Chromosome Painting in Cultivated Bananas and Their Wild Relatives (Musa spp.) Reveals Differences in Chromosome Structure
Abstract
1. Introduction
2. Results
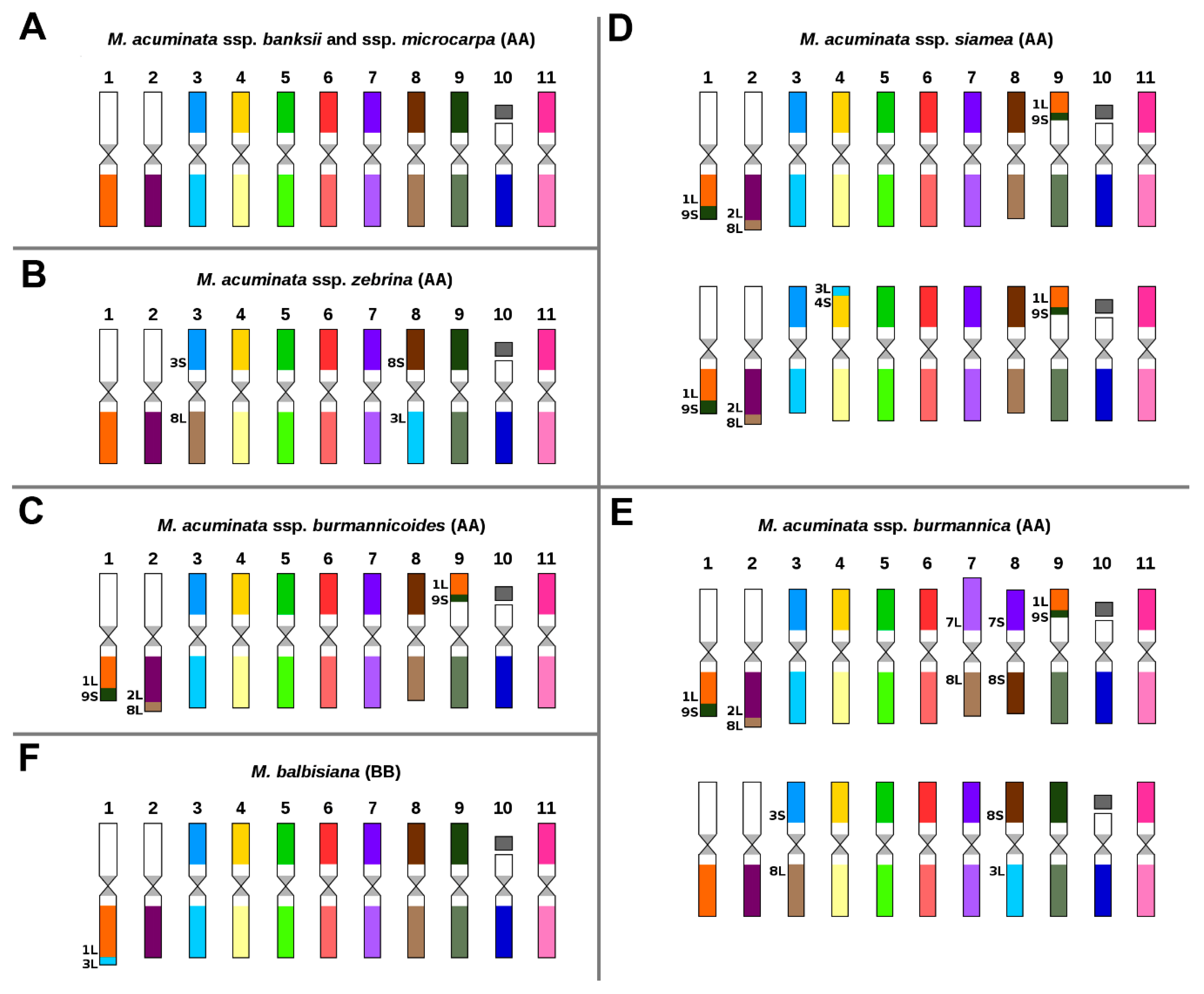
2.1. Karyotype of M. acuminata and M. balbisiana
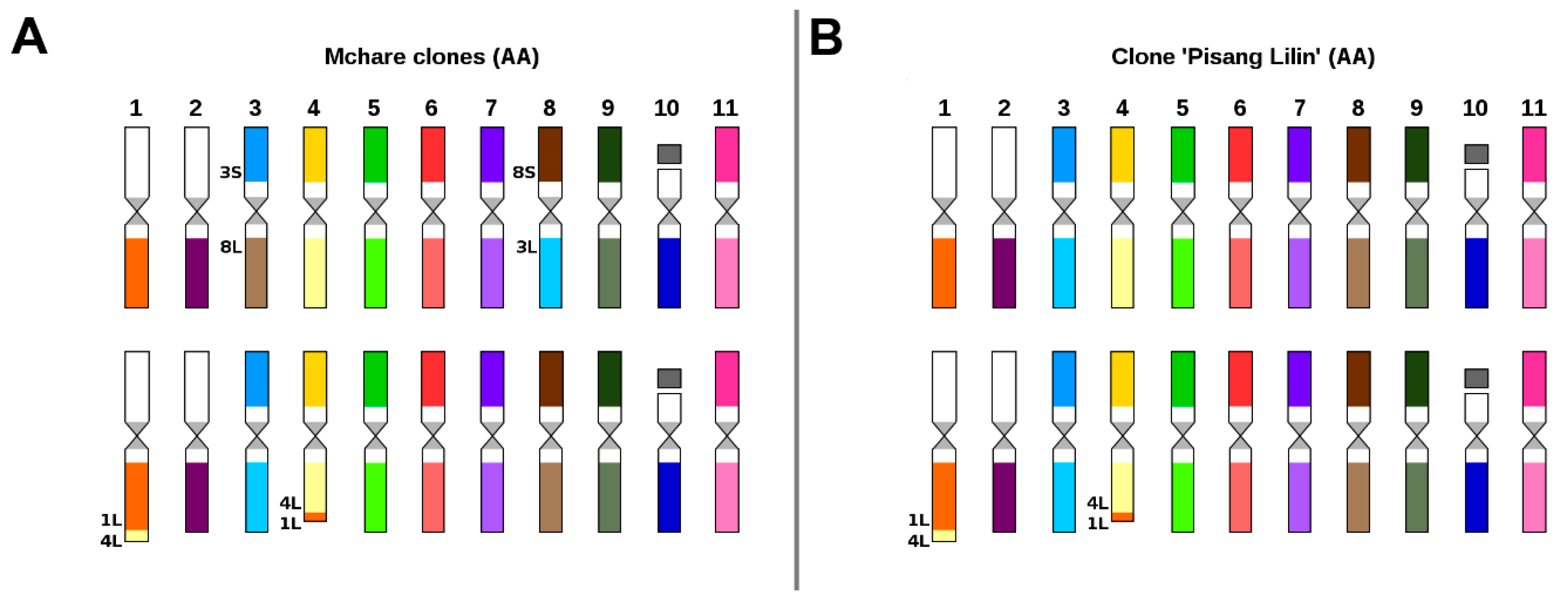
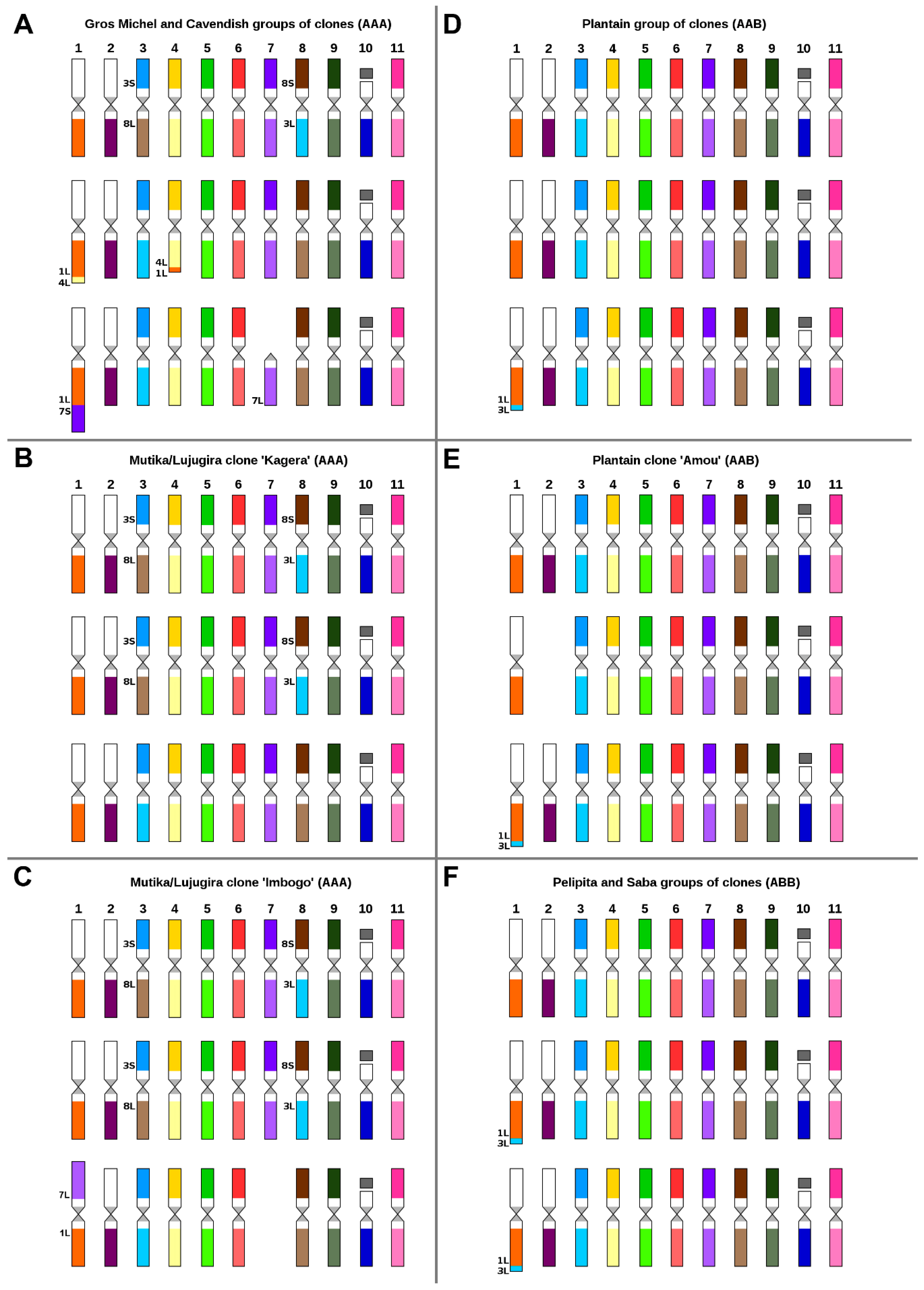
2.2. Karyotype Structure of Edible Clones That Originated as Intraspecific Hybrids
2.3. Karyotype Structure of Interspecific Banana Clones
3. Discussion
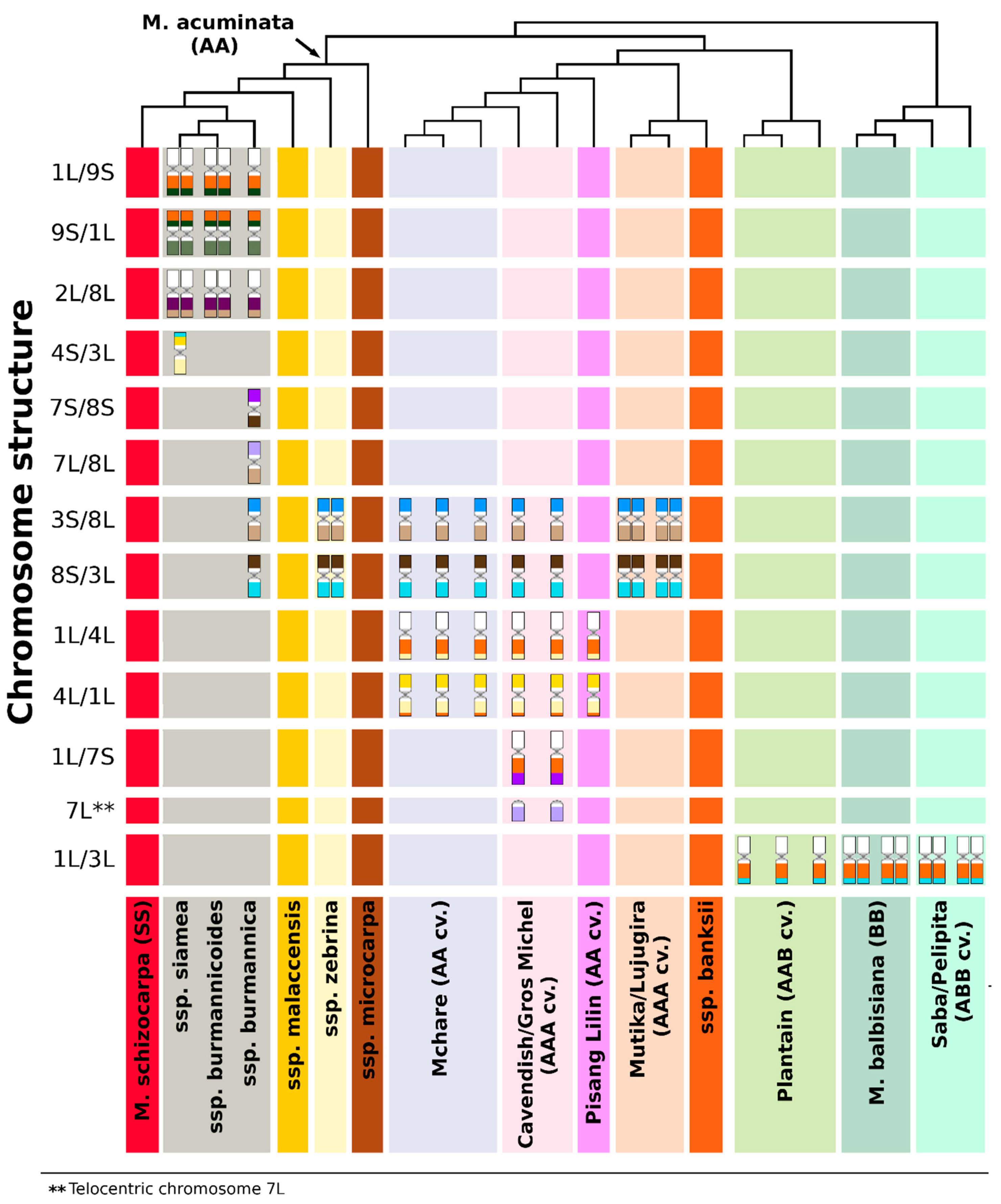
3.1. Structural Genome Variation in Diploid M. acuminata
3.2. Genome Structure and Origin of Cultivated Triploid Musa Clones
3.3. The Origin of Aneuploidy
4. Materials and Methods
4.1. Plant Material and Diversity Tree Construction
4.2. Preparation of Oligo Painting Probes and Mitotic Metaphase Chromosome Spreads
