Comparative Lipidomic Analysis of Extracellular Vesicles Derived from Lactobacillus plantarum APsulloc 331261 Living in Green Tea Leaves Using Liquid Chromatography-Mass Spectrometry
Abstract
1. Introduction
2. Results
2.1. Strain Isolation and Identification
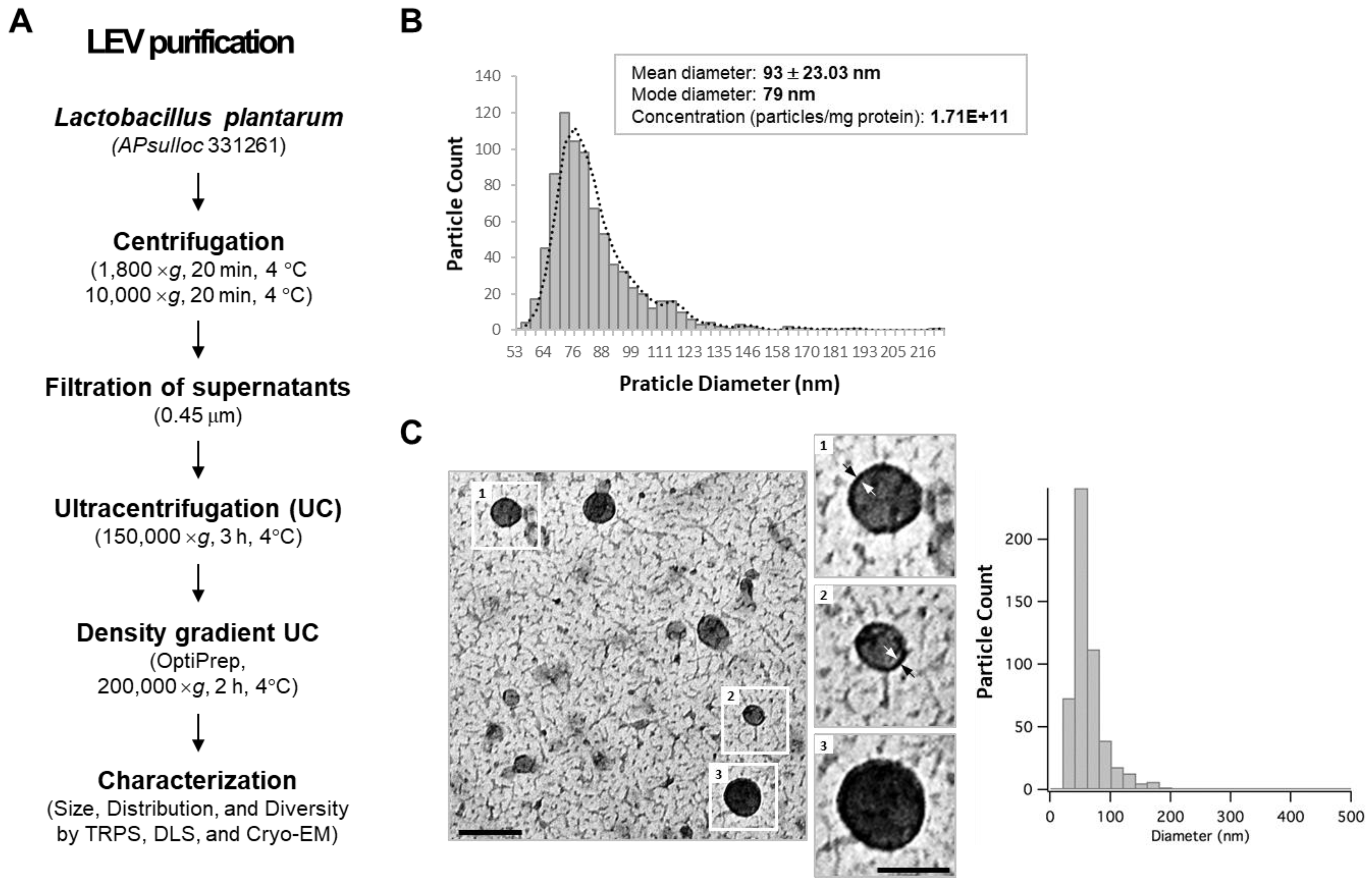
2.2. Spontaneous Release of EVs from L. plantarum APsulloc 331261
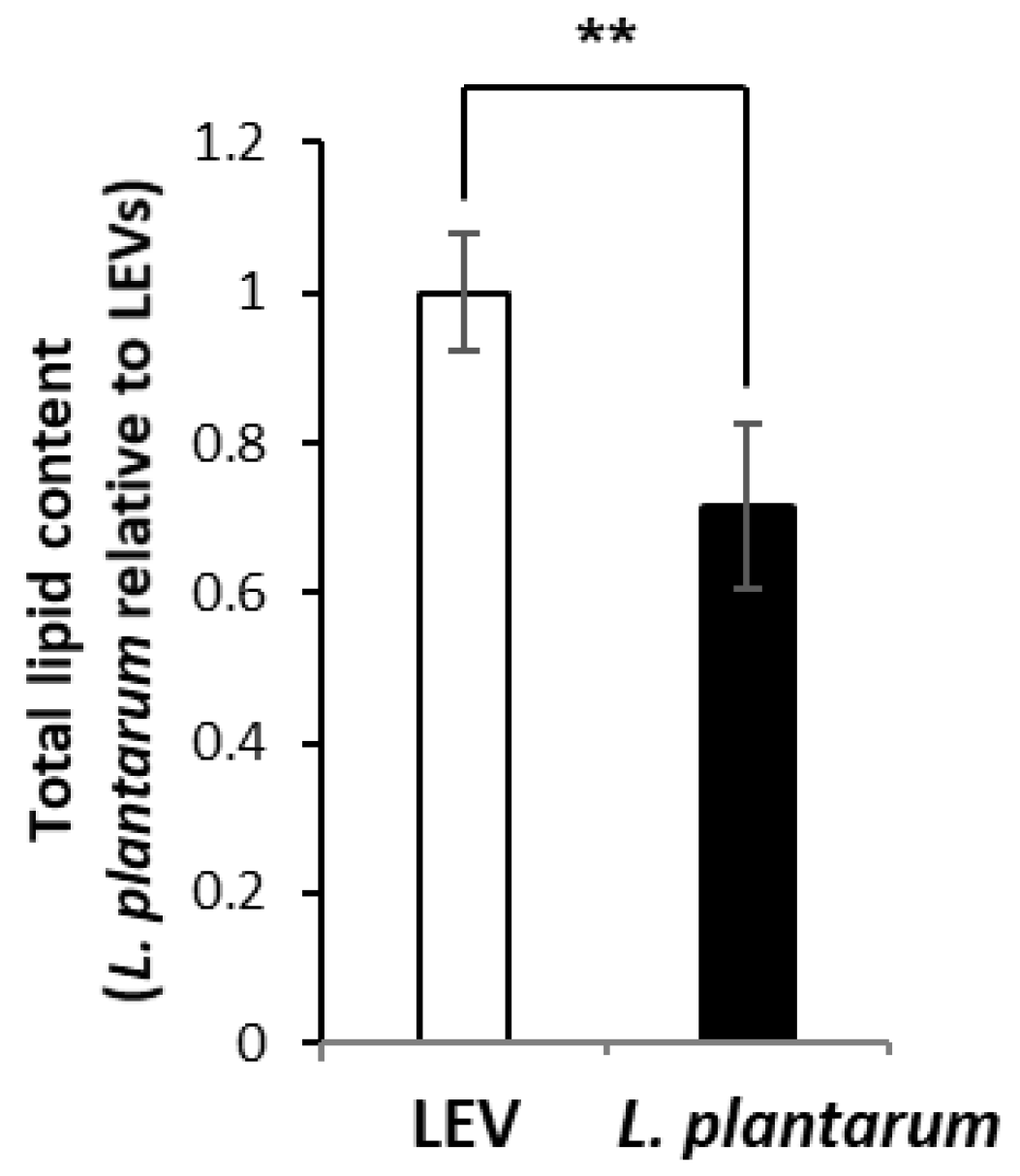
2.3. Lipidomic Profile of LEVs and L. plantarum
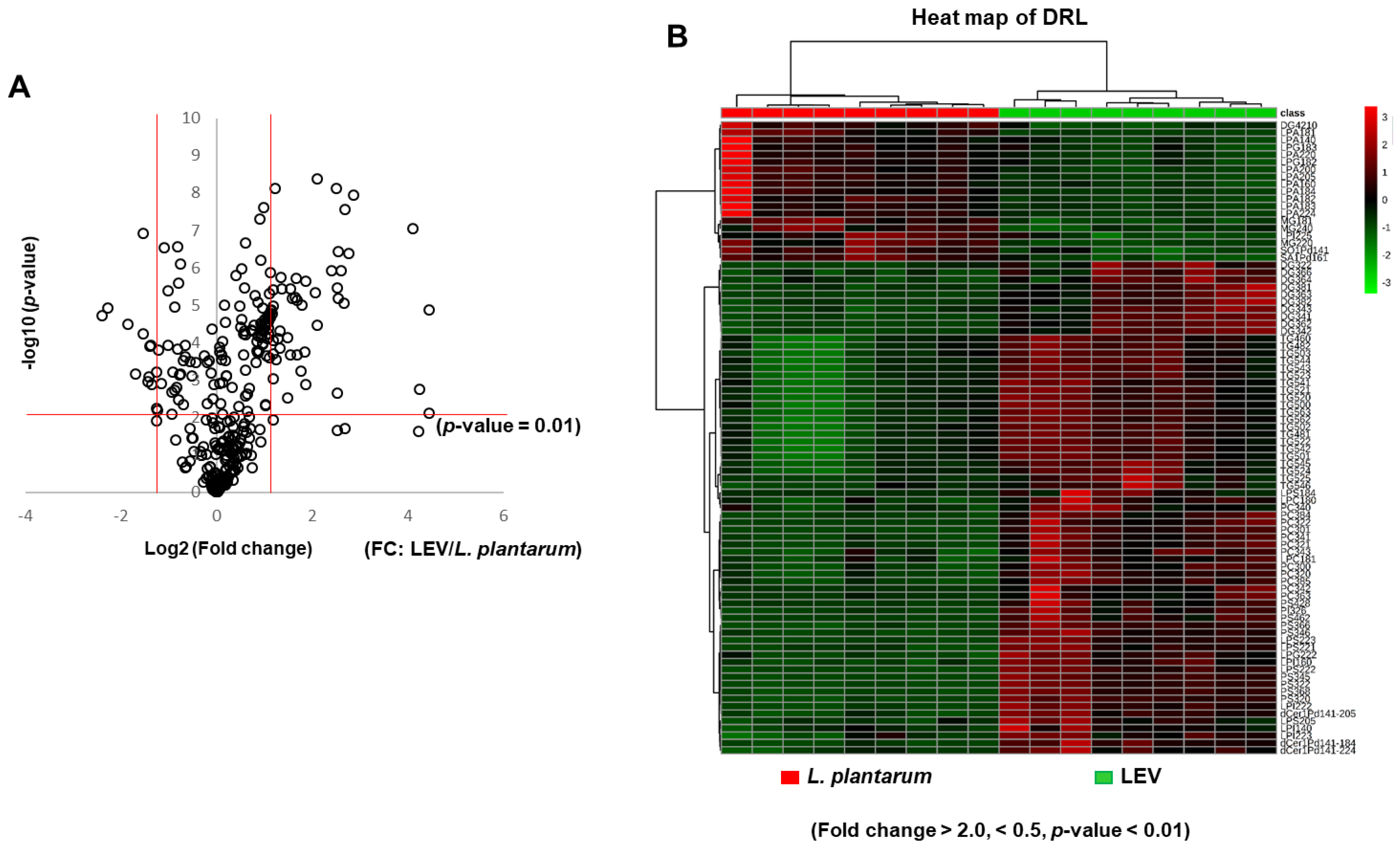
2.4. Differential Enrichement of Lipid Species between LEVs and L. plantarum
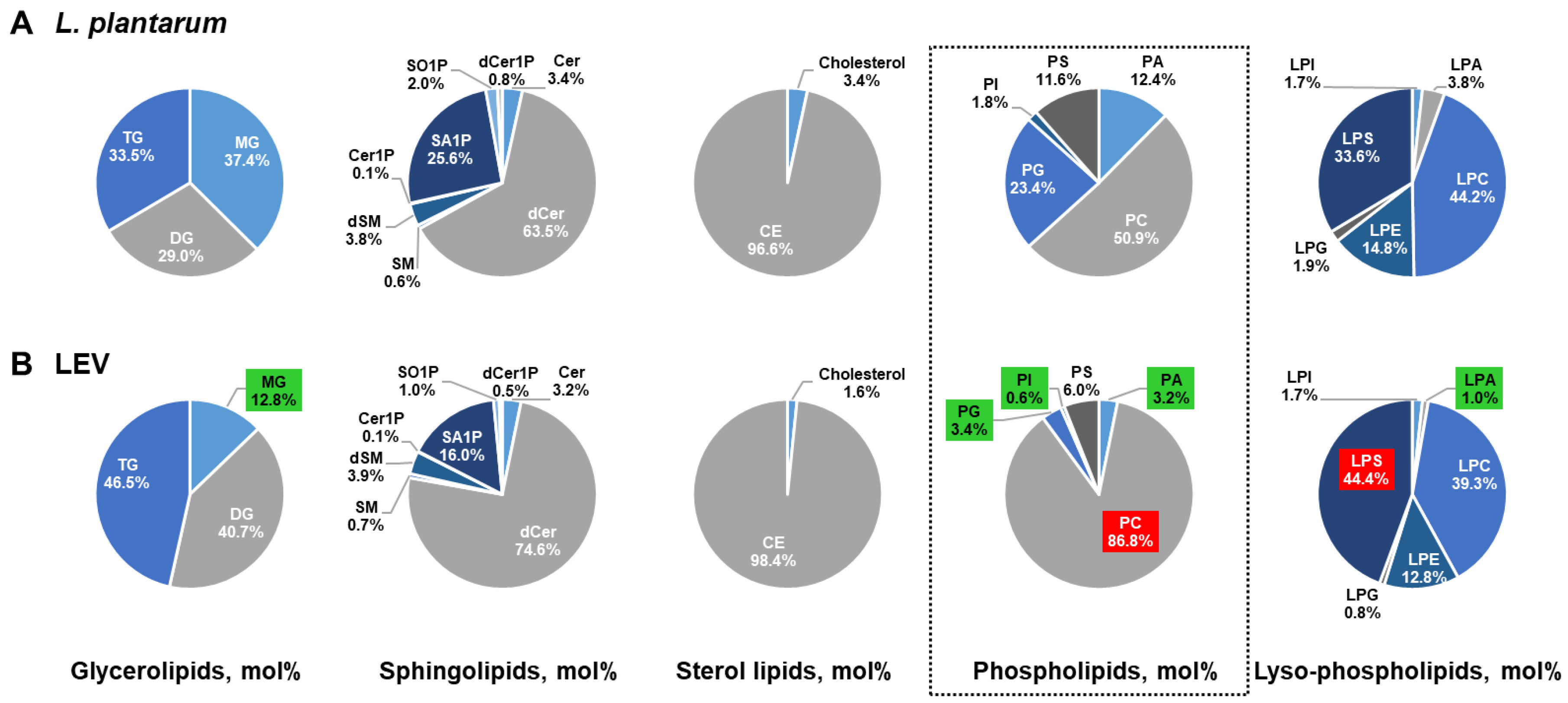
2.5. Comparison of the Lipid Composition in LEVs Versus L. plantarum
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Isolation and Identification of L. plantarum APsulloc
4.3. Isolation of EVs from the Bacterial Culture Medium
4.4. Analysis of LEVs
4.5. Lipid Extraction
4.6. Global Lipid Analysis Using LC-MS
4.7. Processing of Individual Data Obtained by MRM
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- de Vrese, M.; Schrezenmeir, J. Probiotics, prebiotics, and synbiotics. Adv. Biochem. Eng. Biotechnol. 2008, 111, 1–66. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, B.; Delgado, S.; Blanco-Miguez, A.; Lourenco, A.; Gueimonde, M.; Margolles, A. Probiotics, gut microbiota, and their influence on host health and disease. Mol. Nutr. Food Res. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.A.; Blaser, M.J.; Caporaso, J.G.; Jansson, J.K.; Lynch, S.V.; Knight, R. Current understanding of the human microbiome. Nat. Med. 2018, 24, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143. [Google Scholar] [CrossRef] [PubMed]
- Servin, A.L. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol. Rev. 2004, 28, 405–440. [Google Scholar] [CrossRef] [PubMed]
- Marco, M.L.; Pavan, S.; Kleerebezem, M. Towards understanding molecular modes of probiotic action. Curr. Opin. Biotechnol. 2006, 17, 204–210. [Google Scholar] [CrossRef]
- Cataloluk, O.; Gogebakan, B. Presence of drug resistance in intestinal lactobacilli of dairy and human origin in Turkey. FEMS Microbiol. Lett. 2004, 236, 7–12. [Google Scholar] [CrossRef]
- Kaushik, J.K.; Kumar, A.; Duary, R.K.; Mohanty, A.K.; Grover, S.; Batish, V.K. Functional and probiotic attributes of an indigenous isolate of Lactobacillus plantarum. PLoS ONE 2009, 4, e8099. [Google Scholar] [CrossRef]
- Potocnjak, M.; Pusic, P.; Frece, J.; Abram, M.; Jankovic, T.; Gobin, I. Three New Lactobacillus plantarum Strains in the Probiotic Toolbox against Gut Pathogen Salmonella enterica Serotype Typhimurium. Food Technol. Biotechnol. 2017, 55, 48–54. [Google Scholar] [CrossRef]
- Kleerebezem, M.; Boekhorst, J.; van Kranenburg, R.; Molenaar, D.; Kuipers, O.P.; Leer, R.; Tarchini, R.; Peters, S.A.; Sandbrink, H.M.; Fiers, M.W.; et al. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 2003, 100, 1990–1995. [Google Scholar] [CrossRef]
