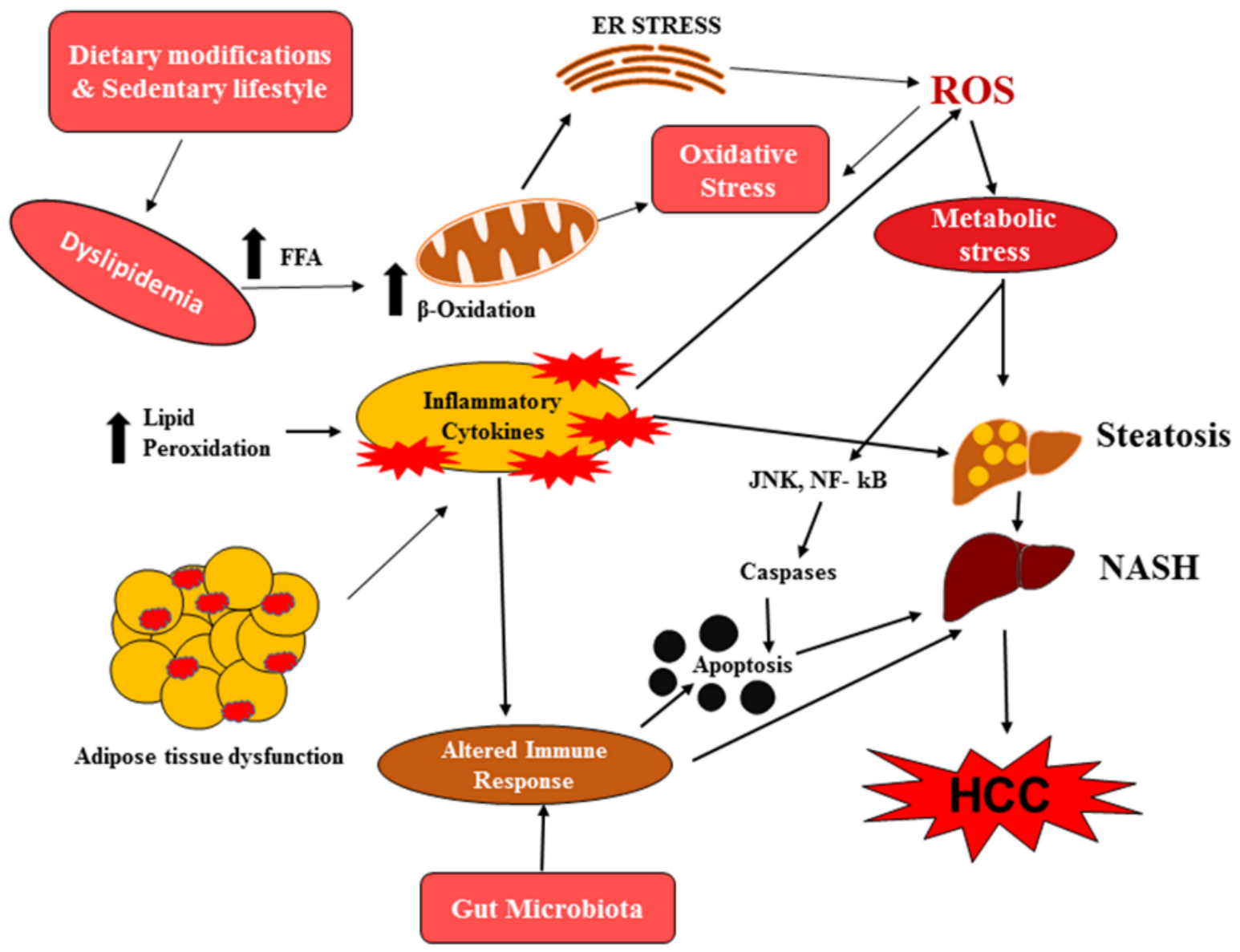
4. Role of Histone Acetylation-/Methylation-Mediated Apoptosis in NAFLD
5. Role of Histone Acetylation-/Methylation-Mediated Apoptosis in HCC
Significance of Histone Acetylation/Methylation Ratio in HCC
6. Epigenetic Therapeutic Implications for HCC
7. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| HATs | Histone acetyltransferases |
| HCC | Hepatocellular carcinoma |
| HDACis | Histone deacetylase inhibitors |
| HDACs | Histone deacetylases |
| HDMs | Histone demethylases |
| HMTs | Histone methyltransferases |
| NAFLD | Nonalcoholic fatty liver disease |
| NASH | Nonalcoholic steatohepatitis |
| PRMTs | Protein arginine methyltransferases |
| PTM | Post-translational modification |
References
- Bird, A. Perceptions of epigenetics. Nature 2007, 447, 396–398. [Google Scholar] [CrossRef]
- Yao, B.; Jin, P. Unlocking epigenetic codes in neurogenesis. Genes Dev. 2014, 28, 1253–1271. [Google Scholar] [CrossRef]
- Blum, R. Stepping inside the realm of epigenetic modifiers. Biomol. Concepts 2015, 6, 119–136. [Google Scholar] [CrossRef]
- Jobe, E.M.; McQuate, A.L.; Zhao, X. Crosstalk among Epigenetic Pathways Regulates Neurogenesis. Front. Behav. Neurosci. 2012, 6, 59. [Google Scholar] [CrossRef] [PubMed]
- Khorasanizadeh, S. The nucleosome: From genomic organization to genomic regulation. Cell 2004, 116, 259–272. [Google Scholar] [CrossRef]
- Côrte-Real, M.; Madeo, F. Yeast Programed Cell Death and Aging. Front. Oncol. 2013, 3, 283. [Google Scholar]
- Dong, Z.-Y.; Zhou, Y.-R.; Wang, L.-X. HDAC1 is indirectly involved in the epigenetic regulation of p38 MAPK that drive the lung cancer progression. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 5980–5986. [Google Scholar]
- Füllgrabe, J.; Hajji, N.; Joseph, B. Cracking the death code: Apoptosis-related histone modifications. Cell Death Differ. 2010, 17, 1238–1243. [Google Scholar] [CrossRef]
- Tan, J.; Yang, X.; Zhuang, L.; Jiang, X.; Chen, W.; Lee, P.L.; Karuturi, R.M.; Tan, P.B.O.; Liu, E.T.; Yu, Q. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007, 21, 1050–1063. [Google Scholar] [CrossRef]
- Cholankeril, G.; Patel, R.; Khurana, S.; Satapathy, S.K. Hepatocellular carcinoma in non-alcoholic steatohepatitis: Current knowledge and implications for management. World J. Hepatol. 2017, 9, 533–543. [Google Scholar] [CrossRef]
- El-Serag, H.B.; Rudolph, K.L. Hepatocellular Carcinoma: Epidemiology and Molecular Carcinogenesis. Gastroenterology 2007, 132, 2557–2576. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.M.; Blissett, D.; Blissett, R.; Henry, L.; Stepanova, M.; Younossi, Y.; Racila, A.; Hunt, S.; Beckerman, R. The economic and clinical burden of nonalcoholic fatty liver disease in the United States and Europe. Hepatology 2016, 64, 1577–1586. [Google Scholar] [CrossRef] [PubMed]
- Michelotti, G.A.; Machado, M.V.; Diehl, A.M. NAFLD, NASH and liver cancer. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Chalasani, N.; Younossi, Z.; LaVine, J.E.; Charlton, M.; Cusi, K.; Rinella, M.; Harrison, S.A.; Brunt, E.M.; Sanyal, A.J. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Hepatology 2018, 67, 328–357. [Google Scholar] [CrossRef] [PubMed]
- Pogribny, I.P.; Rusyn, I. Role of epigenetic aberrations in the development and progression of human hepatocellular carcinoma. Cancer Lett. 2014, 342, 223–230. [Google Scholar] [CrossRef]
- Zucman-Rossi, J.; Villanueva, A.; Nault, J.-C.; Llovet, J.M. Genetic Landscape and Biomarkers of Hepatocellular Carcinoma. Gastroenterology 2015, 149, 1226–1239.e4. [Google Scholar] [CrossRef]
- Lee, J.H.; Friso, S.; Choi, S.-W. Epigenetic Mechanisms Underlying the Link between Non-Alcoholic Fatty Liver Diseases and Nutrition. Nutrients 2014, 6, 3303–3325. [Google Scholar] [CrossRef]
- Sookoian, S.; Rosselli, M.S.; Gemma, C.; Burgueño, A.L.; Gianotti, T.F.; Castaño, G.O.; Pirola, C.J. Epigenetic regulation of insulin resistance in nonalcoholic fatty liver disease: Impact of liver methylation of the peroxisome proliferator-activated receptor γ coactivator 1α promoter. Hepatology 2010, 52, 1992–2000. [Google Scholar] [CrossRef]
- Slomko, H.; Heo, H.J.; Einstein, F.H. Minireview: Epigenetics of Obesity and Diabetes in Humans. Endocrinology 2012, 153, 1025–1030. [Google Scholar] [CrossRef]
- Zimmer, V.; Lammert, F. Genetics and epigenetics in the fibrogenic evolution of chronic liver diseases. Best Pr. Res. Clin. Gastroenterol. 2011, 25, 269–280. [Google Scholar] [CrossRef]
- Chen, Z.J.; Pikaard, C.S. Epigenetic silencing of RNA polymerase I transcription: A role for DNA methylation and histone modification in nucleolar dominance. Genes Dev. 1997, 11, 2124–2136. [Google Scholar] [CrossRef]
- Dorn, E.S.; Cook, J.G. Nucleosomes in the neighborhood: New roles for chromatin modifications in replication origin control. Epigenetics 2011, 6, 552–559. [Google Scholar] [CrossRef] [PubMed]
- Jayani, R.S.; Ramanujam, P.L.; Galande, S. Studying Histone Modifications and Their Genomic Functions by Employing Chromatin Immunoprecipitation and Immunoblotting. Methods Cell Biol. 2010, 98, 35–56. [Google Scholar] [PubMed]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef] [PubMed]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef]
- Berdasco, M.; Esteller, M. Aberrant Epigenetic Landscape in Cancer: How Cellular Identity Goes Awry. Dev. Cell 2010, 19, 698–711. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Krautkramer, K.A.; Feldman, J.L.; Denu, J.M. Metabolic regulation of histone post-translational modifications. ACS Chem. Biol. 2015, 10, 95–108. [Google Scholar] [CrossRef]
