Substrate Specificity and Structural Modeling of Human Carboxypeptidase Z: A Unique Protease with a Frizzled-Like Domain
Abstract
1. Introduction
2. Results
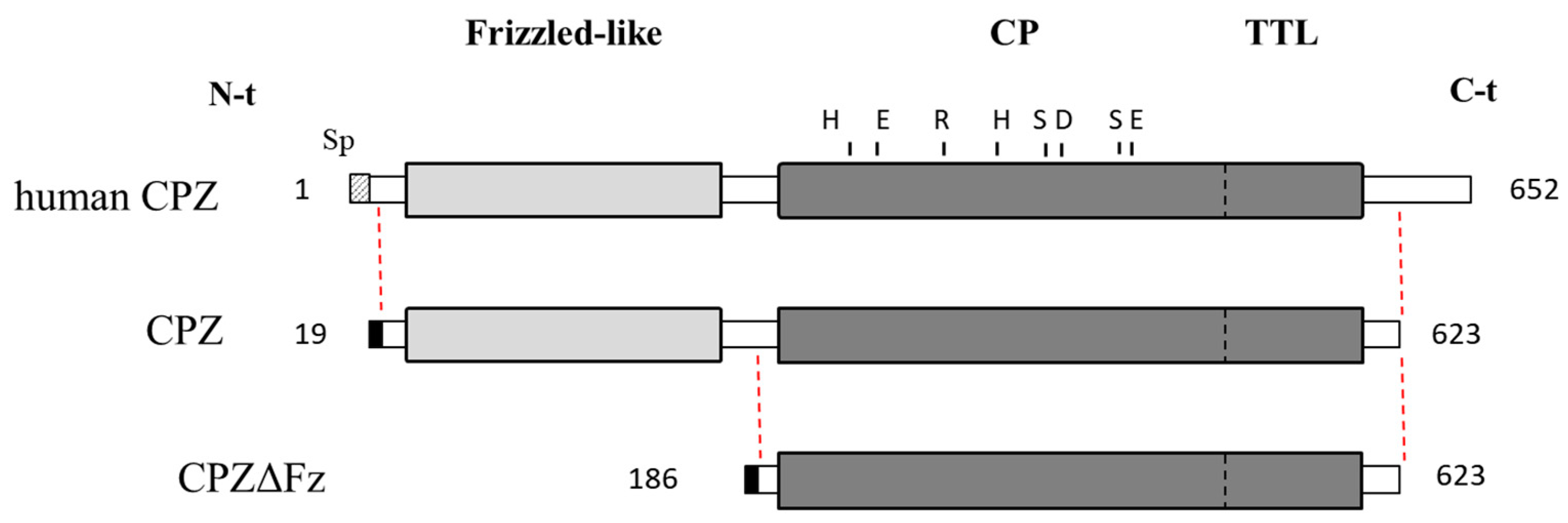
2.1. Recombinant Protein Expression and Purification
2.2. Enzymatic Characterization of CPZ Using Fluorescent Synthetic Substrates
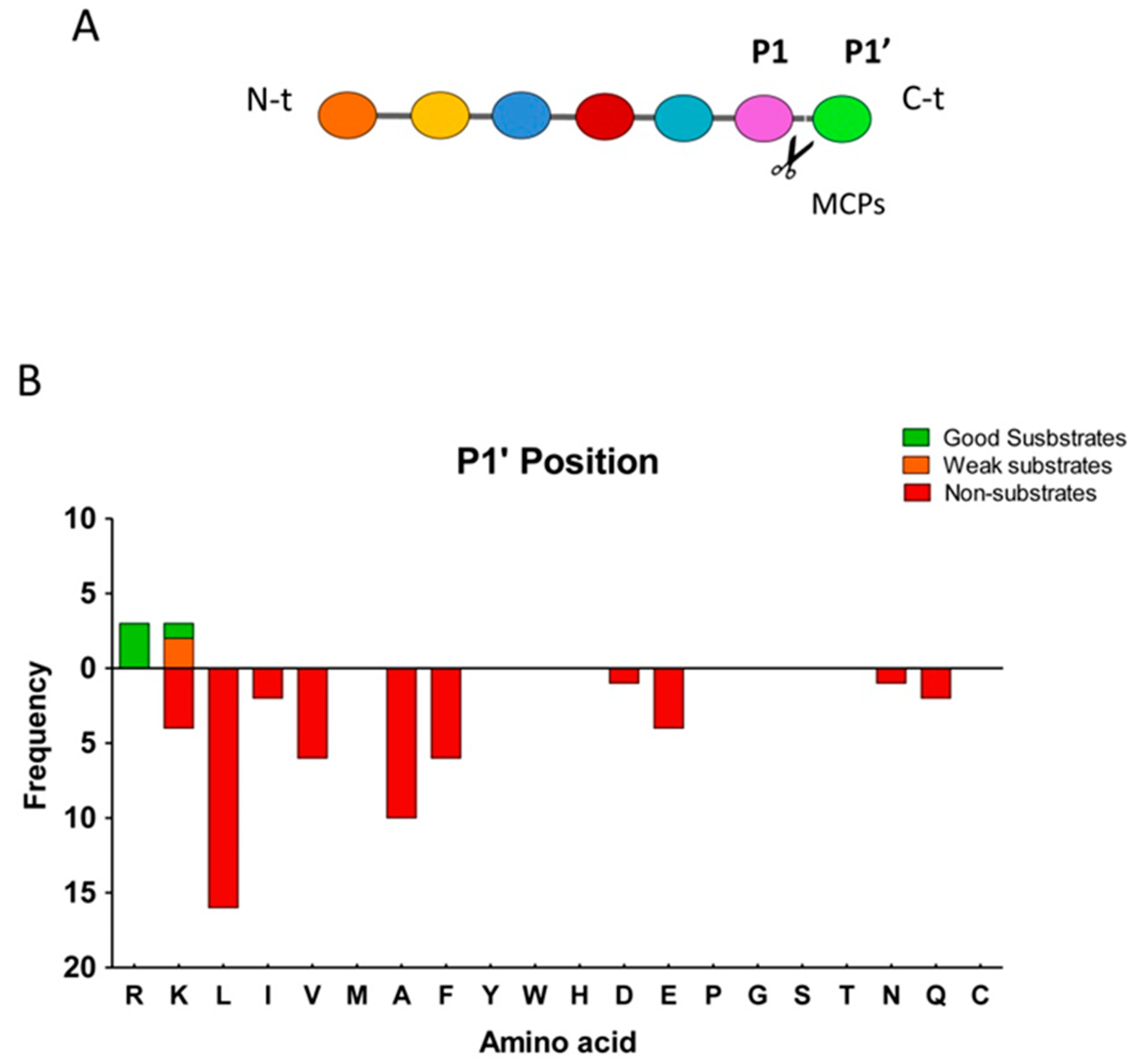
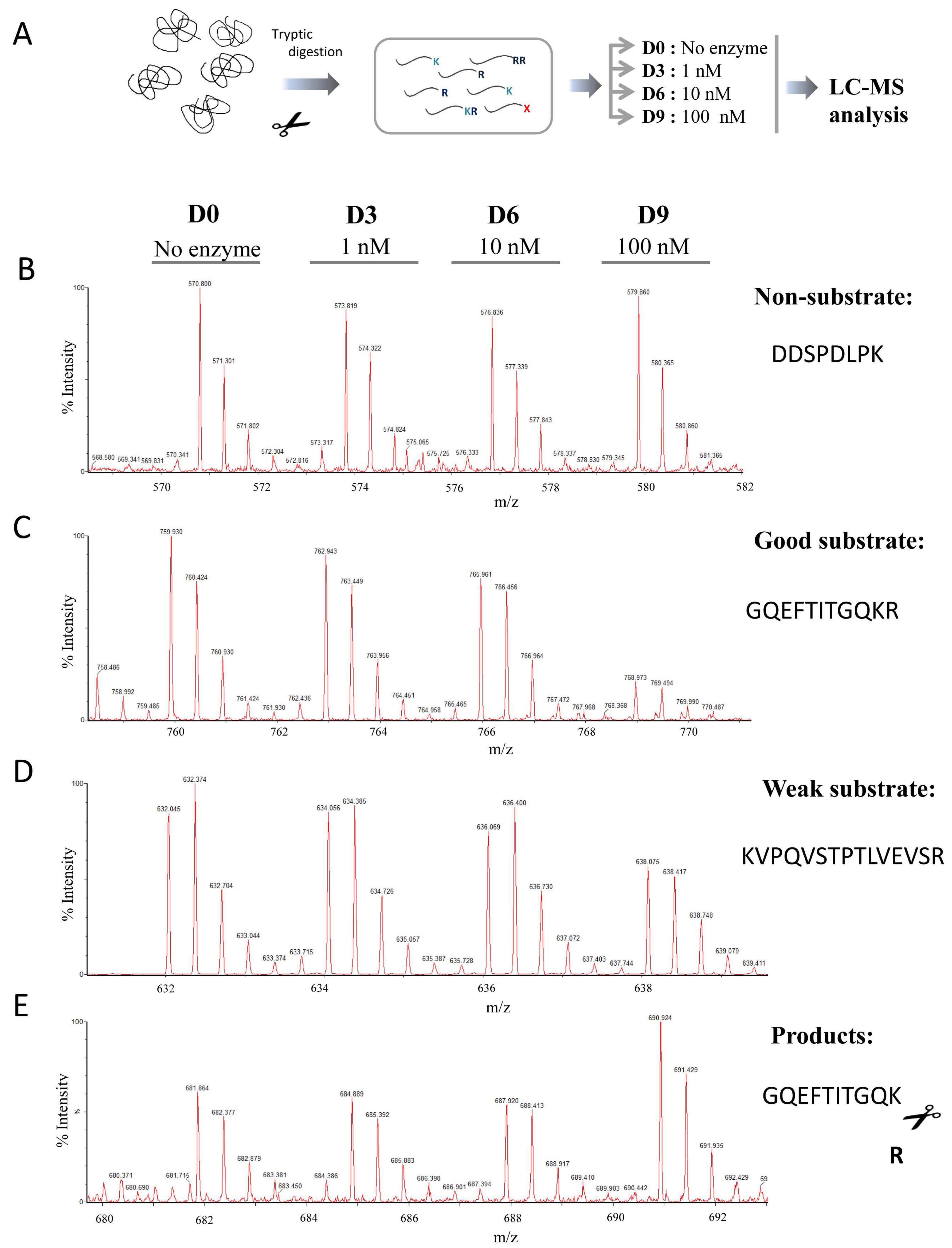
2.3. Substrate Specificity Profiling of Human CPZ by Quantitative Peptidomics Appproaches
2.4. Structural Modeling of the Catalytic and TTL Domains of Human CPZ
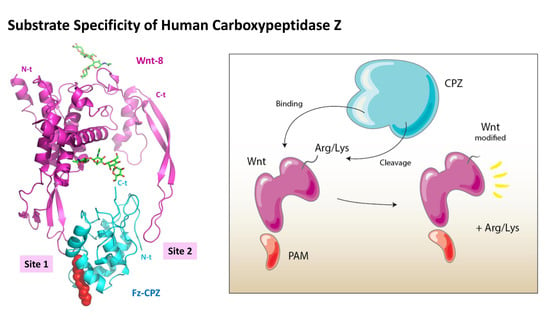
2.5. Structural Modeling of the CPZ Fz Domain: Insights into Its Structure and Wnt Recognition
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Recombinant Protein Production and Purification
4.3. HEK293T Bortezomib Treatment and Peptide Extraction
4.4. Generation of the Tryptic Peptide Library
4.5. Kinetic Measurements Using Fluorescent Synthetic Substrates
4.6. Quantitative Peptidomics Analyses
4.7. MALDI-TOF Mass Spectrometry Experiments
4.8. Sequence Alignment and Structural Modeling
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CP | Carboxypeptidase |
| CPA | Carboxypeptidase A |
| CPA1 | Carboxypeptidase A1 |
| CPD | Carboxypeptidase D |
| CPE | Carboxypeptidase E |
| CPM | Carboxypeptidase M |
| CPN | Carboxypeptidase N |
| CPZ | Carboxypeptidase Z |
| ECM | Extracellular matrix |
| EGF | Epidermal Growth Factor |
| Fz | Frizzled-like |
| LC-MS | Liquid chromatography-mass spectrometry |
| MALDI-TOF MS | Matrix-assisted laser desorption ionization time-of-flight mass spectrometry |
| MCPs | Metallocarboxypeptidases |
| PDB | Protein data bank |
| sFRP | secreted Frizzled-like related protein |
| TMAB | 4-trimethyl-ammoniumbutyrate |
| TTL | Transthyretin-like domain |
| WG | Wingless |
References
- Arolas, J.L.; Vendrell, J.; Aviles, F.X.; Fricker, L.D. Metallocarboxypeptidases: Emerging drug targets in biomedicine. Curr. Pharm. Des. 2007, 13, 349–366. [Google Scholar] [CrossRef] [PubMed]