- Bested, A.C.; Logan, A.C.; Selhub, E.M. Intestinal microbiota, probiotics and mental health: From Metchnikoff to modern advances: Part III—Convergence toward clinical trials. Gut Pathog. 2013, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Troost, F.J.; van Baarlen, P.; Lindsey, P.; Kodde, A.; de Vos, W.M.; Kleerebezem, M.; Brummer, R.J. Identification of the transcriptional response of human intestinal mucosa to Lactobacillus plantarum WCFS1 in vivo. BMC Genom. 2008, 9, 374. [Google Scholar] [CrossRef] [PubMed]
- van Baarlen, P.; Troost, F.J.; van Hemert, S.; van der Meer, C.; de Vos, W.M.; de Groot, P.J.; Hooiveld, G.J.; Brummer, R.J.; Kleerebezem, M. Differential NF-kappaB pathways induction by Lactobacillus plantarum in the duodenum of healthy humans correlating with immune tolerance. Proc. Natl. Acad. Sci. USA 2009, 106, 2371–2376. [Google Scholar] [CrossRef]
- van den Nieuwboer, M.; van Hemert, S.; Claassen, E.; de Vos, W.M. Lactobacillus plantarum WCFS1 and its host interaction: A dozen years after the genome. Microb. Biotechnol. 2016, 9, 452–465. [Google Scholar] [CrossRef]
- Arellano, K.; Vazquez, J.; Park, H.; Lim, J.; Ji, Y.; Kang, H.J.; Cho, D.; Jeong, H.W.; Holzapfel, W.H. Safety Evaluation and Whole-Genome Annotation of Lactobacillus plantarum Strains from Different Sources with Special Focus on Isolates from Green Tea. Probiotics Antimicrob. Proteins 2020, 12, 1057–1070. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Cho, D.; Huang, E.; Seo, J.Y.; Kim, W.G.; Todorov, S.D.; Ji, Y.; Holzapfel, W.H. Amelioration of Alcohol Induced Gastric Ulcers Through the Administration of Lactobacillus plantarum APSulloc 331261 Isolated From Green Tea. Front. Microbiol. 2020, 11, 420. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.Y.; Choi, D.Y.; Kim, D.K.; Kim, J.W.; Park, J.O.; Kim, S.; Kim, S.H.; Desiderio, D.M.; Kim, Y.K.; Kim, K.P.; et al. Gram-positive bacteria produce membrane vesicles: Proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics 2009, 9, 5425–5436. [Google Scholar] [CrossRef]
- Kim, J.H.; Lee, J.; Park, J.; Gho, Y.S. Gram-negative and Gram-positive bacterial extracellular vesicles. Semin. Cell Dev. Biol. 2015, 40, 97–104. [Google Scholar] [CrossRef]
- Brown, L.; Wolf, J.M.; Prados-Rosales, R.; Casadevall, A. Through the wall: Extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 2015, 13, 620–630. [Google Scholar] [CrossRef]
- Colombo, M.; Raposo, G.; Thery, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Choi, E.J.; Lee, H.G.; Bae, I.H.; Kim, W.; Park, J.; Lee, T.R.; Cho, E.G. Propionibacterium acnes-Derived Extracellular Vesicles Promote Acne-Like Phenotypes in Human Epidermis. J. Investig. Dermatol. 2018, 138, 1371–1379. [Google Scholar] [CrossRef]
- Kim, J.; Bin, B.H.; Choi, E.J.; Lee, H.G.; Lee, T.R.; Cho, E.G. Staphylococcus aureus-derived extracellular vesicles induce monocyte recruitment by activating human dermal microvascular endothelial cells in vitro. Clin. Exp. Allergy 2019, 49, 68–81. [Google Scholar] [CrossRef]
