Genes Identification, Molecular Docking and Dynamics Simulation Analysis of Laccases from Amylostereum areolatum Provides Molecular Basis of Laccase Bound to Lignin
Abstract
1. Introduction
2. Results
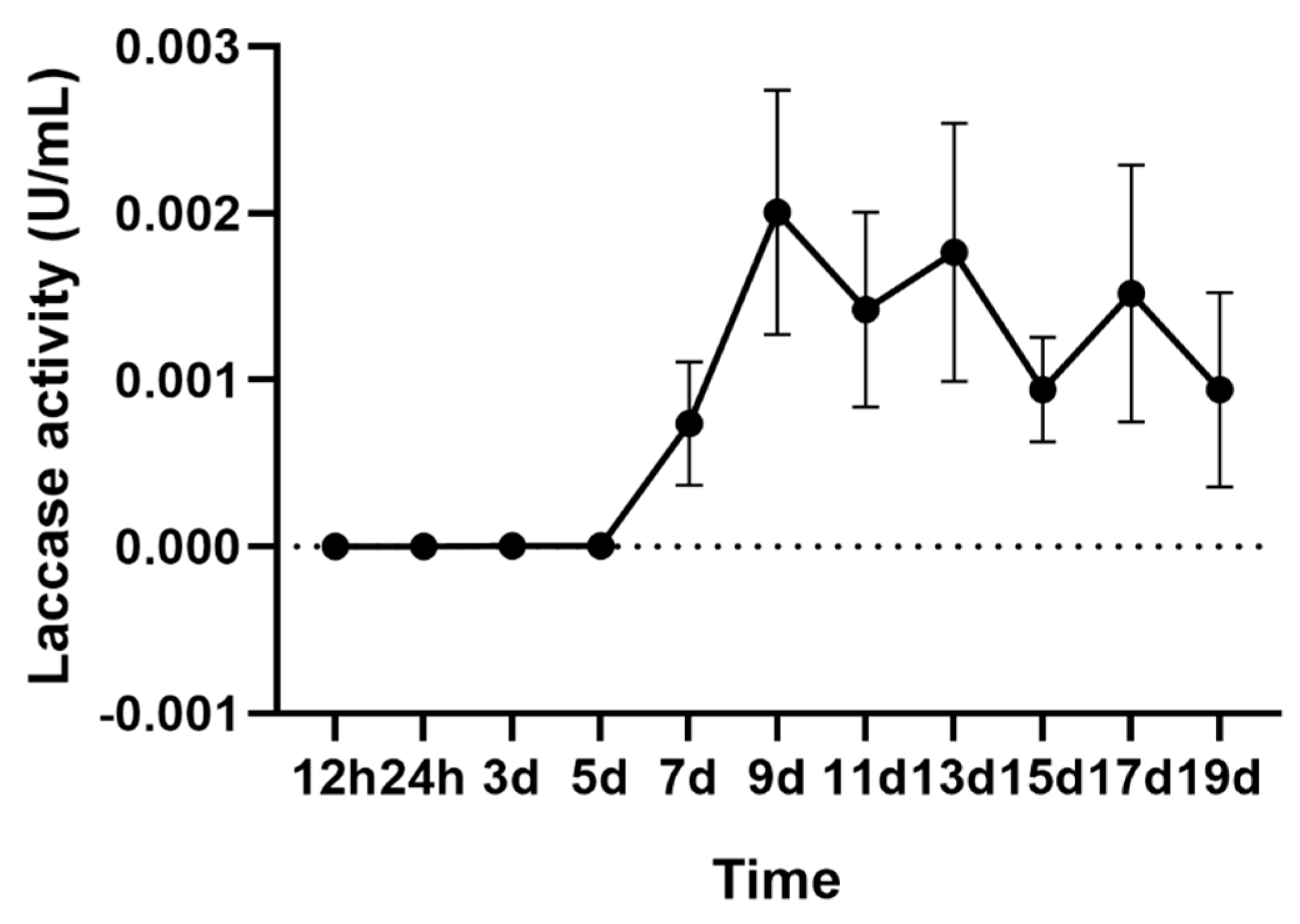
2.1. Laccase Activity of A. areolatum
2.2. Identification and Properties of Laccase Genes in A. areolatum
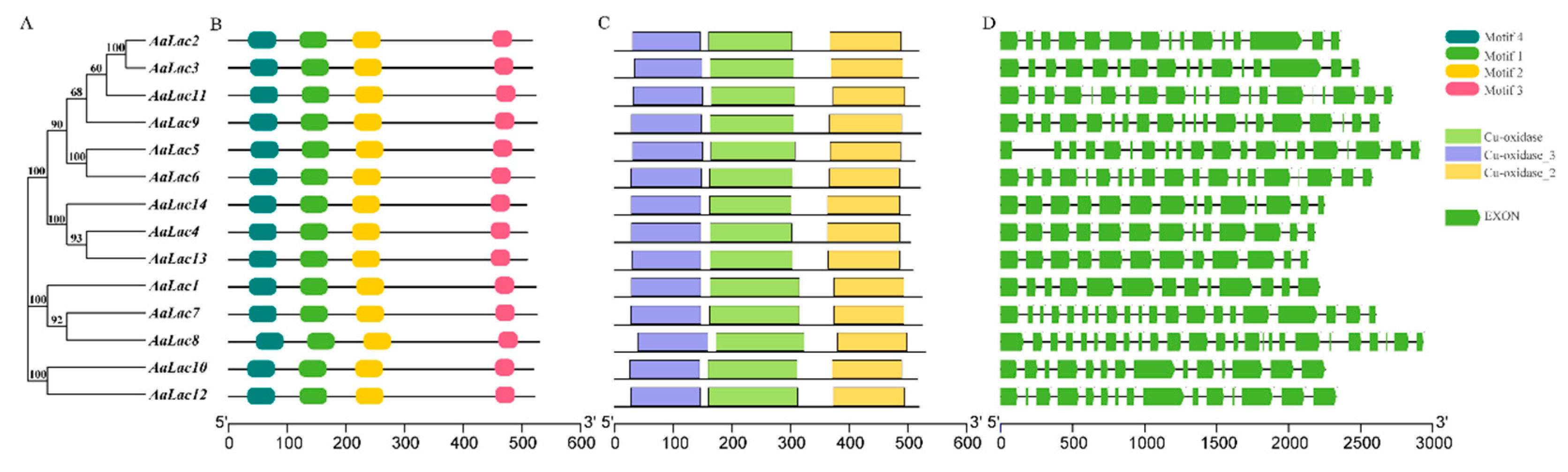
2.3. Gene Structure and Sequence Alignment of Laccase Genes in A. areolatum
2.4. Phylogenetic Analysis of A. areolatum Laccases
2.5. Homology Modeling and Validation
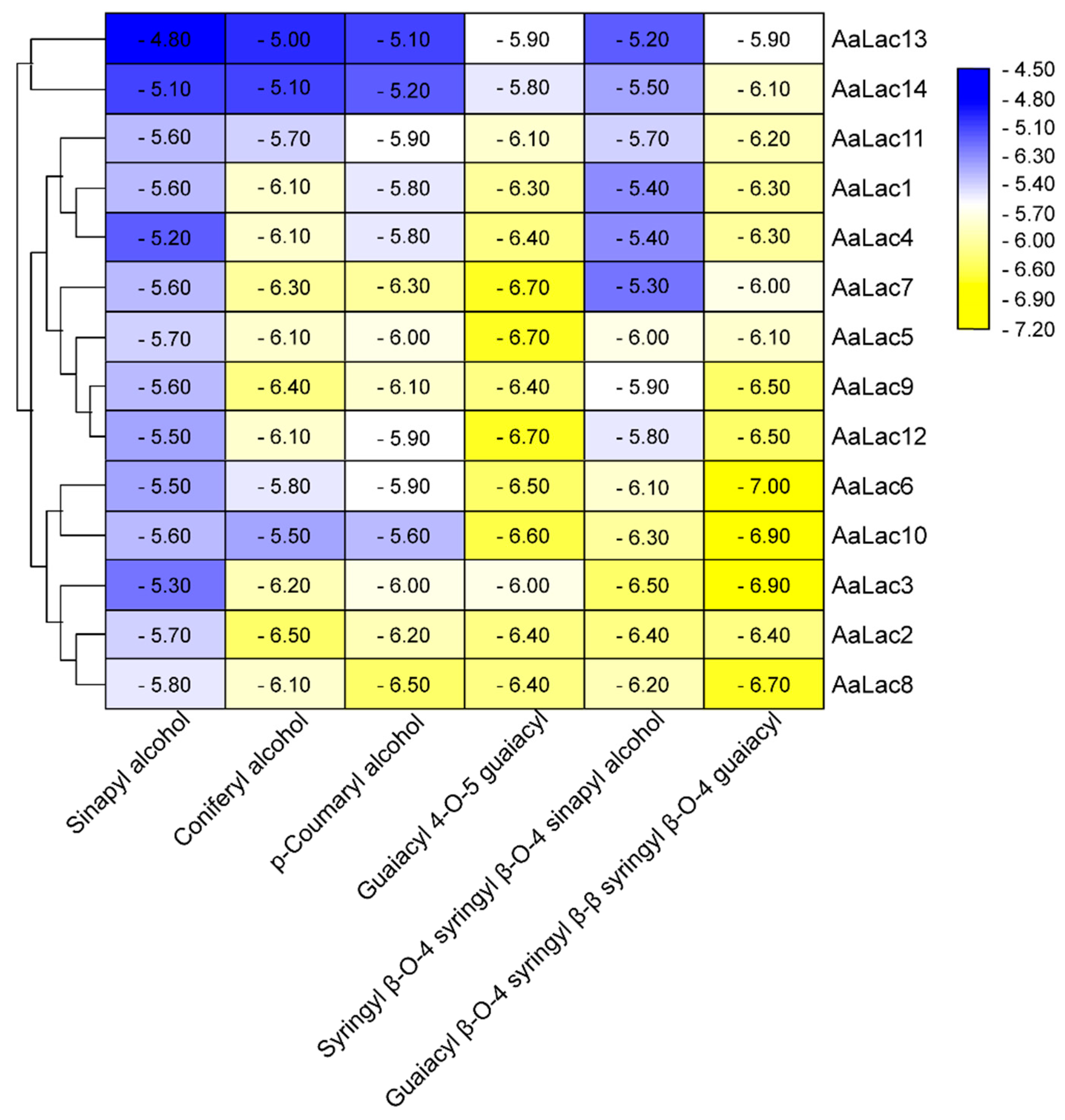
2.6. Molecular Docking of A. areolatum Laccases
2.7. Molecular Dynamics Simulation of Laccase with Lignin Model Compounds
3. Discussion
4. Methods
4.1. Determination of Laccase Activity
4.2. Genome-Wide Identification and Cloning of Laccase Family Genes
4.3. Analysis of the Laccase Sequences
4.4. Phylogenetic Analysis
4.5. Homology Modeling and Validation of Laccase Proteins
4.6. Ligand Preparation and Molecular Docking
4.7. Molecular Dynamics Simulation of Docked Complexes
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| PDB | Potato Dextrose Broth |
| NJ | Neighbor-joining |
| PDA | Potato Dextrose Agar PDA |
| ABTS | 2,2′-azinobis-3-ethylbenzthiazoline-6-sulphonate |
References
- Slippers, B.; De Groot, P.; Wingfield, M.J. The Sirex Woodwasp and Its Fungal Symbiont; Springer: Dordrecht, The Netherlands, 2012. [Google Scholar]
- Ryan, K.; Hurley, B.P. Life History and Biology of Sirex Noctilio; Springer: Dordrecht, The Netherlands, 2012. [Google Scholar]
- Spradbery, J.P.; Kirk, A.A. Aspects of the ecology of siricid woodwasps (Hymenoptera: Siricidae) in Europe, North Africa and Turkey with special reference to the biological control of Sirex noctilio F. in Australia. Bull. Entomol. Res. 1978, 68, 341–359. [Google Scholar]
- Rawlings, G.B. Recent observations on the Sirex noctilio population in Pinus radiata forest in New Zealand. N. Z. J. For. 1948, 5, 411–421. [Google Scholar]
- Iede, E.T.; Bisol, J.; Penteado, S.d.R.C. Primeiro Registro de Ataque de Sirex noctilio em Pinus taeda no Brasil; EMBRAPA-CNPF: Brasilia, Brazil, 1988. [Google Scholar]
- Hurley, B.P.; Slippers, B.; Wingfield, M.J. A comparison of control results for the alien invasive woodwasp, Sirex noctilio, in the southern hemisphere. Agric. For. Entomol. 2007, 9, 159–171. [Google Scholar]
- Neumann, F.; Minko, G. The Sirex wood wasp in Australian radiata pine plantations. Aust. For. 1981, 44, 46–63. [Google Scholar]
- Li, D.; Shi, J.; Lu, M.; Ren, L.; Zhen, C.; Luo, Y. Detection and identification of the invasive Sirex noctilio (Hymenoptera: Siricidae) fungal symbiont, Amylostereum areolatum (Russulales: Amylostereacea), in China and the stimulating effect of insect venom on laccase production by A. areolatum YQL03. J. Econ. Entomol. 2015, 108, 1136–1147. [Google Scholar]