4.3. Fluorescence In Situ Hybridization and Image Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BAC | bacterial artificial chromosome |
| DH Pahang | doubled haploid Pahang |
| EAHB | East African Highland banana |
| FISH | fluorescence in situ hybridization |
| FITC | fluorescein isothiocyanate |
| NM group | Northern Malayan group |
| SNP | single-nucleotide polymorphism |
| spp. | species |
| ssp. | subspecies |
| SSR | simple sequence repeat |
| ST group | standard translocation group |
| UPGMA | unweighted pair group method with arithmetic mean |
References
- FAOSTAT; Agriculture Organization of the United Nations; FAO. 2017. Available online: http://www.fao.org/home/en/ (accessed on 30 January 2020).
- International Trade Statistics. 2019. Available online: https://www.wto.org/ (accessed on 20 May 2020).
- Price, N.S. The origin and development of banana and plantain cultivation. In Bananas and Plantains, World Crop Series; Gowen, S., Ed.; Springer: Dordrecht, The Netherlands, 1995. [Google Scholar]
- Carreel, F.; Fauré, S.; Gonzalez de Leon, D.; Lagoda, P.J.L.; Perrier, X.; Bakry, F.; Tezenas du Montcel, H.; Lanaud, C.; Horry, J.P. Evaluation of the genetic diversity in diploid bananas (Musa spp.). Genet. Sel. Evol. 1994, 26, 125–136. [Google Scholar] [CrossRef]
- Čížková, J.; Hřibová, E.; Humplíková, L.; Christelová, P.; Suchánková, P.; Doležel, J. Molecular analysis and genomic organization of major DNA satellites in banana (Musa spp.). PLoS ONE 2013, 8, e54808. [Google Scholar] [CrossRef] [PubMed]
- Němečková, A.; Christelová, P.; Čížková, J.; Nyine, M.; van den Houwe, I.; Svačina, R.; Uwimana, B.; Swennen, R.; Doležel, J.; Hřibová, E. Molecular and cytogenetic study of East African Highland Banana. Front. Plant Sci. 2018, 9, 1371. [Google Scholar] [CrossRef] [PubMed]
- Perrier, X.; De Langhe, E.; Donohue, M.; Lentfer, C.; Vrydaghs, L.; Bakry, F.; Carreel, F.; Hippolyte, I.; Horry, J.P.; Jenny, C.; et al. Multidisciplinary perspectives on banana (Musa spp.) domestication. Proc. Natl. Acad. Sci. USA. 2011, 108, 11311–11318. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.; Carreel, F.; Coriton, O.; Hervouet, C.; Cardi, C.; Derouault, P.; Roques, D.; Salmon, F.; Rouard, M.; Sardos, J.; et al. Evolution of the banana genome (Musa acuminata) is impacted by large chromosomal translocations. Mol. Biol. Evol. 2017, 34, 2140–2152. [Google Scholar] [CrossRef]
- WCSP. World Checklist of Selected Plant Families. Facilitated by the Royal Botanic Gardens, Kew. 2018. Available online: http://wcsp.science.kew.org/ (accessed on 30 January 2020).