- Dominguez Rubio, A.P.; Martinez, J.H.; Martinez Casillas, D.C.; Coluccio Leskow, F.; Piuri, M.; Perez, O.E. Lactobacillus casei BL23 Produces Microvesicles Carrying Proteins That Have Been Associated with Its Probiotic Effect. Front. Microbiol. 2017, 8, 1783. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Lee, K.; Hsu, M.; Nau, G.; Mylonakis, E.; Ramratnam, B. Lactobacillus-derived extracellular vesicles enhance host immune responses against vancomycin-resistant enterococci. BMC Microbiol. 2017, 17, 66. [Google Scholar] [CrossRef] [PubMed]
- Grande, R.; Celia, C.; Mincione, G.; Stringaro, A.; Di Marzio, L.; Colone, M.; Di Marcantonio, M.C.; Savino, L.; Puca, V.; Santoliquido, R.; et al. Detection and Physicochemical Characterization of Membrane Vesicles (MVs) of Lactobacillus reuteri DSM 17938. Front. Microbiol. 2017, 8, 1040. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Lee, E.J.; Bae, I.H.; Myoung, K.; Kim, S.T.; Park, P.J.; Lee, K.H.; Pham, A.V.Q.; Ko, J.; Oh, S.H.; et al. Lactobacillus plantarum-derived extracellular vesicles induce anti-inflammatory M2 macrophage polarization in vitro. J. Extracell. Vesicles 2020, 9, 1793514. [Google Scholar] [CrossRef]
- Nagakubo, T.; Nomura, N.; Toyofuku, M. Cracking Open Bacterial Membrane Vesicles. Front. Microbiol. 2019, 10, 3026. [Google Scholar] [CrossRef] [PubMed]
- Toyofuku, M.; Nomura, N.; Eberl, L. Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol. 2019, 17, 13–24. [Google Scholar] [CrossRef]
- Onodi, Z.; Pelyhe, C.; Terezia Nagy, C.; Brenner, G.B.; Almasi, L.; Kittel, A.; Mancek-Keber, M.; Ferdinandy, P.; Buzas, E.I.; Giricz, Z. Isolation of High-Purity Extracellular Vesicles by the Combination of Iodixanol Density Gradient Ultracentrifugation and Bind-Elute Chromatography From Blood Plasma. Front. Physiol. 2018, 9, 1479. [Google Scholar] [CrossRef]
- Harkewicz, R.; Dennis, E.A. Applications of mass spectrometry to lipids and membranes. Annu. Rev. Biochem. 2011, 80, 301–325. [Google Scholar] [CrossRef]
- Broadhurst, D.; Goodacre, R.; Reinke, S.N.; Kuligowski, J.; Wilson, I.D.; Lewis, M.R.; Dunn, W.B. Guidelines and considerations for the use of system suitability and quality control samples in mass spectrometry assays applied in untargeted clinical metabolomic studies. Metabolomics 2018, 14, 72. [Google Scholar] [CrossRef] [PubMed]
- Sangster, T.; Major, H.; Plumb, R.; Wilson, A.J.; Wilson, I.D. A pragmatic and readily implemented quality control strategy for HPLC-MS and GC-MS-based metabonomic analysis. Analyst 2006, 131, 1075–1078. [Google Scholar] [CrossRef] [PubMed]
- Dang, V.D.; Jella, K.K.; Ragheb, R.R.T.; Denslow, N.D.; Alli, A.A. Lipidomic and proteomic analysis of exosomes from mouse cortical collecting duct cells. FASEB J. 2017, 31, 5399–5408. [Google Scholar] [CrossRef] [PubMed]
