Affinity Captured Urinary Extracellular Vesicles Provide mRNA and miRNA Biomarkers for Improved Accuracy of Prostate Cancer Detection: A Pilot Study
Abstract
1. Introduction
2. Results
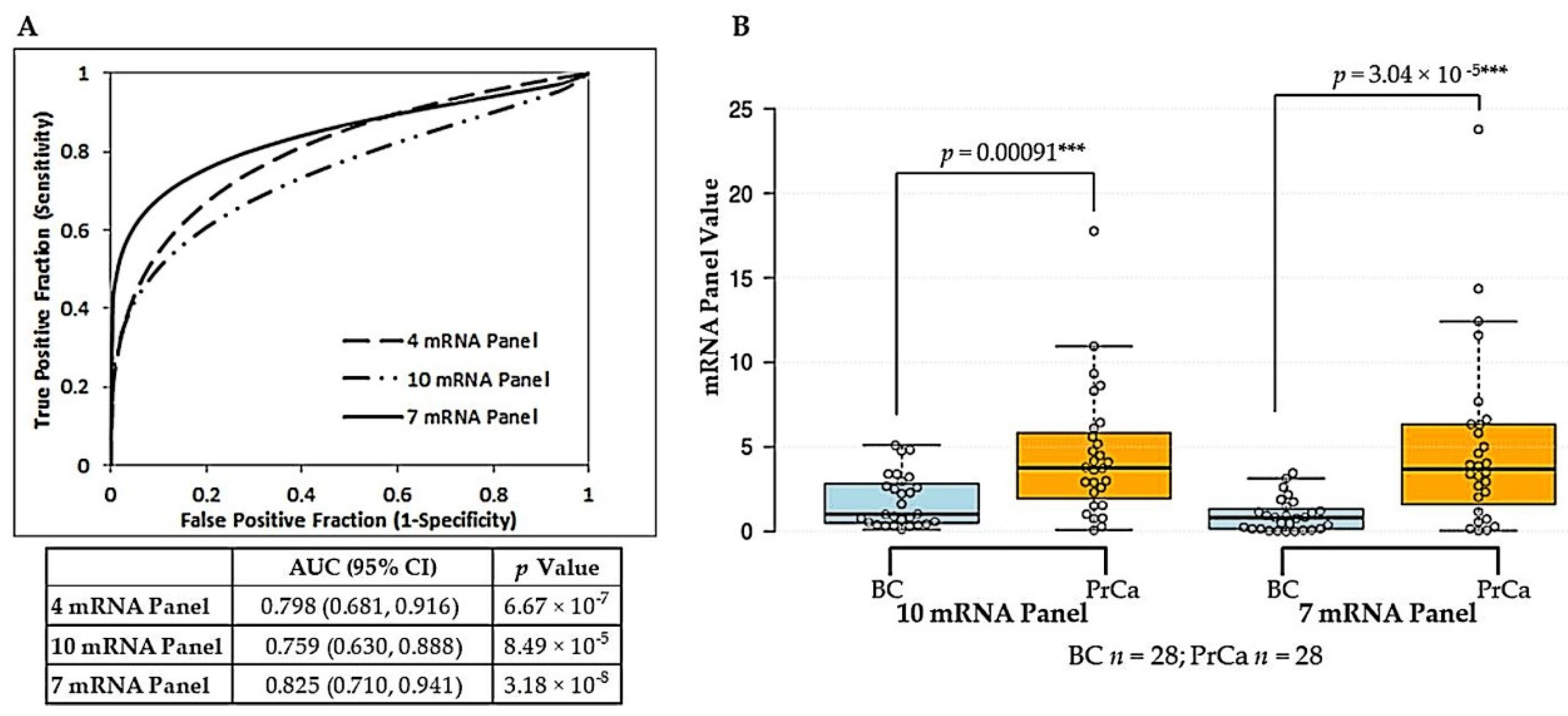
2.1. Derivation of a Vn96-Isolated EV Reference-Free mRNA Panel for Prostate Cancer Diagnosis
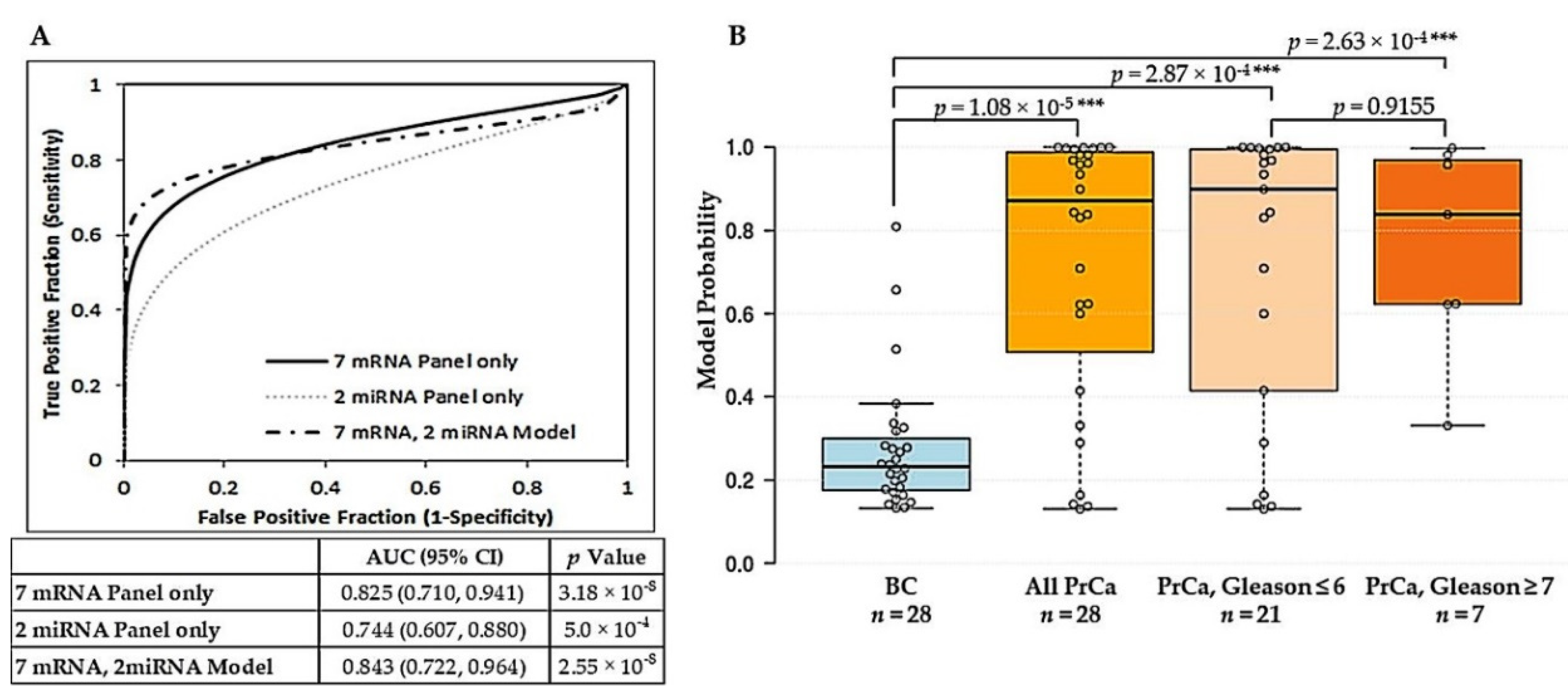
2.2. Derivation of a Vn96-Isolated EV Combined mRNA and miRNA Model for Prostate Cancer Diagnosis
