A Molecular Perspective on Sirtuin Activity
Abstract
1. Introduction
2. Protein Acylation
2.1. N-Terminal Acylation
N-Terminal Acetylation
2.2. Acylation of Internal Lysine Residues
Acetylation of Internal Lysine Residues
2.3. S-Palmitoylation of Cysteine Residues
2.4. The Influence of Protein Acylation in Human Health and Disease
Protein Acylation and “Carbon Stress”
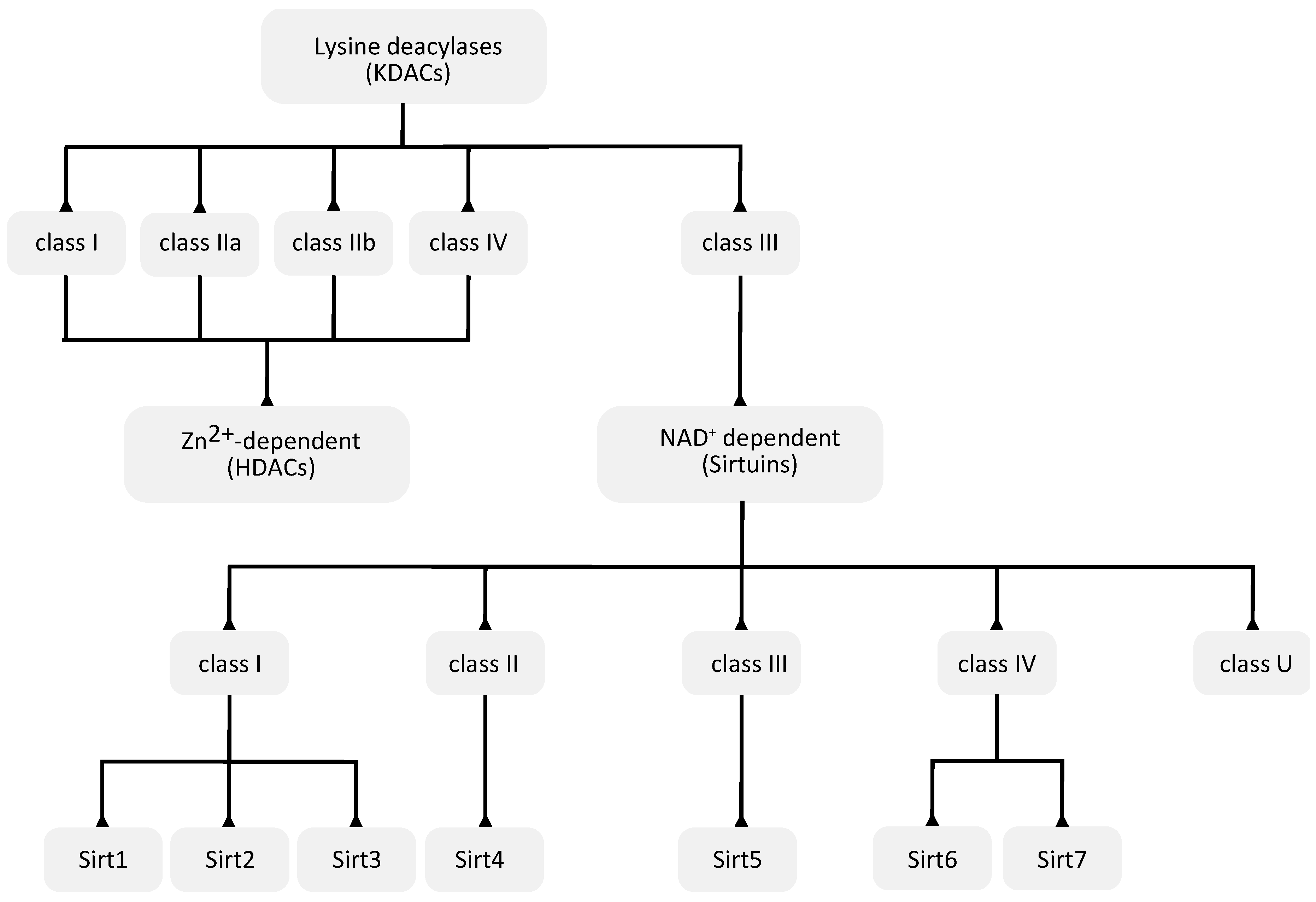
3. Protein Deacylation
3.1. Zn2+-Dependent KDACs
3.2. NAD+-Dependent Sirtuin Deacetylases
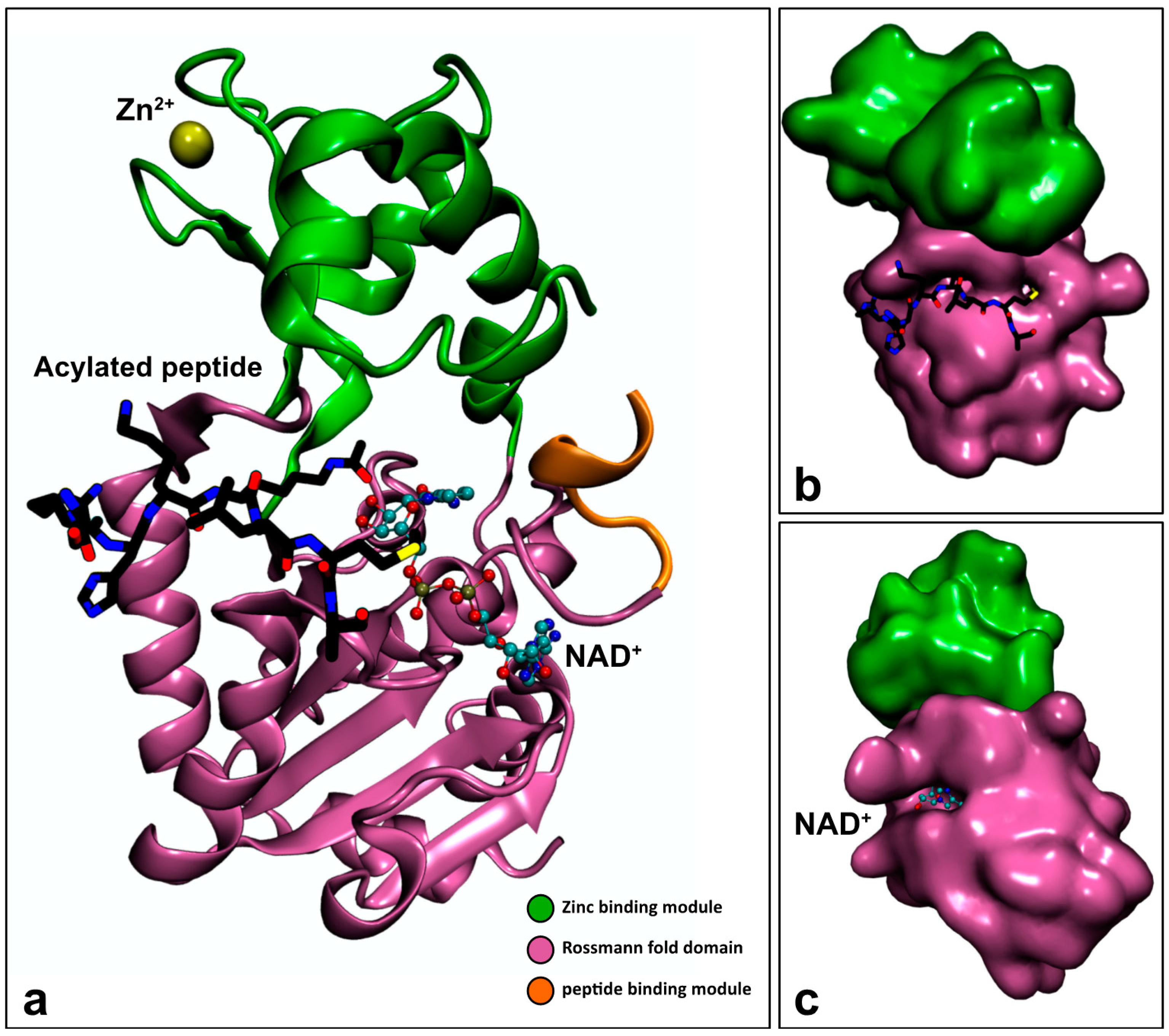
3.2.1. Sirtuin Structure
3.2.2. Substrate Specificity of Sirtuins
The Influence of Sirtuin Structure
The Influence of the Subcellular Localization of Sirtuins
The Influence of the Acylated Residue
The Influence of the Acyl Group
The Influence of Protein Structure
The Influence of Protein Sequence
The Influence of Protein Interactions
3.2.3. Catalytic Mechanism of Sirtuin Deacylation
3.2.4. The Influence of Sirtuins in Human Health and Disease
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature 2004, 431, 931–945. [Google Scholar] [CrossRef]
- Walsh, C.T.; Garneau-Tsodikova, S.; Gatto, G.J., Jr. Protein posttranslational modifications: The chemistry of proteome diversifications. Angew. Chem. Int. Ed. Engl. 2005, 44, 7342–7372. [Google Scholar] [CrossRef]
- Gilbert, W. Why genes in pieces? Nature 1978, 271, 501. [Google Scholar] [CrossRef]
- Roth, M.J.; Forbes, A.J.; Boyne, M.T., 2nd; Kim, Y.B.; Robinson, D.E.; Kelleher, N.L. Precise and parallel characterization of coding polymorphisms, alternative splicing, and modifications in human proteins by mass spectrometry. Mol. Cell. Proteom. 2005, 4, 1002–1008. [Google Scholar] [CrossRef]
- Hunter, T. The age of crosstalk: Phosphorylation, ubiquitination, and beyond. Mol. Cell 2007, 28, 730–738. [Google Scholar] [CrossRef]
- Cantin, G.T.; Yates, J.R., 3rd. Strategies for shotgun identification of post-translational modifications by mass spectrometry. J. Chromatogr. A 2004, 1053, 7–14. [Google Scholar] [CrossRef]
- Nahomi, R.B.; Nandi, S.K.; Rakete, S.; Michel, C.; Fritz, K.S.; Nagaraj, R.H. Lysine malonylation and propionylation are prevalent in human lens proteins. Exp. Eye Res. 2020, 190, 107864. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Tan, M.; Xie, Z.; Dai, L.; Chen, Y.; Zhao, Y. Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 2011, 7, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Qausain, S.; Srinivasan, H.; Jamal, S.; Nasiruddin, M.; Khan, M.K.A. Phosphorylation and Acetylation of Proteins as Posttranslational Modification: Implications in Human Health and Associated Diseases. In Protein Modificomics; Academic Press: Cambridge, MA, USA, 2019; pp. 69–86. [Google Scholar] [CrossRef]
- Brooks, C.L.; Gu, W. Ubiquitination, phosphorylation and acetylation: The molecular basis for p53 regulation. Curr. Opin. Cell Biol. 2003, 15, 164–171. [Google Scholar] [CrossRef]
- Kouzarides, T. Acetylation: A regulatory modification to rival phosphorylation? EMBO J. 2000, 19, 1176–1179. [Google Scholar] [CrossRef]
- Caron, C.; Boyault, C.; Khochbin, S. Regulatory cross-talk between lysine acetylation and ubiquitination: Role in the control of protein stability. Bioessays 2005, 27, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.-J.; Seto, E. Lysine acetylation: Codified crosstalk with other posttranslational modifications. Mol. Cell 2008, 31, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Cheng, Z.; Zhao, Y.; Volchenboum, S.L. Bioinformatic analysis and post-translational modification crosstalk prediction of lysine acetylation. PLoS ONE 2011, 6, e28228. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.C.; Sprung, R.; Chen, Y.; Xu, Y.; Ball, H.; Pei, J.; Cheng, T.; Kho, Y.; Xiao, H.; Xiao, L.; et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 2006, 23, 607–618. [Google Scholar] [CrossRef]
- Lu, J.Y.; Lin, Y.Y.; Sheu, J.C.; Wu, J.T.; Lee, F.J.; Chen, Y.; Lin, M.I.; Chiang, F.T.; Tai, T.Y.; Berger, S.L.; et al. Acetylation of yeast AMPK controls intrinsic aging independently of caloric restriction. Cell 2011, 146, 969–979. [Google Scholar] [CrossRef]
- Chuang, C.; Lin, S.H.; Huang, F.; Pan, J.; Josic, D.; Yu-Lee, L.Y. Acetylation of RNA processing proteins and cell cycle proteins in mitosis. J. Proteome Res. 2010, 9, 4554–4564. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Yang, C.; Xiong, H.; Lin, Y.; Yao, J.; Li, H.; Xie, L.; Zhao, W.; Yao, Y.; et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 2010, 327, 1004–1007. [Google Scholar] [CrossRef]