- Durcin, M.; Fleury, A.; Taillebois, E.; Hilairet, G.; Krupova, Z.; Henry, C.; Truchet, S.; Trotzmuller, M.; Kofeler, H.; Mabilleau, G.; et al. Characterisation of adipocyte-derived extracellular vesicle subtypes identifies distinct protein and lipid signatures for large and small extracellular vesicles. J. Extracell. Vesicles 2017, 6, 1305677. [Google Scholar] [CrossRef]
- Haraszti, R.A.; Didiot, M.C.; Sapp, E.; Leszyk, J.; Shaffer, S.A.; Rockwell, H.E.; Gao, F.; Narain, N.R.; DiFiglia, M.; Kiebish, M.A.; et al. High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J. Extracell. Vesicles 2016, 5, 32570. [Google Scholar] [CrossRef]
- Jung, J.H.; Lee, M.Y.; Choi, D.Y.; Lee, J.W.; You, S.; Lee, K.Y.; Kim, J.; Kim, K.P. Phospholipids of tumor extracellular vesicles stratify gefitinib-resistant nonsmall cell lung cancer cells from gefitinib-sensitive cells. Proteomics 2015, 15, 824–835. [Google Scholar] [CrossRef]
- Llorente, A.; Skotland, T.; Sylvanne, T.; Kauhanen, D.; Rog, T.; Orlowski, A.; Vattulainen, I.; Ekroos, K.; Sandvig, K. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim. Biophys. Acta 2013, 1831, 1302–1309. [Google Scholar] [CrossRef]
- Pienimaeki-Roemer, A.; Kuhlmann, K.; Bottcher, A.; Konovalova, T.; Black, A.; Orso, E.; Liebisch, G.; Ahrens, M.; Eisenacher, M.; Meyer, H.E.; et al. Lipidomic and proteomic characterization of platelet extracellular vesicle subfractions from senescent platelets. Transfusion 2015, 55, 507–521. [Google Scholar] [CrossRef]
- Skotland, T.; Ekroos, K.; Kauhanen, D.; Simolin, H.; Seierstad, T.; Berge, V.; Sandvig, K.; Llorente, A. Molecular lipid species in urinary exosomes as potential prostate cancer biomarkers. Eur. J. Cancer 2017, 70, 122–132. [Google Scholar] [CrossRef]
- Vallejo, M.C.; Nakayasu, E.S.; Longo, L.V.; Ganiko, L.; Lopes, F.G.; Matsuo, A.L.; Almeida, I.C.; Puccia, R. Lipidomic analysis of extracellular vesicles from the pathogenic phase of Paracoccidioides brasiliensis. PLoS ONE 2012, 7, e39463. [Google Scholar] [CrossRef]
- Thorne, K.J. The Phospholipids of Lactobacillus Casei. Biochim. Biophys. Acta 1964, 84, 350–353. [Google Scholar] [CrossRef]
- Geiger, O.; Lopez-Lara, I.M.; Sohlenkamp, C. Phosphatidylcholine biosynthesis and function in bacteria. Biochim. Biophys. Acta 2013, 1831, 503–513. [Google Scholar] [CrossRef]
- Sohlenkamp, C.; Geiger, O. Bacterial membrane lipids: Diversity in structures and pathways. FEMS Microbiol. Rev. 2016, 40, 133–159. [Google Scholar] [CrossRef]
- Sohlenkamp, C.; Lopez-Lara, I.M.; Geiger, O. Biosynthesis of phosphatidylcholine in bacteria. Prog. Lipid Res. 2003, 42, 115–162. [Google Scholar] [CrossRef]
- Li, F.N.; Liao, S.L.; Liu, S.W.; Jin, T.; Sun, C.H. Aeromicrobium endophyticum sp. nov., an endophytic actinobacterium isolated from reed (Phragmites australis). J. Microbiol. 2019, 57, 725–731. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, H.; Zhang, T.; Liu, W.H.; Liu, H.Y.; Yu, L.Y.; Zhang, Y.Q. Desertihabitans aurantiacus gen. nov., sp. nov., a novel member of the family Propionibacteriaceae. Int. J. Syst. Evol. Microbiol. 2019, 69, 2486–2491. [Google Scholar] [CrossRef]
