Circulating Coding and Long Non-Coding RNAs as Potential Biomarkers of Idiopathic Pulmonary Fibrosis
Abstract
1. Introduction
2. Results
2.1. High Throughput Transcriptome Analysis
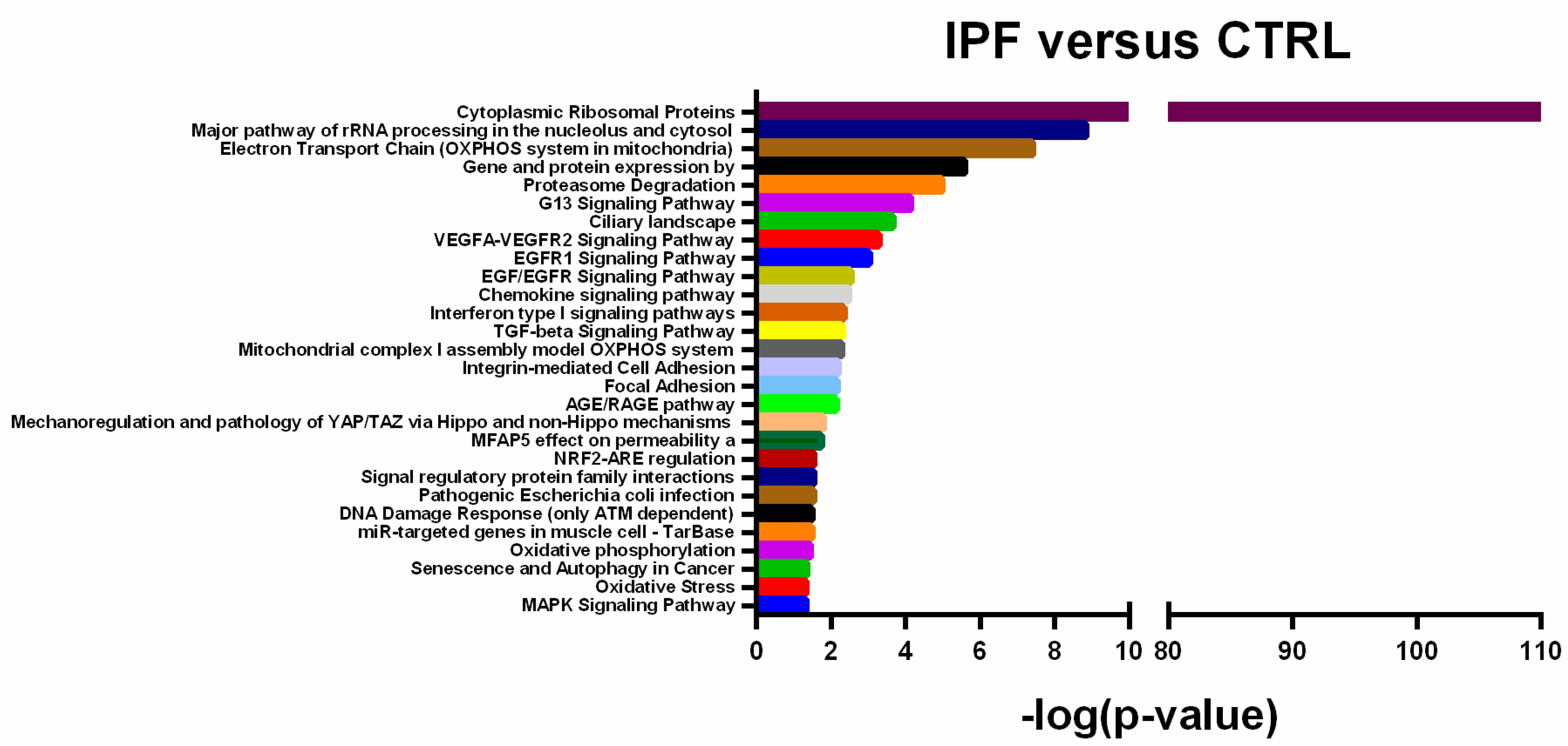
2.2. Differentially Expressed Transcripts are Associated with IPF Molecular Pathways
2.3. Validation of Deregulated Transcripts
2.4. Validated Transcripts are Associated with Lung Function Parameters
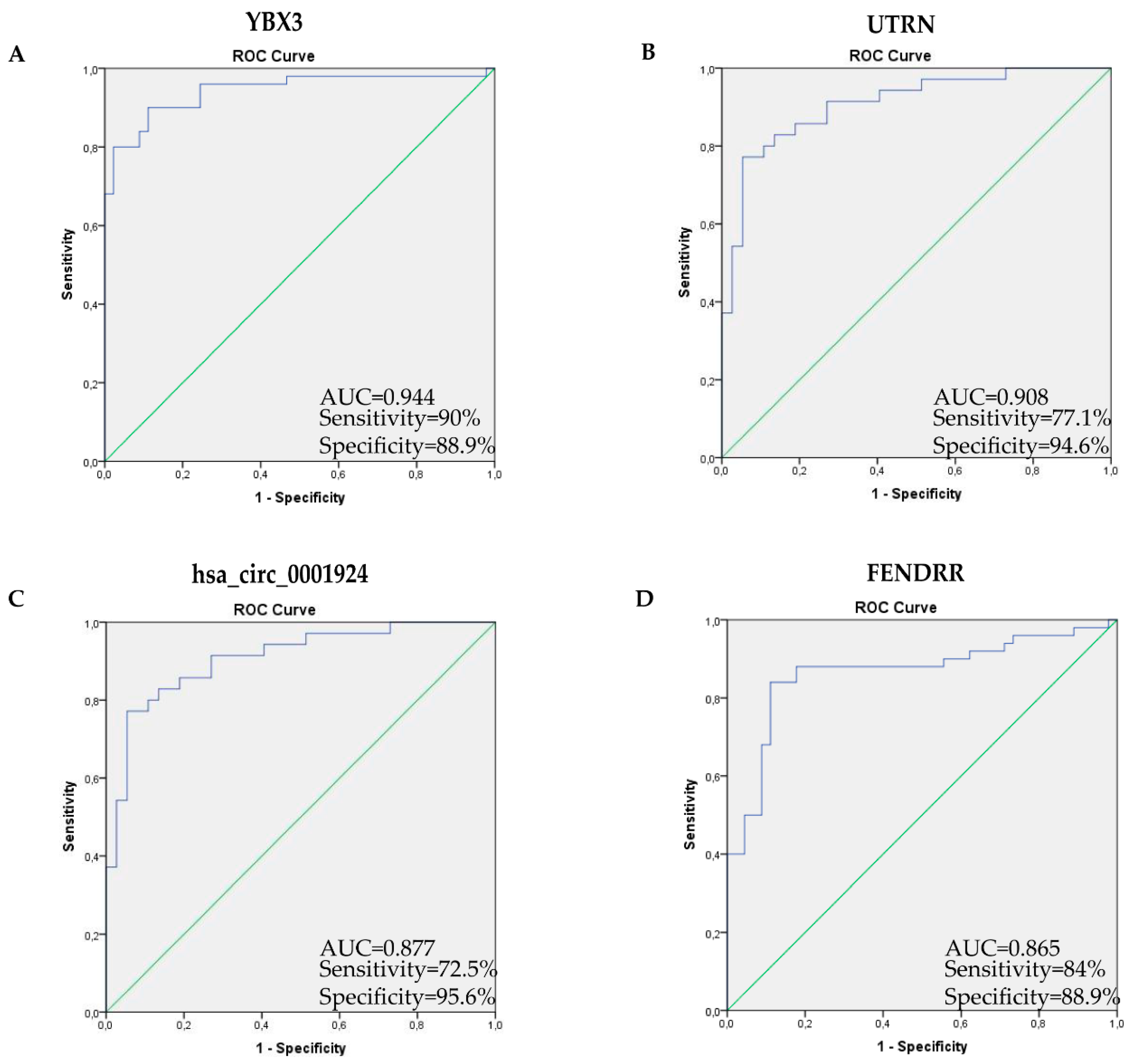
2.5. Receiver Operating Characteristic Curve Analysis Shows High Transcript Diagnostic Performance
3. Discussion
4. Materials and Methods
4.1. Study Subjects
4.2. Sample Processing
4.3. RNA Isolation
4.4. Microarray Analysis
4.5. Computational Analysis
4.6. Single Real-Time PCR Assays
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| IPF | Idiopathic Pulmonary Fibrosis |
| lncRNA | Long Non-Coding RNA |
| FENDRR | FOXF1 adjacent non-coding developmental regulatory RNA |
| UTRN | utrophin |
| YBX3 | Y-box binding protein 3 |
| FVC | Forced Vital Capacity |
| DLCO | Diffusing Capacity of the Lung for carbon monoxide |
| AUC | Area Under the Curve |
| RT | Room Temperature |
| FC | Fold Change |
| FDR | False Discovery Rate |
| DE | Differentially Expressed |
| UPR | Unfolded Protein Response |
| BMI | Body Mass Index |
| FVC | Forced Vital Capacity |
| DLCO | Diffusing Capacity of the Lung for carbon monoxide |
| ROC | Receiver Operating Characteristic |
| AUROC | Area Under the Receiver Operating Characteristics |
| UIP | Usual Interstitial Pneumonia |
| HRCT | High-Resolution Computed Tomography |
| DMD | Duchenne Muscular Dystrophy |
References
- Torrisi, S.E.; Pavone, M.; Vancheri, A.; Vancheri, C. When to start and when to stop antifibrotic therapies. Eur. Respir. Rev. 2017, 26, 170053. [Google Scholar] [CrossRef] [PubMed]
- Fahim, A.; Dettmar, P.W.; Morice, A.H.; Hart, S.P. Gastroesophageal reflux and idiopathic pulmonary fibrosis: A prospective study. Pulm. med. 2011, 47, 28. [Google Scholar] [CrossRef]