- Mellor, K.M.; Brimble, M.A.; Delbridge, L.M. Glucose as an agent of post-translational modification in diabetes—New cardiac epigenetic insights. Life Sci. 2015, 129, 48–53. [Google Scholar] [CrossRef]
- Pena-Altamira, L.; Polazzi, E.; Monti, B.; Peña-Altamira, E. Histone Post-translational Modifications in Huntington’s and Parkinson’s Diseases. Curr. Pharm. Des. 2013, 19, 5085–5092. [Google Scholar] [CrossRef]
- Michalak, E.M.; Burr, M.L.; Bannister, A.J.; Dawson, M.A. The roles of DNA, RNA and histone methylation in ageing and cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 573–589. [Google Scholar] [CrossRef]
- Schreiber, S.L.; Bernstein, B.E. Signaling network model of chromatin. Cell 2002, 111, 771–778. [Google Scholar] [CrossRef]
- Turner, B.M. Cellular memory and the histone code. Cell 2002, 111, 285–291. [Google Scholar] [CrossRef]
- Bannister, A.J.; Schneider, R.; Kouzarides, T. Histone methylation: Dynamic or static? Cell 2002, 109, 801–806. [Google Scholar] [CrossRef]
- Cheung, P.; Lau, P. Epigenetic regulation by histone methylation and histone variants. Mol. Endocrinol. 2005, 19, 563–573. [Google Scholar] [CrossRef]
- Zhang, Y. Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes Dev. 2001, 15, 2343–2360. [Google Scholar] [CrossRef] [PubMed]
- Kaniskan, H.Ü.; Martini, M.L.; Jin, J. Inhibitors of Protein Methyltransferases and Demethylases. Chem. Rev. 2018, 118, 989–1068. [Google Scholar] [CrossRef]
- Deguchi, T.; Barchas, J. Inhibition of transmethylations of biogenic amines by S-adenosylhomocysteine. Enhancement of transmethylation by adenosylhomocysteinase. J. Biol. Chem. 1971, 246, 3175–3181. [Google Scholar] [PubMed]
- Schubeler, D. Function and information content of DNA methylation. Nature 2015, 517, 321–326. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, Z.; Szyf, M. S-adenosyl-methionine (SAM) alters the transcriptome and methylome and specifically blocks growth and invasiveness of liver cancer cells. Oncotarget 2017, 8, 111866–111881. [Google Scholar] [CrossRef]
- McBride, A.E.; Silver, P.A. State of the arg: Protein methylation at arginine comes of age. Cell 2001, 106, 5–8. [Google Scholar] [CrossRef]
- Stallcup, M.R. Role of protein methylation in chromatin remodeling and transcriptional regulation. Oncogene 2001, 20, 3014–3020. [Google Scholar] [CrossRef] [PubMed]
- Bedford, M.T. Arginine methylation at a glance. J. Cell Sci. 2007, 120, 4243–4246. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Sif, S. Interplay between chromatin remodelers and protein arginine methyltransferases. J. Cell Physiol. 2007, 213, 306–315. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.C.; Denu, J.M. Chemical mechanisms of histone lysine and arginine modifications. Biochim. Biophys. Acta 2009, 1789, 45–57. [Google Scholar] [CrossRef]
- Weiss, V.H.; E McBride, A.; A Soriano, M.; Filman, D.J.; A Silver, P.; Hogle, J.M. The structure and oligomerization of the yeast arginine methyltransferase, Hmt1. Nat. Struct. Biol. 2000, 7, 1165–1171. [Google Scholar] [CrossRef]
- Alvarez-Venegas, R.; Avramova, Z. SET-domain proteins of the Su(var)3-9, E(z) and trithorax families. Gene 2002, 285, 25–37. [Google Scholar] [CrossRef]
- Jenuwein, T. The epigenetic magic of histone lysine methylation. FEBS J. 2006, 273, 3121–3135. [Google Scholar] [CrossRef]
- Ng, H.-H.; Xu, R.-M.; Zhang, Y.; Struhl, K. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 2002, 277, 34655–34657. [Google Scholar] [CrossRef]
- Okada, Y.; Feng, Q.; Lin, Y.; Jiang, Q.; Li, Y.; Coffield, V.M.; Su, L.; Xu, G.; Zhang, Y. hDOT1L links histone methylation to leukemogenesis. Cell 2005, 121, 167–178. [Google Scholar] [CrossRef]
- Van Leeuwen, F.; Gafken, P.R.; Gottschling, D.E. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 2002, 109, 745–756. [Google Scholar] [CrossRef]
- Bennani-Baiti, I.M. Integration of ERalpha-PELP1-HER2 signaling by LSD1 (KDM1A/AOF2) offers combinatorial therapeutic opportunities to circumventing hormone resistance in breast cancer. Breast Cancer Res. 2012, 14, 112. [Google Scholar] [CrossRef] [PubMed]
- Hyun, K.; Jeon, J.; Park, K.; Kim, J. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 2017, 49, e324. [Google Scholar] [CrossRef] [PubMed]
- Braunstein, M.; Rose, A.B.; Holmes, S.G.; Allis, C.D.; Broach, J.R. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993, 7, 592–604. [Google Scholar] [CrossRef] [PubMed]
- Kristjuhan, A.; Walker, J.; Suka, N.; Grunstein, M.; Roberts, D.; Cairns, B.R.; Svejstrup, J.Q. Transcriptional inhibition of genes with severe histone h3 hypoacetylation in the coding region. Mol. Cell 2002, 10, 925–933. [Google Scholar] [CrossRef]
- Frankel, A.; Yadav, N.; Lee, J.; Branscombe, T.L.; Clarke, S.; Bedford, M.T. The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J. Biol. Chem. 2002, 277, 3537–3543. [Google Scholar] [CrossRef]
- Chen, D.; Ma, H.; Hong, H.; Koh, S.S.; Huang, S.-M.; Schurter, B.T.; Aswad, D.W.; Stallcup, M.R. Regulation of transcription by a protein methyltransferase. Science 1999, 284, 2174–2177. [Google Scholar] [CrossRef]
- Branscombe, T.L.; Frankel, A.; Lee, J.-H.; Cook, J.R.; Yang, Z.; Pestka, S.; Clarke, S. PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J. Biol. Chem. 2001, 276, 32971–32976. [Google Scholar] [CrossRef]
- Beck, D.B.; Oda, H.; Shen, S.S.; Reinberg, D. PR-Set7 and H4K20me1: At the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes Dev. 2012, 26, 325–337. [Google Scholar] [CrossRef]
- Huang, J.; Perez-Burgos, L.; Placek, B.J.; Sengupta, R.; Richter, M.; Dorsey, J.A.; Kubicek, S.; Opravil, S.; Jenuwein, T.; Berger, S.L. Repression of p53 activity by Smyd2-mediated methylation. Nature 2006, 444, 629–632. [Google Scholar] [CrossRef]