- Thinon, E.; Hang, H.C. Chemical reporters for exploring protein acylation. Biochem. Soc. Trans. 2015, 43, 253–261. [Google Scholar] [CrossRef]
- Deng, S.; Marmorstein, R. Protein N-Terminal Acetylation: Structural Basis, Mechanism, Versatility, and Regulation. Trends Biochem. Sci. 2020. [Google Scholar] [CrossRef]
- Brown, J.L.; Roberts, W.K. Evidence that approximately eighty per cent of the soluble proteins from Ehrlich ascites cells are Nalpha-acetylated. J. Biol. Chem. 1976, 251, 1009–1014. [Google Scholar]
- Arnesen, T.; Van Damme, P.; Polevoda, B.; Helsens, K.; Evjenth, R.; Colaert, N.; Varhaug, J.E.; Vandekerckhove, J.; Lillehaug, J.R.; Sherman, F.; et al. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc. Natl. Acad. Sci. USA 2009, 106, 8157–8162. [Google Scholar] [CrossRef] [PubMed]
- Aksnes, H.; Hole, K.; Arnesen, T. Molecular, cellular, and physiological significance of N-terminal acetylation. Int. Rev. Cell Mol. Biol. 2015, 316, 267–305. [Google Scholar] [CrossRef] [PubMed]
- Drazic, A.; Myklebust, L.M.; Ree, R.; Arnesen, T. The world of protein acetylation. Biochim. Biophys. Acta 2016, 1864, 1372–1401. [Google Scholar] [CrossRef] [PubMed]
- Starheim, K.K.; Gevaert, K.; Arnesen, T. Protein N-terminal acetyltransferases: When the start matters. Trends Biochem. Sci. 2012, 37, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, C.; Kumar, C.; Gnad, F.; Nielsen, M.L.; Rehman, M.; Walther, T.C.; Olsen, J.V.; Mann, M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 2009, 325, 834–840. [Google Scholar] [CrossRef]
- Costantini, C.; Ko, M.H.; Jonas, M.C.; Puglielli, L. A reversible form of lysine acetylation in the ER and Golgi lumen controls the molecular stabilization of BACE1. Biochem. J. 2007, 407, 383–395. [Google Scholar] [CrossRef]
- Farrugia, M.A.; Puglielli, L. Nepsilon-lysine acetylation in the endoplasmic reticulum—A novel cellular mechanism that regulates proteostasis and autophagy. J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef]
- Wagner, G.R.; Hirschey, M.D. Nonenzymatic Protein Acylation as a Carbon Stress Regulated by Sirtuin Deacylases. Mol. Cell 2014, 54, 5–16. [Google Scholar] [CrossRef]
- Ghanta, S.; Grossmann, R.E.; Brenner, C. Mitochondrial protein acetylation as a cell-intrinsic, evolutionary driver of fat storage: Chemical and metabolic logic of acetyl-lysine modifications. Crit. Rev. Biochem. Mol. Biol. 2013, 48, 561–574. [Google Scholar] [CrossRef]
- Allfrey, V.G.; Faulkner, R.; Mirsky, A.E. Acetylation and Methylation of Histones and Their Possible Role in the Regulation of Rna Synthesis. Proc. Natl. Acad. Sci. USA 1964, 51, 786–794. [Google Scholar] [CrossRef]
- Choudhary, C.; Weinert, B.T.; Nishida, Y.; Verdin, E.; Mann, M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 2014, 15, 536–550. [Google Scholar] [CrossRef] [PubMed]
- Soppa, J. Protein acetylation in archaea, bacteria, and eukaryotes. Archaea 2010, 2010. [Google Scholar] [CrossRef] [PubMed]
- Paik, W.K.; Pearson, D.; Lee, H.W.; Kim, S. Nonenzymatic acetylation of histones with acetyl-CoA. Biochim. Biophys. Acta 1970, 213, 513–522. [Google Scholar] [CrossRef]
- Wagner, G.R.; Payne, R.M. Widespread and enzyme-independent Nepsilon-acetylation and Nepsilon-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J. Biol. Chem. 2013, 288, 29036–29045. [Google Scholar] [CrossRef]
- Allfrey, V.G.; Mirsky, A.E. Structural Modifications of Histones and their Possible Role in the Regulation of RNA Synthesis. Science 1964, 144, 559. [Google Scholar] [CrossRef] [PubMed]
- Baddiley, J.; Kekwick, R.A.; Thain, E.M. A new method for acetylating proteins. Nature 1952, 170, 968–970. [Google Scholar] [CrossRef]
- Weinert, B.T.; Iesmantavicius, V.; Moustafa, T.; Scholz, C.; Wagner, S.A.; Magnes, C.; Zechner, R.; Choudhary, C. Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol. Syst. Biol. 2015, 11, 833. [Google Scholar] [CrossRef]
- Weinert, B.T.; Iesmantavicius, V.; Moustafa, T.; Scholz, C.; Wagner, S.A.; Magnes, C.; Zechner, R.; Choudhary, C. Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol. Syst. Biol. 2014, 10, 716. [Google Scholar] [CrossRef]
- Casey, J.R.; Grinstein, S.; Orlowski, J. Sensors and regulators of intracellular pH. Nat. Rev. Mol. Cell Biol. 2010, 11, 50–61. [Google Scholar] [CrossRef]
- Garland, P.B.; Shepherd, D.; Yates, D.W. Steady-state concentrations of coenzyme A, acetyl-coenzyme A and long-chain fatty acyl-coenzyme A in rat-liver mitochondria oxidizing palmitate. Biochem. J. 1965, 97, 587–594. [Google Scholar] [CrossRef]