- Sun, X.; Tao, J.; Ren, L.; Shi, J.; Luo, Y. Identification of Sirex noctilio (Hymenoptera: Siricidae) using a species-specific cytochrome Coxidase subunit I PCR assay. J. Econ. Entomol. 2016, 109, 1424–1430. [Google Scholar]
- Wang, L.; Ren, L.; Li, C.; Gao, C.; Liu, X.; Wang, M.; Luo, Y. Effects of endophytic fungi diversity in different coniferous species on the colonization of Sirex noctilio (Hymenoptera: Siricidae). Sci. Rep. 2019, 9, 5077. [Google Scholar]
- Talbot, P. The Sirex-Amylostereum-Pinus association. Annu. Rev. Phytopathol. 1977, 15, 41–54. [Google Scholar]
- Coutts, M. The mechanism of pathogenicity of Sirex noctilio on Pinus radiata I. Effects of the symbiotic fungus Amylostereum sp. (Thelophoraceae). Aust. J. Biol. Sci. 1969, 22, 915–924. [Google Scholar]
- Hajek, A.E.; Nielsen, C.; Kepler, R.M.; Long, S.J.; Castrillo, L. Fidelity among Sirex woodwasps and their fungal symbionts. Microb. Ecol. 2013, 65, 753–762. [Google Scholar]
- Coutts, M. The mechanism of pathogenicity of Sirex noctilio on Pinus radiata II. Effects of S. noctilio mucus. Aust. J. Biol. Sci. 1969, 22, 1153–1162. [Google Scholar]
- Coutts, M. Rapid physiological change in Pinus radiata following attack by Sirex noctilio and its associated fungus, Amylostereum sp. J. Sci. 1968, 30, 275–277. [Google Scholar]
- Madden, J.; Coutts, M. The role of fungi in the biology and ecology of woodwasps (Hymenoptera: Siricidae). In Insect-Fungus Symbiosis: Nutrition, Mutualism and Commensalism; Batra, L.R., Ed.; Halsted Press: Montclair, NJ, USA, 1979; pp. 165–174. [Google Scholar]
- Kile, G.; GA, K. The effect of radiata pine resin and resin components on the growth of the Sirex symbiont. Austral. For. Res. 1974, 6, 27–34. [Google Scholar]
- Kukor, J.J.; Martin, M.M. Acquisition of digestive enzymes by siricid woodwasps from their fungal symbiont. Science 1983, 220, 1161–1163. [Google Scholar] [PubMed]
- Yousuf, F.; Carnegie, A.J.; Bashford, R.; Bedding, R.A.; Nicol, H.I.; Gurr, G.M. Bark beetle (Ips grandicollis) disruption of woodwasp (Sirex noctilio) biocontrol: Direct and indirect mechanisms. For. Ecol. Manag. 2014, 323, 98–104. [Google Scholar]
- Thompson, B.M.; Bodart, J.; McEwen, C.; Gruner, D.S. Adaptations for symbiont-mediated external digestion in Sirex noctilio (Hymenoptera: Siricidae). Ann. Entomol. Soc. Am. 2014, 107, 453–460. [Google Scholar]
- Thompson, B.M.; Grebenok, R.J.; Behmer, S.T.; Gruner, D.S. Microbial symbionts shape the sterol profile of the xylem-feeding woodwasp, Sirex noctilio. J. Chem. Ecol. 2013, 39, 129–139. [Google Scholar]