- Kurotaki, N.; Imaizumi, K.; Harada, N.; Masuno, M.; Kondoh, T.; Nagai, T.; Ohashi, H.; Naritomi, K.; Tsukahara, M.; Makita, Y.; et al. Haploinsufficiency of NSD1 causes Sotos syndrome. Nat. Genet. 2002, 30, 365–366. [Google Scholar] [CrossRef]
- Schotta, G.; Sengupta, R.; Kubicek, S.; Malin, S.; Kauer, M.; Callén, E.; Celeste, A.; Pagani, M.; Opravil, S.; De La Rosa-Velazquez, I.A.; et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 2008, 22, 2048–2061. [Google Scholar] [CrossRef] [PubMed]
- Irifuku, T.; Doi, S.; Sasaki, K.; Doi, T.; Nakashima, A.; Ueno, T.; Yamada, K.; Arihiro, K.; Kohno, N.; Masaki, T. Inhibition of H3K9 histone methyltransferase G9a attenuates renal fibrosis and retains klotho expression. Kidney Int. 2016, 89, 147–157. [Google Scholar] [CrossRef]
- Rea, S.; Eisenhaber, F.; O’Carroll, D.; Strahl, B.D.; Sun, Z.-W.; Schmid, M.; Opravil, S.; Mechtler, K.; Ponting, C.P.; Allis, C.D.; et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 2000, 406, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Tachibana, M.; Sugimoto, K.; Fukushima, T.; Shinkai, Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 2001, 276, 25309–25317. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yan, W.; Peng, X.; Jiang, Y.; He, L.; Peng, Y.; Chen, X.; Ye, M.; Zhuo, H. Functional Role of SUV39H1 in Human Renal Tubular Epithelial Cells Under High-glucose Ambiance. Inflammation 2018, 41, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wood, A.; Shilatifard, A. Posttranslational modifications of histones by methylation. Adv. Protein Chem. 2004, 67, 201–222. [Google Scholar] [PubMed]
- Zhang, C.; Li, H.; Wang, Y.; Liu, W.; Zhang, Q.; Zhang, T.; Zhang, X.; Han, B.; Zhou, G. Epigenetic inactivation of the tumor suppressor gene RIZ1 in hepatocellular carcinoma involves both DNA methylation and histone modifications. J. Hepatol. 2010, 53, 889–895. [Google Scholar] [CrossRef] [PubMed]
- Faundes, V.; Newman, W.G.; Bernardini, L.; Canham, N.; Clayton-Smith, J.; Dallapiccola, B.; Davies, S.J.; Demos, M.K.; Goldman, A.; Gill, H.; et al. Histone Lysine Methylases and Demethylases in the Landscape of Human Developmental Disorders. Am. J. Hum. Genet. 2018, 102, 175–187. [Google Scholar] [CrossRef]
- Cieniewicz, A.M.; Moreland, L.; Ringel, A.E.; Mackintosh, S.G.; Raman, A.; Gilbert, T.M.; Wolberger, C.; Tackett, A.J.; Taverna, S.D. The bromodomain of Gcn5 regulates site specificity of lysine acetylation on histone H3. Mol. Cell. Proteom. 2014, 13, 2896–2910. [Google Scholar] [CrossRef]
- Nagy, Z.; Tora, L. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene 2007, 26, 5341–5357. [Google Scholar] [CrossRef]
- McCullough, C.E.; Song, S.; Shin, M.H.; Johnson, F.B.; Marmorstein, R. Structural and Functional Role of Acetyltransferase hMOF K274 Autoacetylation. J. Biol. Chem. 2016, 291, 18190–18198. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Rossetto, D.; Mellert, H.; Dang, W.; Srinivasan, M.; Johnson, J.; Hodawadekar, S.; Ding, E.C.; Speicher, K.; Abshiru, N.; et al. MYST protein acetyltransferase activity requires active site lysine autoacetylation. EMBO J. 2011, 31, 58–70. [Google Scholar] [CrossRef] [PubMed]
- Marmorstein, R.; Trievel, R.C. Trievel, Histone modifying enzymes: Structures, mechanisms, and specificities. Biochim. Biophys. Acta 2009, 1789, 58–68. [Google Scholar] [CrossRef]
- Wapenaar, H.; Dekker, F.J. Histone acetyltransferases: Challenges in targeting bi-substrate enzymes. Clin. Epigenetics 2016, 8, 59. [Google Scholar] [CrossRef]
- Parthun, M.R.; Widom, J.; Gottschling, D.E. The major cytoplasmic histone acetyltransferase in yeast: Links to chromatin replication and histone metabolism. Cell 1996, 87, 85–94. [Google Scholar] [CrossRef]
- Brook, P.O.; Perry, M.M.; Adcock, I.M.; Durham, A.L. Epigenome-modifying tools in asthma. Epigenomics 2015, 7, 1017–1032. [Google Scholar] [CrossRef]
- Hull, E.E.; Montgomery, M.R.; Leyva, K.J. HDAC inhibitors as epigenetic regulators of the immune system: Impacts on cancer therapy and inflammatory diseases. BioMed Res. Int. 2016, 2016, 8797206. [Google Scholar] [CrossRef]
- Khan, N.; Jeffers, M.; Kumar, S.; Hackett, C.; Boldog, F.; Khramtsov, N.; Qian, X.; Mills, E.; Berghs, S.C.; Carey, N.; et al. Determination of the class and isoform selectivity of small-molecule histone deacetylase inhibitors. Biochem. J. 2008, 409, 581–589. [Google Scholar] [CrossRef]
- Sun, W.-J.; Zhou, X.; Zheng, J.-H.; Lu, M.-D.; Nie, J.-Y.; Yang, X.-J.; Zheng, Z.-Q. Histone acetyltransferases and deacetylases: Molecular and clinical implications to gastrointestinal carcinogenesis. Acta Biochim. Biophys. Sin. 2012, 44, 80–91. [Google Scholar] [CrossRef]
- Bottomley, M.J.; Surdo, P.L.; Di Giovine, P.; Cirillo, A.; Scarpelli, R.; Ferrigno, F.; Jones, P.; Neddermann, P.; De Francesco, R.; Steinkühler, C.; et al. Structural and functional analysis of the human HDAC4 catalytic domain reveals a regulatory structural zinc-binding domain. J. Biol. Chem. 2008, 283, 26694–26704. [Google Scholar] [CrossRef]
- Gregoretti, I.V.; Lee, Y.-M.; Goodson, H.V. Molecular evolution of the histone deacetylase family: Functional implications of phylogenetic analysis. J. Mol. Biol. 2004, 338, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, P.M.; Cole, K.E.; Dowling, D.P.; Christianson, D.W. Structure, mechanism, and inhibition of histone deacetylases and related metalloenzymes. Curr. Opin. Struct. Biol. 2011, 21, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Schuetz, A.; Min, J.; Allali-Hassani, A.; Schapira, M.; Shuen, M.; Loppnau, P.; Mazitschek, R.; Kwiatkowski, N.P.; Lewis, T.A.; Maglathin, R.L.; et al. Human HDAC7 harbors a class IIa histone deacetylase-specific zinc binding motif and cryptic deacetylase activity. J. Biol. Chem. 2008, 283, 11355–11363. [Google Scholar] [CrossRef] [PubMed]
