Transcriptomic Changes in Endothelial Cells Triggered by Na,K-ATPase Inhibition: A Search for Upstream Na+i/K+i Sensitive Genes
Abstract
1. Introduction
2. Results
2.1. Intracellular Na+ and K+ Content
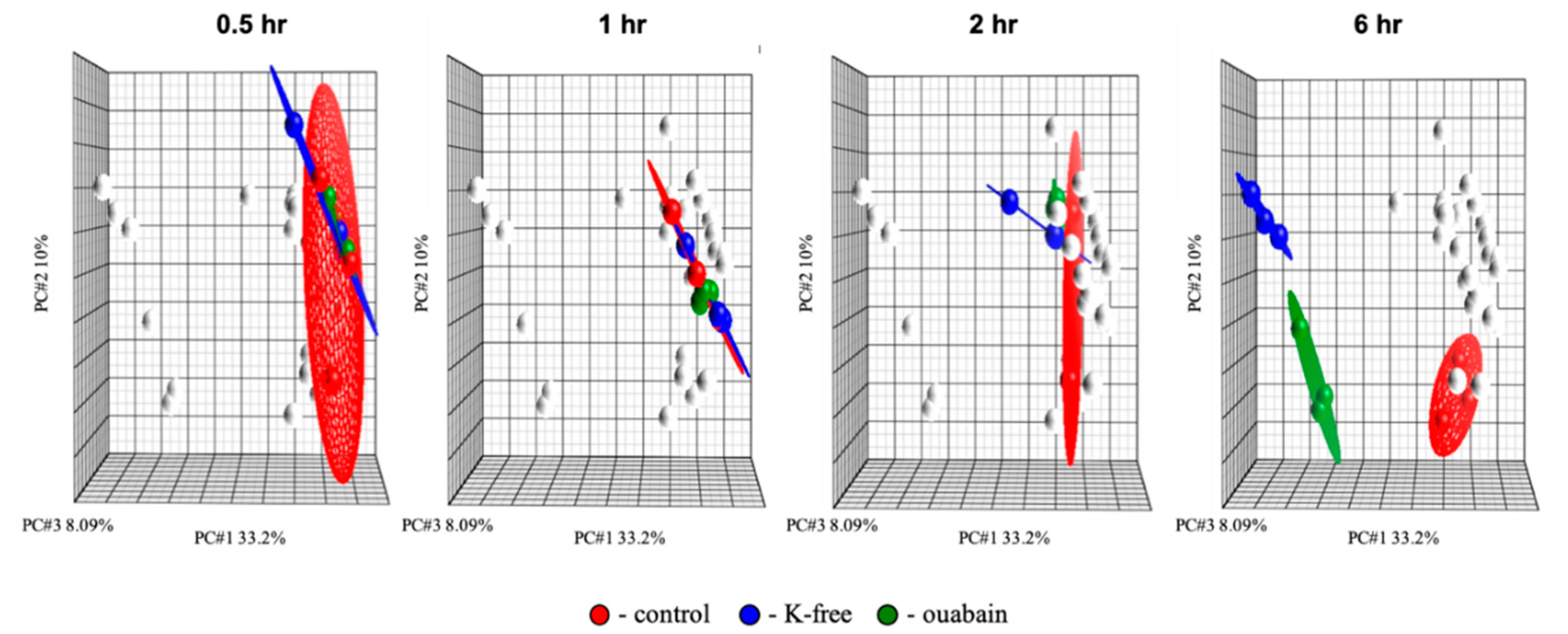
2.2. Transcriptomic Changes Triggered by [Na+]i/[K+]i-Ratio Augmentation
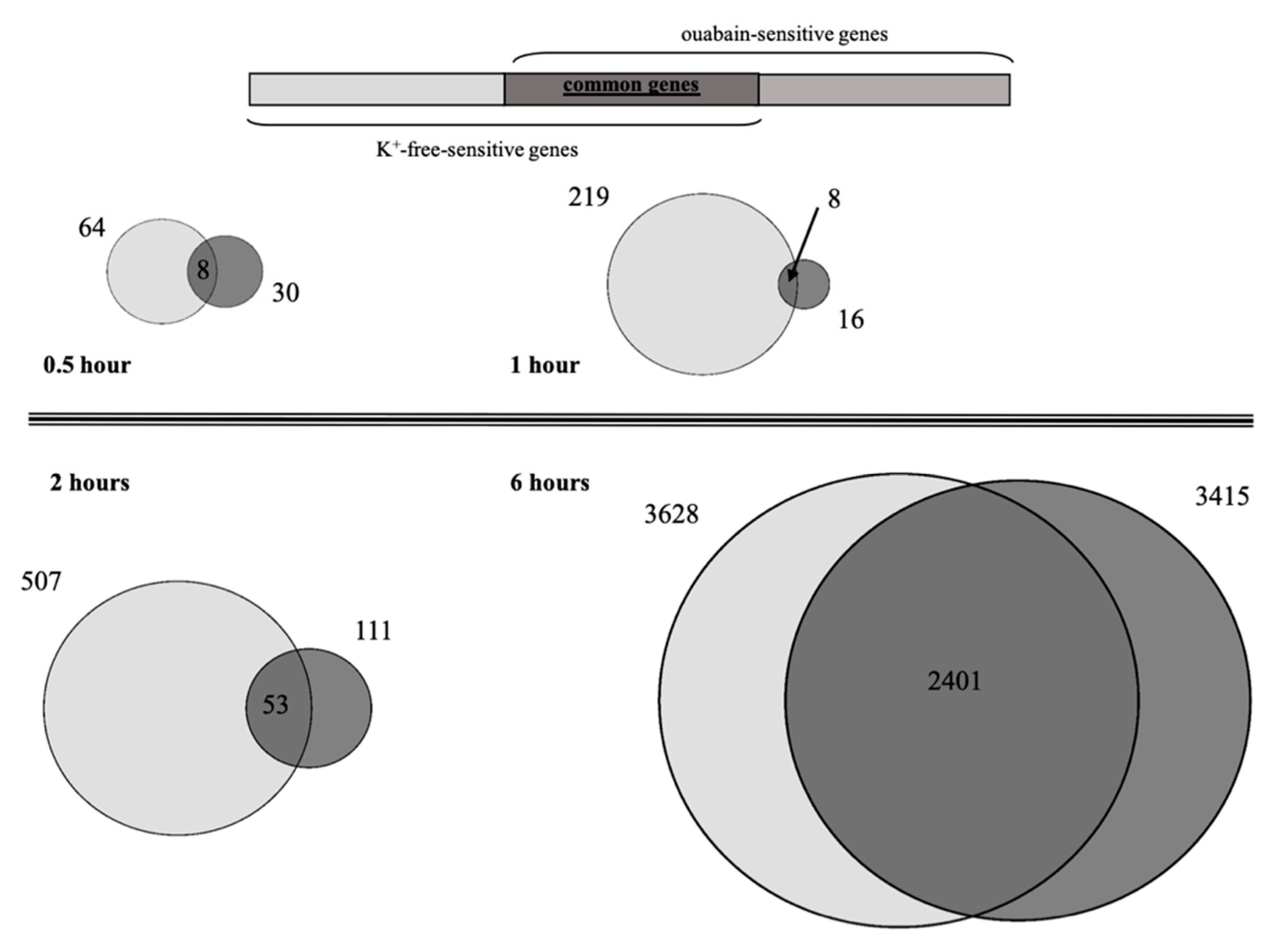
2.3. Analysis of Early Response Transcriptomic Changes Triggered by [Na+]i/[K+]i-Ratio Augmentation
2.4. Analysis of Intermediate and Late Response Transcriptomic Changes Triggered by [Na+]i/[K+]i-Ratio Augmentation
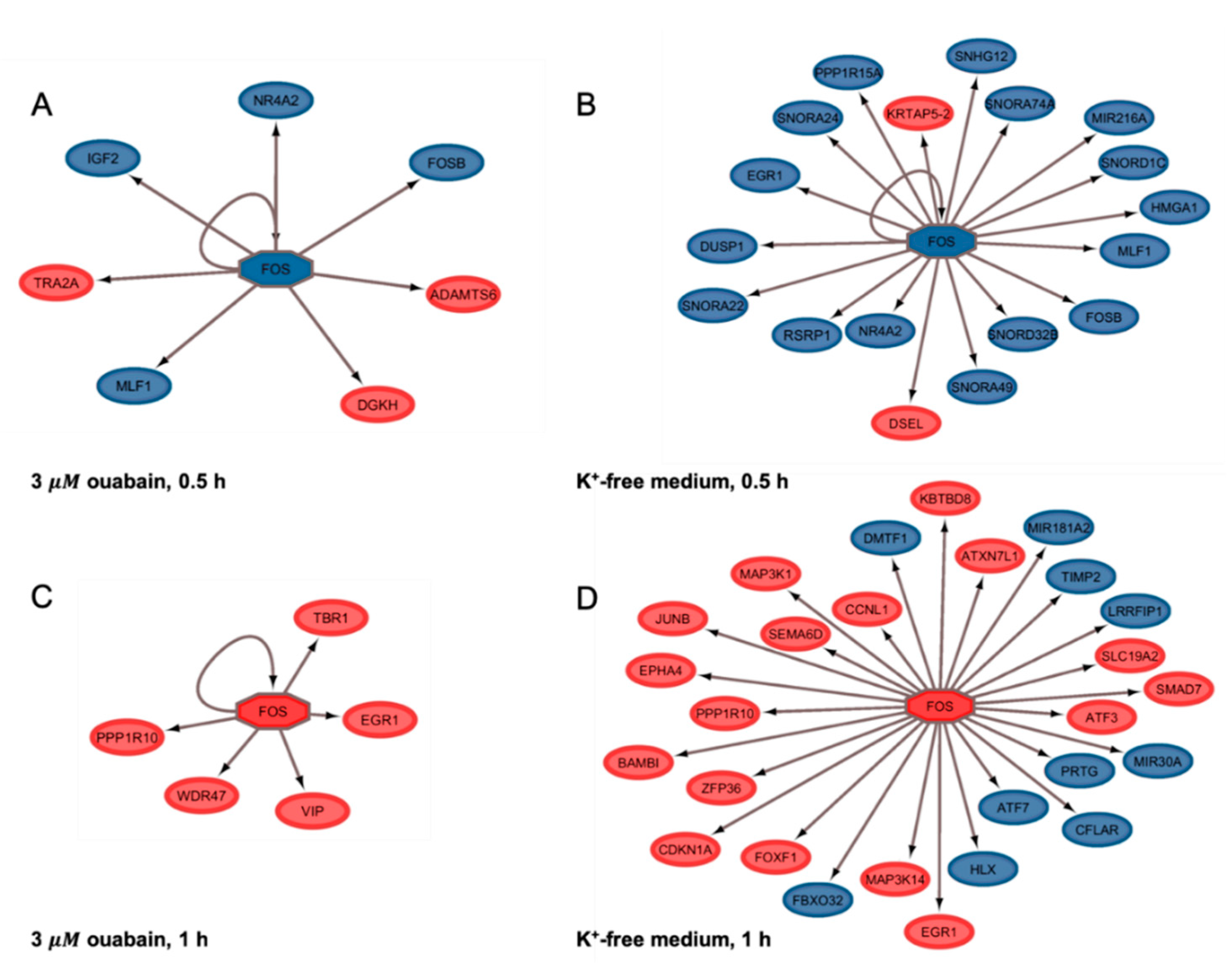
2.5. FOS Is a Conceivable Upstream Na+i/K+i-Sensitive Gene
2.6. Activation of Signaling Pathways Involving Act, CREB, JNK, and ERK
3. Discussions
4. Materials and Methods
4.1. Cell Culture
4.2. Intracellular Na+ and K+ Content
4.3. RNA Isolation
4.4. Gene Chip Expression Analysis
4.5. GO and KEGG Pathways Analysis
4.6. G-Quadruplex-Forming Region Analysis
4.7. Western Blot Analysis
4.8. Chemicals
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Akt | protein kinase B |
| AP-1 | activator protein-1 |
| CaMKI, II or III | Ca2+/calmodulin-dependent protein kinase |
| CBP | CREB-binding protein |
| CRE | Ca2+/cAMP-response element |
| DEGs | differentially expressed genes |
| EGM-2 | endothelial cell growth medium-2 |
| ERG | Early response genes |
| FBS | fetal bovine serum |
| FOXO | forkhead box protein O |
| HeLa | human adenocarcinoma cell line |
| HUVEC | human umbilical vein endothelial cells |
| INFκB | inhibitor of nuclear factor kappa-B |
| IRG | Intermediate response genes |
| LRG | late response genes |
| MAPK | mitogen-activated protein kinases |
| mTOR | mammalian target of rapamycin |
| NFAT | nuclear factor of activated T-cells |
| PCA | principal component analysis |
| PI (3)K | phosphatidyl inositol 3-kinase |
| PKA | cAMP-dependent protein kinase A |
| PKC | cAMP-dependent protein kinase C |
| PtdIns3 ps or PI3P | phosphatidylinositol 3-phosphate |
| RVSM | rat vascular smooth muscle cells |
| SRE | serum response element |
| UTR | 5′-untranslated region |
References
- Komili, S.; Silver, P.A. Coupling and coordination in gene expression processes: A systems biology view. Nat. Rev. Genet. 2008, 9, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Pope, S.D.; Medzhitov, R. Emerging Principles of Gene Expression Programs and Their Regulation. Mol. Cell 2018, 71, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Scheiner-Bobis, G.; Schreiber, S. Glutamic Acid 472 and Lysine 480 of the Sodium Pump α1 Subunit Are Essential for Activity. Their Conservation in Pyrophosphatases Suggests Their Involvement in Recognition of ATP Phosphates. Biochemistry 1999, 38, 9198–9208. [Google Scholar] [CrossRef] [PubMed]