- Rouard, M.; Droc, G.; Martin, G.; Sardos, J.; Hueber, Y.; Guignon, V.; Cenci, A.; Geigle, B.; Hibbins, M.S.; Yahiaoui, N.; et al. Three new genome assemblies support a rapid radiation in Musa acuminata (wild banana). Genome Biol. Evol. 2018, 10, 3129–3140. [Google Scholar] [CrossRef]
- Sharrock, S. Collecting Musa in Papua New Guinea. Identification of genetic diversity in the genus Musa. In International Network for the Improvement of Banana and Plantain; Jarret, R.L., Ed.; INIBAP: Montpellier, France, 1990; pp. 140–157. [Google Scholar]
- Perrier, X.; Bakry, F.; Carreel, F.; Jenny, F.; Horry, J.P.; Lebot, V.; Hippolyte, I. Combining biological approaches to shed light on the evolution of edible bananas. Ethnobot. Res. Appl. 2009, 7, 199–216. [Google Scholar] [CrossRef]
- Cheesman, E.E. Classification of the bananas. Kew Bull. 1948, 3, 145–153. [Google Scholar] [CrossRef]
- De Langhe, E.; Vrydaghs, L.; De Maret, P.; Perrier, X.; Denham, T.P. Why bananas matter: An introduction to the history of banana domestication. Ethnobot. Res. Appl. 2009, 7, 165–177. [Google Scholar] [CrossRef]
- Sand, C. Petite histoire du peuplement de l’Océanie. In Migrations et Identités—Colloque CORAIL, Nouméa (PYF), 1988/11/21-22. Papeete; Ricard, M., Ed.; Université Française du Pacifique, 1989; pp. 39–40. Available online: http://www.documentation.ird.fr/hor/fdi:27803 (accessed on 30 January 2020).
- Denham, T.; Haberle, S.; Lentfer, C. New evidence and revised interpretations of early agriculture in Highland New Guinea. Antiquity 2004, 78, 839–857. [Google Scholar] [CrossRef]
- Denham, T. From domestication histories to regional prehistory: Using plants to re-evaluate early and mid-holocene interaction between New Guinea and Southeast Asia. Food Hist. 2010, 8, 3–22. [Google Scholar] [CrossRef]
- Kagy, V.; Wong, M.; Vandenbroucke, H.; Jenny, C.; Dubois, C.; Ollivier, A.; Cardi, C.; Mournet, P.; Tuia, V.; Roux, N.; et al. Traditional banana diversity in Oceania: An endangered heritage. PLoS ONE 2016, 11, e0151208. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, N.W. The Evolution of the Bananas; Tropical Science Series; Longmans: London, UK, 1962; p. 170. [Google Scholar]
- Raboin, L.M.; Carreel, F.; Noyer, J.L.; Baurens, F.C.; Horry, J.P.; Bakry, F.; Tezenas Du Montcel, H.; Ganry, J.; Lanaud, C.; Lagoda, P.J.L. Diploid ancestors of triploid export banana cultivars: Molecular identification of 2n restitution gamete donors and n gamete donors. Mol. Breed. 2005, 16, 333–341. [Google Scholar] [CrossRef]
- Perrier, X.; Jenny, C.; Bakry, F.; Karamura, D.; Kitavi, M.; Dubois, C.; Hervouet, C.; Philippson, G.; De Langhe, E. East African diploid and triploid bananas: A genetic complex transported from South-East Asia. Ann. Bot. 2019, 123, 19–36. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.; Cardi, C.; Sarah, G.; Ricci, S.; Jenny, C.; Fondi, E.; Perrier, X.; Glaszmann, J.-C.; D’Hont, A.; Yahiaoui, N. Genome ancestry mosaics reveal multiple and cryptic contributors to cultivated banana. Plant. J. 2020, 102, 1008–1025. [Google Scholar] [CrossRef]
- Baurens, F.C.; Martin, G.; Hervouet, C.; Salmon, F.; Yohomé, D.; Ricci, S.; Rouard, M.; Habas, R.; Lemainque, A.; Yahiaoui, N.; et al. Recombination and large structural variations shape interspecific edible bananas genomes. Mol. Biol. Evol. 2019, 36, 97–111. [Google Scholar] [CrossRef]
- De Langhe, E.; Hřibová, E.; Carpentier, S.; Doležel, J.; Swennen, R. Did backcrossing contribute to the origin of hybrid edible bananas? Ann. Bot. 2010, 106, 849–857. [Google Scholar] [CrossRef]