- Bordeaux, J.M. Characterization of Growth Conditions for Production of a Laccase-Like Phenoloxidase by Amylostereum areolatum, a Fungal Pathogen of Pines and Other Conifers; University of Georgia: Athens, GA, USA, 2008. [Google Scholar]
- Giardina, P.; Faraco, V.; Pezzella, C.; Piscitelli, A.; Vanhulle, S.; Sannia, G. Laccases: A never-ending story. Cell. Mol. Life Sci. 2010, 67, 369–385. [Google Scholar]
- Solomon, E.I.; Sundaram, U.M.; Machonkin, T.E. Multicopper oxidases and oxygenases. Chem. Rev. 1996, 96, 2563–2606. [Google Scholar]
- Arora, D.S.; Sharma, R.K. Ligninolytic fungal laccases and their biotechnological applications. Appl. Biochem. Biotechnol. 2010, 160, 1760–1788. [Google Scholar]
- Diamantidis, G.; Effosse, A.; Potier, P.; Bally, R. Purification and characterization of the first bacterial laccase in the rhizospheric bacterium Azospirillum lipoferum. Soil Biol. Biochem. 2000, 32, 919–927. [Google Scholar]
- Cárdenas, W.; Dankert, J.R. Cresolase, catecholase and laccase activities in haemocytes of the red swamp crayfish. Fish Shellfish Immunol. 2000, 10, 33–46. [Google Scholar] [PubMed]
- Thurston, C.F. The structure and function of fungal laccases. Microbiology 1994, 140, 19–26. [Google Scholar]
- Linares, N.C.; Fernández, F.; Loske, A.M.; Gómez-Lim, M.A. Enhanced delignification of lignocellulosic biomass by recombinant fungus phanerochaete chrysosporium overexpressing laccases and peroxidases. J. Mol. Microbiol. Biotechnol. 2018, 28, 1–13. [Google Scholar]
- Lettera, V.; Piscitelli, A.; Leo, G.; Birolo, L.; Pezzella, C.; Sannia, G. Identification of a new member of Pleurotus ostreatus laccase family from mature fruiting body. Fungal Biol. 2010, 114, 724–730. [Google Scholar]
- Temp, U.; Eggert, C. Novel interaction between laccase and cellobiose dehydrogenase during pigment synthesis in the white rot fungus Pycnoporus cinnabarinus. Appl. Environ. Microbiol. 1999, 65, 389–395. [Google Scholar]
- Leonowicz, A.; Cho, N.; Luterek, J.; Wilkolazka, A.; Wojtas-Wasilewska, M.; Matuszewska, A.; Hofrichter, M.; Wesenberg, D.; Rogalski, J. Fungal laccase: Properties and activity on lignin. J. Basic Microbiol. Int. J. Biochem. Physiol. Genet. Morphol. Ecol. Microorg. 2001, 41, 185–227. [Google Scholar]
- Hoegger, P.J.; Kilaru, S.; James, T.Y.; Thacker, J.R.; Kües, U. Phylogenetic comparison and classification of laccase and related multicopper oxidase protein sequences. FEBS J. 2006, 273, 2308–2326. [Google Scholar]
- Bento, I.; Carrondo, M.A.; Lindley, P.F. Reduction of dioxygen by enzymes containing copper. J. Biol. Inorg. Chem. 2006, 11, 539–547. [Google Scholar]