- Reznik, S.E.; Fricker, L.D. Carboxypeptidases from a to Z: Implications in embryonic development and wnt binding. Cell Mol. Life Sci. 2001, 58, 1790–1804. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, N.D. Twenty-five years of nomenclature and classification of proteolytic enzymes. Biochim. Biophys. Acta Proteins Proteom. 2020, 1868, 140345. [Google Scholar] [CrossRef]
- Reverter, D.; Maskos, K.; Tan, F.; Skidgel, R.A.; Bode, W. Crystal structure of human carboxypeptidase M, a membrane-bound enzyme that regulates peptide hormone activity. J. Mol. Biol. 2004, 338, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Pardo, J.; Grana-Montes, R.; Fernandez-Mendez, M.; Ruyra, A.; Roher, N.; Aviles, F.X.; Lorenzo, J.; Ventura, S. Amyloid formation by human carboxypeptidase D transthyretin-like domain under physiological conditions. J. Biol. Chem. 2014, 289, 33783–33796. [Google Scholar] [CrossRef]
- Keil, C.; Maskos, K.; Than, M.; Hoopes, J.T.; Huber, R.; Tan, F.L.; Deddish, P.A.; Erdos, E.G.; Skidgel, R.A.; Bode, W. Crystal structure of the human carboxypeptidase N (kininase I) catalytic domain. J. Mol. Biol. 2007, 366, 504–516. [Google Scholar] [CrossRef]
- Tanco, S.; Arolas, J.L.; Guevara, T.; Lorenzo, J.; Aviles, F.X.; Gomis-Ruth, F.X. Structure-function analysis of the short splicing variant carboxypeptidase encoded by drosophila melanogaster silver. J. Mol. Biol. 2010, 401, 465–477. [Google Scholar] [CrossRef]
- Garcia-Pardo, J.; Tanco, S.; Diaz, L.; Dasgupta, S.; Fernandez-Recio, J.; Lorenzo, J.; Aviles, F.X.; Fricker, L.D. Substrate specificity of human metallocarboxypeptidase D: Comparison of the two active carboxypeptidase domains. PLoS ONE 2017, 12, e0187778. [Google Scholar] [CrossRef]
- Gomis-Ruth, F.X.; Companys, V.; Qian, Y.; Fricker, L.D.; Vendrell, J.; Aviles, F.X.; Coll, M. Crystal structure of avian carboxypeptidase D domain II: A prototype for the regulatory metallocarboxypeptidase subfamily. EMBO J. 1999, 18, 5817–5826. [Google Scholar] [CrossRef]
- Zhang, X.; Che, F.Y.; Berezniuk, I.; Sonmez, K.; Toll, L.; Fricker, L.D. Peptidomics of CPE(fat/fat) mouse brain regions: Implications for neuropeptide processing. J. Neurochem. 2008, 107, 1596–1613. [Google Scholar] [CrossRef]