- Taurin, S.; Dulin, N.O.; Pchejetski, D.; Grygorczyk, R.; Tremblay, J.; Hamet, P.; Orlov, S.N. c-Fos Expression in Ouabain-Treated Vascular Smooth Muscle Cells from rat Aorta: Evidence for an Intracellular-Sodium-Mediated, Calcium-Independent Mechanism. J. Physiol. 2002, 543, 835–847. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yuan, W.; Lu, Z. Effects of ouabain and digoxin on gene expression of sodium pump alpha-subunit isoforms in rat myocardium. Chin. Med. J. 2001, 114, 1055–1059. [Google Scholar]
- Huang, L.; Li, H.; Xie, Z. Ouabain-induced Hypertrophy in Cultured Cardiac Myocytes is Accompanied by Changes in Expression of Several Late Response Genes. J. Mol. Cell. Cardiol. 1997, 29, 429–437. [Google Scholar] [CrossRef]
- Taurin, S.; Seyrantepe, V.; Orlov, S.N.; Tremblay, T.-L.; Thibault, P.; Bennett, M.R.; Hamet, P.; Pshezhetsky, A.V. Proteome Analysis and Functional Expression Identify Mortalin as an Antiapoptotic Gene Induced by Elevation of [Na+]i/[K+]i Ratio in Cultured Vascular Smooth Muscle Cells. Circ. Res. 2002, 91, 915–922. [Google Scholar] [CrossRef]
- Blaustein, M.P.; Lederer, W.J. Sodium/Calcium Exchange: Its Physiological Implications. Physiol. Rev. 1999, 79, 763–854. [Google Scholar] [CrossRef]
- McDonald, T.F.; Pelzer, S.; Trautwein, W.; Pelzer, D.J. Regulation and modulation of calcium channels in cardiac, skeletal, and smooth muscle cells. Physiol. Rev. 1994, 74, 365–507. [Google Scholar] [CrossRef]
- Koltsova, S.V.; Trushina, Y.; Haloui, M.; Akimova, O.A.; Tremblay, J.; Hamet, P.; Orlov, S.N. Ubiquitous [Na+]i/[K+]i-Sensitive Transcriptome in Mammalian Cells: Evidence for Ca2+i-Independent Excitation-Transcription Coupling. PLoS ONE 2012, 7, e38032. [Google Scholar] [CrossRef]
- Medina-Leyte, D.J.; Domínguez-Pérez, M.; Mercado, I.; Villarreal-Molina, M.T.; Jacobo-Albavera, L. Use of Human Umbilical Vein Endothelial Cells (HUVEC) as a Model to Study Cardiovascular Disease: A Review. Appl. Sci. 2020, 10, 938. [Google Scholar] [CrossRef]
- Xie, Z.; Askari, A. Na+/K+-ATPase as a signal transducer. J. Biol. Inorg. Chem. 2002, 269, 2434–2439. [Google Scholar] [CrossRef] [PubMed]
- Garland, C.J.; Hiley, C.R.; A Dora, K. EDHF: Spreading the influence of the endothelium. Br. J. Pharmacol. 2011, 164, 839–852. [Google Scholar] [CrossRef]
- Klimanova, E.A.; Tverskoi, A.M.; Koltsova, S.V.; Sidorenko, S.V.; Lopina, O.D.; Tremblay, J.; Hamet, P.; Kapilevich, L.V.; Orlov, S.N. Time- and dose dependent actions of cardiotonic steroids on transcriptome and intracellular content of Na+ and K+: A comparative analysis. Sci. Rep. 2017, 7, 45403. [Google Scholar] [CrossRef] [PubMed]
- Klimanova, E.A.; Sidorenko, S.V.; Smolyaninova, L.V.; Kapilevich, L.V.; Gusakova, S.V.; Lopina, O.D.; Orlov, S.N. Ubiquitous and cell type-specific transcriptomic changes triggered by dissipation of monovalent cation gradients in rodent cells: Physiological and pathophysiological implications. In Current Topics in Membranes; Elsevier BV: Amsterdam, The Netherlands, 2019; Volume 83, pp. 107–149. [Google Scholar]