- Guardiola, A.R.; Yao, T.-P. Molecular cloning and characterization of a novel histone deacetylase HDAC10. J. Biol. Chem. 2001, 277, 3350–3356. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Faller, D.V. Transcription Regulation by Class. III Histone Deacetylases (HDACs)-Sirtuins. Transl. Oncogenom. 2008, 3, 53–65. [Google Scholar]
- Haberland, M.; Montgomery, R.L.; Olson, E.N. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nat. Rev. Genet. 2009, 10, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.J. Lysine acetylation and the bromodomain: A new partnership for signaling. Bioessays 2004, 26, 1076–1087. [Google Scholar] [CrossRef] [PubMed]
- Kurdistani, S.K.; Grunstein, M. Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell Biol. 2003, 4, 276–284. [Google Scholar] [CrossRef]
- Allis, C.D.; Berger, S.L.; Cote, J.; Dent, S.; Jenuwien, T.; Kouzarides, T.; Pillus, L.; Reinberg, D.; Shi, Y.; Shiekhattar, R.; et al. New nomenclature for chromatin-modifying enzymes. Cell 2007, 131, 633–636. [Google Scholar] [CrossRef]
- Fournier, M.; Tora, L. KAT2-mediated PLK4 acetylation contributes to genomic stability by preserving centrosome number. Mol. Cell Oncol. 2017, 4, e1270391. [Google Scholar] [CrossRef]
- Klein, B.J.; LaLonde, M.-E.; Côté, J.; Yang, X.-J.; Kutateladze, T.G. Crosstalk between epigenetic readers regulates the MOZ/MORF HAT complexes. Epigenetics 2014, 9, 186–193. [Google Scholar] [CrossRef]
- LaLonde, M.-E.; Avvakumov, N.; Glass, K.C.; Joncas, F.-H.; Saksouk, N.; Holliday, M.; Paquet, E.; Yan, K.; Tong, Q.; Klein, B.J.; et al. Exchange of associated factors directs a switch in HBO1 acetyltransferase histone tail specificity. Genes Dev. 2013, 27, 2009–2024. [Google Scholar] [CrossRef]
- Sun, X.-J.; Man, N.; Tan, Y.; Nimer, S.D.; Wang, L. The Role of Histone Acetyltransferases in Normal and Malignant Hematopoiesis. Front. Oncol. 2015, 5, 108. [Google Scholar] [CrossRef]
- Angiolilli, C.; Baeten, D.L.; Radstake, T.R.; Reedquist, K.A. The acetyl code in rheumatoid arthritis and other rheumatic diseases. Epigenomics 2017, 9, 447–461. [Google Scholar] [CrossRef]
- Ceccacci, E.; Minucci, S. Inhibition of histone deacetylases in cancer therapy: Lessons from leukaemia. Br. J. Cancer 2016, 114, 605–611. [Google Scholar] [CrossRef]
- Fierz, B.; Muir, T.W. Chromatin as an expansive canvas for chemical biology. Nat. Chem. Biol. 2012, 8, 417–427. [Google Scholar] [CrossRef]
- Santos-Rosa, H.; Caldas, C. Chromatin modifier enzymes, the histone code and cancer. Eur. J. Cancer 2005, 41, 2381–2402. [Google Scholar] [CrossRef]
- Lawen, A. Apoptosis—An introduction. Bioessays 2003, 25, 888–896. [Google Scholar] [CrossRef]
- Boix-Chornet, M.; Fraga, M.F.; Villar-Garea, A.; Caballero, R.; Espada, J.; Nuñez, A.; Casado-Vela, J.; Largo, C.; Casal, J.I.; Cigudosa, J.C.; et al. Release of hypoacetylated and trimethylated histone H4 is an epigenetic marker of early apoptosis. J. Biol. Chem. 2006, 281, 13540–13547. [Google Scholar] [CrossRef]
- Hendzel, M.J.; Nishioka, W.K.; Raymond, Y.; Allis, C.D.; Bazett-Jones, D.P.; Th’ng, J.P. Chromatin condensation is not associated with apoptosis. J. Biol. Chem. 2006, 281, 13540–13547. [Google Scholar] [CrossRef]
- Wu, D.; Ingram, A.; Lahti, J.H.; Mazza, B.; Grenet, J.; Kapoor, A.; Liu, L.; Kidd, V.J.; Tang, D. Apoptotic release of histones from nucleosomes. J. Biol. Chem. 2002, 277, 12001–12008. [Google Scholar] [CrossRef]
- Brochier, C.; Dennis, G.; Rivieccio, M.A.; McLaughlin, K.; Coppola, G.; Ratan, R.R.; Langley, B. Specific acetylation of p53 by HDAC inhibition prevents DNA damage-induced apoptosis in neurons. J. Neurosci. 2013, 33, 8621–8632. [Google Scholar] [CrossRef]
- Duan, H.; Heckman, C.A.; Boxer, L.M. Histone deacetylase inhibitors down-regulate bcl-2 expression and induce apoptosis in t(14;18) lymphomas. Mol. Cell. Biol. 2005, 25, 1608–1619. [Google Scholar] [CrossRef]
- Gong, P.; Wang, Y.; Jing, Y. Apoptosis Induction byHistone Deacetylase Inhibitors in Cancer Cells: Role of Ku70. Int. J. Mol. Sci. 2019, 20, 1601. [Google Scholar] [CrossRef]
- Roy, S.; Packman, K.; Jeffrey, R.; Tenniswood, M. Histone deacetylase inhibitors differentially stabilize acetylated p53 and induce cell cycle arrest or apoptosis in prostate cancer cells. Cell Death Differ. 2005, 12, 482–491. [Google Scholar] [CrossRef]
- Weidle, U.H.; Grossmann, A. Inhibition of histone deacetylases: A new strategy to target epigenetic modifications for anticancer treatment. Anticancer Res. 2000, 20, 1471–1485. [Google Scholar]
- Liu, T.; Kuljaca, S.; Tee, A.; Marshall, G.M. Histone deacetylase inhibitors: Multifunctional anticancer agents. Cancer Treat. Rev. 2006, 32, 157–165. [Google Scholar] [CrossRef]
- Hajji, N.; Wallenborg, K.; Vlachos, P.; Nyman, U.; Hermanson, O.; Joseph, B. Combinatorial action of the HDAC inhibitor trichostatin A and etoposide induces caspase-mediated AIF-dependent apoptotic cell death in non-small cell lung carcinoma cells. Oncogene 2007, 27, 3134–3144. [Google Scholar] [CrossRef]
- Li, Y.; Yuan, Z.; Liu, B.; Sailhamer, E.A.; Shults, C.; Velmahos, G.C.; DeMoya, M.; Alam, H.B. Prevention of hypoxia-induced neuronal apoptosis through histone deacetylase inhibition. J. Trauma 2008, 64, 863–870, discussion 870–871. [Google Scholar] [CrossRef]