- Deiteren, K.; Hendriks, D.; Scharpe, S.; Lambeir, A.M. Carboxypeptidase M: Multiple alliances and unknown partners. Clin. Chim. Acta 2009, 399, 24–39. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Tan, F.; Skidgel, R.A. Carboxypeptidase M is a positive allosteric modulator of the kinin b1 receptor. J. Biol. Chem. 2013, 288, 33226–33240. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Pierce, S.E.; Li, A.; Spees, K.; Anderson, G.R.; Seoane, J.A.; Lo, Y.H.; Dubreuil, M.; Olivas, M.; Kamber, R.A.; et al. CRISPR screens in cancer spheroids identify 3D growth-specific vulnerabilities. Nature 2020, 580, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Novikova, E.; Fricker, L.D.; Reznik, S.E. Metallocarboxypeptidase Z is dynamically expressed in mouse development. Mech. Dev. 2001, 102, 259–262. [Google Scholar] [CrossRef]
- Novikova, E.G.; Fricker, L.D. Purification and characterization of human metallocarboxypeptidase Z. Biochem. Biophys. Res. Commun. 1999, 256, 564–568. [Google Scholar] [CrossRef]
- Novikova, E.G.; Reznik, S.E.; Varlamov, O.; Fricker, L.D. Carboxypeptidase Z is present in the regulated secretory pathway and extracellular matrix in cultured cells and in human tissues. J. Biol. Chem. 2000, 275, 4865–4870. [Google Scholar] [CrossRef]
- Song, L.; Fricker, L.D. Cloning and expression of human carboxypeptidase Z, a novel metallocarboxypeptidase. J. Biol. Chem. 1997, 272, 10543–10550. [Google Scholar] [CrossRef]
- Rehn, M.; Pihlajaniemi, T.; Hofmann, K.; Bucher, P. The frizzled motif: In how many different protein families does it occur? Trends Biochem. Sci. 1998, 23, 415–417. [Google Scholar] [CrossRef]
- Reya, T.; Clevers, H. Wnt signalling in stem cells and cancer. Nature 2005, 434, 843–850. [Google Scholar] [CrossRef]
- Clevers, H.; Nusse, R. Wnt/beta-catenin signaling and disease. Cell 2012, 149, 1192–1205. [Google Scholar] [CrossRef]
- Anastas, J.N.; Moon, R.T. Wnt signalling pathways as therapeutic targets in cancer. Nat. Rev. Cancer 2013, 13, 11–26. [Google Scholar] [CrossRef]
- Koni, M.; Pinnaro, V.; Brizzi, M.F. The Wnt signalling pathway: A tailored target in cancer. Int. J. Mol. Sci. 2020, 21, 7697. [Google Scholar] [CrossRef]
- Leimeister, C.; Bach, A.; Gessler, M. Developmental expression patterns of mouse sFRP genes encoding members of the secreted frizzled related protein family. Mech. Dev. 1998, 75, 29–42. [Google Scholar] [CrossRef]