- Manavalan, T.; Manavalan, A.; Heese, K. Characterization of lignocellulolytic enzymes from white-rot fungi. Curr. Microbiol. 2015, 70, 485–498. [Google Scholar]
- Eggert, C.; Temp, U.; Eriksson, K.E.L. The Ligninolytic System of the White Rot Fungus Pycnoporus cinnabarinus: Purification and Characterization of the Laccase. Appl. Environ. Microbiol. 1996, 62, 1151–1158. [Google Scholar] [PubMed]
- Bourbonnais, R.; Paice, M.G.; Freiermuth, B.; Bodie, E.; Borneman, S. Reactivities of various mediators and laccases with kraft pulp and lignin model compounds. Appl. Environ. Microbiol. 1997, 63, 4627–4632. [Google Scholar] [PubMed]
- Camarero, S.; Ibarra, D.; Martínez, M.J.; Martínez, Á.T. Lignin-derived compounds as efficient laccase mediators for decolorization of different types of recalcitrant dyes. Appl. Environ. Microbiol. 2005, 71, 1775–1784. [Google Scholar] [PubMed]
- Rencoret, J.; Pereira, A.; José, C.; Martínez, A.T.; Gutiérrez, A. Laccase-mediator pretreatment of wheat straw degrades lignin and improves saccharification. Bioenergy Res. 2016, 9, 917–930. [Google Scholar]
- Munk, L.; Sitarz, A.K.; Kalyani, D.C.; Mikkelsen, J.D.; Meyer, A.S. Can laccases catalyze bond cleavage in lignin? Biotechnol. Adv. 2015, 33, 13–24. [Google Scholar]
- Kameshwar, A.K.S.; Barber, R.; Qin, W. Comparative modeling and molecular docking analysis of white, brown and soft rot fungal laccases using lignin model compounds for understanding the structural and functional properties of laccases. J. Mol. Graph. Model. 2018, 79, 15–26. [Google Scholar]
- Tamboli, A.S.; Rane, N.R.; Patil, S.M.; Biradar, S.P.; Pawar, P.K.; Govindwar, S.P. Physicochemical characterization, structural analysis and homology modeling of bacterial and fungal laccases using in silico methods. Netw. Modeling Anal. Health Inform. Bioinform. 2015, 4, 17. [Google Scholar]
- Chen, M.; Zeng, G.; Lai, C.; Li, J.; Xu, P.; Wu, H. Molecular basis of laccase bound to lignin: Insight from comparative studies on the interaction of Trametes versicolor laccase with various lignin model compounds. RSC Adv. 2015, 5, 52307–52313. [Google Scholar]
- Fu, N.; Wang, M.; Wang, L.; Luo, Y.; Ren, L. Genome Sequencing and Analysis of the Fungal Symbiont of Sirex noctilio, Amylostereum areolatum: Revealing the Biology of Fungus-Insect Mutualism. Msphere 2020, 5. [Google Scholar] [CrossRef]
- Bertrand, T.; Jolivalt, C.; Briozzo, P.; Caminade, E.; Joly, N.; Madzak, C.; Mougin, C. Crystal structure of a four-copper laccase complexed with an arylamine: Insights into substrate recognition and correlation with kinetics. Biochemistry 2002, 41, 7325–7333. [Google Scholar]
- Ducros, V.; Brzozowski, A.M.; Wilson, K.S.; Brown, S.H.; Østergaard, P.; Schneider, P.; Yaver, D.S.; Pedersen, A.H.; Davies, G.J. Crystal structure of the type-2 Cu depleted laccase from Coprinus cinereus at 2.2 Å resolution. Nat. Struct. Biol. 1998, 5, 310–316. [Google Scholar] [PubMed]