- Desai, O.; Winkler, J.; Minasyan, M.; Herzog, E.L. The Role of Immune and Inflammatory Cells in Idiopathic Pulmonary Fibrosis. Front. Med. 2018, 5, 43. [Google Scholar] [CrossRef] [PubMed]
- Sgalla, G.; Iovene, B.; Calvello, M.; Ori, M.; Varone, F.; Richeldi, L. Idiopathic pulmonary fibrosis: Pathogenesis and management. Respir. Res. 2018, 19, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.; Wang, J.; Shan, B.; Peng, Z.; Dong, Y.; Shi, W.; He, D.; Cheng, Y.; Zhao, W.; Zhang, C.; et al. Diagnostic and Prognostic Potential of Circulating Long Non-Coding RNAs in Non Small Cell Lung Cancer. Cell. Physiol. Biochem. 2018, 49, 816–827. [Google Scholar] [CrossRef]
- Yuan, Y.; Peng, W.; Liu, Y.; Xu, Z.-S. Circulating miR-130 and its target PPAR-γ may be potential biomarkers in patients of coronary artery disease with type 2 diabetes mellitus. Mol. Genet. Genom. Med. 2019, 7, e909. [Google Scholar] [CrossRef]
- Di Mauro, S.; Scamporrino, A.; Petta, S.; Urbano, F.; Filippello, A.; Ragusa, M.; Di Martino, M.T.; Scionti, F.; Grimaudo, S.; Pipitone, R.M.; et al. Serum coding and non-coding RNAs as biomarkers of NAFLD and fibrosis severity. Liver int. off. J. Int. Assoc. Study Liver 2019, 39, 1742–1754. [Google Scholar] [CrossRef]
- Santos, M.C.D.L.; Dragomir, M.P.; Calin, G.A. The role of exosomal long non-coding RNAs in cancer drug resistance. Cancer Drug Resist. 2019, 2, 1178–1192. [Google Scholar] [CrossRef]
- Zhang, Y.; Luo, G.; Zhang, Y.; Zhang, M.; Zhou, J.; Gao, W.; Xuan, X.; Yang, X.; Yang, D.; Tian, Z.; et al. Critical effects of long non-coding RNA on fibrosis diseases. Exp. Mol. Med. 2018, 50, e428. [Google Scholar] [CrossRef]
- Wetmore, B.A.; Brees, D.J.; Singh, R.; Watkins, P.B.; Andersen, M.E.; Loy, J.; Thomas, R.S. Quantitative analyses and transcriptomic profiling of circulating messenger RNAs as biomarkers of rat liver injury. Hepatology 2010, 51, 2127–2139. [Google Scholar] [CrossRef]