- Williams, G.T.; Farzaneh, F. Are snoRNAs and snoRNA host genes new players in cancer? Nat. Rev. Cancer 2012, 12, 84–88. [Google Scholar] [CrossRef]
- Yoneda-Kato, N.; Tomoda, K.; Umehara, M.; Arata, Y.; Kato, J.-Y. Myeloid leukemia factor 1 regulates p53 by suppressing COP1 via COP9 signalosome subunit 3. EMBO J. 2005, 24, 1739–1749. [Google Scholar] [CrossRef]
- Ronchetti, D.; Mosca, L.; Cutrona, G.; Tuana, G.; Gentile, M.; Fabris, S.; Agnelli, L.; Ciceri, G.; Matis, S.; Massucco, C.; et al. Small nucleolar RNAs as new biomarkers in chronic lymphocytic leukemia. BMC Med. Genom. 2013, 6, 27. [Google Scholar] [CrossRef]
- Williams, J.H.; Daly, L.N.; Ingley, E.; Beaumont, J.G.; Tilbrook, P.A.; LaLonde, J.; Stillitano, J.P.; Klinken, S.P. HLS7, a hemopoietic lineage switch gene homologous to the leukemia-inducing gene MLF1. EMBO J. 1999, 18, 5559–5566. [Google Scholar] [CrossRef][Green Version]
- Sun, W.; Zhang, K.; Zhang, X.; Lei, W.; Xiao, T.; Ma, J.; Guo, S.; Shao, S.; Zhang, H.; Liu, Y.; et al. Identification of differentially expressed genes in human lung squamous cell carcinoma using suppression subtractive hybridization. Cancer Lett. 2004, 212, 83–93. [Google Scholar] [CrossRef]
- Ding, L.; Ni, J.; Yang, F.; Huang, L.; Deng, H.; Wu, Y.; Ding, X.; Tang, J. Promising therapeutic role of miR-27b in tumor. Tumor Biol. 2017, 39, 1010428317691657. [Google Scholar] [CrossRef]
- Cheetham, S.W.; Faulkner, G.J.; Dinger, M.E. Overcoming challenges and dogmas to understand the functions of pseudogenes. Nat. Rev. Genet. 2019, 21, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Kharel, P.; Balaratnam, S.; Beals, N.; Basu, S. The role of RNA G-quadruplexes in human diseases and therapeutic strategies. Wiley Interdiscip. Rev. RNA 2019, 11, e1568. [Google Scholar] [CrossRef] [PubMed]
- Shiyan, A.A.; Sidorenko, S.V.; Fedorov, D.; Klimanova, E.A.; Smolyaninova, L.V.; Kapilevich, L.V.; Grygorczyk, R.; Orlov, S.N. Elevation of Intracellular Na+ Contributes to Expression of Early Response Genes Triggered by Endothelial Cell Shrinkage. Cell. Physiol. Biochem. 2019, 53, 638–647. [Google Scholar] [PubMed]
- Sidorenko, S.V.; Klimanova, E.A.; Milovanova, K.G.; Lopina, O.D.; Kapilevich, L.V.; Chibalin, A.V.; Orlov, S.N. Transcriptomic changes in C2C12 myotubes triggered by electrical stimulation: Role of Ca2+i-mediated and Ca2+i-independent signaling and elevated [Na+]i/[K+]i ratio. Cell Calcium 2018, 76, 72–86. [Google Scholar] [CrossRef] [PubMed]
- Haloui, M.; Taurin, S.; Akimova, O.A.; Guo, D.-F.; Tremblay, J.; Dulin, N.O.; Hamet, P.; Orlov, S.N. [Na+]i-induced c-Fos expression is not mediated by activation of the 5′-promoter containing known transcriptional elements. FEBS J. 2007, 274, 3557–3567. [Google Scholar] [CrossRef] [PubMed]