- Matera, I.; Gullotto, A.; Tilli, S.; Ferraroni, M.; Scozzafava, A.; Briganti, F. Crystal structure of the blue multicopper oxidase from the white-rot fungus Trametes trogii complexed with p-toluate. Inorg. Chim. Acta 2008, 361, 4129–4137. [Google Scholar]
- Ferraroni, M.; Myasoedova, N.M.; Schmatchenko, V.; Leontievsky, A.A.; Golovleva, L.A.; Scozzafava, A.; Briganti, F. Crystal structure of a blue laccase from Lentinus tigrinus: Evidences for intermediates in the molecular oxygen reductive splitting by multicopper oxidases. BMC Struct. Biol. 2007, 7, 60. [Google Scholar]
- Polyakov, K.M.; Fedorova, T.V.; Stepanova, E.V.; Cherkashin, E.A.; Kurzeev, S.A.; Strokopytov, B.V.; Lamzin, V.S.; Koroleva, O.V. Structure of native laccase from Trametes hirsuta at 1.8 Å resolution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009, 65, 611–617. [Google Scholar]
- Kumar, S.S.; Phale, P.S.; Durani, S.; Wangikar, P.P. Combined sequence and structure analysis of the fungal laccase family. Biotechnol. Bioeng. 2003, 83, 386–394. [Google Scholar]
- Jiao, X.; Li, G.; Wang, Y.; Nie, F.; Cheng, X.; Abdullah, M.; Lin, Y.; Cai, Y. Systematic analysis of the Pleurotus ostreatus laccase gene (PoLac) family and functional characterization of PoLac2 involved in the degradation of cotton-straw lignin. Molecules 2018, 23, 880. [Google Scholar]
- Larrondo, L.F.; Salas, L.; Melo, F.; Vicuña, R.; Cullen, D. A novel extracellular multicopper oxidase from Phanerochaete chrysosporium with ferroxidase activity. Appl. Environ. Microbiol. 2003, 69, 6257–6263. [Google Scholar]
- Hakulinen, N.; Kiiskinen, L.-L.; Kruus, K.; Saloheimo, M.; Paananen, A.; Koivula, A.; Rouvinen, J. Crystal structure of a laccase from Melanocarpus albomyces with an intact trinuclear copper site. Nat. Struct. Biol. 2002, 9, 601–605. [Google Scholar]
- Kilaru, S.; Hoegger, P.J.; Kües, U. The laccase multi-gene family in Coprinopsis cinerea has seventeen different members that divide into two distinct subfamilies. Curr. Genet. 2006, 50, 45–60. [Google Scholar]
- Gilbert, W. The exon theory of genes. Proc. Cold Spring Harb Symp. Quant. Biol. 1987, 52, 901–905. [Google Scholar]
- Bradshaw, R.A.J.T.i.b.s. Protein translocation and turnover in eukaryotic cells. Trends Biochem. Sci. 1989, 14, 276–279. [Google Scholar] [PubMed]
- Nakashima, H.; Nishikawa, K. Discrimination of intracellular and extracellular proteins using amino acid composition and residue-pair frequencies. J. Mol. Biol. 1994, 238, 54–61. [Google Scholar] [PubMed]