- Hasselmann, D.O.; Rappl, G.; Tilgen, W.; Reinhold, U. Extracellular Tyrosinase mRNA within Apoptotic Bodies Is Protected from Degradation in Human Serum. Clin. Chem. 2001, 47, 1488–1489. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Gao, G.; Cao, Y. Long Noncoding RNAs as Novel Biomarkers Have a Promising Future in Cancer Diagnostics. Dis. Markers 2016, 2016, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kishikawa, T.; Otsuka, M.; Ohno, M.; Yoshikawa, T.; Takata, A.; Koike, K. Circulating RNAs as new biomarkers for detecting pancreatic cancer. World J. Gastroenterol. 2015, 21, 8527–8540. [Google Scholar] [CrossRef] [PubMed]
- Yuan, T.; Huang, X.; Woodcock, M.; Du, M.; Dittmar, R.; Wang, Y.; Tsai, S.; Kohli, M.; Boardman, L.A.; Patel, T.; et al. Plasma extracellular RNA profiles in healthy and cancer patients. Sci. Rep. 2016, 6, srep19413. [Google Scholar] [CrossRef]
- Yang, G.; Yang, L.; Wang, W.; Wang, J.; Wang, J.; Xu, Z. Discovery and validation of extracellular/circulating microRNAs during idiopathic pulmonary fibrosis disease progression. Gene 2015, 562, 138–144. [Google Scholar] [CrossRef]
- Huang, C.; Liang, Y.; Zeng, X.; Yang, X.; Xu, D.; Gou, X.; Sathiaseelan, R.; Senavirathna, L.K.; Wang, P.; Liu, L. Long Noncoding RNA FENDRR Exhibits Antifibrotic Activity in Pulmonary Fibrosis. Am. J. Respir. Cell Mol. Biol. 2020, 62, 440–453. [Google Scholar] [CrossRef]
- Senavirathna, L.K.; Huang, C.; Pushparaj, S.; Xu, D.; Liu, L. Hypoxia and transforming growth factor beta1 regulation of long non-coding RNA transcriptomes in human pulmonary fibroblasts. Phys. Rep. 2020, 8, e14343. [Google Scholar]
- Cooke, A.; Schwarzl, T.; Huppertz, I.; Kramer, G.; Mantas, P.; Alleaume, A.-M.; Huber, W.; Krijgsveld, J.; Hentze, M.W. The RNA-Binding Protein YBX3 Controls Amino Acid Levels by Regulating SLC mRNA Abundance. Cell Rep. 2019, 27, 3097–3106.e5. [Google Scholar] [CrossRef]
- Yilermi, S. Claudins in lung diseases. Respir. Res. 2011, 12, 70. [Google Scholar]
- Levi, O.; Genin, O.; Angelini, C.; Halevy, O.; Pines, M. Inhibition of muscle fibrosis results in increases in both utrophin levels and the number of revertant myofibers in Duchenne muscular dystrophy. Oncotarget 2015, 6, 23249–23260. [Google Scholar] [CrossRef]