- D’Alayer, J.; Expert-Bezancon, N.; Beguin, P. Time- and temperature-dependent acetylation of the chemokine RANTES produced in recombinant Escherichia coli. Protein Expr. Purif. 2007, 55, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Hentschel, A.; Zahedi, R.P.; Ahrends, R. Protein lipid modifications-More than just a greasy ballast. Proteomics 2016, 16, 759–782. [Google Scholar] [CrossRef] [PubMed]
- Lanyon-Hogg, T.; Faronato, M.; Serwa, R.A.; Tate, E.W. Dynamic Protein Acylation: New Substrates, Mechanisms, and Drug Targets. Trends Biochem. Sci. 2017, 42, 566–581. [Google Scholar] [CrossRef] [PubMed]
- Chamberlain, L.H.; Shipston, M.J. The physiology of protein S-acylation. Physiol. Rev. 2015, 95, 341–376. [Google Scholar] [CrossRef] [PubMed]
- Fukata, M.; Fukata, Y.; Adesnik, H.; Nicoll, R.A.; Bredt, D.S. Identification of PSD-95 palmitoylating enzymes. Neuron 2004, 44, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Roth, A.F.; Feng, Y.; Chen, L.; Davis, N.G. The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J. Cell Biol. 2002, 159, 23–28. [Google Scholar] [CrossRef]
- Politis, E.G.; Roth, A.F.; Davis, N.G. Transmembrane topology of the protein palmitoyl transferase Akr1. J. Biol. Chem. 2005, 280, 10156–10163. [Google Scholar] [CrossRef]
- Narita, T.; Weinert, B.T.; Choudhary, C. Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 156–174. [Google Scholar] [CrossRef]
- Cohen, T.J.; Hwang, A.W.; Restrepo, C.R.; Yuan, C.X.; Trojanowski, J.Q.; Lee, V.M. An acetylation switch controls TDP-43 function and aggregation propensity. Nat. Commun. 2015, 6, 5845. [Google Scholar] [CrossRef]
- Min, S.W.; Chen, X.; Tracy, T.E.; Li, Y.; Zhou, Y.; Wang, C.; Shirakawa, K.; Minami, S.S.; Defensor, E.; Mok, S.A.; et al. Critical role of acetylation in tau-mediated neurodegeneration and cognitive deficits. Nat. Med. 2015, 21, 1154–1162. [Google Scholar] [CrossRef]
- Clayton-Smith, J.; O’ullivan, J.; Daly, S.; Bhaskar, S.; Day, R.; Anderson, B.; Voss, A.K.; Thomas, T.; Biesecker, L.G.; Smith, P.; et al. Whole-exome-sequencing identifies mutations in histone acetyltransferase gene KAT6B in individuals with the Say-Barber-Biesecker variant of Ohdo syndrome. Am. J. Hum. Genet. 2011, 89, 675–681. [Google Scholar] [CrossRef] [PubMed]
- Simpson, M.A.; Deshpande, C.; Dafou, D.; Vissers, L.E.; Woollard, W.J.; Holder, S.E.; Gillessen-Kaesbach, G.; Derks, R.; White, S.M.; Cohen-Snuijf, R.; et al. De novo mutations of the gene encoding the histone acetyltransferase KAT6B cause Genitopatellar syndrome. Am. J. Hum. Genet. 2012, 90, 290–294. [Google Scholar] [CrossRef] [PubMed]
- Campeau, P.M.; Kim, J.C.; Lu, J.T.; Schwartzentruber, J.A.; Abdul-Rahman, O.A.; Schlaubitz, S.; Murdock, D.M.; Jiang, M.M.; Lammer, E.J.; Enns, G.M.; et al. Mutations in KAT6B, encoding a histone acetyltransferase, cause Genitopatellar syndrome. Am. J. Hum. Genet. 2012, 90, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Tham, E.; Lindstrand, A.; Santani, A.; Malmgren, H.; Nesbitt, A.; Dubbs, H.A.; Zackai, E.H.; Parker, M.J.; Millan, F.; Rosenbaum, K.; et al. Dominant mutations in KAT6A cause intellectual disability with recognizable syndromic features. Am. J. Hum. Genet. 2015, 96, 507–513. [Google Scholar] [CrossRef]
- Arboleda, V.A.; Lee, H.; Dorrani, N.; Zadeh, N.; Willis, M.; Macmurdo, C.F.; Manning, M.A.; Kwan, A.; Hudgins, L.; Barthelemy, F.; et al. De novo nonsense mutations in KAT6A, a lysine acetyl-transferase gene, cause a syndrome including microcephaly and global developmental delay. Am. J. Hum. Genet. 2015, 96, 498–506. [Google Scholar] [CrossRef]
- Pasqualucci, L.; Dominguez-Sola, D.; Chiarenza, A.; Fabbri, G.; Grunn, A.; Trifonov, V.; Kasper, L.H.; Lerach, S.; Tang, H.; Ma, J.; et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature 2011, 471, 189–195. [Google Scholar] [CrossRef]
- Houtkooper, R.H.; Pirinen, E.; Auwerx, J. Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol. 2012, 13, 225–238. [Google Scholar] [CrossRef]
- Lasko, L.M.; Jakob, C.G.; Edalji, R.P.; Qiu, W.; Montgomery, D.; Digiammarino, E.L.; Hansen, T.M.; Risi, R.M.; Frey, R.; Manaves, V.; et al. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 2017, 550, 128–132. [Google Scholar] [CrossRef]
- Baell, J.B.; Leaver, D.J.; Hermans, S.J.; Kelly, G.L.; Brennan, M.S.; Downer, N.L.; Nguyen, N.; Wichmann, J.; McRae, H.M.; Yang, Y.; et al. Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth. Nature 2018, 560, 253–257. [Google Scholar] [CrossRef]
- Salaun, C.; Greaves, J.; Chamberlain, L.H. The intracellular dynamic of protein palmitoylation. J. Cell Biol. 2010, 191, 1229–1238. [Google Scholar] [CrossRef]