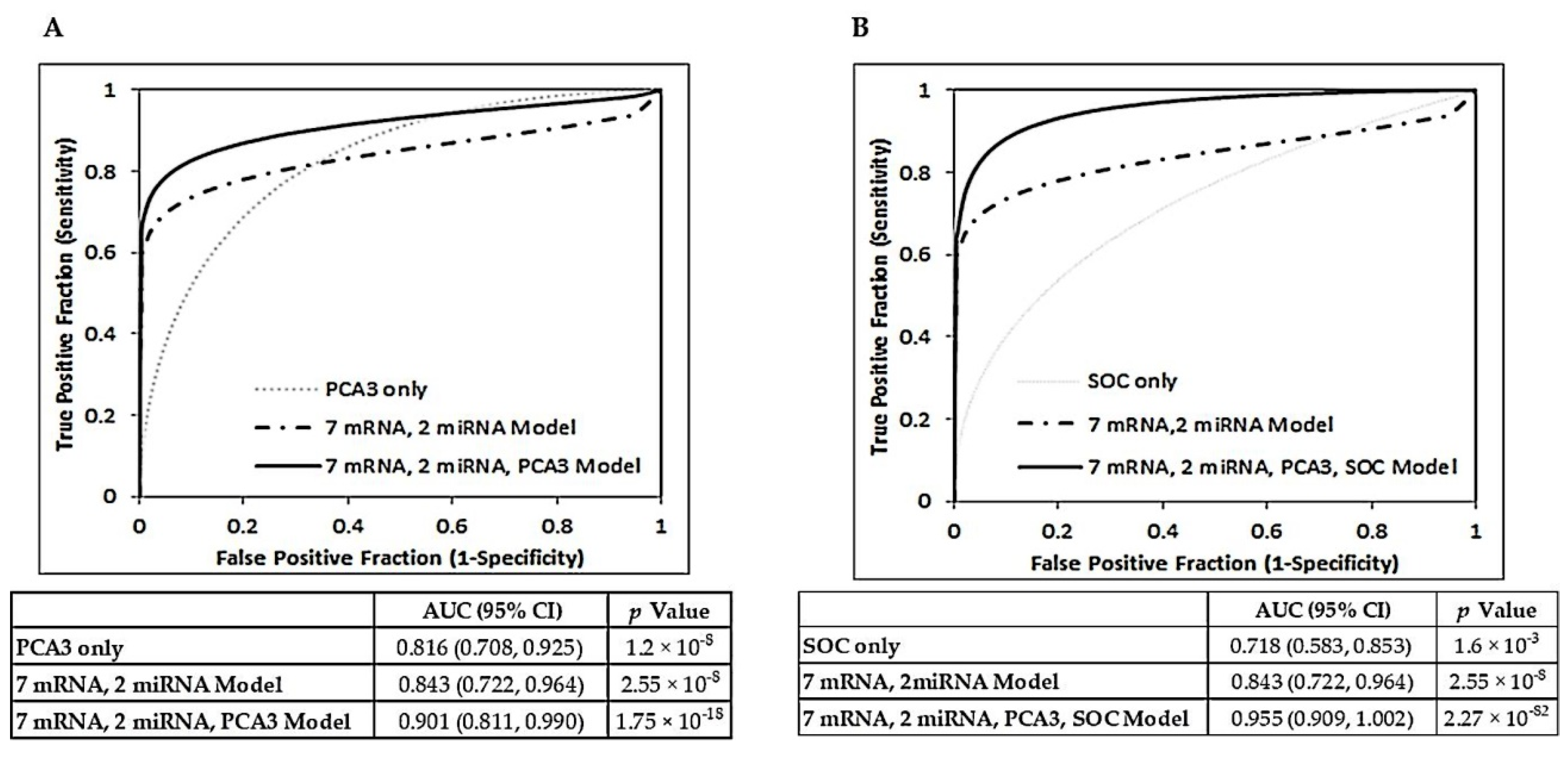
2.3. Additive Value of Combining Vn96-Isolated EV mRNA and miRNA Panels with PCA3 and Clinical Characteristics
3. Discussion
4. Materials and Methods
4.1. Study Participants
4.2. Urine Collection and Processing
4.3. Isolation of Extracellular Vesicles Using the Vn96 Peptide
4.4. RNA Extraction from Urinary EVs
4.5. Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
geomean(REχ1,…,REχη), where χ = individual mRNA of interest.