- Bovolenta, P.; Esteve, P.; Ruiz, J.M.; Cisneros, E.; Lopez-Rios, J. Beyond Wnt inhibition: New functions of secreted Frizzled-related proteins in development and disease. J. Cell. Sci. 2008, 121, 737–746. [Google Scholar] [CrossRef]
- Moeller, C.; Swindell, E.C.; Kispert, A.; Eichele, G. Carboxypeptidase Z (CPZ) modulates Wnt signaling and regulates the development of skeletal elements in the chicken. Development 2003, 130, 5103–5111. [Google Scholar] [CrossRef]
- Wang, L.; Shao, Y.Y.; Ballock, R.T. Carboxypeptidase Z (CPZ) links thyroid hormone and Wnt signaling pathways in growth plate chondrocytes. J. Bone Miner. Res. 2009, 24, 265–273. [Google Scholar] [CrossRef]
- Garcia-Pardo, J.; Tanco, S.; Arolas, J.L.; Aviles, F.X.; Lorenzo, J. Expression and Funcional Characterization of Six Recombinant Variants of Carboxypeptidase Z, a New Carboxypeptidase Involved in the Wnt Signaling Pathway; Institut de Biotecnologia i Biomedicina and Departament de Bioquimica i Biologia Molecular, Universitat Autònoma de Barcelona: Bellaterra (Barcelona), Spain, 2012. [Google Scholar]
- Fricker, L.D.; Devi, L. Comparison of a spectrophotometric, a fluorometric, and a novel radiometric assay for carboxypeptidase E (ec 3.4.17.10) and other carboxypeptidase b-like enzymes. Anal. Biochem. 1990, 184, 21–27. [Google Scholar] [CrossRef]
- Gelman, J.S.; Sironi, J.; Berezniuk, I.; Dasgupta, S.; Castro, L.M.; Gozzo, F.C.; Ferro, E.S.; Fricker, L.D. Alterations of the intracellular peptidome in response to the proteasome inhibitor bortezomib. PLoS ONE 2013, 8, e53263. [Google Scholar] [CrossRef]
- Morano, C.; Zhang, X.; Fricker, L.D. Multiple isotopic labels for quantitative mass spectrometry. Anal. Chem. 2008, 80, 9298–9309. [Google Scholar] [CrossRef]
- Fricker, L.D. Limitations of mass spectrometry-based peptidomic approaches. J. Am. Soc. Mass Spectrom. 2015, 26, 1981–1991. [Google Scholar] [CrossRef]
- Schechter, I.; Berger, A. On the size of the active site in proteases I. Papain. Biochem. Biophys. Res. Commun. 1967, 27, 157–162. [Google Scholar] [CrossRef]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef]