- Chamberlain, C.A.; Hatch, M.; Garrett, T.J. Metabolomic profiling of oxalate-degrading probiotic Lactobacillus acidophilus and Lactobacillus gasseri. PLoS ONE 2019, 14, e0222393. [Google Scholar] [CrossRef]
- Sandoval-Calderon, M.; Nguyen, D.D.; Kapono, C.A.; Herron, P.; Dorrestein, P.C.; Sohlenkamp, C. Plasticity of Streptomyces coelicolor Membrane Composition Under Different Growth Conditions and During Development. Front. Microbiol. 2015, 6, 1465. [Google Scholar] [CrossRef]
- Wei, J.H.; Yin, X.; Welander, P.V. Sterol Synthesis in Diverse Bacteria. Front. Microbiol. 2016, 7, 990. [Google Scholar] [CrossRef]
- Holert, J.; Brown, K.; Hashimi, A.; Eltis, L.D.; Mohn, W.W. Steryl Ester Formation and Accumulation in Steroid-Degrading Bacteria. Appl. Environ. Microbiol. 2020, 86. [Google Scholar] [CrossRef]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Lorenzen, W.; Bozhuyuk, K.A.; Cortina, N.S.; Bode, H.B. A comprehensive insight into the lipid composition of Myxococcus xanthus by UPLC-ESI-MS. J. Lipid Res. 2014, 55, 2620–2633. [Google Scholar] [CrossRef]
- Jeon, J.; Park, S.C.; Her, J.; Lee, J.W.; Han, J.K.; Kim, Y.K.; Kim, K.P.; Ban, C. Comparative lipidomic profiling of the human commensal bacterium Propionibacterium acnes and its extracellular vesicles. RSC Adv. 2018, 8, 15241–15247. [Google Scholar] [CrossRef]
- Heung, L.J.; Luberto, C.; Del Poeta, M. Role of sphingolipids in microbial pathogenesis. Infect. Immun. 2006, 74, 28–39. [Google Scholar] [CrossRef]
- Morita, S.Y.; Tsuji, T.; Terada, T. Protocols for Enzymatic Fluorometric Assays to Quantify Phospholipid Classes. Int. J. Mol. Sci. 2020, 21, 1032. [Google Scholar] [CrossRef]
- Yao, J.; Rock, C.O. Exogenous fatty acid metabolism in bacteria. Biochimie 2017, 141, 30–39. [Google Scholar] [CrossRef]
- Brinster, S.; Lamberet, G.; Staels, B.; Trieu-Cuot, P.; Gruss, A.; Poyart, C. Type II fatty acid synthesis is not a suitable antibiotic target for Gram-positive pathogens. Nature 2009, 458, 83–86. [Google Scholar] [CrossRef]
- Muller, J.A.; Ross, R.P.; Sybesma, W.F.; Fitzgerald, G.F.; Stanton, C. Modification of the technical properties of Lactobacillus johnsonii NCC 533 by supplementing the growth medium with unsaturated fatty acids. Appl. Environ. Microbiol. 2011, 77, 6889–6898. [Google Scholar] [CrossRef]
- Benesch, M.G.; Ko, Y.M.; McMullen, T.P.; Brindley, D.N. Autotaxin in the crosshairs: Taking aim at cancer and other inflammatory conditions. FEBS Lett. 2014, 588, 2712–2727. [Google Scholar] [CrossRef]
- Benesch, M.G.K.; MacIntyre, I.T.K.; McMullen, T.P.W.; Brindley, D.N. Coming of Age for Autotaxin and Lysophosphatidate Signaling: Clinical Applications for Preventing, Detecting and Targeting Tumor-Promoting Inflammation. Cancers 2018, 10, 73. [Google Scholar] [CrossRef]