4.6. miRNA Detection and Quantification
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ACTB | Actin Beta |
| ANXA3 | Annexin A3 |
| AUC | Area Under the Curve |
| BC | Benign Control |
| BPH | Benign Prostate Hyperplasia |
| CFD | Complement Factor D |
| DNA | Deoxyribonucleic acid |
| dNTP | Deoxynucleoside triphosphate |
| DRE | Digital Rectal Examination |
| DTT | Dithiothreitol |
| ERG | Erythroblast Transformation-Specific (ETS) Transcription Factor ERG |
| EV | Extracellular Vesicle |
| FOLH1 | Folate Hydrolase 1 |
| GOLM1 | Golgi Membrane Protein 1 |
| GOLPH2 | Golgi phosphoprotein 2 |
| GSTM4 | Glutathione S-Transferase Mu 4 |
| HPN | Hepsin |
| Hsp/c70 | Heat shock protein/cognate 70 |
| ITSN1 | Intersectin 1 |
| PIN | Prostatic Intraepithelial Neoplasia |
| KLK3 | Kallikrein Related Peptidase 3 |
| LTBP4 | Latent Transforming Growth Factor Beta Binding Protein 4 |
| mRNA | Messenger ribonucleic acid |
| NELL2 | Neural EGFL Like 2 |
| PBS | Phosphate-buffered saline |
| PCA3 | Prostate Cancer Associated 3 |
| PDCD6IP | Programmed Cell Death 6 Interacting Protein |
| PrCa | Prostate Cancer |
| PSCA | Prostate Stem Cell Antigen |
| qPCR | Quantitative Polymerase Chain Reaction |
| ROC | Receiver Operator Characteristic |
| SDS-PAGE | Sodium dodecyl sulphate-polyacrylamide gel electrophoresis |
| SLC45A3 | Solute Carrier Family 45 Member 3 |
| SNORD44 | Small Nucleolar RNA, C/D Box 44 |
| SPINK1 | Serine Peptidase Inhibitor Kazal Type 1 |
| sPSA | Serum Prostate Specific Antigen |
| TMN | Tumour, nodes, metastasis |
| TMPRSS2 | Transmembrane Serine Protease 2 |
| TRUS | Transrectal Ultrasound |
| XBP1 | X-Box Binding Protein 1 |
References
- Bray, F.; Me, J.F.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Rendon, R.A.; Mason, R.J.; Marzouk, K.; Finelli, A.; Saad, F.; So, A.; Violette, P.D.; Breau, R.H. Canadian Urological Association recommendations on prostate cancer screening and early diagnosis. Can. Urol. Assoc. J. 2017, 11, 298–309. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fenton, J.J.; Weyrich, M.S.; Durbin, S.; Liu, Y.; Bang, H.; Melnikow, J. Prostate-Specific Antigen–Based Screening for Prostate Cancer. JAMA 2018, 319, 1914–1931. [Google Scholar] [CrossRef] [PubMed]
- Berman, D.M.; Epstein, J.I. When is Prostate Cancer Really Cancer? Urol. Clin. N. Am. 2014, 41, 339–346. [Google Scholar] [CrossRef]
- Ilic, D.; Djulbegovic, M.; Jung, J.H.; Hwang, E.C.; Zhou, Q.; Cleves, A.; Agoritsas, T.; Dahm, P. Prostate cancer screening with prostate-specific antigen (PSA) test: A systematic review and meta-analysis. BMJ 2018, 362, k3519. [Google Scholar] [CrossRef] [PubMed]
- Draisma, G.; Etzioni, R.; Tsodikov, A.; Mariotto, A.; Wever, E.; Gulati, R.; Feuer, E.; De Koning, H. Lead Time and Overdiagnosis in Prostate-Specific Antigen Screening: Importance of Methods and Context. J. Natl. Cancer Inst. 2009, 101, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Prensner, J.R.; Rubin, M.A.; Wei, J.T.; Chinnaiyan, A.M. Beyond PSA: The Next Generation of Prostate Cancer Biomarkers. Sci. Transl. Med. 2012, 4, 127rv3. [Google Scholar] [CrossRef]
- Blute, M.L.; Abel, E.J.; Downs, T.M.; Kelcz, F.; Jarrard, D.F. Addressing the need for repeat prostate biopsy: New technology and approaches. Nat. Rev. Urol. 2015, 12, 435–444. [Google Scholar] [CrossRef]
- Thompson, I.M.; Pauler, D.K.; Goodman, P.J.; Tangen, C.M.; Lucia, M.S.; Parnes, H.L.; Minasian, L.M.; Ford, L.G.; Lippman, S.M.; Crawford, E.D.; et al. Prevalence of Prostate Cancer among Men with a Prostate-Specific Antigen Level ≤4.0 ng per Milliliter. N. Engl. J. Med. 2004, 350, 2239–2246. [Google Scholar] [CrossRef]
- Esserman, L.J.; Thompson, I.M.; Reid, B.J. Overdiagnosis and Overtreatment in Cancer. JAMA 2013, 310, 797–798. [Google Scholar] [CrossRef] [PubMed]
- Loeb, S.; Vellekoop, A.; Ahmed, H.U.; Catto, J.; Emberton, M.; Nam, R.; Rosario, D.J.; Scattoni, V.; Lotan, Y. Systematic Review of Complications of Prostate Biopsy. Eur. Urol. 2013, 64, 876–892. [Google Scholar] [CrossRef]
- Van Der Leest, M.; Cornel, E.; Israël, B.; Hendriks, R.; Padhani, A.R.; Hoogenboom, M.; Zamecnik, P.; Bakker, D.; Setiasti, A.Y.; Veltman, J.; et al. Head-to-head Comparison of Transrectal Ultrasound-guided Prostate Biopsy Versus Multiparametric Prostate Resonance Imaging with Subsequent Magnetic Resonance-guided Biopsy in Biopsy-naïve Men with Elevated Prostate-specific Antigen: A Large Prospective Multicenter Clinical Study. Eur. Urol. 2019, 75, 570–578. [Google Scholar] [CrossRef]
- Omer, A.; Lamb, A.D. Optimizing prostate biopsy techniques. Curr. Opin. Urol. 2019, 29, 578–586. [Google Scholar] [CrossRef]
- Bratulic, S.; Gatto, F.; Nielsen, J.C. The Translational Status of Cancer Liquid Biopsies. Regen. Eng. Transl. Med. 2019, 1–41. [Google Scholar] [CrossRef]
- Mattox, A.K.; Bettegowda, C.; Zhou, S.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B. Applications of liquid biopsies for cancer. Sci. Transl. Med. 2019, 11, eaay1984. [Google Scholar] [CrossRef]
- Di Meo, A.; Bartlett, J.; Cheng, Y.; Pasic, M.D.; Yousef, G.M. Liquid biopsy: A step forward towards precision medicine in urologic malignancies. Mol. Cancer 2017, 16, 1–14. [Google Scholar] [CrossRef]