- Cooper, H.; Spillane, C.; Hodgkin, T. Broadening the Genetic Bases of Crop. Production; FAO: Rome, Italy, 2001. [Google Scholar]
- Tugume, A.K.; Lubega, G.W.; Rubaihayo, P.R. Genetic diversity of East African Highland bananas using AFLP. Infomusa 2003, 11, 28–32. [Google Scholar]
- Kitavi, M.; Downing, T.; Lorenzen, J.; Karamura, D.; Onyango, M.; Nyine, M.; Ferguson, M.; Spillane, C. The triploid East African Highland Banana (EAHB) genepool is genetically uniform arising from a single ancestral clone that underwent population expansion by vegetative propagation. Theor. Appl. Genet. 2016, 129, 547–561. [Google Scholar] [CrossRef]
- Christelová, P.; De Langhe, E.; Hřibová, E.; Čížková, J.; Sardos, J.; Hušáková, M.; van den Houwe, I.; Sutanto, A.; Kepler, A.K.; Swennen, R.; et al. Molecular and cytological characterization of the global Musa germplasm collection provides insights into the treasure of banana diversity. Biodivers. Conserv. 2017, 26, 801–824. [Google Scholar] [CrossRef]
- Burke, J.M.; Arnold, M.L. Genetics and the fitness of hybrids. Annu. Rev. Genet. 2001, 35, 31–52. [Google Scholar] [CrossRef] [PubMed]
- Batte, M.; Swennen, R.; Uwimana, B.; Akech, V.; Brown, A.; Tumuhimbise, R.; Hovmalm, H.P.; Geleta, M.; Ortiz, R. Crossbreeding East African Highland Bananas: Lessons learnt relevant to the botany of the crop after 21 years of genetic enhancement. Front. Plant. Sci. 2019, 10, 81. [Google Scholar] [CrossRef] [PubMed]
- Bakry, F.; Horry, J.P. Tetraploid hybrids from interploid 3×/2× crosses in cooking bananas. Fruits 1992, 47, 641–655. [Google Scholar]
- Tomepke, K.; Jenny, C.; Escalant, J.-V. A review of conventional improvement strategies for Musa. InfoMusa 2004, 13, 2–6. [Google Scholar]
- Ortiz, R. Conventional banana and plantain breeding. Acta Hortic. 2013, 986, 177–194. [Google Scholar] [CrossRef]
- Nyine, M.; Uwimana, B.; Swennen, R.; Batte, M.; Brown, A.; Christelová, P.; Hřibová, E.; Lorenzen, J.; Doležel, J. Trait variation and genetic diversity in a banana genomic selection training population. PLoS ONE 2017, 12, e0178734. [Google Scholar] [CrossRef]
- Amorim, E.P.; Silva, S.O.; Amorim, V.B.O.; Pillay, M. Quality improvement of cultivated Musa. In Banana Breeding: Progress and Challenges; Pillay, M., Tenkouano, A., Eds.; CRC Press: New York, NY, USA, 2011; pp. 252–280. [Google Scholar]
- Amorim, E.P.; dos Santos-Serejo, J.A.; Amorim, V.B.O.; Ferreira, C.F.; Silva, S.O. Banana breeding at embrapa cassava and fruits. Acta Hortic. 2013, 968, 171–176. [Google Scholar] [CrossRef]
- Tenkouano, A.; Vuylsteke, D.; Agogbua, J.U.; Makumbi, D.; Swennen, R.; Ortiz, R. Diploid banana hybrids TMB2x5105-1 and TMB2x9128-3 with good combining ability, resistance to black sigatoga and nematodes. HortScience 2003, 38, 468–472. [Google Scholar] [CrossRef]
- Šimoníková, D.; Němečková, A.; Karafiátová, M.; Uwimana, B.; Swennen, R.; Doležel, J.; Hřibová, E. Chromosome painting facilitates anchoring reference genome sequence to chromosomes in situ and integrated karyotyping in banana (Musa spp.). Front. Plant. Sci. 2019, 10, 1503. [Google Scholar] [CrossRef]
- De Langhe, E.; Perrier, X.; Donohue, M.; Denham, T. The original Banana Split: Multi-disciplinary implications of the generation of African and Pacific Plantains in Island Southeast Asia. Ethnobot. Res. Appl. 2015, 14, 299–312. [Google Scholar] [CrossRef]