- Liu, P.; Jiao, B.; Zhang, R.; Zhao, H.; Zhang, C.; Wu, M.; Li, D.; Zhao, X.; Qiu, Q.; Li, J.; et al. Palmitoylacyltransferase Zdhhc9 inactivation mitigates leukemogenic potential of oncogenic Nras. Leukemia 2016, 30, 1225–1228. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.-X.; Du, Z.-M.; Jiao, L.; Shao, Q.; Fu, S.; Shao, J.-Y.; Zhu, X.-F.; Ernberg, I.; Li, Y.-H. Inhibition of MiR-155 suppresses cell migration in nasopharyngeal carcinoma through targeting ZDHHC2. Int. J. Clin. Exp. Med. 2015, 8, 8472–8484. [Google Scholar] [PubMed]
- Cho, E.; Park, M. Palmitoylation in Alzheimer’s disease and other neurodegenerative diseases. Pharmacol. Res. 2016, 111, 133–151. [Google Scholar] [CrossRef] [PubMed]
- Carafa, V.; Nebbioso, A.; Altucci, L. Sirtuins and disease: The road ahead. Front. Pharm. 2012, 3, 4. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Xu, W.; Jiang, W.; Yu, W.; Lin, Y.; Zhang, T.; Yao, J.; Zhou, L.; Zeng, Y.; Li, H.; et al. Regulation of cellular metabolism by protein lysine acetylation. Science 2010, 327, 1000–1004. [Google Scholar] [CrossRef]
- Finkel, T.; Deng, C.X.; Mostoslavsky, R. Recent progress in the biology and physiology of sirtuins. Nature 2009, 460, 587–591. [Google Scholar] [CrossRef]
- Brownlee, M. Biochemistry and molecular cell biology of diabetic complications. Nature 2001, 414, 813–820. [Google Scholar] [CrossRef]
- Fukami, K.; Yamagishi, S.; Okuda, S. Role of AGEs-RAGE system in cardiovascular disease. Curr. Pharm. Des. 2014, 20, 2395–2402. [Google Scholar] [CrossRef]
- Houtkooper, R.H.; Mouchiroud, L.; Ryu, D.; Moullan, N.; Katsyuba, E.; Knott, G.; Williams, R.W.; Auwerx, J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature 2013, 497, 451–457. [Google Scholar] [CrossRef]
- Gottlieb, S.; Esposito, R.E. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 1989, 56, 771–776. [Google Scholar] [CrossRef]
- Bennett, C.B.; Snipe, J.R.; Westmoreland, J.W.; Resnick, M.A. SIR functions are required for the toleration of an unrepaired double-strand break in a dispensable yeast chromosome. Mol. Cell. Biol. 2001, 21, 5359–5373. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brachmann, C.B.; Sherman, J.M.; Devine, S.E.; Cameron, E.E.; Pillus, L.; Boeke, J.D. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 1995, 9, 2888–2902. [Google Scholar] [CrossRef] [PubMed]
- Araki, T.; Sasaki, Y.; Milbrandt, J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science 2004, 305, 1010–1013. [Google Scholar] [CrossRef] [PubMed]
- Picard, F.; Kurtev, M.; Chung, N.; Topark-Ngarm, A.; Senawong, T.; Machado De Oliveira, R.; Leid, M.; McBurney, M.W.; Guarente, L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature 2004, 429, 771–776. [Google Scholar] [CrossRef]
- Brunet, A.; Sweeney, L.B.; Sturgill, J.F.; Chua, K.F.; Greer, P.L.; Lin, Y.; Tran, H.; Ross, S.E.; Mostoslavsky, R.; Cohen, H.Y.; et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 2004, 303, 2011–2015. [Google Scholar] [CrossRef]
- Luo, J.; Nikolaev, A.Y.; Imai, S.; Chen, D.; Su, F.; Shiloh, A.; Guarente, L.; Gu, W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 2001, 107, 137–148. [Google Scholar] [CrossRef]
- Lin, S.J.; Defossez, P.A.; Guarente, L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 2000, 289, 2126–2128. [Google Scholar] [CrossRef]
- Kaeberlein, M.; McVey, M.; Guarente, L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev 1999, 13, 2570–2580. [Google Scholar] [CrossRef]
- Du, J.; Zhou, Y.; Su, X.; Yu, J.J.; Khan, S.; Jiang, H.; Kim, J.; Woo, J.; Kim, J.H.; Choi, B.H.; et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 2011, 334, 806–809. [Google Scholar] [CrossRef]
- Peng, C.; Lu, Z.; Xie, Z.; Cheng, Z.; Chen, Y.; Tan, M.; Luo, H.; Zhang, Y.; He, W.; Yang, K.; et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol. Cell. Proteom. 2011, 10. [Google Scholar] [CrossRef]
- Tan, M.; Peng, C.; Anderson, K.A.; Chhoy, P.; Xie, Z.; Dai, L.; Park, J.; Chen, Y.; Huang, H.; Zhang, Y.; et al. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 2014, 19, 605–617. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.A.; Huynh, F.K.; Fisher-Wellman, K.; Stuart, J.D.; Peterson, B.S.; Douros, J.D.; Wagner, G.R.; Thompson, J.W.; Madsen, A.S.; Green, M.F.; et al. SIRT4 Is a Lysine Deacylase that Controls Leucine Metabolism and Insulin Secretion. Cell Metab. 2017, 25, 838–855.e15. [Google Scholar] [CrossRef] [PubMed]