- Hendriks, R.J.; Van Oort, I.M.; Schalken, J.A. Blood-based and urinary prostate cancer biomarkers: A review and comparison of novel biomarkers for detection and treatment decisions. Prostate Cancer Prostatic Dis. 2016, 20, 12–19. [Google Scholar] [CrossRef]
- Boerrigter, E.; Groen, L.N.; Van Erp, N.P.; Verhaegh, G.W.; Schalken, J.A. Clinical utility of emerging biomarkers in prostate cancer liquid biopsies. Expert Rev. Mol. Diagn. 2019, 20, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Raja, N.; Russell, C.M.; George, A.K. Urinary markers aiding in the detection and risk stratification of prostate cancer. Transl. Androl. Urol. 2018, 7, S436–S442. [Google Scholar] [CrossRef]
- Chevli, K.K.; Duff, M.; Walter, P.; Yu, C.; Capuder, B.; Elshafei, A.; Malczewski, S.; Kattan, M.W.; Jones, J.S. Urinary PCA3 as a Predictor of Prostate Cancer in a Cohort of 3,073 Men Undergoing Initial Prostate Biopsy. J. Urol. 2014, 191, 1743–1748. [Google Scholar] [CrossRef] [PubMed]
- Tomlins, S.A.; Day, J.R.; Lonigro, R.J.; Hovelson, D.H.; Siddiqui, J.; Kunju, L.P.; Dunn, R.L.; Meyer, S.; Hodge, P.; Groskopf, J.; et al. Urine TMPRSS2:ERG Plus PCA3 for Individualized Prostate Cancer Risk Assessment. Eur. Urol. 2016, 70, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Laxman, B.; Morris, D.S.; Yu, J.; Siddiqui, J.; Cao, J.; Mehra, R.; Lonigro, R.J.; Tsodikov, A.; Wei, J.T.; Tomlins, S.A.; et al. A First-Generation Multiplex Biomarker Analysis of Urine for the Early Detection of Prostate Cancer. Cancer Res. 2008, 68, 645–649. [Google Scholar] [CrossRef] [PubMed]
- Groskopf, J.; Aubin, S.M.; Deras, I.L.; Blase, A.; Bodrug, S.; Clark, C.; Brentano, S.; Mathis, J.; Pham, J.; Meyer, T.; et al. APTIMA PCA3 Molecular Urine Test: Development of a Method to Aid in the Diagnosis of Prostate Cancer. Clin. Chem. 2006, 52, 1089–1095. [Google Scholar] [CrossRef] [PubMed]
- De La Taille, A.; Irani, J.; Graefen, M.; Chun, F.; De Reijke, T.; Kil, P.; Gontero, P.; Mottaz, A.; Haese, A. Clinical Evaluation of the PCA3 Assay in Guiding Initial Biopsy Decisions. J. Urol. 2011, 185, 2119–2125. [Google Scholar] [CrossRef]
- Van Neste, L.; Hendriks, R.J.; Dijkstra, S.; Trooskens, G.; Cornel, E.B.; Jannink, S.A.; De Jong, H.; Hessels, D.; Smit, F.P.; Melchers, W.J.G.; et al. Detection of High-grade Prostate Cancer Using a Urinary Molecular Biomarker–Based Risk Score. Eur. Urol. 2016, 70, 740–748. [Google Scholar] [CrossRef]
- McKiernan, J.; Donovan, M.J.; O’Neill, V.; Bentink, S.; Noerholm, M.; Belzer, S.; Skog, J.; Kattan, M.W.; Partin, A.; Andriole, G.; et al. A Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer at Initial Biopsy. JAMA Oncol. 2016, 2, 882–889. [Google Scholar] [CrossRef]
- Hessels, D.; Smit, F.P.; Verhaegh, G.W.; Witjes, J.; Cornel, E.B.; Schalken, J.A. Detection of TMPRSS2-ERG Fusion Transcripts and Prostate Cancer Antigen 3 in Urinary Sediments May Improve Diagnosis of Prostate Cancer. Clin. Cancer Res. 2007, 13, 5103–5108. [Google Scholar] [CrossRef] [PubMed]
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef]
- Dong, L.; Zieren, R.C.; Wang, Y.; De Reijke, T.M.; Xue, W.; Pienta, K.J. Recent advances in extracellular vesicle research for urological cancers: From technology to application. Biochim. Et Biophys. Acta (BBA)-Bioenerg. 2019, 1871, 342–360. [Google Scholar] [CrossRef]
- Linxweiler, J.; Junker, K. Extracellular vesicles in urological malignancies: An update. Nat. Rev. Urol. 2019, 17, 1–17. [Google Scholar] [CrossRef]
- Barreiro, K.; Holthofer, H. Urinary extracellular vesicles. A promising shortcut to novel biomarker discoveries. Cell Tissue Res. 2017, 369, 217–227. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ghosh, A.; Davey, M.; Chute, I.C.; Griffiths, S.G.; Lewis, S.; Chacko, S.; Barnett, D.; Crapoulet, N.; Fournier, S.; Joy, A.; et al. Rapid Isolation of Extracellular Vesicles from Cell Culture and Biological Fluids Using a Synthetic Peptide with Specific Affinity for Heat Shock Proteins. PLoS ONE 2014, 9, e110443. [Google Scholar] [CrossRef]
- Bijnsdorp, I.V.; Maxouri, O.; Kardar, A.; Schelfhorst, T.; Piersma, S.R.; Pham, T.V.; Vis, A.; Van Moorselaar, R.J.; Jimenez, C.R. Feasibility of urinary extracellular vesicle proteome profiling using a robust and simple, clinically applicable isolation method. J. Extracell. Vesicles 2017, 6, 1313091. [Google Scholar] [CrossRef]
- Saucier, D.; Wajnberg, G.; Roy, J.; Beauregard, A.-P.; Chacko, S.; Crapoulet, N.; Fournier, S.; Ghosh, A.; Lewis, S.M.; Marrero, A.; et al. Identification of a circulating miRNA signature in extracellular vesicles collected from amyotrophic lateral sclerosis patients. Brain Res. 2019, 1708, 100–108. [Google Scholar] [CrossRef]
- Joy, A.P.; Ayre, D.C.; Chute, I.C.; Beauregard, A.-P.; Wajnberg, G.; Ghosh, A.; Lewis, S.M.; Ouellette, R.J.; Barnett, D.A. Proteome profiling of extracellular vesicles captured with the affinity peptide Vn96: Comparison of Laemmli and TRIzol© protein-extraction methods. J. Extracell. Vesicles 2018, 7, 1438727. [Google Scholar] [CrossRef] [PubMed]
- Stokman, M.F.; Bijnsdorp, I.V.; Schelfhorst, T.; Pham, T.V.; Piersma, S.R.; Knol, J.C.; Giles, R.H.; Bongers, E.M.; Knoers, N.V.; Lilien, M.R.; et al. Changes in the urinary extracellular vesicle proteome are associated with nephronophthisis-related ciliopathies. J. Proteom. 2019, 192, 27–36. [Google Scholar] [CrossRef]