- Zhu, W.; Hammad, L.A.; Hsu, F.; Mao, Y.; Luo, Z.Q. Induction of caspase 3 activation by multiple Legionella pneumophila Dot/Icm substrates. Cell Microbiol. 2013, 15, 1783–1795. [Google Scholar] [CrossRef]
- Hong, S.W.; Kim, M.R.; Lee, E.Y.; Kim, J.H.; Kim, Y.S.; Jeon, S.G.; Yang, J.M.; Lee, B.J.; Pyun, B.Y.; Gho, Y.S.; et al. Extracellular vesicles derived from Staphylococcus aureus induce atopic dermatitis-like skin inflammation. Allergy 2011, 66, 351–359. [Google Scholar] [CrossRef]
- Yoon, J.H.; Lee, S.T.; Park, Y.H. Inter- and intraspecific phylogenetic analysis of the genus Nocardioides and related taxa based on 16S rDNA sequences. Int. J. Syst. Bacteriol. 1998, 48 Pt 1, 187–194. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Breil, C.; Abert Vian, M.; Zemb, T.; Kunz, W.; Chemat, F. “Bligh and Dyer” and Folch Methods for Solid-Liquid-Liquid Extraction of Lipids from Microorganisms. Comprehension of Solvatation Mechanisms and towards Substitution with Alternative Solvents. Int. J. Mol. Sci. 2017, 18, 708. [Google Scholar] [CrossRef]
- Lee, J.W.N.S.; Yoshida, M.; Fukusaki, E.; Bamba, T. Simultaneous profiling of polar lipids by supercritical fluid chromatography/tandem mass spectrometry with methylation. J. Chromatogr. A 2013, 1279, 98–107. [Google Scholar] [CrossRef]
- Lee, J.W.; Mok, H.J.; Lee, D.Y.; Park, S.C.; Ban, M.S.; Choi, J.; Park, C.G.; Ahn, Y.S.; Kim, K.P.; Kim, H.D. UPLC-MS/MS based profiling of Eicosanoids in RAW264.7 cells treated with Lipopolysaccharide. Int. J. Mol. Sci. 2016, 17, 508. [Google Scholar] [CrossRef]

| Lipid Class | Lipid Species Quantitated |
|---|---|
| Phosphatidic acid (PA) | 50 |
| Triacylglycerol (TG) | 43 |
| Diacylglycerol (DG) | 23 |
| Phosphatidylserine (PS) | 23 |
| Phosphatidylcholine (PC) | 20 |
| Monoacylglycerol (MG) | 17 |
| Lysophosphatidic acid (LPA) | 17 |
| Cholesterylester (CE) | 16 |
| Lysophosphatidylserine (LPS) | 16 |
| Lysophosphatidylglycerol (LPG) | 13 |
| Dihydroceramide (dCer) | 11 |
| Phosphatidylglycerol (PG) | 11 |
| Lysophosphatidylinocitol (LPI) | 11 |
| Lysophosphatidylcholine (LPC) | 8 |
| Ceramide (Cer) | 7 |
| Phosphatidylinositol (PI) | 7 |
| Dihydroceramide-1-phosphate (dCer1P) | 6 |
| Ceramide-1-phosphate (Cer1P) | 5 |
| Dihydrosphingomyelin (DSM) | 4 |
| Sphingomyeline (SM) | 3 |
| Sphinganine-1-phosphate (SA1P) | 3 |
| Sphingosine-1-phosphate (SO1P) | 3 |
| Lysophosphatidylethanolamine (LPE) | 2 |
| Cholesterol | 1 |
| Total | 320 |
| Lipid Category | Lipid Class | Fold Change a (Mean ± S.D.) | p-Value |
|---|---|---|---|
| Glycerolipids | MG | 0.505 ± 0.0898 | 4 × 10−9 |
| DG | 2.04 ± 0.476 | 1 × 10−4 | |
| TG | 2.26 ± 0.625 | 1 × 10−5 | |
| Sphingolipids | Cer | 1.039 ± 0.0627 | 3 × 10−1 |
| dCer | 1.36 ± 0.188 | 2 × 10−2 | |
| SM | 1.194 ± 0.148 | 7 × 10−3 | |
| dSM | 1.15 ± 0.062 | 2 × 10−2 | |
| Cer1P | 0.91 ± 0.0496 | 6 × 10−2 | |
| SA1P | 0.685 ± 0.0877 | 1 × 10−3 | |
| SO1P | 0.536 ± 0.0824 | 3 × 10−6 | |
| dCer1P | 0.682 ± 0.285 | 2 × 10−1 | |