- Eggert, C.; Temp, U.; Eriksson, K.-E.L. Laccase is essential for lignin degradation by the white-rot fungus Pycnoporus cinnabarinus. FEBS Lett. 1997, 407, 89–92. [Google Scholar] [PubMed]
- Morozova, O.; Shumakovich, G.; Gorbacheva, M.; Shleev, S.; Yaropolov, A. “Blue” laccases. J. Biochem. 2007, 72, 1136–1150. [Google Scholar]
- Xu, F. Oxidation of phenols, anilines, and benzenethiols by fungal laccases: Correlation between activity and redox potentials as well as halide inhibition. J. Biochem. 1996, 35, 7608–7614. [Google Scholar]
- Chen, M.; Zeng, G.; Tan, Z.; Jiang, M.; Li, H.; Liu, L.; Zhu, Y.; Yu, Z.; Wei, Z.; Liu, Y. Understanding lignin-degrading reactions of ligninolytic enzymes: Binding affinity and interactional profile. PLoS ONE 2011, 6, e25647. [Google Scholar]
- Hongyan, L.; Zexiong, Z.; Shiwei, X.; He, X.; Yinian, Z.; Haiyun, L.; Zhongsheng, Y. Study on transformation and degradation of bisphenol A by Trametes versicolor laccase and simulation of molecular docking. Chemosphere 2019, 224, 743–750. [Google Scholar]
- Awasthi, M.; Jaiswal, N.; Singh, S.; Pandey, V.P.; Dwivedi, U.N. Molecular docking and dynamics simulation analyses unraveling the differential enzymatic catalysis by plant and fungal laccases with respect to lignin biosynthesis and degradation. J. Biomol. Struct. Dyn. 2015, 33, 1835–1849. [Google Scholar]
- Kallio, J.; Auer, S.; Jänis, J.; Andberg, M.; Kruus, K.; Rouvinen, J.; Koivula, A.; Hakulinen, N. Structure-function studies of a Melanocarpus albomyces laccase suggest a pathway for oxidation of phenolic compounds. J. Mol. Biol. 2009, 392, 895–909. [Google Scholar]
- Bourbonnais, R.; Paice, M.G. Demethylation and delignification of kraft pulp by Trametes versicolor laccase in the presence of 2,2′-azinobis-(3-ethylbenzthiazoline-6-sulphonate). Appl. Microbiol. Biotechnol. 1992, 36, 823–827. [Google Scholar]
- Kues, U.; Ruhl, M. Multiple multi-copper oxidase gene families in basidiomycetes-what for? Curr. Genom. 2011, 12, 72–94. [Google Scholar]
- Lalitha, S. Primer premier 5. Biotech Softw. Internet Rep. Comput. Softw. J. Sci. 2000, 1, 270–272. [Google Scholar]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Springer: Cham, Switzerland, 2005; pp. 571–607. [Google Scholar]
- Armenteros, J.J.A.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar]
- Ferrè, F.; Clote, P. DiANNA 1.1: An extension of the DiANNA web server for ternary cysteine classification. Nucleic Acids Res. 2006, 34, W182–W185. [Google Scholar]
- Geourjon, C.; Deleage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 1995, 11, 681–684. [Google Scholar]
- Gupta, R.; Jung, E.; Brunak, S. Prediction of N-Glycosylation Sites in Human Proteins. 2004. Available online: http://www.cbs.dtu.dk/services/NetNGlyc/ (accessed on 1 August 2020).