- Haberland, M.; Montgomery, R.L.; Olson, E.N. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nat. Rev. Genet. 2009, 10, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Altamura, S.; De Francesco, R.; Gallinari, P.; Lahm, A.; Neddermann, P.; Rowley, M.; Serafini, S.; Steinkuhler, C. Probing the elusive catalytic activity of vertebrate class IIa histone deacetylases. Bioorg Med. Chem. Lett. 2008, 18, 1814–1819. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.J.; Seto, E. The Rpd3/Hda1 family of lysine deacetylases: From bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 2008, 9, 206–218. [Google Scholar] [CrossRef]
- Jackson, M.D.; Denu, J.M. Structural identification of 2′- and 3′-O-acetyl-ADP-ribose as novel metabolites derived from the Sir2 family of beta -NAD+-dependent histone/protein deacetylases. J. Biol. Chem. 2002, 277, 18535–18544. [Google Scholar] [CrossRef]
- Rine, J.; Herskowitz, I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics 1987, 116, 9–22. [Google Scholar]
- Frye, R.A. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 2000, 273, 793–798. [Google Scholar] [CrossRef]
- Sanders, B.D.; Jackson, B.; Marmorstein, R. Structural basis for sirtuin function: What we know and what we don’t. Biochim. Biophys. Acta 2010, 1804, 1604–1616. [Google Scholar] [CrossRef]
- Hawse, W.F.; Wolberger, C. Structure-based mechanism of ADP-ribosylation by sirtuins. J. Biol. Chem. 2009, 284, 33654–33661. [Google Scholar] [CrossRef]
- Tanner, K.G.; Landry, J.; Sternglanz, R.; Denu, J.M. Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl. Acad. Sci. USA 2000, 97, 14178–14182. [Google Scholar] [CrossRef] [PubMed]
- Bheda, P.; Jing, H.; Wolberger, C.; Lin, H. The Substrate Specificity of Sirtuins. Ann. Rev. Biochem. 2016, 85, 405–429. [Google Scholar] [CrossRef] [PubMed]
- Avalos, J.L.; Celic, I.; Muhammad, S.; Cosgrove, M.S.; Boeke, J.D.; Wolberger, C. Structure of a Sir2 enzyme bound to an acetylated p53 peptide. Mol. Cell 2002, 10, 523–535. [Google Scholar] [CrossRef]
- Avalos, J.L.; Boeke, J.D.; Wolberger, C. Structural basis for the mechanism and regulation of Sir2 enzymes. Mol. Cell 2004, 13, 639–648. [Google Scholar] [CrossRef]
- Min, J.; Landry, J.; Sternglanz, R.; Xu, R.M. Crystal structure of a SIR2 homolog-NAD complex. Cell 2001, 105, 269–279. [Google Scholar] [CrossRef]
- Hoff, K.G.; Avalos, J.L.; Sens, K.; Wolberger, C. Insights into the Sirtuin Mechanism from Ternary Complexes Containing NAD+ and Acetylated Peptide. Structure 2006, 14, 1231–1240. [Google Scholar] [CrossRef]
- Rauh, D.; Fischer, F.; Gertz, M.; Lakshminarasimhan, M.; Bergbrede, T.; Aladini, F.; Kambach, C.; Becker, C.F.; Zerweck, J.; Schutkowski, M.; et al. An acetylome peptide microarray reveals specificities and deacetylation substrates for all human sirtuin isoforms. Nat. Commun. 2013, 4, 2327. [Google Scholar] [CrossRef]
- De Boor, S.; Knyphausen, P.; Kuhlmann, N.; Wroblowski, S.; Brenig, J.; Scislowski, L.; Baldus, L.; Nolte, H.; Kruger, M.; Lammers, M. Small GTP-binding protein Ran is regulated by posttranslational lysine acetylation. Proc. Natl. Acad. Sci. USA 2015, 112, E3679–E3688. [Google Scholar] [CrossRef]
- Kuhlmann, N.; Wroblowski, S.; Knyphausen, P.; de Boor, S.; Brenig, J.; Zienert, A.Y.; Meyer-Teschendorf, K.; Praefcke, G.J.; Nolte, H.; Kruger, M.; et al. Structural and Mechanistic Insights into the Regulation of the Fundamental Rho Regulator RhoGDIalpha by Lysine Acetylation. J. Biol. Chem. 2016, 291, 5484–5499. [Google Scholar] [CrossRef]
- Zhao, K.; Chai, X.; Marmorstein, R. Structure and substrate binding properties of cobB, a Sir2 homolog protein deacetylase from Escherichia coli. J. Mol. Biol. 2004, 337, 731–741. [Google Scholar] [CrossRef]
- Tanno, M.; Sakamoto, J.; Miura, T.; Shimamoto, K.; Horio, Y. Nucleocytoplasmic Shuttling of the NAD+-dependent Histone Deacetylase SIRT1. J. Biol. Chem. 2007, 282, 6823–6832. [Google Scholar] [CrossRef] [PubMed]
- Gomes, P.; Fleming Outeiro, T.; Cavadas, C. Emerging Role of Sirtuin 2 in the Regulation of Mammalian Metabolism. Trends Pharmacol. Sci. 2015, 36, 756–768. [Google Scholar] [CrossRef] [PubMed]
- Ansari, A.; Rahman, M.S.; Saha, S.K.; Saikot, F.K.; Deep, A.; Kim, K.H. Function of the SIRT3 mitochondrial deacetylase in cellular physiology, cancer, and neurodegenerative disease. Aging Cell 2017, 16, 4–16. [Google Scholar] [CrossRef] [PubMed]
- Michan, S.; Sinclair, D. Sirtuins in mammals: Insights into their biological function. Biochem. J. 2007, 404, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Nishida, Y.; Rardin, M.J.; Carrico, C.; He, W.; Sahu, A.K.; Gut, P.; Najjar, R.; Fitch, M.; Hellerstein, M.; Gibson, B.W.; et al. SIRT5 Regulates both Cytosolic and Mitochondrial Protein Malonylation with Glycolysis as a Major Target. Mol. Cell 2015, 59, 321–332. [Google Scholar] [CrossRef]