- Cuperlovic-Culf, M.; Belacel, N.; Davey, M.; Ouellette, R.J. Multi-gene biomarker panel for reference free prostate cancer diagnosis: Determination and independent validation. Biomarkers 2010, 15, 693–706. [Google Scholar] [CrossRef]
- Endzeliņš, E.; Melne, V.; Kalniņa, Z.; Lietuvietis, V.; Riekstiņa, U.; Llorente, A.; Linē, A. Diagnostic, prognostic and predictive value of cell-free miRNAs in prostate cancer: A systematic review. Mol. Cancer 2016, 15, 1–13. [Google Scholar] [CrossRef]
- Paiva, R.M.; Zauli, D.A.G.; Neto, B.S.; Brum, I.S. Urinary microRNAs expression in prostate cancer diagnosis: A systematic review. Clin. Transl. Oncol. 2020, 22, 2061–2073. [Google Scholar] [CrossRef]
- Movahedpour, A.; Ahmadi, N.; Ghasemi, Y.; Savardashtaki, A.; Shabaninejad, Z. Circulating microRNAs as potential diagnostic biomarkers and therapeutic targets in prostate cancer: Current status and future perspectives. J. Cell. Biochem. 2019, 120, 16316–16329. [Google Scholar] [CrossRef]
- Cochetti, G.; De Vermandois, J.A.R.; Maulà, V.; Giulietti, M.; Cecati, M.; Del Zingaro, M.; Cagnani, R.; Suvieri, C.; Paladini, A.; Mearini, E. Role of miRNAs in prostate cancer: Do we really know everything? Urol. Oncol. Semin. Orig. Investig. 2020, 38, 623–635. [Google Scholar] [CrossRef]
- Bin Riaz, I.; Wang, L.; Kohli, M. Liquid biopsy approach in the management of prostate cancer. Transl. Res. 2018, 201, 60–70. [Google Scholar] [CrossRef]
- Lu, Y.-T.; Delijani, K.; Mecum, A.; Goldkorn, A. Current status of liquid biopsies for the detection and management of prostate cancer. Cancer Manag. Res. 2019, 11, 5271–5291. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Nonomura, N. Urinary biomarkers of prostate cancer. Int. J. Urol. 2018, 25, 770–779. [Google Scholar] [CrossRef]
- Eskra, J.N.; Rabizadeh, D.; Pavlovich, C.P.; Catalona, W.J.; Luo, J. Approaches to urinary detection of prostate cancer. Prostate Cancer Prostatic Dis. 2019, 22, 362–381. [Google Scholar] [CrossRef]
- Vlaeminck-Guillem, V. Extracellular Vesicles in Prostate Cancer Carcinogenesis, Diagnosis, and Management. Front. Oncol. 2018, 8, 222. [Google Scholar] [CrossRef]
- Xu, R.; Rai, A.; Chen, M.; Suwakulsiri, W.; Greening, D.W.; Simpson, R.J. Extracellular vesicles in cancer—Implications for future improvements in cancer care. Nat. Rev. Clin. Oncol. 2018, 15, 617–638. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Sun, X.; Scicluna, B.J.; Coleman, B.M.; Hill, A.F. Characterization and deep sequencing analysis of exosomal and non-exosomal miRNA in human urine. Kidney Int. 2014, 86, 433–444. [Google Scholar] [CrossRef]
- Anand, S.; Samuel, M.; Kumar, S.; Mathivanan, S. Ticket to a bubble ride: Cargo sorting into exosomes and extracellular vesicles. Biochim. Et Biophys. Acta (BBA)-Proteins Proteom. 2019, 1867, 140203. [Google Scholar] [CrossRef]
- Becker, A.; Thakur, B.K.; Weiss, J.M.; Kim, H.S.; Peinado, H.; Lyden, D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell 2016, 30, 836–848. [Google Scholar] [CrossRef]
- Groot, M.; Lee, H. Sorting Mechanisms for MicroRNAs into Extracellular Vesicles and Their Associated Diseases. Cells 2020, 9, 1044. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Underwood, P.; Hughes, S.J. From tumor microenvironment communicants to biomarker discovery: Selectively packaged extracellular vesicular cargoes in pancreatic cancer. Cytokine Growth Factor Rev. 2020, 51, 61–68. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef]
- Villarroya-Beltri, C.; Baixauli, F.; Gutiérrez-Vázquez, C.; Sánchez-Madrid, F.; Mittelbrunn, M. Sorting it out: Regulation of exosome loading. Semin. Cancer Biol. 2014, 28, 3–13. [Google Scholar] [CrossRef]
- Musante, L.; Tataruch, D.; Gu, D.; Benito-Martin, A.; Calzaferri, G.; Aherne, S.; Holthofer, H. A Simplified Method to Recover Urinary Vesicles for Clinical Applications and Sample Banking. Sci. Rep. 2014, 4, 7532. [Google Scholar] [CrossRef]
- Musante, L.; Tataruch-Weinert, D.; Kerjaschki, D.; Henry, M.; Meleady, P.; Holthofer, H. Residual urinary extracellular vesicles in ultracentrifugation supernatants after hydrostatic filtration dialysis enrichment. J. Extracell. Vesicles 2016, 6, 1267896. [Google Scholar] [CrossRef]
- Dijkstra, S.; Birker, I.L.; Smit, F.P.; Leyten, G.H.J.M.; De Reijke, T.M.; Van Oort, I.M.; Mulders, P.F.A.; Jannink, S.A.; Schalken, J.A. Prostate Cancer Biomarker Profiles in Urinary Sediments and Exosomes. J. Urol. 2014, 191, 1132–1138. [Google Scholar] [CrossRef]
- Pellegrini, K.L.; Patil, D.; Douglas, K.J.; Lee, G.; Wehrmeyer, K.; Torlak, M.; Clark, J.; Cooper, C.S.; Moreno, C.S.; Sanda, M. Detection of prostate cancer-specific transcripts in extracellular vesicles isolated from post-DRE urine. Prostate 2017, 77, 990–999. [Google Scholar] [CrossRef]
- Hessels, D.; Gunnewiek, J.M.K.; Van Oort, I.; Karthaus, H.F.; Van Leenders, G.J.; Van Balken, B.; Kiemeney, L.A.; Witjes, J.; Schalken, J.A. DD3PCA3-based Molecular Urine Analysis for the Diagnosis of Prostate Cancer. Eur. Urol. 2003, 44, 8–16. [Google Scholar] [CrossRef]