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [PubMed]
- Ge, H.; Gao, Y.; Hong, Y.; Zhang, M.; Xiao, Y.; Teng, M.; Niu, L. Structure of native laccase B from Trametes sp. AH28-2. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 254–258. [Google Scholar] [PubMed]
- Polyakov, K.M.; Gavryushov, S.; Ivanova, S.; Fedorova, T.V.; Glazunova, O.A.; Popov, A.N.; Koroleva, O.V. Structural study of the X-ray-induced enzymatic reduction of molecular oxygen to water by Steccherinum murashkinskyi laccase: Insights into the reaction mechanism. Acta Crystallogr. 2017, 73, 388–401. [Google Scholar]
- Glazunova, O.A.; Polyakov, K.M.; Moiseenko, K.V.; Kurzeev, S.A.; Fedorova, T.V. Structure-function study of two new middle-redox potential laccases from basidiomycetes Antrodiella faginea and Steccherinum murashkinskyi. Int. J. Biol. Macromol. 2018, 118 Pt A, 406–418. [Google Scholar]
- Wu, M.H.; Lee, C.C.; Hsiao, A.S.; Yu, S.M.; Wang, A.H.; Ho, T.D. Kinetic analysis and structural studies of a high-efficiency laccase from Cerrena sp. RSD1. FEBS Open Bio 2018, 8, 1230–1246. [Google Scholar] [PubMed]
- Polyakov, K.M.; Gavryushov, S.; Fedorova, T.V.; Glazunova, O.A.; Popov, A.N. The subatomic resolution study of laccase inhibition by chloride and fluoride anions using single-crystal serial crystallography: Insights into the enzymatic reaction mechanism. Acta Crystallogr. Sect. D 2019, 75, 804–816. [Google Scholar]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar]
- Heo, L.; Park, H.; Seok, C. GalaxyRefine: Protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 2013, 41, W384–W388. [Google Scholar]
- Pontius, J.; Richelle, J.; Wodak, S.J. Deviations from standard atomic volumes as a quality measure for protein crystal structures. J. Mol. Biol. 1996, 264, 121–136. [Google Scholar]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [PubMed]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple Ligand–Protein Interaction Diagrams for Drug Discovery; ACS Publications: Washington, DC, USA, 2011. [Google Scholar]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. Ccp4 Newsl. Protein Crystallogr. 2002, 40, 82–92. [Google Scholar]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar]




| Gene Name | DNA Length (bp) * | Intron # | Mature Protein (aa) | MW (kDa) | pI | Signal Peptide (aa) | Cysteine Residues # | Predicted S-S Bonds |
|---|---|---|---|---|---|---|---|---|
| AaLac1 | 2223 | 12 | 523 | 56.96001 | 4.70 | 20–21 | 5 | 104–512, 136–474 |
| AaLac2 | 2353 | 14 | 517 | 56.48112 | 6.12 | 18–19 | 5 | 103–470, 135–224 |
| AaLac3 | 2489 | 15 | 518 | 56.37469 | 5.04 | / | 5 | 106–473, 138–226 |
| AaLac4 | 2182 | 12 | 509 | 54.40684 | 4.90 | 20–21 | 5 | 104–467, 136–223 |
| AaLac5 | 2910 | 19 | 520 | 55.87515 | 4.52 | 22–23 | 5 | 107–471, 139–229 |
| AaLac6 | 2579 | 18 | 520 | 56.39443 | 5.56 | 20–21 | 5 | / |
| AaLac7 | 2604 | 19 | 524 | 57.36822 | 4.81 | 20–21 | 5 | 104–475, 136–230 |
| AaLac8 | 2934 | 23 | 529 | 57.82204 | 5.04 | 32–33 | 5 | 116–242, 148–480 |
| AaLac9 | 2630 | 19 | 525 | 56.82742 | 4.93 | 19–20 | 5 | 105–509, 137–226 |
| AaLac10 | 2253 | 13 | 520 | 56.42250 | 4.53 | 19–20 | 3 | 102–474 |
| AaLac11 | 2717 | 19 | 523 | 57.07710 | 5.72 | 22–23 | 5 | 107–511, 228–476 |
| AaLac12 | 2332 | 14 | 521 | 56.41483 | 4.45 | 19–20 | 3 | 102–475 |