- Lin, H. The Enzymatic Activities of Sirtuins. In Introductory Review on Sirtuins in Biology, Aging, and Disease; Academic Press: Cambridge, MA, USA, 2018; pp. 45–62. [Google Scholar] [CrossRef]
- Michishita, E.; Park, J.Y.; Burneskis, J.M.; Barrett, J.C.; Horikawa, I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell 2005, 16, 4623–4635. [Google Scholar] [CrossRef]
- Feldman, J.L.; Baeza, J.; Denu, J.M. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J. Biol. Chem. 2013, 288, 31350–31356. [Google Scholar] [CrossRef]
- Jiang, H.; Khan, S.; Wang, Y.; Charron, G.; He, B.; Sebastian, C.; Du, J.; Kim, R.; Ge, E.; Mostoslavsky, R.; et al. SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature 2013, 496, 110–113. [Google Scholar] [CrossRef]
- Li, L.; Shi, L.; Yang, S.; Yan, R.; Zhang, D.; Yang, J.; He, L.; Li, W.; Yi, X.; Sun, L.; et al. SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nat. Commun. 2016, 7, 12235. [Google Scholar] [CrossRef]
- Wei, L.; Hu, F.; Shen, Y.; Chen, Z.; Yu, Y.; Lin, C.C.; Wang, M.C.; Min, W. Live-cell imaging of alkyne-tagged small biomolecules by stimulated Raman scattering. Nat. Methods 2014, 11, 410–412. [Google Scholar] [CrossRef]
- Khan, A.N.; Lewis, P.N. Unstructured conformations are a substrate requirement for the Sir2 family of NAD-dependent protein deacetylases. J. Biol. Chem. 2005, 280, 36073–36078. [Google Scholar] [CrossRef] [PubMed]
- Knyphausen, P.; de Boor, S.; Kuhlmann, N.; Scislowski, L.; Extra, A.; Baldus, L.; Schacherl, M.; Baumann, U.; Neundorf, I.; Lammers, M. Insights into Lysine Deacetylation of Natively Folded Substrate Proteins by Sirtuins. J. Biol. Chem. 2016, 291, 14677–14694. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, M.S.; Bever, K.; Avalos, J.L.; Muhammad, S.; Zhang, X.; Wolberger, C. The structural basis of sirtuin substrate affinity. Biochemistry 2006, 45, 7511–7521. [Google Scholar] [CrossRef] [PubMed]
- Vaziri, H.; Dessain, S.K.; Eaton, E.N.; Imai, S.-I.; Frye, R.A.; Pandita, T.K.; Guarente, L.; Weinberg, R.A. hSIR2SIRT1 Functions as an NAD-Dependent p53 Deacetylase. Cell 2001, 107, 149–159. [Google Scholar] [CrossRef]
- Schwer, B.; Bunkenborg, J.; Verdin, R.O.; Andersen, J.S.; Verdin, E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc. Natl. Acad. Sci. USA 2006, 103, 10224–10229. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, T.L.A.; Michishita, E.; Adler, A.S.; Damian, M.; Berber, E.; Lin, M.; McCord, R.A.; Ongaigui, K.C.L.; Boxer, L.D.; Chang, H.Y.; et al. SIRT6 Links Histone H3 Lysine 9 Deacetylation to NF-κB-Dependent Gene Expression and Organismal Life Span. Cell 2009, 136, 62–74. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; D’Urso, A.; Toiber, D.; Sebastian, C.; Henry, R.E.; Vadysirisack, D.D.; Guimaraes, A.; Marinelli, B.; Wikstrom, J.D.; Nir, T.; et al. The Histone Deacetylase Sirt6 Regulates Glucose Homeostasis via Hif1α. Cell 2010, 140, 280–293. [Google Scholar] [CrossRef]
- Barber, M.F.; Michishita-Kioi, E.; Xi, Y.; Tasselli, L.; Kioi, M.; Moqtaderi, Z.; Tennen, R.I.; Paredes, S.; Young, N.L.; Chen, K.; et al. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature 2012, 487, 114–118. [Google Scholar] [CrossRef]
- Sauve, A.A. Sirtuin chemical mechanisms. Biochim. Biophys. Acta 2010, 1804, 1591–1603. [Google Scholar] [CrossRef]
- Shi, Y.; Zhou, Y.; Wang, S.; Zhang, Y. Sirtuin Deacetylation Mechanism and Catalytic Role of the Dynamic Cofactor Binding Loop. J. Phys. Chem. Lett. 2013, 4, 491–495. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, H.; He, B.; Du, J.; Lin, H.; Cerione, R.A.; Hao, Q. The bicyclic intermediate structure provides insights into the desuccinylation mechanism of human sirtuin 5 (SIRT5). J. Biol. Chem. 2012, 287, 28307–28314. [Google Scholar] [CrossRef] [PubMed]
- Sauve, A.A.; Celic, I.; Avalos, J.; Deng, H.; Boeke, J.D.; Schramm, V.L. Chemistry of gene silencing: The mechanism of NAD+-dependent deacetylation reactions. Biochemistry 2001, 40, 15456–15463. [Google Scholar] [CrossRef] [PubMed]
- Tanny, J.C.; Moazed, D. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: Evidence for acetyl transfer from substrate to an NAD breakdown product. Proc. Natl. Acad. Sci. USA 2001, 98, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Bosch-Presegue, L.; Vaquero, A. Sirtuin-dependent epigenetic regulation in the maintenance of genome integrity. FEBS J. 2015, 282, 1745–1767. [Google Scholar] [CrossRef]