- Dann, C.E.; Hsieh, J.C.; Rattner, A.; Sharma, D.; Nathans, J.; Leahy, D.J. Insights into Wnt binding and signalling from the structures of two Frizzled cysteine-rich domains. Nature 2001, 412, 86–90. [Google Scholar] [CrossRef]
- Janda, C.Y.; Waghray, D.; Levin, A.M.; Thomas, C.; Garcia, K.C. Structural basis of Wnt recognition by Frizzled. Science 2012, 337, 59–64. [Google Scholar] [CrossRef]
- Dang, L.T.; Miao, Y.; Ha, A.; Yuki, K.; Park, K.; Janda, C.Y.; Jude, K.M.; Mohan, K.; Ha, N.; Vallon, M.; et al. Receptor subtype discrimination using extensive shape complementary designed interfaces. Nat. Struct. Mol. Biol. 2019, 26, 407–414. [Google Scholar] [CrossRef]
- Chang, T.H.; Hsieh, F.L.; Zebisch, M.; Harlos, K.; Elegheert, J.; Jones, E.Y. Structure and functional properties of norrin mimic Wnt for signalling with frizzled4, lrp5/6, and proteoglycan. Elife 2015, 4, e06554. [Google Scholar] [CrossRef]
- Xin, X.N.; Day, R.; Dong, W.J.; Lei, Y.H.; Fricker, L.D. Cloning, sequence analysis, and distribution of rat metallocarboxypeptidase z. DNA Cell Biol. 1998, 17, 311–319. [Google Scholar] [CrossRef]
- Skalka, N.; Caspi, M.; Caspi, E.; Loh, Y.P.; Rosin-Arbesfeld, R. Carboxypeptidase E: A negative regulator of the canonical Wnt signaling pathway. Oncogene 2013, 32, 2836–2847. [Google Scholar] [CrossRef]
- Skalka, N.; Caspi, M.; Lahav-Ariel, L.; Loh, Y.P.; Hirschberg, K.; Rosin-Arbesfeld, R. Carboxypeptidase e (cpe) inhibits the secretion and activity of wnt3a. Oncogene 2016, 35, 6416–6428. [Google Scholar] [CrossRef]
- Sharma, S.; Schiller, M.R. The carboxy-terminus, a key regulator of protein function. Crit. Rev. Biochem. Mol. Biol. 2019, 54, 85–102. [Google Scholar] [CrossRef]
- Tanco, S.; Tort, O.; Demol, H.; Aviles, F.X.; Gevaert, K.; Van Damme, P.; Lorenzo, J. C-terminomics screen for natural substrates of cytosolic carboxypeptidase 1 reveals processing of acidic protein C termini. Mol. Cell. Proteom. 2015, 14, 177–190. [Google Scholar] [CrossRef]
- Tanco, S.; Gevaert, K.; Van Damme, P. C-terminomics: Targeted analysis of natural and posttranslationally modified protein and peptide c-termini. Proteomics 2015, 15, 903–914. [Google Scholar] [CrossRef]
- Calnan, D.P.; Fagbemi, A.; Berlanga-Acosta, J.; Marchbank, T.; Sizer, T.; Lakhoo, K.; Edwards, A.D.; Playford, R.J. Potency and stability of C terminal truncated human epidermal growth factor. Gut 2000, 47, 622–627. [Google Scholar] [CrossRef]