| AaLac13 | 2134 | 11 | 509 | 54.50674 | 4.63 | 20–21 | 5 | 104–467, 136–223 |
| AaLac14 | 2249 | 13 | 508 | 54.90668 | 4.20 | 20–21 | 5 | 104–467, 136–223 |
| Protein Name | Template | Sequence Identity | Coverage | Verify 3D | ERRAT | G-Factors | LGscore | MaxSub | Z-Score | GMQE | QMEAN | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dihedrals | Covalent | Overall | |||||||||||
| AaLac1 | 3kw7.1.A | 53.88% | 0.91 | 90.30% | 86.5263 | 0.12 | −0.41 | −0.07 | 4.598 | 0.313 | −7.33 | 0.75 | −3.74 |
| AaLac2 | 5mhw.1.A | 58.37% | 0.95 | 91.89% | 83.9248 | 0.17 | −0.37 | −0.03 | 4.995 | 0.342 | −8.45 | 0.81 | −0.24 |
| AaLac3 | 5ehf.1.A | 57.99% | 0.94 | 92.67% | 86.8476 | 0.18 | −0.31 | 0 | 4.742 | 0.322 | −8.27 | 0.8 | 0.08 |
| AaLac4 | 5mej.1.A | 63.39% | 0.94 | 92.07% | 80.9829 | 0.12 | −0.38 | −0.06 | 4.544 | 0.319 | −7.94 | 0.81 | −0.98 |
| AaLac5 | 5mej.1.A | 63.69% | 0.95 | 97.17% | 82.5 | 0.16 | −0.38 | −0.05 | 5.087 | 0.343 | −7.17 | 0.81 | −0.23 |
| AaLac6 | 5mhw.1.A | 63.37% | 0.93 | 93.25% | 89.0985 | 0.16 | −0.42 | −0.06 | 5.022 | 0.355 | −7.16 | 0.81 | 0.49 |
| AaLac7 | 5mew.1.A | 52.62% | 0.91 | 92.48% | 88.843 | 0.14 | −0.4 | −0.06 | 4.832 | 0.328 | −8.62 | 0.77 | −1.67 |
| AaLac8 | 5mej.1.A | 56.05% | 0.89 | 90.31% | 84.8936 | 0.16 | −0.44 | −0.06 | 4.999 | 0.324 | −8.16 | 0.76 | −1.93 |
| AaLac9 | 5z1x.1.A | 60.25% | 0.93 | 95.77% | 84.7107 | 0.14 | −0.39 | −0.06 | 4.995 | 0.345 | −7.85 | 0.79 | −1.61 |
| AaLac10 | 6rhh.1.A | 45.67% | 0.91 | 92.02% | 85.8672 | 0.12 | −0.42 | −0.08 | 3.891 | 0.208 | −7.4 | 0.74 | −1.52 |
| AaLac11 | 5ehf.1.A | 61.54% | 0.92 | 96.93% | 85.2083 | 0.14 | −0.42 | −0.06 | 5.089 | 0.36 | −7.94 | 0.8 | −0.51 |
| AaLac12 | 2xyb.1.A | 42.86% | 0.93 | 92.46% | 83.8057 | 0.13 | −0.44 | −0.08 | 4.179 | 0.233 | −7.08 | 0.72 | −3.5 |
| AaLac13 | 5mew.1.A | 65.06% | 0.94 | 93.11% | 88.0952 | 0.14 | −0.36 | −0.04 | 4.245 | 0.29 | −7.96 | 0.81 | −0.27 |
| AaLac14 | 5mew.1.A | 62.21% | 0.94 | 95.82% | 89.0792 | 0.14 | −0.35 | −0.04 | 4.450 | 0.31 | −8.18 | 0.82 | 0.28 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, N.; Li, J.; Wang, M.; Ren, L.; Luo, Y. Genes Identification, Molecular Docking and Dynamics Simulation Analysis of Laccases from Amylostereum areolatum Provides Molecular Basis of Laccase Bound to Lignin. Int. J. Mol. Sci. 2020, 21, 8845. https://doi.org/10.3390/ijms21228845
Fu N, Li J, Wang M, Ren L, Luo Y. Genes Identification, Molecular Docking and Dynamics Simulation Analysis of Laccases from Amylostereum areolatum Provides Molecular Basis of Laccase Bound to Lignin. International Journal of Molecular Sciences. 2020; 21(22):8845. https://doi.org/10.3390/ijms21228845
Chicago/Turabian StyleFu, Ningning, Jiaxing Li, Ming Wang, Lili Ren, and Youqing Luo. 2020. "Genes Identification, Molecular Docking and Dynamics Simulation Analysis of Laccases from Amylostereum areolatum Provides Molecular Basis of Laccase Bound to Lignin" International Journal of Molecular Sciences 21, no. 22: 8845. https://doi.org/10.3390/ijms21228845
APA StyleFu, N., Li, J., Wang, M., Ren, L., & Luo, Y. (2020). Genes Identification, Molecular Docking and Dynamics Simulation Analysis of Laccases from Amylostereum areolatum Provides Molecular Basis of Laccase Bound to Lignin. International Journal of Molecular Sciences, 21(22), 8845. https://doi.org/10.3390/ijms21228845