- Aubin, S.M.; Reid, J.; Sarno, M.J.; Blase, A.; Aussie, J.; Rittenhouse, H.; Rittmaster, R.; Andriole, G.L.; Groskopf, J. PCA3 Molecular Urine Test for Predicting Repeat Prostate Biopsy Outcome in Populations at Risk: Validation in the Placebo Arm of the Dutasteride REDUCE Trial. J. Urol. 2010, 184, 1947–1952. [Google Scholar] [CrossRef]
- Salagierski, M.; Schalken, J.A. Molecular Diagnosis of Prostate Cancer: PCA3 and TMPRSS2: ERG Gene Fusion. J. Urol. 2012, 187, 795–801. [Google Scholar] [CrossRef]
- Margolis, L.; Sadovsky, Y. The biology of extracellular vesicles: The known unknowns. PLoS Biol. 2019, 17, e3000363. [Google Scholar] [CrossRef] [PubMed]
- Rigau, M.; Ortega, I.; Mir, M.C.; Ballesteros, C.; Garcia, M.; Llauradó, M.; Colás, E.; Pedrola, N.; Montes, M.; Sequeiros, T.; et al. A Three-Gene panel on urine increases PSA specificity in the detection of prostate cancer. Prostate 2011, 71, 1736–1745. [Google Scholar] [CrossRef]
- Talesa, V.N.; Antognelli, C.; Del Buono, C.; Stracci, F.; Serva, M.R.; Cottini, E.; Mearini, E. Diagnostic potential in prostate cancer of a panel of urinary molecular tumor markers. Cancer Biomark. 2009, 5, 241–251. [Google Scholar] [CrossRef]
- Stephan, C.; Yousef, G.M.; Scorilas, A.; Jung, K.; Jung, M.; Kristiansen, G.; Hauptmann, S.; Kishi, T.; Nakamura, T.; Loening, S.A.; et al. Hepsin is Highly Over Expressed in and a New Candidate for a Prognostic Indicator in Prostate Cancer. J. Urol. 2004, 171, 187–191. [Google Scholar] [CrossRef]
- Schostak, M.; Krause, H.; Miller, K.; Schrader, M.; Weikert, S.; Christoph, F.; Kempkensteffen, C.; Kollermann, J. Quantitative real-time RT-PCR of CD24 mRNA in the detection of prostate cancer. BMC Urol. 2006, 6, 7. [Google Scholar] [CrossRef][Green Version]
- Quek, S.-I.; Ho, M.E.; Loprieno, M.A.; Ellis, W.J.; Elliott, N.; Liu, A.Y. A Multiplex Assay to Measure RNA Transcripts of Prostate Cancer in Urine. PLoS ONE 2012, 7, e45656. [Google Scholar] [CrossRef][Green Version]
- Sanguedolce, F.; Cormio, A.; Brunelli, M.; D’Amuri, A.; Carrieri, G.; Bufo, P.; Cormio, L. Urine TMPRSS2: ERG Fusion Transcript as a Biomarker for Prostate Cancer: Literature Review. Clin. Genitourin. Cancer 2016, 14, 117–121. [Google Scholar] [CrossRef]
- Song, C.; Chen, H. Predictive significance of TMRPSS2-ERG fusion in prostate cancer: A meta-analysis. Cancer Cell Int. 2018, 18, 177. [Google Scholar] [CrossRef]
- Hunter, M.P.; Russo, A.; O’Bryan, J.P. Emerging Roles for Intersectin (ITSN) in Regulating Signaling and Disease Pathways. Int. J. Mol. Sci. 2013, 14, 7829–7852. [Google Scholar] [CrossRef]
- Cao, D.-L.; Ye, D.-W.; Zhang, H.-L.; Zhu, Y.; Wang, Y.-X.; Yao, X.-D. A multiplex model of combining gene-based, protein-based, and metabolite-based with positive and negative markers in urine for the early diagnosis of prostate cancer. Prostate 2010, 71, 700–710. [Google Scholar] [CrossRef]
- Schostak, M.; Schwall, G.P.; Poznanovic, S.; Groebe, K.; Müller, M.; Messinger, D.; Miller, K.; Krause, H.; Pelzer, A.; Horninger, W.; et al. Annexin A3 in Urine: A Highly Specific Noninvasive Marker for Prostate Cancer Early Detection. J. Urol. 2009, 181, 343–353. [Google Scholar] [CrossRef]
- Hernández, S.; Font-Tello, A.; Juanpere, N.; De Muga, S.; Lorenzo, M.; Salido, M.; Fumadó, L.; Serrano, L.; Cecchini, L.; Serrano, S.; et al. ConcurrentTMPRSS2-ERGandSLC45A3-ERGrearrangements plusPTENloss are not found in low grade prostate cancer and define an aggressive tumor subset. Prostate 2016, 76, 854–865. [Google Scholar] [CrossRef]
- Rickman, D.S.; Pflueger, R.; Moss, B.; Van Doren, V.; Chen, C.X.; De La Taille, A.; Kuefer, R.; Tewari, A.K.; Setlur, S.R.; Demichelis, F.; et al. SLC45A3-ELK4 is a novel and frequent erythroblast transformation-specific fusion transcript in prostate cancer. Cancer Res. 2009, 69, 2734–2738. [Google Scholar] [CrossRef]
- Gouin, K.; Peck, K.; Antes, T.; Johnson, J.L.; Li, C.; Vaturi, S.D.; Middleton, R.; De Couto, G.; Walravens, A.-S.; Rodriguez-Borlado, L.; et al. A comprehensive method for identification of suitable reference genes in extracellular vesicles. J. Extracell. Vesicles 2017, 6, 1347019. [Google Scholar] [CrossRef]
- Gunasekaran, P.M.; Luther, J.M.; Byrd, J.B. For what factors should we normalize urinary extracellular mRNA biomarkers? Biomol. Detect. Quantif. 2019, 17, 100090. [Google Scholar] [CrossRef]
- Mateescu, B.; Kowal, E.J.K.; Van Balkom, B.W.M.; Bartel, S.; Bhattacharyya, S.N.; Buzás, E.I.; Buck, A.H.; De Candia, P.; Chow, F.W.N.; Das, S.; et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA–An ISEV position paper. J. Extracell. Vesicles 2017, 6, 1286095. [Google Scholar] [CrossRef] [PubMed]
- Bryant, R.J.; Pawlowski, T.; Catto, J.W.F.; Marsden, G.; Vessella, R.L.; Rhees, B.; Kuslich, C.; Visakorpi, T.; Hamdy, F.C. Changes in circulating microRNA levels associated with prostate cancer. Br. J. Cancer 2012, 106, 768–774. [Google Scholar] [CrossRef]
- Endzeliņš, E.; Berger, A.; Melne, V.; Bajo-Santos, C.; Soboļevska, K.; Ābols, A.; Rodriguez, M.; Šantare, D.; Rudņickiha, A.; Lietuvietis, V.; et al. Detection of circulating miRNAs: Comparative analysis of extracellular vesicle-incorporated miRNAs and cell-free miRNAs in whole plasma of prostate cancer patients. BMC Cancer 2017, 17, 1–13. [Google Scholar] [CrossRef]