- Morgoulis, D.; Berenstein, P.; Cazacu, S.; Kazimirsky, G.; Dori, A.; Barnea, E.R.; Brodie, C. sPIF promotes myoblast differentiation and utrophin expression while inhibiting fibrosis in Duchenne muscular dystrophy via the H19/miR-675/let-7 and miR-21 pathways. Cell Death Dis. 2019, 10, 82. [Google Scholar] [CrossRef] [PubMed]
- Di Mauro, S.; Ragusa, M.; Urbano, F.; Filippello, A.; Di Pino, A.; Scamporrino, A.; Pulvirenti, A.; Ferro, A.; Rabuazzo, A.; Purrello, R.; et al. Intracellular and extracellular miRNome deregulation in cellular models of NAFLD or NASH: Clinical implications. Nutr. Metab. Cardiovasc. Dis. 2016, 26, 1129–1139. [Google Scholar] [CrossRef] [PubMed]
- Scicali, R.; Di Pino, A.; Pavanello, C.; Ossoli, A.; Strazzella, A.; Alberti, A.; Di Mauro, S.; Scamporrino, A.; Urbano, F.; Filippello, A.; et al. Analysis of HDL-microRNA panel in heterozygous familial hypercholesterolemia subjects with LDL receptor null or defective mutation. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Filippello, A.; Urbano, F.; Di Mauro, S.; Scamporrino, A.; Di Pino, A.; Scicali, R.; Rabuazzo, A.M.; Purrello, F.; Piro, S. Chronic Exposure to Palmitate Impairs Insulin Signaling in an Intestinal L-cell Line: A Possible Shift from GLP-1 to Glucagon Production. Int. J. Mol. Sci. 2018, 19, 3791. [Google Scholar] [CrossRef]


| Clinical Characteristics | Control (n = 45) | IPF (n = 50) | p-Value |
|---|---|---|---|
| Gender (% M) | 60 | 78 | 0.216 |
| Age (years) | 68.24 ± 8.53 | 71.32 ± 8.28 | 0.078 |
| Weight (Kg) | 74.59 ± 14.98 | 73.12 ± 13.25 | 0.617 |
| Height (cm) | 167 ± 9.19 | 164 ± 9.79 | 0.131 |
| BMI (kg/m2) | 27.06 ± 4.02 | 27.56 ± 3.54 | 0.525 |
| Pack-years | 15 ± 25.70 | 20 ± 33.83 | 0.790 |
| FVC% | 82.02 ± 19.08 | ||
| DLCO% | 55 ± 14.76 |
| PEARSON CORRELATION | ||
| FVC % | ||
| p-value | R2 | |
| FENDRR | 0.035 | 0.07 |
| hsa_circ_0001924 | 0.033 | 0.09 |
| SPEARMAN CORRELATION | ||
| DLCO % | PACK YEARS | |