| Glycero-phospholipids | PA | 1.09 ± 0.0318 | 5 × 10−4 |
| PC | 6.92 ± 1.44 | 3 × 10−4 | |
| PG | 0.573 ± 0.282 | 8 × 10−2 | |
| LPA | 0.491 ± 0.0811 | 6 × 10−4 | |
| LPC | 1.63 ± 0.215 | 9 × 10−3 | |
| LPE | 2.41 ± 2.696 | 2 × 10−1 | |
| LPG | 0.812 ± 0.215 | 1 × 10−1 | |
| PI | 1.52 ± 0.122 | 6 × 10−6 | |
| LPI | 1.77 ± 0.167 | 3 × 10−4 | |
| PS | 2.22 ± 0.204 | 8 × 10−7 | |
| LPS | 2.23 ± 0.763 | 2 × 10−3 | |
| Sterol lipids | Cholesterol | 0.553 ± 0.193 | 2 × 10−3 |
| CE | 1.26 ± 0.134 | 9 × 10−2 |
| No. | Lipid | Fold Change a (Mean ± S.D.) | p-Value |
|---|---|---|---|
| 1 | LPS(18:4) | 21.66 ± 13.15 | 8 × 10−3 |
| 2 | PC(32:2) | 21.64 ± 3.35 | 1 × 10−5 |
| 3 | PC(34:2) | 18.84 ± 4.08 | 2 × 10−3 |
| 4 | PC(30:1) | 17.21 ± 1.35 | 9 × 10−8 |
| 5 | PS(32:2) | 7.32 ± 0.94 | 1 × 10−8 |
| 6 | LPI(22:2) | 6.79 ± 0.54 | 4 × 10−7 |
| 7 | LPS(22:2) | 6.43 ± 0.81 | 3 × 10−8 |
| 8 | PC(34:1) | 6.29 ± 0.96 | 9 × 10−6 |
| 9 | DG(36:2) | 6.04 ± 1.13 | 1 × 10−6 |
| 10 | PC(32:1) | 5.85 ± 0.5 | 6 × 10−6 |
| 11 | LPS(22:3) | 5.84 ± 0.84 | 4 × 10−7 |
| 12 | PC(36:3) | 5.75 ± 1.33 | 2 × 10−3 |
| 13 | dCer1P(d14:1–18:4) | 5.74 ± 0.75 | 3 × 10−6 |
| 14 | PS(36:8) | 5.69 ± 0.66 | 8 × 10−9 |
| 15 | dCer1P(d14:1–20:5) | 5.26 ± 1.86 | 1 × 10−6 |
| 16 | DG(36:3) | 4.3 ± 1.2 | 3 × 10−5 |
| 17 | PS(34:5) | 4.3 ± 0.65 | 4 × 10−9 |
| 18 | DG(34:1) | 4.17 ± 0.81 | 5 × 10−6 |
| 19 | TG(52:5) | 3.64 ± 1.21 | 1 × 10−3 |
| 20 | LPS(22:1) | 3.62 ± 0.36 | 2 × 10−6 |
| 21 | dCer1P(d14:1–22:4) | 3.6 ± 3.31 | 2 × 10−4 |
| 22 | DG(34:2) | 3.46 ± 0.71 | 1 × 10−5 |
| 23 | LPC(18:1) | 3.41 ± 2.7 | 6 × 10−4 |
| 24 | LPC(18:0) | 3.23 ± 0.88 | 2 × 10−4 |
| 25 | LPG(22:2) | 3.2 ± 0.45 | 7 × 10−6 |
| 26 | PC(38:4) | 3.18 ± 0.5 | 2 × 10−6 |
| 27 | PC(30:0) | 3.1 ± 0.42 | 6 × 10−6 |
| 28 | PI(32:6) | 2.97 ± 0.55 | 6 × 10−6 |
| 29 | DG(38:1) | 2.91 ± 0.54 | 2 × 10−4 |
| 30 | PC(32:0) | 2.89 ± 0.29 | 4 × 10−6 |
| 31 | TG(54:6) | 2.8 ± 0.63 | 3 × 10−3 |
| 32 | DG(34:3) | 2.78 ± 0.37 | 7 × 10−5 |
| 33 | PC(38:5) | 2.58 ± 0.61 | 4 × 10−6 |
| 34 | LPI(16:0) | 2.55 ± 0.23 | 2 × 10−6 |
| 35 | DG(38:2) | 2.51 ± 0.57 | 1 × 10−4 |
| 36 | TG(54:5) | 2.5 ± 0.28 | 5 × 10−5 |
| 37 | PS(32:0) | 2.44 ± 0.18 | 1 × 10−9 |
| 38 | TG(52:4) | 2.44 ± 0.63 | 8 × 10−5 |
| 39 | LPI(22:3) | 2.37 ± 0.94 | 3 × 10−4 |
| 40 | PS(36:6) | 2.32 ± 0.13 | 7 × 10−9 |
| 41 | PS(42:8) | 2.28 ± 0.68 | 2 × 10−4 |
| 42 | TG(56:3) | 2.27 ± 0.46 | 4 × 10−6 |
| 43 | LPS(20:5) | 2.26 ± 0.45 | 9 × 10−4 |
| 44 | DG(36:6) | 2.26 ± 0.35 | 9 × 10−5 |
| 45 | TG(54:3) | 2.26 ± 0.62 | 1 × 10−5 |
| 46 | TG(52:1) | 2.23 ± 0.53 | 2 × 10−5 |
| 47 | TG(52:3) | 2.2 ± 0.66 | 1 × 10−5 |
| 48 | PS(46:2) | 2.2 ± 0.53 | 1 × 10−5 |
| 49 | PS(34:6) | 2.18 ± 0.28 | 1 × 10−6 |
| 50 | TG(52:0) | 2.18 ± 0.61 | 2 × 10−5 |
| 51 | TG(54:4) | 2.15 ± 0.61 | 2 × 10−5 |
| 52 | TG(56:2) | 2.15 ± 0.72 | 5 × 10−6 |
| 53 | DG(32:2) | 2.14 ± 0.42 | 2 × 10−4 |
| 54 | TG(54:2) | 2.14 ± 0.74 | 2 × 10−5 |