- Panosa, C.; Tebar, F.; Ferrer-Batalle, M.; Fonge, H.; Seno, M.; Reilly, R.M.; Massaguer, A.; De Llorens, R. Development of an epidermal growth factor derivative with EGFR blocking activity. PLoS ONE 2013, 8, e69325. [Google Scholar] [CrossRef]
- Garcia-Guerrero, M.C.; Garcia-Pardo, J.; Berenguer, E.; Fernandez-Alvarez, R.; Barfi, G.B.; Lyons, P.J.; Aviles, F.X.; Huber, R.; Lorenzo, J.; Reverter, D. Crystal structure and mechanism of human carboxypeptidase o: Insights into its specific activity for acidic residues. Proc. Natl. Acad. Sci. USA 2018, 115, E3932–E3939. [Google Scholar] [CrossRef]
- Lyons, P.J.; Callaway, M.B.; Fricker, L.D. Characterization of carboxypeptidase A6, an extracellular matrix peptidase. J. Biol. Chem. 2008, 283, 7054–7063. [Google Scholar] [CrossRef]
- Wernersson, S.; Pejler, G. Mast cell secretory granules: Armed for battle. Nat. Rev. Immunol. 2014, 14, 478–494. [Google Scholar] [CrossRef]
- Fricker, L.D. Methods for studying carboxypeptidase E. Pep. Neuropept. Process. 1995, 23, 237–250. [Google Scholar]
- Motulsky, H.J. Graphpad Prism Version 6.01; GraphPad Sofware Inc.: San Diego, CA, USA, 2012. [Google Scholar]
- Lyons, P.J.; Fricker, L.D. Substrate specificity of human carboxypeptidase A6. J. Biol. Chem. 2010, 285, 38234–38242. [Google Scholar] [CrossRef]
- Tanco, S.; Zhang, X.; Morano, C.; Aviles, F.X.; Lorenzo, J.; Fricker, L.D. Characterization of the substrate specificity of human carboxypeptidase A4 and implications for a role in extracellular peptide processing. J. Biol. Chem. 2010, 285, 18385–18396. [Google Scholar] [CrossRef]
- Ye, Y.; Godzik, A. FATCAT: A web server for flexible structure comparison and structure similarity searching. Nucleic Acids Res. 2004, 32, W582–W585. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Drozdetskiy, A.; Cole, C.; Procter, J.; Barton, G.J. Jpred4: A protein secondary structure prediction server. Nucleic Acids Res. 2015, 43, W389–W394. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK- A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- DeLano, W.L. The Pymol Molecular Graphics System; DeLano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]