- Allera, C.; Lazzarini, G.; Patrone, E.; Alberti, I.; Barboro, P.; Sanna, P.; Melchiori, A.; Parodi, S.; Balbi, C. The condensation of chromatin in apoptotic thymocytes shows a specific structural change. J. Biol. Chem. 1997, 272, 10817–10822. [Google Scholar] [CrossRef]
- Shogren-Knaak, M.; Ishii, H.; Sun, J.-M.; Pazin, M.J.; Davie, J.R.; Peterson, C.L. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 2006, 311, 844–847. [Google Scholar] [CrossRef] [PubMed]
- Hajji, N.; Wallenborg, K.; Vlachos, P.; Füllgrabe, J.; Hermanson, O.; Joseph, B. Opposing effects of hMOF and SIRT1 on H4K16 acetylation and the sensitivity to the topoisomerase II inhibitor etoposide. Oncogene 2010, 29, 2192–2204. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.-F.; Lee, C.-H.; Hsia, K.-T.; Huang, G.-S.; Lee, H.-S. Methylation of histone H3 lysine 27 associated with apoptosis in osteosarcoma cells induced by staurosporine. Histol. Histopathol. 2009, 24, 1105–1111. [Google Scholar] [PubMed]
- Greer, E.L.; Maures, T.J.; Hauswirth, A.G.; Green, E.M.; Leeman, D.S.; Maro, G.S.; Han, S.; Banko, M.R.; Gozani, O.; Brunet, A. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature 2010, 466, 383–387. [Google Scholar] [CrossRef]
- Chen, C.-C.; Carson, J.J.; Feser, J.; Tamburini, B.; Zabaronick, S.; Linger, J.; Tyler, J.K. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell 2008, 134, 231–243. [Google Scholar] [CrossRef]
- Farooq, Z.; Banday, S.; Pandita, T.K.; Altaf, M. The many faces of histone H3K79 methylation. Mutat. Res. Mutat. Res. 2016, 768, 46–52. [Google Scholar] [CrossRef]
- Fernandez-Capetillo, O.; Allis, C.D.; Nussenzweig, A. Phosphorylation of histone H2B at DNA double-strand breaks. J. Exp. Med. 2004, 199, 1671–1677. [Google Scholar] [CrossRef]
- Liu, W.; Deng, L.; Song, Y.; Redell, M.S. DOT1L inhibition sensitizes MLL-rearranged AML to chemotherapy. PLoS ONE 2014, 9, e98270. [Google Scholar] [CrossRef]
- Prokhorova, E.A.; Zamaraev, A.V.; Kopeina, G.S.; Zhivotovsky, B.; Lavrik, I.N. Role of the nucleus in apoptosis: Signaling and execution. Cell. Mol. Life Sci. 2015, 72, 4593–4612. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, Y.; Liu, M.; Lan, M.S.; Fei, J.; Fan, W.; Gao, X.; Lu, D.-R. Hypermethylation of hepatic glucokinase and L-type pyruvate kinase promoters in high-fat diet-induced obese rats. Endocrinology 2011, 152, 1284–1289. [Google Scholar] [CrossRef]
- Pirola, C.J.; Gianotti, T.F.; Burgueño, A.L.; Rey-Funes, M.; Loidl, C.F.; Mallardi, P.; Martino, J.S.; Castaño, G.; Sookoian, S. Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease. Gut 2012, 62, 1356–1363. [Google Scholar] [CrossRef] [PubMed]
- Sookoian, S.; Pirola, C.J. DNA methylation and hepatic insulin resistance and steatosis. Curr. Opin. Clin. Nutr. Metab. Care 2012, 15, 350–356. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Fan, J.; Qiao, L. Potential epigenetic mechanism in non-alcoholic Fatty liver disease. Int. J. Mol. Sci. 2015, 16, 5161–5179. [Google Scholar] [CrossRef] [PubMed]
- Dawson, M.A.; Kouzarides, T. Cancer epigenetics: From mechanism to therapy. Cell 2012, 150, 12–27. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Wong, V.W.-S.; Chan, H.L.-Y.; Cheng, A.S.-L. Epigenetic regulation of hepatocellular carcinoma in non-alcoholic fatty liver disease. Semin. Cancer Biol. 2013, 23, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Brenner, C.; Galluzzi, L.; Kepp, O.; Kroemer, G. Decoding cell death signals in liver inflammation. J. Hepatol. 2013, 59, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Kanda, T.; Matsuoka, S.; Yamazaki, M.; Shibata, T.; Nirei, K.; Takahashi, H.; Kaneko, T.; Fujisawa, M.; Higuchi, T.; Nakamura, H.; et al. Apoptosis and non-alcoholic fatty liver diseases. World J. Gastroenterol. 2018, 24, 2661–2672. [Google Scholar] [CrossRef]
- Ibrahim, S.; Kohli, R.; Gores, G.J. Gores, Mechanisms of lipotoxicity in NAFLD and clinical implications. Pediatr. Gastroenterol. Nutr. 2011, 53, 131–140. [Google Scholar]
- Kim, J.H.; Jung, D.Y.; Nagappan, A.; Jung, M.H. Histone H3K9 demethylase JMJD2B induces hepatic steatosis through upregulation of PPARgamma2. Sci. Rep. 2018, 8, 13734. [Google Scholar]
- Wieckowska, A.; Zein, N.N.; Yerian, L.M.; Lopez, A.R.; McCullough, A.J.; Feldstein, A.E. In vivo assessment of liver cell apoptosis as a novel biomarker of disease severity in nonalcoholic fatty liver disease. Hepatology 2006, 44, 27–33. [Google Scholar] [CrossRef]
- Akazawa, Y.; Nakao, K. To die or not to die: Death signaling in nonalcoholic fatty liver disease. J. Gastroenterol. 2018, 53, 893–906. [Google Scholar] [CrossRef] [PubMed]
- Lomba, A.; Milagro, F.I.; Garcia-Diaz, D.F.; Marti, A.; Campion, J.; Martínez, J.A. Obesity induced by a pair-fed high fat sucrose diet: Methylation and expression pattern of genes related to energy homeostasis. Lipids Health Dis. 2010, 9, 60. [Google Scholar] [CrossRef] [PubMed]
- Paternain, L.; Batlle, M.; De La Garza, A.; Milagro, F.I.; Martinez, J.; Campion, J. Transcriptomic and epigenetic changes in the hypothalamus are involved in an increased susceptibility to a high-fat-sucrose diet in prenatally stressed female rats. Neuroendocrinology 2012, 96, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Chan, H.M.; La Thangue, N.B. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 2001, 114, 2363–2373. [Google Scholar]
- Bricambert, J.; Miranda, J.; Benhamed, F.; Girard, J.; Postic, C.; Dentin, R. Salt-inducible kinase 2 links transcriptional coactivator p300 phosphorylation to the prevention of ChREBP-dependent hepatic steatosis in mice. J. Clin. Investig. 2010, 120, 4316–4331. [Google Scholar] [CrossRef]