- Curran, T.; Franza, B. Fos and jun: The AP-1 connection. Cell 1988, 55, 395–397. [Google Scholar] [CrossRef]
- Whitmarsh, A.J.; Davis, R.J. Transcription factor AP-1 regulation by mitogen-activated protein kinase signal transduction pathways. J. Mol. Med. 1996, 74, 589–607. [Google Scholar] [CrossRef]
- Piechaczyk, M.; Blanchard, J.-M. c-fos proto-oncogene regulation and function. Crit. Rev. Oncol. 1994, 17, 93–131. [Google Scholar] [CrossRef]
- Papa, S.; Choy, P.M.; Bubici, C. The ERK and JNK pathways in the regulation of metabolic reprogramming. Oncogene 2018, 38, 2223–2240. [Google Scholar] [CrossRef]
- Lopachev, A.V.; Lopacheva, O.M.; Osipova, E.A.; Vladychenskaya, E.A.; Smolyaninova, L.V.; Fedorova, T.N.; Koroleva, O.V.; Akkuratov, E.E. Ouabain-induced changes in MAP kinase phosphorylation in primary culture of rat cerebellar cells. Cell Biochem. Funct. 2016, 34, 367–377. [Google Scholar] [CrossRef]
- Akimova, O.A.; Hamet, P.; Orlov, S.N. [Na+] i /[K+] i -independent death of ouabain-treated renal epithelial cells is not mediated by Na+,K+-ATPase internalization and de novo gene expression. Pflug. Arch. Eur. J Phy. 2007, 455, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Kolkhof, P.; Geerts, A.; Schäfer, S.; Torzewski, J. Cardiac glycosides potently inhibit C-reactive protein synthesis in human hepatocytes. Biochem. Biophys. Res. Commun. 2010, 394, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Smolyaninova, L.V.; Shiyan, A.A.; Kapilevich, L.V.; Lopachev, A.V.; Fedorova, T.N.; Klementieva, T.S.; Moskovtsev, A.A.; Kubatiev, A.A.; Orlov, S.N. Transcriptomic changes triggered by ouabain in rat cerebellum granule cells: Role of α3- and α1-Na+,K+-ATPase-mediated signaling. PLoS ONE 2019, 14, e0222767. [Google Scholar] [CrossRef]
- Liang, G.H.; Kim, J.A.; Seol, G.H.; Choi, S.; Suh, S.H. The Na+/Ca2+ exchanger inhibitor KB-R7943 activates large-conductance Ca2+-activated K+ channels in endothelial and vascular smooth muscle cells. Eur. J. Pharmacol. 2008, 582, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Coulon, V.; Blanchard, J. Flux calciques et expression génique. Médecine/Sciences 2001, 17, 969. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, D.; McQuade, J.S.; Behbehani, M.; Tsien, J.Z.; Xu, M. c-Fos regulates neuronal excitability and survival. Nat. Genet. 2002, 30, 416–420. [Google Scholar] [CrossRef]
- Kapilevich, L.V.; Kironenko, T.A.; Zaharova, A.N.; Kotelevtsev, Y.; Dulin, N.O.; Orlov, S.N. Skeletal muscle as an endocrine organ: Role of [Na+]i/[K+]i-mediated excitation-transcription coupling. Genes Dis. 2015, 2, 328–336. [Google Scholar] [CrossRef]