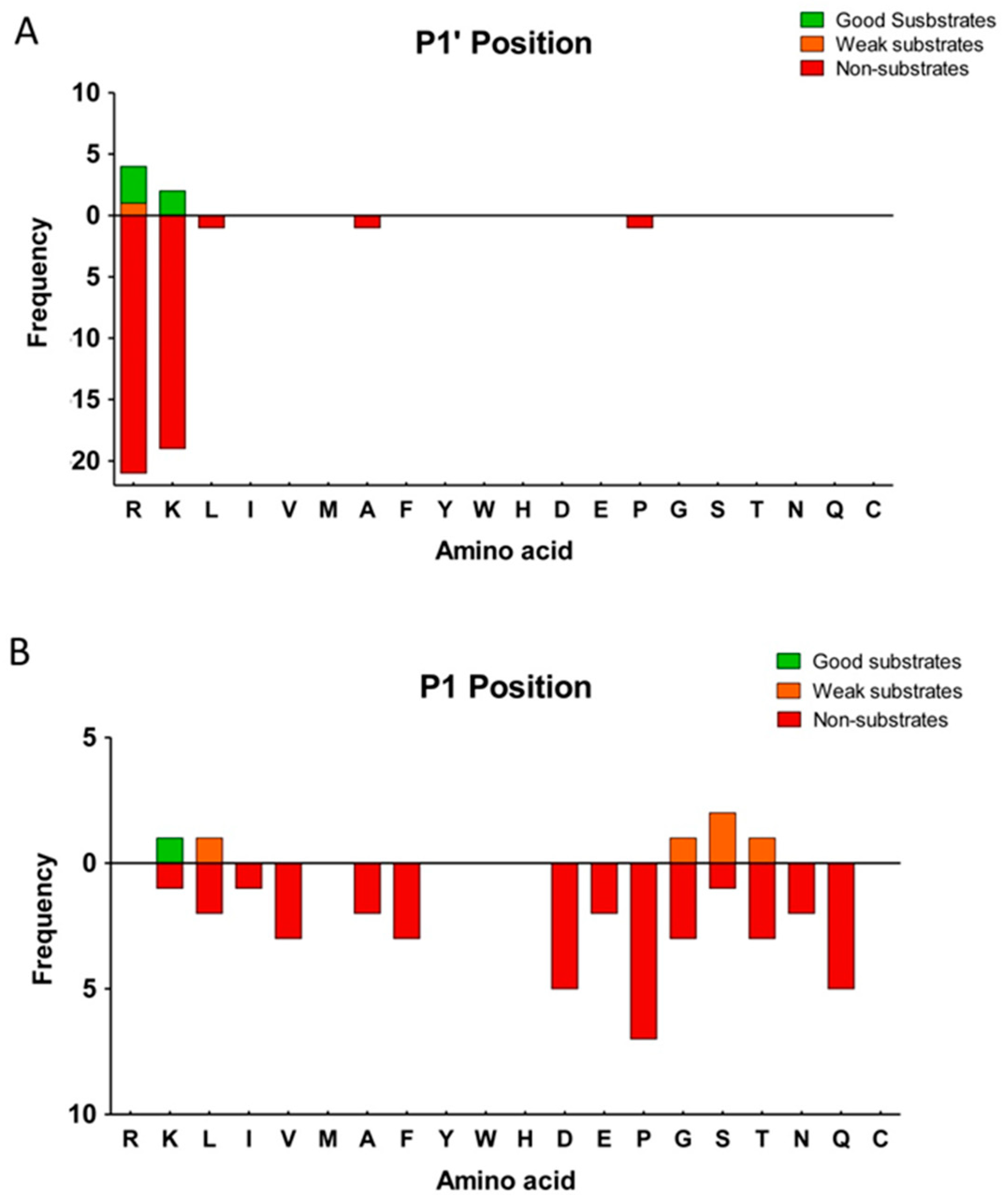
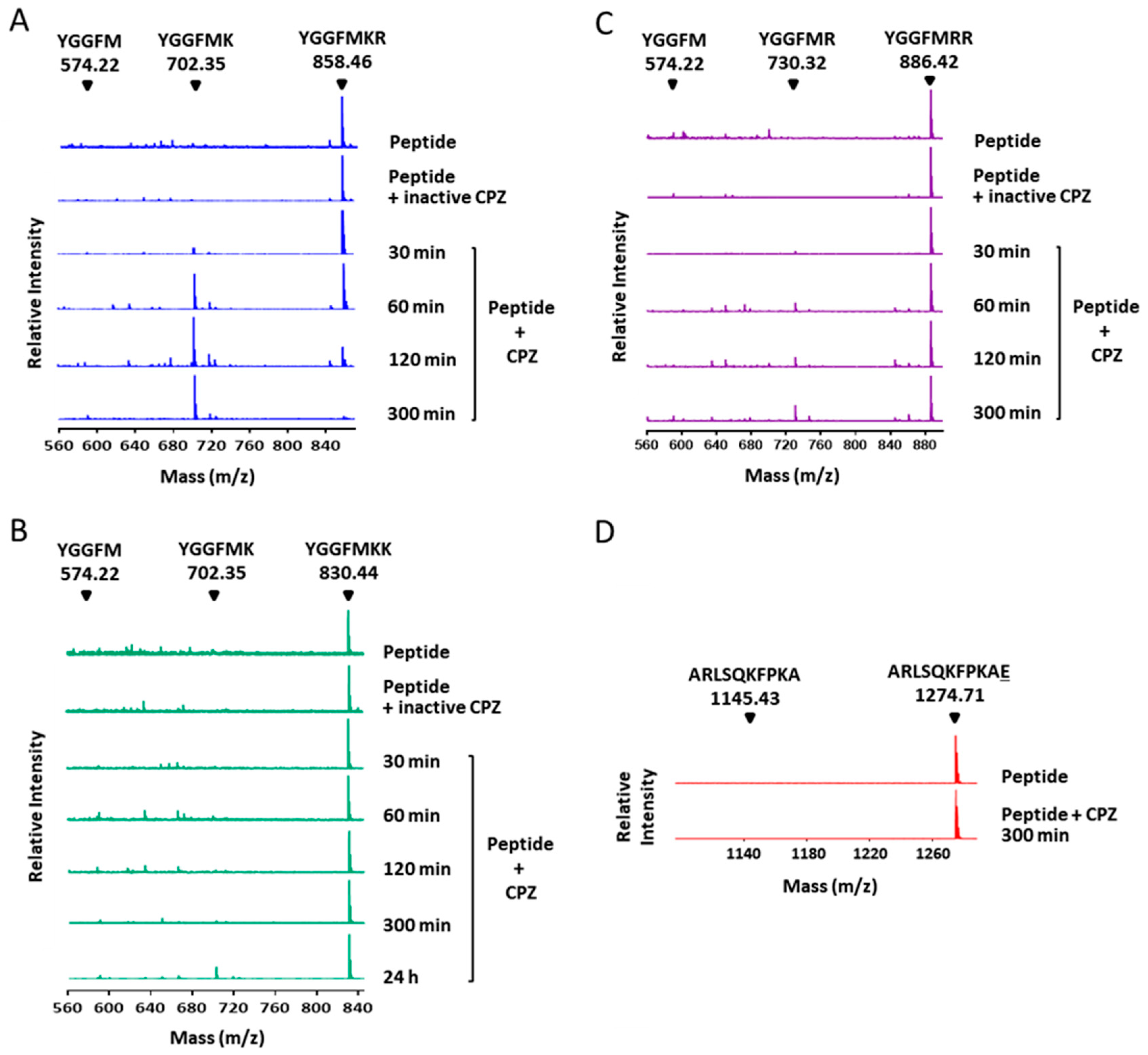


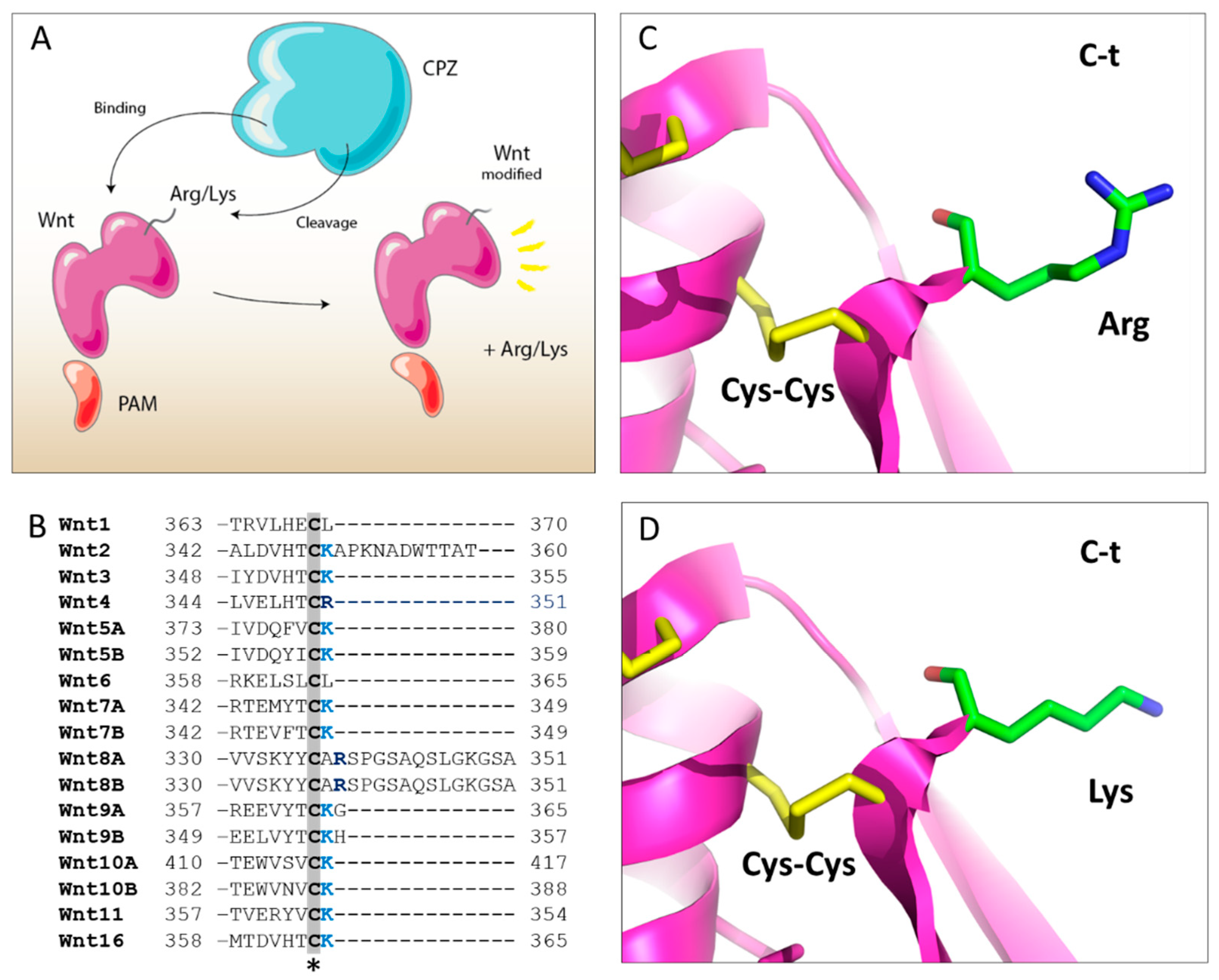
| Enzyme | Km (µM) | kcat (s−1) | kcat/Km (µM−1 s−1) |
|---|---|---|---|
| CPZ | 1905 ± 360 | 5.3 ± 0.6 | 0.0028 ± 0.0008 |
| CPZΔFz | 1667 ± 385 | 6.2 ± 0.8 | 0.0039 ± 0.0014 |
| CPD domain I (a) | 319 ± 37 | 8.5 ± 0.5 | 0.027 ± 0.003 |
| CPD domain II (a) | 844 ± 139 | 7.0 ± 0.9 | 0.008 ± 0.001 |
| CPE (b) | 34 | 13 | 0.38 |
| Ratio CPZ/No Enzyme | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Type | Protein Precursor | Peptide Sequence | Z | T | Obs M | Theor M | ppm | 100 nM | 10 nM | 1 nM | 0.1 nM |
| Good | Histidine triad nucleotide-binding protein 1 | Ac-ADEIAKAQVAR | 2 | 1 | 1212.66 | 1212.65 | 14 | 0.02 | 0.94 | 1.08 | 1.00 |
| Good | Eukaryotic translation initiation factor 5A | SAMoxTEEAAVAIKAMAK | 3 | 3 | 1636.81 | 1636.82 | −5 | 0.07 | 0.15 | 0.95 | 0.95 |
| Good | Vimentin | AELEQLKGQGKSR | 4 | 3 | 1442.78 | 1442.78 | −3 | 0.18 | 1.00 | 1.03 | 0.95 |
| Good | Hematological and neurological expressed 1 protein | Ac-TTTTTFKGVDPNSRNSSR | 3 | 1 | 2010.00 | 2009.98 | 13 | 0.31 | 0.89 | 1.24 | 1.07 |
| Weak | Eukaryotic translation initiation factor 5A | NMDVPNIKR | 3 | 2 | 1085.56 | 1085.57 | −6 | 0.45 | n.d. | n.d. | 0.82 |
| Weak | Ubiquitin-60S ribosomal protein L40 | IIEPSLR | 2 | 1 | 826.49 | 826.49 | 0 | 0.60 | 1.00 | 1.13 | 1.04 |
| Protein Precursor | Sequence | Cleaved aa | Z | T | Obs M | Theor M | ppm | Ratio CPZ/No Enzyme | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 100 nM | 10 nM | 1 nM | 0.1 nM | ||||||||
| Heat shock 10 kDa protein 1 | AVGSGSKGKGGEIQPVSV | K | 3 | 3 | 1655.89 | 1655.89 | 2 | 1.30 | 1.16 | 1.28 | 0.93 |
| Peptidylprolyl isomerase A | ADKVPKTAENF | R | 3 | 3 | 1218.62 | 1218.62 | −3 | 1.36 | 1.09 | 1.13 | 1.09 |
| Nucleophosmin | EKTPKTPKGPSSVEDIKA | K | 5 | 5 | 1911.00 | 1911.03 | −18 | 1.46 | 1.00 | 1.23 | 1.15 |
| FK506 Binding Protein | VFDVELL | K | 1 | 1 | 833.45 | 833.45 | 2 | 1.50 | 1.28 | ND | 1.25 |
| Ratio CPZ/No Enzyme | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Type | Protein Precursor | Peptide Sequence | Cleaved aa | Z | T | Obs M | Theor M | ppm | 100 nM | 10 nM | 1 nM |