- Li, Y.; Reddy, M.A.; Miao, F.; Shanmugam, N.; Yee, J.K.; Hawkins, D.; Ren, B.; Natarajan, R. Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-kappaB-dependent inflammatory genes. Relevance to diabetes and inflammation. J. Biol. Chem. 2008, 283, 26771–26781. [Google Scholar] [CrossRef]
- Pathak, P.; Li, T.; Chiang, J.Y.L. Retinoic acid-related orphan receptor alpha regulates diurnal rhythm and fasting induction of sterol 12alpha-hydroxylase in bile acid synthesis. J. Biol. Chem. 2013, 288, 37154–37165. [Google Scholar] [CrossRef]
- Mikula, M.; Majewska, A.; Ledwon, J.K.; Dzwonek, A.; Ostrowski, J. Obesity increases histone H3 lysine 9 and 18 acetylation at Tnfa and Ccl2 genes in mouse liver. Int. J. Mol. Med. 2014, 34, 1647–1654. [Google Scholar] [CrossRef]
- Jun, H.J.; Kim, J.; Hoang, M.H.; Lee, S.J. Hepatic lipid accumulation alters global histone h3 lysine 9 and 4 trimethylation in the peroxisome proliferator-activated receptor alpha network. PLoS ONE 2012, 7, e44345. [Google Scholar] [CrossRef]
- Çolak, Y.; Ozturk, O.; Senates, E.; Tuncer, I.; Yorulmaz, E.; Adali, G.; Doganay, L.; Enc, F.Y. SIRT1 as a potential therapeutic target for treatment of nonalcoholic fatty liver disease. Med. Sci. Monit. 2011, 17, HY5–HY9. [Google Scholar] [CrossRef]
- Hirschey, M.D.; Shimazu, T.; Jing, E.; Grueter, C.A.; Collins, A.M.; Aouizerat, B.; Stančáková, A.; Goetzman, E.; Lam, M.M.; Schwer, B.; et al. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol. Cell 2011, 44, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Schwer, B.; Verdin, E. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 2008, 7, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Raza, S.; Rajak, S.; Anjum, B.; Sinha, R.A. Molecular links between non-alcoholic fatty liver disease and hepatocellular carcinoma. Hepatoma Res. 2019, 2019, 42. [Google Scholar] [CrossRef] [PubMed]
- Labi, V.; Erlacher, M. How cell death shapes cancer. Cell Death Dis. 2015, 6, e1675. [Google Scholar] [CrossRef]
- Yang, Y.M.; Kim, S.Y.; Seki, E. Inflammation and Liver Cancer: Molecular Mechanisms and Therapeutic Targets. Semin. Liver Dis. 2019, 39, 26–42. [Google Scholar] [CrossRef]
- Heinrichsdorff, J.; Luedde, T.; Perdiguero, E.; Nebreda, A.R.; Pasparakis, M. p38 alpha MAPK inhibits JNK activation and collaborates with IkappaB kinase 2 to prevent endotoxin-induced liver failure. EMBO Rep. 2008, 9, 1048–1054. [Google Scholar] [CrossRef]
- Pahl, H.L. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene 1999, 18, 6853–6866. [Google Scholar] [CrossRef]
- Tang, G.; Minemoto, Y.; Dibling, B.; Purcell, N.H.; Li, Z.; Karin, M.; Lin, A. Inhibition of JNK activation through NF-kappaB target genes. Nature 2001, 414, 313–317. [Google Scholar] [CrossRef]
- Beerheide, W.; Tan, Y.-J.; Teng, E.; Ting, A.E.; Jedpiyawongse, A.; Srivatanakul, P. Downregulation of proapoptotic proteins Bax and Bcl-X(S) in p53 overexpressing hepatocellular carcinomas. Biochem. Biophys. Res. Commun. 2000, 273, 54–61. [Google Scholar] [CrossRef]
- Ito, T.; Shiraki, K.; Sugimoto, K.; Yamanaka, T.; Fujikawa, K.; Ito, M.; Takase, K.; Moriyama, M.; Kawano, H.; Hayashida, M.; et al. Survivin promotes cell proliferation in human hepatocellular carcinoma. Hepatology 2000, 31, 1080–1085. [Google Scholar] [CrossRef]
- Zhu, H.; Chen, X.-P.; Zhang, W.-G.; Luo, S.-F.; Zhang, B.-X. Expression and significance of new inhibitor of apoptosis protein survivin in hepatocellular carcinoma. World J. Gastroenterol. 2005, 11, 3855–3859. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Kesarwala, A.H.; Eggert, T.; Medina-Echeverz, J.; Kleiner, D.E.; Jin, P.; Stroncek, P.J.D.F.; Terabe, M.; Kapoor, V.; Elgindi, M.; et al. NAFLD causes selective CD4(+) T lymphocyte loss and promotes hepatocarcinogenesis. Nature 2016, 531, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Marquardt, J.U.; Edlich, F. Predisposition to Apoptosis in Hepatocellular Carcinoma: From Mechanistic Insights to Therapeutic Strategies. Front. Oncol. 2019, 9, 1421. [Google Scholar] [CrossRef] [PubMed]
- Bayo, J.; Fiore, E.J.; Dominguez, L.M.; Real, A.; Malvicini, M.; Rizzo, M.; Atorrasagasti, C.; García, M.G.; Argemi, J.; Martinez, E.D.; et al. A comprehensive study of epigenetic alterations in hepatocellular carcinoma identifies potential therapeutic targets. J. Hepatol. 2019, 71, 78–90. [Google Scholar] [CrossRef] [PubMed]
- Salerno, D.; Chiodo, L.; Alfano, V.; Floriot, O.; Cottone, G.; Paturel, A.; Pallocca, M.; Plissonnier, M.-L.; Jeddari, S.; Belloni, L.; et al. Hepatitis B protein HBx binds the DLEU2 lncRNA to sustain cccDNA and host cancer-related gene transcription. Gut 2020, 69, 2016–2024. [Google Scholar] [CrossRef] [PubMed]
- Karanth, A.V.; Maniswami, R.R.; Prashanth, S.; Govindaraj, H.; Padmavathy, R.; Jegatheesan, S.K.; Mullangi, R.; Rajagopal, S. Emerging role of SETDB1 as a therapeutic target. Expert Opin. Ther. Targets 2017, 21, 319–331. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, J.; Li, Q.; Chen, K.; Liang, Y.; Zhan, Z.; Ye, F.; Ni, W.; Chen, L.; Ding, Y. Histone methyltransferase SETDB1 promotes cells proliferation and migration by interacting withTiam1 in hepatocellular carcinoma. BMC Cancer 2018, 18, 539. [Google Scholar] [CrossRef]
- Han, T.-S.; Ban, H.S.; Hur, K.; Cho, H.-S. The Epigenetic Regulation of HCC Metastasis. Int. J. Mol. Sci. 2018, 19, 3978. [Google Scholar] [CrossRef]
- Wei, L.; Chiu, D.K.-C.; Tsang, F.H.-C.; Law, C.-T.; Cheng, C.L.-H.; Au, S.L.-K.; Lee, J.M.-F.; Wong, C.C.-L.; Kung-Chun, C.D.; Wong, C.-M. Histone methyltransferase G9a promotes liver cancer development by epigenetic silencing of tumor suppressor gene RARRES3. J. Hepatol. 2017, 67, 758–769. [Google Scholar] [CrossRef]