- Nakagawa, Y.; Rivera, V.; Larner, A.C. A role for the Na/K-ATPase in the control of human c-fos and c-jun transcription. J. Biol. Chem. 1992, 267, 8785–8788. [Google Scholar]
- Nakagawa, Y.; Petricoin, E.F.; Akai, H.; Grimley, P.M.; Rupp, B.; Larner, A.C. Interferon-alpha-induced gene expression: Evidence for a selective effect of ouabain on activation of the ISGF3 transcription complex. Virology 1992, 190, 210–220. [Google Scholar] [CrossRef]
- Choudhury, G.G. Akt Serine Threonine Kinase Regulates Platelet-derived Growth Factor-induced DNA Synthesis in Glomerular Mesangial Cells. J. Biol. Chem. 2001, 276, 35636–35643. [Google Scholar] [CrossRef]
- He, H.; Ping, F. The SIE, SRE, CRE, and FAP-1 four intracellular signal pathways between stimulus and the expression of c-fos promoter. J. Cell. Biochem. 2009, 106, 764–768. [Google Scholar] [CrossRef]
- Orlov, S.N.; Hamet, P. Intracellular Monovalent Ions as Second Messengers. J. Membr. Biol. 2006, 210, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Klimanova, E.A.; Sidorenko, S.V.; Tverskoi, A.M.; Shiyan, A.A.; Smolyaninova, L.V.; Kapilevich, L.V.; Gusakova, S.V.; Maksimov, G.V.; Lopina, O.D.; Orlov, S.N. Search for Intracellular Sensors Involved in the Functioning of Monovalent Cations as Secondary Messengers. Biochemistry 2019, 84, 1280–1295. [Google Scholar] [CrossRef]
- Lopina, O.D.; Tverskoi, A.M.; Klimanova, E.A.; Sidorenko, S.V.; Orlov, S.N. Ouabain-Induced Cell Death and Survival. Role of α1-Na,K-ATPase-Mediated Signaling and [Na+]i/[K+]i-Dependent Gene Expression. Front. Physiol. 2020, 11, 1060. [Google Scholar] [CrossRef]
- Huppert, J.L. Four-stranded DNA: Cancer, gene regulation and drug development. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2007, 365, 2969–2984. [Google Scholar] [CrossRef]
- Sen, D.; Gilbert, W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nat. Cell Biol. 1990, 344, 410–414. [Google Scholar] [CrossRef]
- Pandith, A.; Siddappa, R.G.; Seo, Y.-J. Recent developments in novel blue/green/red/NIR small fluorescent probes for in cellulo tracking of RNA/DNA G-quadruplexes. J. Photochem. Photobiol. C Photochem. Rev. 2019, 40, 81–116. [Google Scholar] [CrossRef]
- Spiegel, J.; Adhikari, S.; Balasubramanian, S. The Structure and Function of DNA G-Quadruplexes. Trends Chem. 2020, 2, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, E.; Richter, S.N. G-quadruplexes and G-quadruplex ligands: Targets and tools in antiviral therapy. Nucleic Acids Res. 2018, 46, 3270–3283. [Google Scholar] [CrossRef] [PubMed]
- Mijatovic, T.; Van Quaquebeke, E.; Delest, B.; Debeir, O.; Darro, F.; Kiss, R. Cardiotonic steroids on the road to anti-cancer therapy. Biochim. Biophys. Acta (BBA)—Bioenerg. 2007, 1776, 32–57. [Google Scholar] [CrossRef]