| 55 | PC(34:3) | 2.13 ± 0.31 | 4 × 10−5 |
| 56 | TG(50:3) | 2.12 ± 0.62 | 2 × 10−5 |
| 57 | TG(54:1) | 2.11 ± 0.58 | 6 × 10−5 |
| 58 | TG(52:2) | 2.11 ± 0.84 | 3 × 10−5 |
| 59 | TG(50:2) | 2.07 ± 0.66 | 2 × 10−5 |
| 60 | TG(50:0) | 2.07 ± 0.82 | 8 × 10−5 |
| 61 | DG(36:4) | 2.07 ± 0.4 | 2 × 10−4 |
| 62 | TG(50:1) | 2.06 ± 0.65 | 3 × 10−5 |
| 63 | TG(48:1) | 2.05 ± 0.6 | 3 × 10−5 |
| 64 | LPI(14:0) | 2.03 ± 0.44 | 5 × 10−3 |
| 65 | PC(34:0) | 2.02 ± 0.33 | 5 × 10−3 |
| 66 | TG(46:0) | 2 ± 0.6 | 3 × 10−5 |
| 67 | TG(48:2) | 2 ± 0.53 | 2 × 10−5 |
| No. | Lipid | Fold Change a (Mean ± S.D.) | p-Value |
|---|---|---|---|
| 1 | SO1P(d14:1) | 0.49 ± 0.05 | 4 × 10−6 |
| 2 | DG(42:10) | 0.49 ± 0.05 | 1 × 10−4 |
| 3 | MG(18:1) | 0.46 ± 0.02 | 3 × 10−7 |
| 4 | LPA(18:3) | 0.44 ± 0.09 | 1 × 10−3 |
| 5 | LPI(22:5) | 0.43 ± 0.11 | 2 × 10−4 |
| 6 | LPG(18:3) | 0.42 ± 0.11 | 6 × 10−3 |
| 7 | LPA(22:4) | 0.42 ± 0.15 | 6 × 10−4 |
| 8 | LPA(14:0) | 0.41 ± 0.08 | 6 × 10−3 |
| 9 | LPA(22:0) | 0.39 ± 0.08 | 1 × 10−3 |
| 10 | LPA(20:0) | 0.38 ± 0.05 | 1 × 10−4 |
| 11 | LPA(18:2) | 0.38 ± 0.06 | 1 × 10−4 |
| 12 | LPA(16:0) | 0.37 ± 0.07 | 8 × 10−4 |
| 13 | LPG(18:2) | 0.36 ± 0.09 | 1 × 10−3 |
| 14 | SA1P(d16:1) | 0.34 ± 0.05 | 1 × 10−7 |
| 15 | LPA(20:5) | 0.34 ± 0.05 | 6 × 10−5 |
| 16 | LPA(18:4) | 0.3 ± 0.06 | 7 × 10−4 |
| 17 | MG(24:0) | 0.27 ± 0.07 | 3 × 10−5 |
| 18 | MG(22:0) | 0.2 ± 0.02 | 1 × 10−5 |
| 19 | LPA(18:1) | 0.19 ± 0.05 | 2 × 10−5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.; Kim, M.; Myoung, K.; Kim, W.; Ko, J.; Kim, K.P.; Cho, E.-G. Comparative Lipidomic Analysis of Extracellular Vesicles Derived from Lactobacillus plantarum APsulloc 331261 Living in Green Tea Leaves Using Liquid Chromatography-Mass Spectrometry. Int. J. Mol. Sci. 2020, 21, 8076. https://doi.org/10.3390/ijms21218076
Kim H, Kim M, Myoung K, Kim W, Ko J, Kim KP, Cho E-G. Comparative Lipidomic Analysis of Extracellular Vesicles Derived from Lactobacillus plantarum APsulloc 331261 Living in Green Tea Leaves Using Liquid Chromatography-Mass Spectrometry. International Journal of Molecular Sciences. 2020; 21(21):8076. https://doi.org/10.3390/ijms21218076
Chicago/Turabian StyleKim, Hyoseon, Minjung Kim, Kilsun Myoung, Wanil Kim, Jaeyoung Ko, Kwang Pyo Kim, and Eun-Gyung Cho. 2020. "Comparative Lipidomic Analysis of Extracellular Vesicles Derived from Lactobacillus plantarum APsulloc 331261 Living in Green Tea Leaves Using Liquid Chromatography-Mass Spectrometry" International Journal of Molecular Sciences 21, no. 21: 8076. https://doi.org/10.3390/ijms21218076
APA StyleKim, H., Kim, M., Myoung, K., Kim, W., Ko, J., Kim, K. P., & Cho, E.-G. (2020). Comparative Lipidomic Analysis of Extracellular Vesicles Derived from Lactobacillus plantarum APsulloc 331261 Living in Green Tea Leaves Using Liquid Chromatography-Mass Spectrometry. International Journal of Molecular Sciences, 21(21), 8076. https://doi.org/10.3390/ijms21218076