- De Conti, A.; Dreval, K.; Tryndyak, V.; Orisakwe, O.E.; Ross, S.A.; Beland, F.A.; Pogribny, I.P. Inhibition of the Cell Death Pathway in Nonalcoholic Steatohepatitis (NASH)-Related Hepatocarcinogenesis Is Associated with Histone H4 lysine 16 Deacetylation. Mol. Cancer Res. 2017, 15, 1163–1172. [Google Scholar] [CrossRef]
- Ji, X.; Jin, S.; Qu, X.; Li, K.; Wang, H.; He, H.; Guo, F.; Dong, L. Lysine-specific demethylase 5C promotes hepatocellular carcinoma cell invasion through inhibition BMP7 expression. BMC Cancer 2015, 15, 801. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Qi, G.; Tang, F.; Yuan, S.; Wang, Z.; Liang, X.; Li, B.; Yu, S.; Liu, J.; Huang, Q.; et al. JARID1B promotes metastasis and epithelial-mesenchymal transition via PTEN/AKT signaling in hepatocellular carcinoma cells. Oncotarget 2015, 6, 12723–12739. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wang, X.; Liu, R.; Chen, L.; Yi, J.; Qi, B.; Shuang, Z.; Liu, M.; Li, X.; Li, S.; et al. KDM4B-mediated epigenetic silencing of miRNA-615-5p augments RAB24 to facilitate malignancy of hepatoma cells. Oncotarget 2016, 8, 17712–17725. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.-P.; Wang, X.-L.; Zhao, W.; Qi, B.; Yang, Q.; Wan, H.-Y.; Shuang, Z.-Y.; Liu, M.; Li, X.; Li, S.-P.; et al. DNA methylation-mediated repression of miR-941 enhances lysine (K)-specific demethylase 6B expression in hepatoma cells. J. Biol. Chem. 2014, 289, 24724–24735. [Google Scholar] [CrossRef]
- Zheng, X.; Gai, X.; Ding, F.; Lu, Z.; Tu, K.; Yao, Y.; Liu, Q. Histone acetyltransferase PCAF up-regulated cell apoptosis in hepatocellular carcinoma via acetylating histone H4 and inactivating AKT signaling. Mol. Cancer 2013, 12, 96. [Google Scholar] [CrossRef]
- Coradini, D.; Speranza, A. Histone deacetylase inhibitors for treatment of hepatocellular carcinoma. Acta Pharmacol. Sin. 2005, 26, 1025–1033. [Google Scholar] [CrossRef]
- Coradini, D.; Zorzet, S.; Rossin, R.; Scarlata, I.; Pellizzaro, C.; Turrin, C.-O.; Bello, M.; Cantoni, S.; Speranza, A.; Sava, G.; et al. Inhibition of hepatocellular carcinomas in vitro and hepatic metastases in vivo in mice by the histone deacetylase inhibitor HA-But. Clin. Cancer Res. 2004, 10, 4822–4830. [Google Scholar] [CrossRef]
- Lee, Y.-H.; Seo, D.; Choi, K.-J.; Andersen, J.B.; Won, M.-A.; Kitade, M.; Gómez-Quiroz, L.E.; Judge, A.D.; Marquardt, J.U.; Raggi, C.; et al. Antitumor effects in hepatocarcinoma of isoform-selective inhibition of HDAC2. Cancer Res. 2014, 74, 4752–4761. [Google Scholar] [CrossRef]
- Ler, S.Y.; Leung, C.H.W.; Khin, L.W.; Lu, G.-D.; Salto-Tellez, M.; Hartman, M.; Iau, P.T.C.; Yap, C.T.; Hooi, S.C. HDAC1 and HDAC2 independently predict mortality in hepatocellular carcinoma by a competing risk regression model in a Southeast. Asian population. Oncol. Rep. 2015, 34, 2238–2250. [Google Scholar] [CrossRef]
- Yang, J.; Jin, X.; Yan, Y.; Shao, Y.; Pan, Y.; Roberts, L.R.; Zhang, J.; Huang, H.; Jiang, J. Inhibiting histone deacetylases suppresses glucose metabolism and hepatocellular carcinoma growth by restoring FBP1 expression. Sci. Rep. 2017, 7, 43864. [Google Scholar] [CrossRef]
- Semenza, G.L. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene 2010, 29, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Wu, L.; Liang, T.; Liu, Z.; Li, J.; Li, N.; Xie, H.; Yin, S.; Yu, J.; Lin, Q.; et al. Overexpression of myocyte enhancer factor 2 and histone hyperacetylation in hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 2008, 134, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.-Y.; Hou, J.-H.; Rao, H.-L.; Luo, R.-Z.; Li, M.; Pei, X.-Q.; Lin, M.C.; Guan, X.-Y.; Kung, H.-F.; Zeng, Y.-X.; et al. High expression of H3K27me3 in human hepatocellular carcinomas correlates closely with vascular invasion and predicts worse prognosis in patients. Mol. Med. 2010, 17, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Shi, Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Hałasa, M.; Wawruszak, A.; Przybyszewska, A.; Jaruga, A.; Guz, M.; Kalafut, J.; Stepulak, A.; Cybulski, M. H3K18Ac as a Marker of Cancer Progression and Potential Target of Anti-Cancer Therapy. Cells 2019, 8, 485. [Google Scholar]
- He, C.; Xu, J.; Zhang, J.; Xie, D.; Ye, H.; Xiao, Z.; Cai, M.; Xu, K.; Zeng, Y.; Li, H.; et al. High expression of trimethylated histone H3 lysine 4 is associated with poor prognosis in hepatocellular carcinoma. Hum. Pathol. 2012, 43, 1425–1435. [Google Scholar] [CrossRef]
- Heintzman, N.D.; Hon, G.C.; Hawkins, R.D.; Kheradpour, P.; Stark, A.; Harp, L.F.; Ye, Z.; Lee, L.K.; Stuart, R.K.; Ching, C.W.; et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 2009, 459, 108–112. [Google Scholar] [CrossRef]
- Ong, C.-T.; Corces, V.G. Enhancer function: New insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 2011, 12, 283–293. [Google Scholar] [CrossRef]
- Seligson, D.B.; Horvath, S.; McBrian, M.A.; Mah, V.; Yu, H.; Tze, S.; Wang, Q.; Chia, D.; Goodglick, L.; Kurdistani, S.K. Global levels of histone modifications predict prognosis in different cancers. Am. J. Pathol. 2009, 174, 1619–1628. [Google Scholar] [CrossRef]
- Varier, R.A.; Timmers, H.M. Histone lysine methylation and demethylation pathways in cancer. Biochim. Biophys. Acta 2011, 1815, 75–89. [Google Scholar] [CrossRef]