- Do Amaral, C.M.; de Almeida dos Santos-Serejo, J.; de Oliveira e Silva, S.; da Silva Ledo, C.A.; Amorim, E.P. Agronomic characterization of autotetraploid banana plants derived from ‘Pisang Lilin’ (AA) obtained through chromosome doubling. Euphytica 2015, 202, 435–443. [Google Scholar] [CrossRef]
- Robertson, W.R.B. Chromosome studies. I. Taxonomic relationships shown in the chromosomes of Tettigidae and Acrididae: V-shaped chromosomes and their significance in Acrididae, Locustidae, and Gryllidae: Chromosomes and variation. J. Morphol. 1916, 27, 179–331. [Google Scholar] [CrossRef]
- Schubert, I.; Fransz, P.F.; Fuchs, J.; De Jong, J.H. Chromosome painting in plants. Meth. Cell Sci. 2001, 23, 57–69. [Google Scholar] [CrossRef]
- Jiang, J. Fluorescence in situ hybridization in plants: Recent developments and future applications. Chromosome Res. 2019, 27, 153–165. [Google Scholar] [CrossRef]
- Lysák, M.A.; Fransz, P.F.; Ali, H.B.M.; Schubert, I. Chromosome painting in A. thaliana. Plant. J. 2001, 28, 689–697. [Google Scholar] [CrossRef]
- Idziak, D.; Hazuka, I.; Poliwczak, B.; Wiszynska, A.; Wolny, E.; Hasterok, R. Insight into the karyotype evolution of Brachypodium species using comparative chromosome barcoding. PLoS ONE 2014, 9, e93503. [Google Scholar] [CrossRef]
- Han, Y.; Zhang, T.; Thammapichai, P.; Weng, Y.; Jiang, J. Chromosome-specific painting in Cucumis species using bulked oligonucleotides. Genetics 2015, 200, 771–779. [Google Scholar] [CrossRef]
- Qu, M.; Li, K.; Han, Y.; Chen, L.; Li, Z.; Han, Y. Integrated karyotyping of woodland strawberry (Fragaria vesca) with oligopaint FISH probes. Cytogenet. Genome Res. 2017, 153, 158–164. [Google Scholar] [CrossRef]
- Braz, G.T.; He, L.; Zhao, H.; Zhang, T.; Semrau, K.; Rouillard, J.M.; Torres, G.A.; Jiang, J. Comparative oligo-FISH mapping: An efficient and powerful methodology to reveal karyotypic and chromosomal evolution. Genetics 2018, 208, 513–523. [Google Scholar] [CrossRef]
- He, L.; Braz, G.T.; Torres, G.A.; Jiang, J.M. Chromosome painting in meiosis reveals pairing of specific chromosomes in polyploid Solanum species. Chromosoma 2018, 127, 505–513. [Google Scholar] [CrossRef]
- Machado, M.A.; Pieczarka, J.C.; Silva, F.H.R.; O’Brien, P.C.M.; Ferguson-Smith, M.A.; Nagamachi, C.Y. Extensive karyotype reorganization in the fish Gymnotus arapaima (Gymnotiformes, Gymnotidae) highlighted by zoo-FISH analysis. Front. Genet. 2018, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Albert, P.S.; Zhang, T.; Semrau, K.; Rouillard, J.M.; Kao, Y.H.; Wang, C.J.R.; Danilova, T.V.; Jiang, J.; Birchler, J.A. Whole-chromosome paints in maize reveal rearrangements, nuclear domains, and chromosomal relationships. Proc. Natl. Acad. Sci. USA 2019, 116, 1679–1685. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Zhao, Q.; Yan, W.; Li, M.; Liu, Y.; Cheng, C.; Zhang, L.; Yu, X.; Li, J.; Qian, C.; et al. Flexible chromosome painting based on multiplex PCR of oligonucleotides and its application for comparative chromosome analyses in Cucumis. Plant. J. 2020, 102, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, K. Cytogenetics of the Genus Musa; International Network for the Improvement of Banana and Plantain: Montpellier, France, 1999; ISBN 2-910810-25-9. [Google Scholar]
- Fauré, S.; Noyer, J.L.; Horry, J.P.; Bakry, F.; Lanaud, C.; de León, D.G. A molecular marker-based linkage map of diploid bananas (Musa acuminata). Theor. Appl. Genet. 1993, 87, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Hippolyte, I.; Bakry, F.; Seguin, M.; Gardes, L.; Rivallan, R.; Risterucci, A.M.; Jenny, C.; Perrier, X.; Carreel, F.; Argout, X.; et al. A saturated SSR/DArT linkage map of Musa acuminata addressing genome rearrangements among bananas. BMC Plant. Biol. 2010, 10, 65. [Google Scholar] [CrossRef] [PubMed]