- Masri, S.; Sassone-Corsi, P. Sirtuins and the circadian clock: Bridging chromatin and metabolism. Sci. Signal. 2014, 7, re6. [Google Scholar] [CrossRef]
- Guarente, L. Sirtuins, aging, and metabolism. Cold Spring Harb Symp. Quant. Biol. 2011, 76, 81–90. [Google Scholar] [CrossRef]
- Haigis, M.C.; Sinclair, D.A. Mammalian sirtuins: Biological insights and disease relevance. Annu. Rev. Pathol. 2010, 5, 253–295. [Google Scholar] [CrossRef]
- Longo, V.D.; Antebi, A.; Bartke, A.; Barzilai, N.; Brown-Borg, H.M.; Caruso, C.; Curiel, T.J.; Cabo, R.; Franceschi, C.; Gems, D.; et al. Interventions to Slow Aging in Humans: Are We Ready? Aging Cell 2015, 14, 497–510. [Google Scholar] [CrossRef]
- Watroba, M.; Szukiewicz, D. The role of sirtuins in aging and age-related diseases. Adv. Med. Sci. 2016, 61, 52–62. [Google Scholar] [CrossRef]
- Chen, D.; Steele, A.D.; Lindquist, S.; Guarente, L. Increase in activity during calorie restriction requires Sirt1. Science 2005, 310, 1641. [Google Scholar] [CrossRef]
- Boily, G.; Seifert, E.L.; Bevilacqua, L.; He, X.H.; Sabourin, G.; Estey, C.; Moffat, C.; Crawford, S.; Saliba, S.; Jardine, K.; et al. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS ONE 2008, 3, e1759. [Google Scholar] [CrossRef]
- Grabowska, W.; Sikora, E.; Bielak-Zmijewska, A. Sirtuins, a promising target in slowing down the ageing process. Biogerontology 2017, 18, 447–476. [Google Scholar] [CrossRef]
- Chung, J.H.; Manganiello, V.; Dyck, J.R.B. Resveratrol as a calorie restriction mimetic: Therapeutic implications. Trends Cell Biol. 2012, 22, 546–554. [Google Scholar] [CrossRef]
- Bosch-Presegue, L.; Vaquero, A. The dual role of sirtuins in cancer. Genes Cancer 2011, 2, 648–662. [Google Scholar] [CrossRef]
- Deng, C.-X. SIRT1, Is It a Tumor Promoter or Tumor Suppressor? Int. J. Biol. Sci. 2009, 5, 147–152. [Google Scholar] [CrossRef]
- El-Rifai, W.; Zhang, T.; Rong, N.; Chen, J.; Zou, C.; Jing, H.; Zhu, X.; Zhang, W. SIRT1 Expression Is Associated with the Chemotherapy Response and Prognosis of Patients with Advanced NSCLC. PLoS ONE 2013, 8, e79162. [Google Scholar] [CrossRef]
- McGlynn, L.M.; Zino, S.; MacDonald, A.I.; Curle, J.; Reilly, J.E.; Mohammed, Z.M.A.; McMillan, D.C.; Mallon, E.; Payne, A.P.; Edwards, J.; et al. SIRT2: Tumour suppressor or tumour promoter in operable breast cancer? Eur. J. Cancer 2014, 50, 290–301. [Google Scholar] [CrossRef]
- Ozden, O.; Park, S.-H.; Wagner, B.A.; Yong Song, H.; Zhu, Y.; Vassilopoulos, A.; Jung, B.; Buettner, G.R.; Gius, D. SIRT3 deacetylates and increases pyruvate dehydrogenase activity in cancer cells. Free Radic. Biol. Med. 2014, 76, 163–172. [Google Scholar] [CrossRef]
- Miyo, M.; Yamamoto, H.; Konno, M.; Colvin, H.; Nishida, N.; Koseki, J.; Kawamoto, K.; Ogawa, H.; Hamabe, A.; Uemura, M.; et al. Tumour-suppressive function of SIRT4 in human colorectal cancer. Br. J. Cancer 2015, 113, 492–499. [Google Scholar] [CrossRef]
- Lu, W.; Zuo, Y.; Feng, Y.; Zhang, M. SIRT5 facilitates cancer cell growth and drug resistance in non-small cell lung cancer. Tumour. Biol. 2014, 35, 10699–10705. [Google Scholar] [CrossRef]
- Desantis, V.; Lamanuzzi, A.; Vacca, A. The role of SIRT6 in tumors. Haematologica 2018, 103, 1–4. [Google Scholar] [CrossRef]
- Elibol, B.; Kilic, U. High Levels of SIRT1 Expression as a Protective Mechanism Against Disease-Related Conditions. Front. Endocrinol. 2018, 9, 614. [Google Scholar] [CrossRef]
- Lutz, M.I.; Milenkovic, I.; Regelsberger, G.; Kovacs, G.G. Distinct patterns of sirtuin expression during progression of Alzheimer’s disease. Neuromol. Med. 2014, 16, 405–414. [Google Scholar] [CrossRef]
- Chan, S.H.; Hung, C.H.; Shih, J.Y.; Chu, P.M.; Cheng, Y.H.; Lin, H.C.; Tsai, K.L. SIRT1 inhibition causes oxidative stress and inflammation in patients with coronary artery disease. Redox Biol. 2017, 13, 301–309. [Google Scholar] [CrossRef]
- Costa, C.d.S.; Hammes, T.O.; Rohden, F.; Margis, R.; Bortolotto, J.W.; Padoin, A.V.; Mottin, C.C.; Guaragna, R.M. SIRT1 Transcription Is Decreased in Visceral Adipose Tissue of Morbidly Obese Patients with Severe Hepatic Steatosis. Obes. Surg. 2009, 20, 633–639. [Google Scholar] [CrossRef]
- Braidy, N.; Jayasena, T.; Poljak, A.; Sachdev, P.S. Sirtuins in cognitive ageing and Alzheimer’s disease. Curr. Opin. Psychiatry 2012, 25, 226–230. [Google Scholar] [CrossRef]
- Singh, P.; Hanson, P.S.; Morris, C.M. SIRT1 ameliorates oxidative stress induced neural cell death and is down-regulated in Parkinson’s disease. BMC Neurosci. 2017, 18, 1–13. [Google Scholar] [CrossRef]