- Foj, L.; Ferrer, F.; Serra, M.; Arévalo, A.; Gavagnach, M.; Giménez, N.; Filella, X. Exosomal and Non-Exosomal Urinary miRNAs in Prostate Cancer Detection and Prognosis. Prostate 2016, 77, 573–583. [Google Scholar] [CrossRef]
- Huang, X.; Yuan, T.; Liang, M.; Du, M.; Xia, S.; Dittmar, R.; Wang, D.; See, W.; Costello, B.A.; Quevedo, F.; et al. Exosomal miR-1290 and miR-375 as Prognostic Markers in Castration-resistant Prostate Cancer. Eur. Urol. 2015, 67, 33–41. [Google Scholar] [CrossRef]
- Koppers-Lalic, D.; Hackenberg, M.; De Menezes, R.; Misovic, B.; Wachalska, M.; Geldof, A.; Zini, N.; De Reijke, T.; Wurdinger, T.; Vis, A.; et al. Non-invasive prostate cancer detection by measuring miRNA variants (isomiRs) in urine extracellular vesicles. Oncotarget 2016, 7, 22566–22578. [Google Scholar] [CrossRef]
- Kumar, B.; Rosenberg, A.Z.; Choi, S.M.; Fox-Talbot, K.; De Marzo, A.M.; Nonn, L.; Brennen, W.N.; Marchionni, L.; Halushka, M.K.; Lupold, S.E. Cell-type specific expression of oncogenic and tumor suppressive microRNAs in the human prostate and prostate cancer. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef]
- Samsonov, R.; Shtam, T.; Burdakov, V.; Glotov, A.S.; Tsyrlina, E.; Berstein, L.; Nosov, A.; Evtushenko, V.; Filatov, M.; Malek, A. Lectin-induced agglutination method of urinary exosomes isolation followed by mi-RNA analysis: Application for prostate cancer diagnostic. Prostate 2015, 76, 68–79. [Google Scholar] [CrossRef]
- Sharma, N.; Baruah, M.M. The microRNA signatures: Aberrantly expressed miRNAs in prostate cancer. Clin. Transl. Oncol. 2018, 21, 126–144. [Google Scholar] [CrossRef]
- Laxman, B.; Tomlins, S.A.; Mehra, R.; Morris, D.S.; Wang, L.; Helgeson, B.E.; Shah, R.B.; Rubin, M.; Wei, J.T.; Chinnaiyan, A.M. Noninvasive Detection of TMPRSS2:ERG Fusion Transcripts in the Urine of Men with Prostate Cancer1. Neoplasia 2006, 8, 885–888. [Google Scholar] [CrossRef]
- Descotes, J.-L. Diagnosis of prostate cancer. Asian J. Urol. 2019, 6, 129–136. [Google Scholar] [CrossRef]
- Epstein, J.I. An Update of the Gleason Grading System. J. Urol. 2010, 183, 433–440. [Google Scholar] [CrossRef]
- Sobin, L.H.; Gospodarowicz, M.K.; Wittekind, C.; International Union against Cancer. TNM Classification of Malignant Tumours; Wiley-Blackwell: Chichester, West Sussex, UK; Hoboken, NJ, USA, 2009. [Google Scholar]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Goksuluk, D.; Korkmaz, S.; Zararsiz, G.; Karaagaoglu, A.E. easyROC: An Interactive Web-tool for ROC Curve Analysis Using R Language Environment. R J. 2016, 8, 213–230. [Google Scholar] [CrossRef]
- Spitzer, M.; Wildenhain, J.; Rappsilber, J.; Tyers, M. BoxPlotR: A web tool for generation of box plots. Nat. Methods 2014, 11, 121–122. [Google Scholar] [CrossRef]

| Patient Group | |||
|---|---|---|---|
| Total | Benign Controls (BC) | Patients with Prostate Cancer (PrCa) | |
| Number (n) | 56 | 28 | 28 |
| Age (years) | |||
| Mean (±SD) | 67.6 (±6.8) | 67.3 (±7.4) | 68.1 (±6.3) |
| Range | 48–79 | 50–79 | 48–79 |
| Serum PSA (ng/mL) | |||
| Mean (±SD) | 4.6 (±2.6) | 4.5 (±3.0) | 4.8 (±2.1) |
| Range | 0.6–14.7 | 0.6–14.7 | 0.9–9.1 |
| 0–4 ng/mL | 22 (39%) | 13 (46%) | 9 (32%) |
| 4–10 ng/mL | 32 (57%) | 13 (46%) | 19 (68%) |
| >10 ng/mL | 1(1.8%) | 1 (3.6%) | 0 |
| Unknown | 1 | 1 | 0 |
| Other prostate conditions | |||
| BPH * | 16 | 7 | 9 |
| PIN † | 13 | 5 | 8 |
| Both BPH and PIN | 7 | 1 | 6 |
| Nodule(s) | 11 | 5 | 6 |
| LUTS †† | 6 | 1 | 5 |
| Firm to touch | 13 | 7 | 6 |
| Increased volume | 22 | 13 | 9 |
| Lobe Asymmetry | 11 | 3 | 8 |
| Gleason grade at diagnosis | |||
| ≤6 (3 + 3) | NA | NA | 21 |
| 7 (3 + 4) | NA | NA | 4 |
| 7 (4 + 3) or higher | NA | NA | 3 |
| Clinical Stage | |||
| T1 | NA | NA | 9 |
| T2 | NA | NA | 11 |
| T3 | NA | NA | 1 |
| Not Available | NA | NA | 7 |
| mRNA Panel Variable | Logistic Regression Analysis | ROC Curve Analysis | ||
|---|---|---|---|---|
| OR (95% CI) | p Value | AUC (95% CI) | p Value | |
| 8 mRNA Panel | 1.0396 (1.0062, 1.0741) | 0.0199 | 0.695 (0.553, 0.837) | 0.0071 |
| LTBP4 and NELL2 removed | 1.2386 (1.0360, 1.4809) | 0.0189 | 0.694 (0.555, 0.832) | 0.0061 |
| 5 mRNA Panels | ||||
| FOLH1 removed | 1.0488 (0.9439, 1.1655) | 0.3755 | 0.651 (0.502, 0.799) | 0.0473 |
| HPN removed | 1.4259 (1.0146, 2.0040) | 0.041 | 0.667 (0.523, 0.811) | 0.023 |
| XBP1 removed | 1.1272 (1.0416, 1.2199) | 0.003 | 0.761 (0.636, 0.887) | <0.0001 |
| ITSN1 removed | 1.0154 (0.9616, 1.0722) | 0.5817 | 0.67 (0.525, 0.814) | 0.0212 |
| GSTM4 removed | 1.2439 (1.0482, 1.4762) | 0.0124 | 0.751 (0.623, 0.880) | 0.0001 |
| CFD removed | 1.1995 (0.9935, 1.4481) | 0.0584 | 0.662 (0.519, 0.805) | 0.026 |
| 4 mRNA Panel | ||||
| (FOLH1, HPN, ITSN1, CFD) | 1.1359 (1.0479, 1.2313) | 0.002 | 0.798 (0.681, 0.916) | <0.0001 |
| mRNA Panel Variable | Logistic Regression Analysis | ROC Curve Analysis | ||
|---|---|---|---|---|
| OR (95% CI) | p Value | AUC (95% CI) | p Value | |
| 4 mRNA Panel | ||||
| (FOLH1, HPN, ITSN1, CFD) | 1.1359 (1.0479, 1.2313) | 0.002 | 0.798 (0.681, 0.916) | 6.67 × 10−7 |
| New 6 mRNA Panel | ||||
| (GOLM1, ANXA3, CD24, TMPRSS2-ERG, PSCA, SLC45A3) | 4.4649 (1.3686, 14.5668) | 0.0131 | 0.725 (0.585, 0.865) | 1.62 × 10−3 |
| Combined 10 mRNA Panel | 1.6359 (1.1808, 2.2663) | 0.0031 | 0.759 (0.630, 0.888) | 8.49 × 10−5 |