| Good | Thyroglobulin | GQEFTITGQKR | - | 2 | 2 | 1263.66 | 1263.60 | −2 | 0.38 | 1.13 | 0.97 |
| Weak | Thyroglobulin | ALEQATR | - | 2 | 1 | 787.42 | 787.42 | 3 | 0.57 | 1.04 | 1.00 |
| Weak | Thyroglobulin | AVKQFEESQGR | - | 3 | 2 | 1277.64 | 1277.64 | 0 | 0.68 | 0.86 | 0.95 |
| Weak | Bovine serum albumin | KVPQVSTPTLVEVSR | - | 3 | 2 | 1638.93 | 1638.93 | −2 | 0.70 | 0.90 | 0.95 |
| Weak | Bovine serum albumin | KQTALVELLK | - | 3 | 3 | 1141.69 | 1141.71 | −22 | 0.73 | 0.80 | 0.89 |
| Weak | Thyroglobulin | LPESK | - | 2 | 2 | 572.31 | 572.32 | −24 | 0.77 | 0.83 | 0.83 |
| Product | Thyroglobulin | GQEFTITGQK | R | 2 | 2 | 1107.56 | 1107.56 | −4 | 1.52 | 0.78 | 1.04 |
| Product | Thyroglobulin | LF | R | 1 | 1 | 278.16 | 278.15 | 14 | 1.56 | 0.88 | 0.88 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garcia-Pardo, J.; Tanco, S.; Garcia-Guerrero, M.C.; Dasgupta, S.; Avilés, F.X.; Lorenzo, J.; Fricker, L.D. Substrate Specificity and Structural Modeling of Human Carboxypeptidase Z: A Unique Protease with a Frizzled-Like Domain. Int. J. Mol. Sci. 2020, 21, 8687. https://doi.org/10.3390/ijms21228687
Garcia-Pardo J, Tanco S, Garcia-Guerrero MC, Dasgupta S, Avilés FX, Lorenzo J, Fricker LD. Substrate Specificity and Structural Modeling of Human Carboxypeptidase Z: A Unique Protease with a Frizzled-Like Domain. International Journal of Molecular Sciences. 2020; 21(22):8687. https://doi.org/10.3390/ijms21228687
Chicago/Turabian StyleGarcia-Pardo, Javier, Sebastian Tanco, Maria C. Garcia-Guerrero, Sayani Dasgupta, Francesc Xavier Avilés, Julia Lorenzo, and Lloyd D. Fricker. 2020. "Substrate Specificity and Structural Modeling of Human Carboxypeptidase Z: A Unique Protease with a Frizzled-Like Domain" International Journal of Molecular Sciences 21, no. 22: 8687. https://doi.org/10.3390/ijms21228687
APA StyleGarcia-Pardo, J., Tanco, S., Garcia-Guerrero, M. C., Dasgupta, S., Avilés, F. X., Lorenzo, J., & Fricker, L. D. (2020). Substrate Specificity and Structural Modeling of Human Carboxypeptidase Z: A Unique Protease with a Frizzled-Like Domain. International Journal of Molecular Sciences, 21(22), 8687. https://doi.org/10.3390/ijms21228687