- Dai, H.; Sinclair, D.A.; Ellis, J.L.; Steegborn, C. Sirtuin activators and inhibitors: Promises, achievements, and challenges. Pharmacol. Ther. 2018, 188, 140–154. [Google Scholar] [CrossRef]
- Dompierre, J.P.; Godin, J.D.; Charrin, B.C.; Cordelieres, F.P.; King, S.J.; Humbert, S.; Saudou, F. Histone deacetylase 6 inhibition compensates for the transport deficit in Huntington’s disease by increasing tubulin acetylation. J. Neurosci. 2007, 27, 3571–3583. [Google Scholar] [CrossRef]
- Govindarajan, N.; Rao, P.; Burkhardt, S.; Sananbenesi, F.; Schluter, O.M.; Bradke, F.; Lu, J.; Fischer, A. Reducing HDAC6 ameliorates cognitive deficits in a mouse model for Alzheimer’s disease. EMBO Mol. Med. 2013, 5, 52–63. [Google Scholar] [CrossRef]
- Saba, N.F.; Magliocca, K.R.; Kim, S.; Muller, S.; Chen, Z.; Owonikoko, T.K.; Sarlis, N.J.; Eggers, C.; Phelan, V.; Grist, W.J.; et al. Acetylated tubulin (AT) as a prognostic marker in squamous cell carcinoma of the head and neck. Head Neck Pathol. 2014, 8, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Boggs, A.E.; Vitolo, M.I.; Whipple, R.A.; Charpentier, M.S.; Goloubeva, O.G.; Ioffe, O.B.; Tuttle, K.C.; Slovic, J.; Lu, Y.; Mills, G.B.; et al. alpha-Tubulin acetylation elevated in metastatic and basal-like breast cancer cells promotes microtentacle formation, adhesion, and invasive migration. Cancer Res. 2015, 75, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Ge, J.; Li, H. Lysine acetyltransferases and lysine deacetylases as targets for cardiovascular disease. Nat. Rev. Cardiol. 2019, 17, 96–115. [Google Scholar] [CrossRef] [PubMed]
- McLendon, P.M.; Ferguson, B.S.; Osinska, H.; Bhuiyan, M.S.; James, J.; McKinsey, T.A.; Robbins, J. Tubulin hyperacetylation is adaptive in cardiac proteotoxicity by promoting autophagy. Proc. Natl. Acad. Sci. USA 2014, 111, E5178–E5186. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, D.A.; Guarente, L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell 1997, 91, 1033–1042. [Google Scholar] [CrossRef]
- Satoh, A.; Stein, L.; Imai, S. The role of mammalian sirtuins in the regulation of metabolism, aging, and longevity. Handb. Exp. Pharm. 2011, 206, 125–162. [Google Scholar] [CrossRef]
- Chakraborty, C.; Doss, C.G. Sirtuins family--recent development as a drug target for aging, metabolism, and age related diseases. Curr. Drug Targets 2013, 14, 666–675. [Google Scholar] [CrossRef]
- Kane, A.E.; Sinclair, D.A. Sirtuins and NAD(+) in the Development and Treatment of Metabolic and Cardiovascular Diseases. Circ. Res. 2018, 123, 868–885. [Google Scholar] [CrossRef]
- Satoh, A.; Brace, C.S.; Rensing, N.; Cliften, P.; Wozniak, D.F.; Herzog, E.D.; Yamada, K.A.; Imai, S. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab. 2013, 18, 416–430. [Google Scholar] [CrossRef]
- Kanfi, Y.; Naiman, S.; Amir, G.; Peshti, V.; Zinman, G.; Nahum, L.; Bar-Joseph, Z.; Cohen, H.Y. The sirtuin SIRT6 regulates lifespan in male mice. Nature 2012, 483, 218–221. [Google Scholar] [CrossRef]
- Mitchell, S.J.; Martin-Montalvo, A.; Mercken, E.M.; Palacios, H.H.; Ward, T.M.; Abulwerdi, G.; Minor, R.K.; Vlasuk, G.P.; Ellis, J.L.; Sinclair, D.A.; et al. The SIRT1 activator SRT1720 extends lifespan and improves health of mice fed a standard diet. Cell Rep. 2014, 6, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Mercken, E.M.; Mitchell, S.J.; Martin-Montalvo, A.; Minor, R.K.; Almeida, M.; Gomes, A.P.; Scheibye-Knudsen, M.; Palacios, H.H.; Licata, J.J.; Zhang, Y.; et al. SRT2104 extends survival of male mice on a standard diet and preserves bone and muscle mass. Aging Cell 2014, 13, 787–796. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teixeira, C.S.S.; Cerqueira, N.M.F.S.A.; Gomes, P.; Sousa, S.F. A Molecular Perspective on Sirtuin Activity. Int. J. Mol. Sci. 2020, 21, 8609. https://doi.org/10.3390/ijms21228609
Teixeira CSS, Cerqueira NMFSA, Gomes P, Sousa SF. A Molecular Perspective on Sirtuin Activity. International Journal of Molecular Sciences. 2020; 21(22):8609. https://doi.org/10.3390/ijms21228609
Chicago/Turabian StyleTeixeira, Carla S. S., Nuno M. F. S. A. Cerqueira, Pedro Gomes, and Sérgio F. Sousa. 2020. "A Molecular Perspective on Sirtuin Activity" International Journal of Molecular Sciences 21, no. 22: 8609. https://doi.org/10.3390/ijms21228609
APA StyleTeixeira, C. S. S., Cerqueira, N. M. F. S. A., Gomes, P., & Sousa, S. F. (2020). A Molecular Perspective on Sirtuin Activity. International Journal of Molecular Sciences, 21(22), 8609. https://doi.org/10.3390/ijms21228609