| GOLM1 removed | 1.5596 (1.1723, 2.0748) | 0.0023 | 0.797 (0.677, 0.917) | 1.19 × 10−6 |
| ANXA3 removed | 1.0827 (0.9981, 1.1745) | 0.0557 | 0.709 (0.570, 0.849) | 3.33 × 10−3 |
| CD24 removed | 1.7905 (1.2128, 2.6433) | 0.0034 | 0.769 (0.643, 0.896) | 3.08 × 10−5 |
| TMPRSS2-ERG removed | 1.5597 (1.0128, 2.4017) | 0.0436 | 0.704 (0.563, 0.845) | 4.52 × 10−3 |
| PSCA removed | 1.5355 (1.1665, 2.0213) | 0.0022 | 0.781 (0.656, 0.905) | 1.03 × 10−5 |
| SLC45A3 removed | 1.1223 (0.9776, 1.2885) | 0.1013 | 0.723 (0.584, 0.862) | 1.66 × 10−3 |
| FOLH1 removed | 1.0836 (0.9653, 1.2164) | 0.1733 | 0.699 (0.558, 0.840) | 5.54 × 10−3 |
| HPN removed | 1.5006 (1.1264, 1.9991) | 0.0055 | 0.741 (0.608, 0.875) | 4.00 × 10−4 |
| ITSN1 removed | 2.1580 (1.2820, 3.6325) | 0.0038 | 0.749 (0.617, 0.880) | 2.05 × 10−4 |
| CFD removed | 3.1347 (1.5581, 6.3064) | 0.0014 | 0.787 (0.663, 0.911) | 5.82 × 10−6 |
| 7 mRNA Panel | ||||
| (ANXA3, CD24, TMPRSS2-ERG, SLC45A3, FOLH1, HPN, ITSN1) | 2.2371 (1.4036, 3.5656) | 0.0007 | 0.825 (0.710, 0.941) | 3.18 × 10−8 |
| miRNA or Combined Variable | Logistic Regression Analysis | ROC Curve Analysis | ||
|---|---|---|---|---|
| Univariate | OR (95% CI) | p Value | AUC (95% CI) | p Value |
| miR-141-3p | 1.6604 (1.0373, 2.6578) | 0.0346 | 0.645 (0.492, 0.797) | 0.0629 |
| miR-375-3p | 1.1049 (1.0130, 1.2051) | 0.0243 | 0.744 (0.603, 0.885) | 0.0007 |
| miR-574-3p | 1.7572 (1.0882, 2.8373) | 0.0211 | 0.733 (0.599, 0.866) | 0.0006 |
| miR-21-3p | 1.2562 (1.0154, 1.5540) | 0.0357 | 0.698 (0.553, 0.843) | 0.0073 |
| 4 miRNA Panel | 1.5012 (1.0761, 2.0942) | 0.0168 | 0.719 (0.574, 0.865) | 0.0031 |
| 3 miRNA Panel (miR-141-3p, miR-375-3p, miR-574-3p) | 1.4851 (1.0815, 2.0393) | 0.0145 | 0.704 (0.559, 0.849) | 0.0059 |
| 3 miRNA Panel (miR-375-3p, miR-574-3p, miR-21-3p) | 1.3990 (1.0750, 1.8206) | 0.0125 | 0.737 (0.597, 0.876) | 0.0009 |
| 2 miRNA Panel (miR-375-3p, miR-574-3p) | 1.3390 (1.0592, 1.6928) | 0.0147 | 0.744 (0.607, 0.880) | 0.0005 |
| Multivariable | ||||
| 7 Gene Score + 2 miRNA Model | NA | NA | 0.843 (0.722, 0.964) | 2.55 × 10−8 |
| 7 mRNA Panel | 1.8463 (1.1375, 2.9968) | 0.0131 | NA | NA |
| 2 miRNA Panel | 1.1822 (0.9581, 1.4587) | 0.1822 | NA | NA |
| Variable | Logistic Regression Analysis | ROC Curve Analysis | ||
|---|---|---|---|---|
| Univariate | OR (95% CI) | p Value | AUC (95% CI) | p Value |
| Urinary Vn96-EV PCA3 | 1.0960 (1.0298, 1.1665) | 0.0039 | 0.816 (0.708, 0.925) | 1.20 × 10−8 |
| Age at collection | 1.0181 (0.9420, 1.1004) | 0.6509 | 0.543 (0.387, 0.700) | 0.5864 |
| Serum PSA at collection | 1.0742 (0.8671, 1.3308) | 0.5124 | 0.594 (0.440, 0.749) | 0.2303 |
| Prostate Volume at collection | 0.9726 (0.9421, 1.0041) | 0.0874 | 0.656 (0.508, 0.805) | 0.0391 |
| DRE Result | 0.8627 (0.2972, 2.5041) | 0.7860 | 0.518 (0.387, 0.649) | 0.7895 |
| Multivariable | ||||
| Model 1 (SOC † Only) | NA | NA | 0.718 (0.583, 0.853) | 0.0016 |
| Age | 1.0746 (0.9785, 1.1802) | 0.1322 | NA | NA |
| Serum PSA | 1.2333 (0.9385, 1.6209) | 0.1325 | NA | NA |
| Prostate Volume | 0.9427 (0.9001, 0.9873) | 0.0123 | NA | NA |
| DRE Result | 0.4799 (0.1327, 1.7351) | 0.2629 | NA | NA |
| Model 2 †† | NA | NA | 0.901 (0.811, 0.990) | 1.75 × 10−18 |
| 7 mRNA Panel | 1.7053 (1.0026, 2.9005) | 0.0489 | NA | NA |
| 2 miRNA Panel | 1.1670 (0.9429, 1.4445) | 0.1088 | NA | NA |
| PCA3 Value | 1.0615 (1.0047, 1.1216) | 0.0335 | NA | NA |
| Model 3 ††† | NA | NA | 0.955 (0.909, 1.002) | 2.27 × 10−82 |
| 7 mRNA Panel | 1.4312 (0.7581, 2.7021) | 0.2688 | NA | NA |
| 2 miRNA Panel | 1.4024 (0.9767, 2.0138) | 0.0669 | NA | NA |
| PCA3 Value | 1.0810 (0.9984, 1.1705) | 0.0548 | NA | NA |
| Age | 1.0921 (0.8962, 1.3309) | 0.3823 | NA | NA |
| Serum PSA | 1.5220 (0.8605, 2.6921) | 0.1489 | NA | NA |
| Prostate Volume | 0.8741 (0.7764, 0.9841) | 0.0261 | NA | NA |
| DRE Results | 0.1393 (0.0117, 1.6573) | 0.1187 | NA | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Davey, M.; Benzina, S.; Savoie, M.; Breault, G.; Ghosh, A.; Ouellette, R.J. Affinity Captured Urinary Extracellular Vesicles Provide mRNA and miRNA Biomarkers for Improved Accuracy of Prostate Cancer Detection: A Pilot Study. Int. J. Mol. Sci. 2020, 21, 8330. https://doi.org/10.3390/ijms21218330
Davey M, Benzina S, Savoie M, Breault G, Ghosh A, Ouellette RJ. Affinity Captured Urinary Extracellular Vesicles Provide mRNA and miRNA Biomarkers for Improved Accuracy of Prostate Cancer Detection: A Pilot Study. International Journal of Molecular Sciences. 2020; 21(21):8330. https://doi.org/10.3390/ijms21218330
Chicago/Turabian StyleDavey, Michelle, Sami Benzina, Marc Savoie, Guy Breault, Anirban Ghosh, and Rodney J. Ouellette. 2020. "Affinity Captured Urinary Extracellular Vesicles Provide mRNA and miRNA Biomarkers for Improved Accuracy of Prostate Cancer Detection: A Pilot Study" International Journal of Molecular Sciences 21, no. 21: 8330. https://doi.org/10.3390/ijms21218330
APA StyleDavey, M., Benzina, S., Savoie, M., Breault, G., Ghosh, A., & Ouellette, R. J. (2020). Affinity Captured Urinary Extracellular Vesicles Provide mRNA and miRNA Biomarkers for Improved Accuracy of Prostate Cancer Detection: A Pilot Study. International Journal of Molecular Sciences, 21(21), 8330. https://doi.org/10.3390/ijms21218330