- Pessôa, M.T.C.; Cortes, V.F.; Barbosa, L.A. Na, K-ATPase Cell Signaling Pathways and Cancer. In Regulation of Membrane Na+-K+ ATPase; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2015; pp. 51–61. [Google Scholar]
- Bagrov, A.Y.; Shapiro, J.I.; Fedorova, O.V. Endogenous Cardiotonic Steroids: Physiology, Pharmacology, and Novel Therapeutic Targets. Pharmacol. Rev. 2009, 61, 9–38. [Google Scholar] [CrossRef]
- Amarelle, L.; Katzen, J.; Shigemura, M.; Welch, L.C.; Cajigas, H.; Peteranderl, C.; Celli, D.; Herold, S.; Lecuona, E.; Sznajder, J.I. Cardiac glycosides decrease influenza virus replication by inhibiting cell protein translational machinery. Am. J. Physiol. Cell. Mol. Physiol. 2019, 316, L1094–L1106. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.W.; Balachandran, A.; Ostrowski, M.A.; Cochrane, A. Digoxin Suppresses HIV-1 Replication by Altering Viral RNA Processing. PLoS ONE Pathog. 2013, 9, e1003241. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.W.; Lingwood, C.A.; Ostrowski, M.A.; Cabral, T.; Cochrane, A. Cardiac glycoside/aglycones inhibit HIV-1 gene expression by a mechanism requiring MEK1/2-ERK1/2 signaling. Sci. Rep. 2018, 8, 850. [Google Scholar] [CrossRef]
- Grosso, F.; Stoilov, P.; Lingwood, C.; Brown, M.; Cochrane, A. Suppression of Adenovirus Replication by Cardiotonic Steroids. J. Virol. 2016, 91, 91. [Google Scholar] [CrossRef]
- Dvela, M.; Rosen, H.; Feldmann, T.; Nesher, M.; Lichtstein, D. Diverse biological responses to different cardiotonic steroids. Pathophysiology 2007, 14, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Triana-Martínez, F.; Picallos-Rabina, P.; Da Silva-Álvarez, S.; Pietrocola, F.; Llanos, S.; Rodilla, V.; Soprano, E.; Pedrosa, P.; Ferreirós, A.; Barradas, M.; et al. Identification and characterization of Cardiac Glycosides as senolytic compounds. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Pearson, K. LIII. On lines and planes of closest fit to systems of points in space. Lond. Edinb. Dublin Philos. Mag. J. Sci. 1901, 2, 559–572. [Google Scholar] [CrossRef]
- Ülgen, E.; Ozisik, O.; Sezerman, O.U. pathfindR: An R Package for Comprehensive Identification of Enriched Pathways in Omics Data Through Active Subnetworks. Front. Genet. 2019, 10, 858. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009. [Google Scholar]
- Doluca, O. G4Catchall: A G-quadruplex prediction approach considering atypical features. J. Theor. Biol. 2019, 463, 92–98. [Google Scholar] [CrossRef]
- Bennett, G.F. Lowry’s handbook of right-to-know emergency planning. J. Hazard. Mater. 1992, 30, 361–362. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nat. Cell Biol. 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]



| Ouabain | K+-Free Medium | |||||||
|---|---|---|---|---|---|---|---|---|
| 0.5 h | 1 h | 2 h | 6 h | 0.5 h | 1 h | 2 h | 6 h | |
| Number of differentially expressed genes | 30 | 16 | 111 | 3415 | 64 | 219 | 507 | 3628 |