- Zhou, X.; Sun, H.; Chen, H.; Zavadil, J.; Kluz, T.; Arita, A.; Costa, M. Hypoxia induces trimethylated H3 lysine 4 by inhibition of JARID1A demethylase. Cancer Res. 2010, 70, 4214–4221. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, A.; Yamauchi, N.; Shibahara, J.; Kimura, H.; Morikawa, T.; Ishikawa, S.; Nagae, G.; Nishi, A.; Sakamoto, Y.; Kokudo, N.; et al. Concurrent activation of acetylation and tri-methylation of H3K27 in a subset of hepatocellular carcinoma with aggressive behavior. PLoS ONE 2014, 9, e91330. [Google Scholar] [CrossRef]
- Ji, H.; Zhou, Y.; Zhuang, X.; Zhu, Y.; Wu, Z.; Lu, Y.; Li, S.; Zeng, Y.; Lu, Q.R.; Huo, Y.; et al. HDAC3 Deficiency Promotes Liver Cancer through a Defect in H3K9ac/H3K9me3 Transition. Cancer Res. 2019, 79, 3676–3688. [Google Scholar] [CrossRef] [PubMed]
- Kelly, T.K.; De Carvalho, D.D.; A Jones, P. Epigenetic modifications as therapeutic targets. Nat. Biotechnol. 2010, 28, 1069–1078. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, N.U.; Altaf, M. Epigenetics: An emerging field in the pathogenesis of nonalcoholic fatty liver disease. Mutat. Res. Mutat. Res. 2018, 778, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Cazaly, E.; Charlesworth, J.C.; Dickinson, J.L.; Holloway, A.F. Genetic Determinants of Epigenetic Patterns: Providing Insight into Disease. Mol. Med. 2015, 21, 400–409. [Google Scholar] [CrossRef]
- Toh, T.B.; Lim, J.J.; Chow, E.K. Epigenetics of hepatocellular carcinoma. Clin. Transl. Med. 2019, 8, 13. [Google Scholar] [CrossRef]
- Prince, H.M.; Bishton, M.; Johnstone, R.W. Panobinostat (LBH589): A potent pan-deacetylase inhibitor with promising activity against hematologic and solid tumors. Futur. Oncol. 2009, 5, 601–612. [Google Scholar] [CrossRef]
- Lachenmayer, A.; Toffanin, S.; Cabellos, L.; Alsinet, C.; Hoshida, Y.; Villanueva, A.; Minguez, B.; Tsai, H.-W.; Ward, S.C.; Thung, S.; et al. Combination therapy for hepatocellular carcinoma: Additive preclinical efficacy of the HDAC inhibitor panobinostat with sorafenib. J. Hepatol. 2012, 56, 1343–1350. [Google Scholar] [CrossRef]
- Gailhouste, L.; Liew, L.C.; Yasukawa, K.; Hatada, I.; Tanaka, Y.; Nakagama, H.; Ochiya, T. Differentiation Therapy by Epigenetic Reconditioning Exerts Antitumor Effects on Liver Cancer Cells. Mol. Ther. 2018, 26, 1840–1854. [Google Scholar] [CrossRef]
- Hardy, T.; A Mann, D. Epigenetics in liver disease: From biology to therapeutics. Gut 2016, 65, 1895–1905. [Google Scholar] [CrossRef] [PubMed]

| Histone Modifications | Biological Effects in NAFLD/HCC | References |
|---|---|---|
| HAT activity of p300/SIK2 | Transcriptional activation of lipogenic and glycolytic genes | [135] |
| HMT activity of SET7/9 on H3K4 | NF-κB-induced inflammation | [136] |
| Histone acetylation of Sterol 12α-hydroxylase (CYP8B1) by RORα | Dyslipidemia-associated inflammatory changes and regulation of bile acid synthesis and cholesterol levels | [137] |
| H3K9 and H3K18 acetylation of TNF-α and CCL2 | Obesity and fatty liver | [138] |
| H3K4 and H3K9 trimethylation in PPARα and lipid catabolism-related genes | Hepatic steatosis and NASH progression | [139] |
| Sirtuins | Mediate adaptive responses to metabolic stress and regulate adipogenesis and insulin secretion in NAFLD | [140,141,142] |
| H3K27 trimethylation by EZH2 | Cell cycle arrest and apoptosis in HCC | [155] |
| Methylation/demethylation of histone H3-K4 and K36 of mesenchymal-epithelial transition (MET)-related genes | Malignancy progression to metastasis in HCC | [24] |
| SETDB1-mediated histone H3K9 methylation | Downregulation of T-lymphoma invasion and metastasis gene (Tiam1) | [156,157] |
| H3K9 methylation by G9a, EHMT2 and SUV39H1 | Malignant clinicopathological features of HCC | [158,159] |
| Loss of histone H4K20 trimethylation and deacetylation of H4K16 | Alteration in cell death pathway | [160] |
| Demethylation of H3K4 by KDM5c and JARID1B | Suppression of gene expression in HCC | [161,162] |
| Demethylation of histone H3K9 (KDM4B) and K27 (KDM6B) | HCC cell proliferation and migration | [163,164] |
| Histone H4 acetylation by P300/CBP-associated factor (PCAF) | Promote cell apoptosis and inhibits tumor growth | [165] |
| Activity of HDAC1 and HDAC2 on metabolic enzymes | Regulate cell proliferation and cell death in HCC pathogenesis | [94,166,167,168,169,170] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rajan, P.K.; Udoh, U.-A.; Sanabria, J.D.; Banerjee, M.; Smith, G.; Schade, M.S.; Sanabria, J.; Sodhi, K.; Pierre, S.; Xie, Z.; et al. The Role of Histone Acetylation-/Methylation-Mediated Apoptotic Gene Regulation in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2020, 21, 8894. https://doi.org/10.3390/ijms21238894
Rajan PK, Udoh U-A, Sanabria JD, Banerjee M, Smith G, Schade MS, Sanabria J, Sodhi K, Pierre S, Xie Z, et al. The Role of Histone Acetylation-/Methylation-Mediated Apoptotic Gene Regulation in Hepatocellular Carcinoma. International Journal of Molecular Sciences. 2020; 21(23):8894. https://doi.org/10.3390/ijms21238894
Chicago/Turabian StyleRajan, Pradeep Kumar, Utibe-Abasi Udoh, Juan D. Sanabria, Moumita Banerjee, Gary Smith, Mathew Steven Schade, Jacqueline Sanabria, Komal Sodhi, Sandrine Pierre, Zijian Xie, and et al. 2020. "The Role of Histone Acetylation-/Methylation-Mediated Apoptotic Gene Regulation in Hepatocellular Carcinoma" International Journal of Molecular Sciences 21, no. 23: 8894. https://doi.org/10.3390/ijms21238894
APA StyleRajan, P. K., Udoh, U.-A., Sanabria, J. D., Banerjee, M., Smith, G., Schade, M. S., Sanabria, J., Sodhi, K., Pierre, S., Xie, Z., Shapiro, J. I., & Sanabria, J. (2020). The Role of Histone Acetylation-/Methylation-Mediated Apoptotic Gene Regulation in Hepatocellular Carcinoma. International Journal of Molecular Sciences, 21(23), 8894. https://doi.org/10.3390/ijms21238894