| p-value | p-value | |
| FENDRR | 0.008 | 0.139 |
| hsa_circ_0001924 | 0.016 | 0.454 |
| UTRN | 0.228 | 0.038 |
| YBX3 | 0.388 | 0.034 |
| Forward | Reverse | |
|---|---|---|
| hsa_circ_0001924 | ACCATGGCGAGTGGAAGAAAT | TTTAATCTAGGTGAAATGCCCCTT |
| FENDRR | CACGAAAGGTGGTGCCGAGA | TCCGTTTGCATCCAACATTGTCT |
| UTRN | CCTGACAATGGGCAGAACGA | CCCACTCTTTGAAAATCGAGCA |
| YBX3 | AGCGAAGACGCGGAGAAAAA | TCCATTTCTGACGTTGAACCA |
| GAPDH | TGCACCACCAACTGCTTAGC | GGCATGGACTGTGGTCATGAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Mauro, S.; Scamporrino, A.; Fruciano, M.; Filippello, A.; Fagone, E.; Gili, E.; Scionti, F.; Purrazzo, G.; Di Pino, A.; Scicali, R.; et al. Circulating Coding and Long Non-Coding RNAs as Potential Biomarkers of Idiopathic Pulmonary Fibrosis. Int. J. Mol. Sci. 2020, 21, 8812. https://doi.org/10.3390/ijms21228812
Di Mauro S, Scamporrino A, Fruciano M, Filippello A, Fagone E, Gili E, Scionti F, Purrazzo G, Di Pino A, Scicali R, et al. Circulating Coding and Long Non-Coding RNAs as Potential Biomarkers of Idiopathic Pulmonary Fibrosis. International Journal of Molecular Sciences. 2020; 21(22):8812. https://doi.org/10.3390/ijms21228812
Chicago/Turabian StyleDi Mauro, Stefania, Alessandra Scamporrino, Mary Fruciano, Agnese Filippello, Evelina Fagone, Elisa Gili, Francesca Scionti, Giacomo Purrazzo, Antonino Di Pino, Roberto Scicali, and et al. 2020. "Circulating Coding and Long Non-Coding RNAs as Potential Biomarkers of Idiopathic Pulmonary Fibrosis" International Journal of Molecular Sciences 21, no. 22: 8812. https://doi.org/10.3390/ijms21228812
APA StyleDi Mauro, S., Scamporrino, A., Fruciano, M., Filippello, A., Fagone, E., Gili, E., Scionti, F., Purrazzo, G., Di Pino, A., Scicali, R., Di Martino, M. T., Malaguarnera, R., Malatino, L., Purrello, F., Vancheri, C., & Piro, S. (2020). Circulating Coding and Long Non-Coding RNAs as Potential Biomarkers of Idiopathic Pulmonary Fibrosis. International Journal of Molecular Sciences, 21(22), 8812. https://doi.org/10.3390/ijms21228812