| Number of up-regulated genes | 22 | 15 | 64 | 1231 | 6 | 133 | 280 | 1011 |
| Maximal fold of activation | 1.56 | 2.04 | 2.73 | 14.49 | 1.48 | 2.55 | 17.22 | 24.57 |
| Number of down-regulated genes | 8 | 1 | 44 | 2184 | 58 | 86 | 227 | 2617 |
| Maximal fold of inhibition | −1.73 | 1.93 | −2.20 | −7.49 | −2.47 | −3.44 | −4.99 | −12.42 |
| Hours | Gene Symbol | 3 μM Ouabain | K+-Free Medium | ||
|---|---|---|---|---|---|
| FC | p-Value | FC | p-Value | ||
| 0.5 | RNU6–447P | 1.44 | 1.23 × 103 | 1.36 | 5.26 × 103 |
| RNU6–747P | 1.36 | 1.48 × 103 | 1.32 | 5.26 × 103 | |
| SNORD32B | −1.29 | 1.21 × 102 | −1.26 | 2.43 × 102 | |
| MLF1 | −1.29 | 6.26 × 103 | −1.27 | 9.23 × 103 | |
| SNORA20 | −1.45 | 476 × 102 | −1.86 | 6.49 × 104 | |
| NR4A2 | −1.51 | 6.26 × 103 | −2.47 | 7.12 × 108 | |
| FOSB | −1.73 | 3.11 × 103 | −2.42 | 5.16 × 106 | |
| FOS | −1.73 | 3.71 × 102 | −2.17 | 2.72 × 103 | |
| 1 | EGR1 | 2.04 | 1.90 × 103 | 1.69 | 8.07 × 103 |
| FOS | 1.96 | 1.19 × 103 | 2.52 | 3.65 × 105 | |
| LSMEM1 | 1.63 | 1.41 × 103 | 1.32 | 2.92 × 102 | |
| SNORA67 | 1.47 | 5.42 × 103 | 1.78 | 3.33 × 106 | |
| SNORA21 | 1.35 | 1.06 × 102 | 1.26 | 2.23 × 102 | |
| PPP1R10 | 1.29 | 1.90 × 103 | 1.21 | 6.48 × 103 | |
| WDR47 | 1.29 | 3.80 × 102 | 1.36 | 7.36 × 104 | |
| MIR27B | −1.93 | 1.49 × 103 | −1.75 | 9.21 × 104 | |
| Hours | Gene Symbol | 3 µM Ouabain | K+-Free Medium | ||
|---|---|---|---|---|---|
| FC | p-Value | FC | p-Value | ||
| 2 | RN7SL600P | 2.71 | 8.26 × 106 | 8.81 | 9.24 × 1013 |
| RN7SL473P | 2.45 | 1.03 × 107 | 7.58 | 1.26 × 1015 | |
| RN7SL849P | 1.53 | 6.66 × 103 | 6.33 | 6.76 × 1014 | |
| IL1A | 2.21 | 8.31 × 109 | 5.83 | 1.18 × 1016 | |
| KITLG | 1.53 | 5.75 × 105 | 5.36 | 1.11 × 1016 | |
| FLJ35409 | −2.13 | 2.02 × 103 | −2.85 | 5.98 × 106 | |
| AC002350.1 | −1.78 | 3.43 × 103 | −3.16 | 4.24 × 108 | |
| LOC100130713 | −2.14 | 9.38 × 107 | −3.49 | 1.12 × 1011 | |
| SNORD52 | −2.20 | 1.72 × 106 | −3.96 | 7.64 × 1012 | |
| DEPP1 | −1.69 | 8.84 × 106 | −4.99 | 9.57 × 1016 | |
| 6 | FLJ43390 | 14.33 | 2.91 × 1017 | 2.74 | 3.40 × 109 |
| TFPI2 | 9.07 | 4.76 × 1018 | 14.21 | 8.28 × 1020 | |
| HIVEP2 | 6.83 | 9.42 × 1019 | 13.52 | 1.80 × 1021 | |
| PLA2G4C | 6.48 | 1.63 × 1018 | 8.03 | 1.19 × 1019 | |
| TRAF1 | 3.85 | 2.23 × 1015 | 20.12 | 3.62 × 1022 | |
| AGGF1 | −5.68 | 7.59 × 1021 | −5.94 | 2.60 × 1021 | |
| GCNT1 | −5.78 | 2.51 × 1016 | −6.16 | 8.77 × 1017 | |
| NMI | −5.86 | 5.04 × 1019 | −6.64 | 8.28 × 1020 | |
| SAMD9L | −6.92 | 1.61 × 1019 | −5.01 | 3.19 × 1018 | |
| GIMAP2 | −7.38 | 2.77 × 1019 | −12.42 | 2.03 × 1021 | |
| Sequence Definition | NCBI Reference Sequence | Beginning of G4 Sequence (First Nucleotide = 0) | End of G4 Sequence | Length | G4-Forming Sequence | G4HScore |
|---|---|---|---|---|---|---|
| FOS | NC_000014.9 | 341 | 368 | 27 | ggggccgggggcttggggtcgcggagg | 2 |
| FOS | NC_000014.9 | 1405 | 1427 | 22 | gggaatgtgggggctgggtggg | 2.136 |
| FOS | NC_000014.9 | 2755 | 2778 | 23 | gtgagggggcagggaaggggagg | 2.174 |
| FOSB | NC_000019.10 | 2242 | 2265 | 23 | ggggtgggggtggggtgttgtgg | 2.522 |
| FOSB | NC_000019.10 | 3910 | 3930 | 20 | gggaggtagagagggagggg | 2 |
| FOSB | NC_000019.10 | 5323 | 5348 | 25 | ggggatgggtggggaggggggcggg | 2.92 |
| LSMEM1 | NC_000007.14 | 5835 | 5858 | 23 | gggtgtggtggagggggaggggg | 2.522 |
| LSMEM1 | NC_000007.14 | 6220 | 6244 | 24 | gggttggagaaagggggtgggggg | 2.417 |
| LSMEM1 | NC_000007.14 | 9144 | 9167 | 23 | gggttgggactgggagggagggg | 2.217 |
| PPP1R10 | NC_000006.12 | 3449 | 3473 | 24 | ggggtgtggggggggggttgcagg | 2.542 |
| PPP1R10 | NC_000006.12 | 4975 | 4995 | 20 | ggagcagttgggtggggggg | 2 |
| WDR47 | NC_000001.11 | 61981 | 62015 | 34 | gggtggggtgggaaggggtagggatggatggtttaggg | 2 |
| MLF1 | NC_000003.12 | 2170 | 2225 | 55 | ggggcggggcggggggaggggggcgggaggagggaggagggaggagggcggcgggggggggggggcggcgggggggggggggtgtgtgtg | 2.633 |
| MLF1 | NC_000003.12 | 27114 | 27155 | 41 | ggtggggggacaggagggaggtgtgggggttgggggtaagg | 2.171 |
| Hours | 3 µM Ouabain | K+-Free Medium | ||
|---|---|---|---|---|
| FC | p-Value | FC | p-Value | |
| 0.5 | −1.73 | 3.71 × 102 | −2.17 | 2.72 × 103 |
| 1 | 1.96 | 1.19 × 102 | 2.52 | 3.65 × 105 |
| 2 | 2.47 | 1.80 × 105 | 18.07 | 1.00 × 1015 |
| 6 | 8.14 | 8.44 × 1014 | 5.68 | 2.64 × 1012 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klimanova, E.A.; Sidorenko, S.V.; Abramicheva, P.A.; Tverskoi, A.M.; Orlov, S.N.; Lopina, O.D. Transcriptomic Changes in Endothelial Cells Triggered by Na,K-ATPase Inhibition: A Search for Upstream Na+i/K+i Sensitive Genes. Int. J. Mol. Sci. 2020, 21, 7992. https://doi.org/10.3390/ijms21217992
Klimanova EA, Sidorenko SV, Abramicheva PA, Tverskoi AM, Orlov SN, Lopina OD. Transcriptomic Changes in Endothelial Cells Triggered by Na,K-ATPase Inhibition: A Search for Upstream Na+i/K+i Sensitive Genes. International Journal of Molecular Sciences. 2020; 21(21):7992. https://doi.org/10.3390/ijms21217992
Chicago/Turabian StyleKlimanova, Elizaveta A., Svetlana V. Sidorenko, Polina A. Abramicheva, Artem M. Tverskoi, Sergei N. Orlov, and Olga D. Lopina. 2020. "Transcriptomic Changes in Endothelial Cells Triggered by Na,K-ATPase Inhibition: A Search for Upstream Na+i/K+i Sensitive Genes" International Journal of Molecular Sciences 21, no. 21: 7992. https://doi.org/10.3390/ijms21217992
APA StyleKlimanova, E. A., Sidorenko, S. V., Abramicheva, P. A., Tverskoi, A. M., Orlov, S. N., & Lopina, O. D. (2020). Transcriptomic Changes in Endothelial Cells Triggered by Na,K-ATPase Inhibition: A Search for Upstream Na+i/K+i Sensitive Genes. International Journal of Molecular Sciences, 21(21), 7992. https://doi.org/10.3390/ijms21217992