- Mbanjo, E.G.N.; Tchoumbougnang, F.; Mouelle, A.S.; Oben, J.E.; Nyine, M.; Dochez, C.; Ferguson, M.E.; Lorenzen, J. Molecular marker-based genetic linkage map of a diploid banana population (Musa acuminata Colla). Euphytica 2012, 188, 369–386. [Google Scholar] [CrossRef]
- Noumbissié, G.B.; Chabannes, M.; Bakry, F.; Ricci, S.; Cardi, C.; Njembele, J.C.; Yohoume, D.; Tomekpe, K.; Iskra-Caruana, M.L.; D’Hont, A.; et al. Chromosome segregation in an allotetraploid banana hybrid (AAAB) suggests a translocation between the A and B genomes and results in eBSV-free offsprings. Mol. Breed. 2016, 36, 38–52. [Google Scholar] [CrossRef]
- Carreel, F.; Gonzalez de Leon, D.; Lagoda, P.; Lanaud, C.; Jenny, C.; Horry, J.; Tezenas du Montcel, H. Ascertaining maternal and paternal lineage within Musa by chloroplast and mitochondrial DNA RFLP analyses. Genome 2002, 45, 679–692. [Google Scholar] [CrossRef]
- Hřibová, E.; Čížková, J.; Christelová, P.; Taudien, S.; De Langhe, E.; Doležel, J. The ITS-5.8S-ITS2 sequence region in the Musaceae: Structure, diversity and use in molecular phylogeny. PLoS ONE 2011, 6, e17863. [Google Scholar] [CrossRef]
- Dupouy, M.; Baurens, F.C.; Derouault, P.; Hervouet, C.; Cardi, C.; Cruaud, C.; Istace, B.; Labadie, K.; Guiougou, C.; Toubi, L.; et al. Two large reciprocal translocations characterized in the disease resistance-rich burmannica genetic group of Musa acuminata. Ann. Bot. 2019, 124, 31–329. [Google Scholar] [CrossRef]
- Martin, G.; Baurens, F.C.; Droc, G.; Rouard, M.; Cenci, A.; Kilian, A.; Hastie, A.; Doležel, J.; Aury, J.M.; Alberti, A.; et al. Improvement of the banana “Musa acuminata” reference sequence using NGS data and semi-automated bioinformatics methods. BMC Genomics 2016, 17, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Horry, J.P. Chimiotaxonomie et organisation génétique dans le genre Musa (III). Fruits 1989, 44, 573–578. [Google Scholar]
- Lebot, V.; Aradhya, M.K.; Manshardt, R.M.; Meilleur, B.A. Genetic relationships among cultivated bananas and plantains from Asia and the Pacific. Euphytica 1993, 67, 163–175. [Google Scholar] [CrossRef]
- Hippolyte, I.; Jenny, C.; Gardes, L.; Bakry, F.; Rivallan, R.; Pomies, V.; Cubry, P.; Tomekpe, K.; Risterucci, A.M.; Roux, N.; et al. Foundation characteristics of edible Musa triploids revealed from allelic distribution of SSR markers. Ann. Bot. 2012, 109, 937–951. [Google Scholar] [CrossRef]
- Li, L.F.; Wang, H.Y.; Zhang, C.; Wang, X.F.; Shi, F.X.; Chen, W.N.; Ge, X.J. Origins and domestication of cultivated banana inferred from chloroplast and nuclear genes. PLoS ONE 2013, 8, e80502. [Google Scholar] [CrossRef]
- Sandoval, J.A.; Côte, F.X.; Escoute, J. Chromosome number variations in micropropagated true-to-type and off-type banana plants (Musa AAA Grande Naine cv.). In Vitro Cell. Dev. Biol. Plant 1996, 32, 14–17. [Google Scholar] [CrossRef]
- Shepherd, K.; Da Silva, K.M. Mitotic instability in banana varieties. Aberrations in conventional triploid plants. Fruits 1996, 51, 99–103. [Google Scholar]
- Bartoš, J.; Alkhimova, O.; Doleželová, M.; De Langhe, E.; Doležel, J. Nuclear genome size and genomic distribution of ribosomal DNA in Musa and Ensete (Musaceae): Taxonomic implications. Cytogenet. Genome Res. 2005, 109, 50–57. [Google Scholar] [CrossRef]
- Čížková, J.; Hřibová, E.; Christelová, P.; van den Houwe, I.; Häkkinen, M.; Roux, N.; Swennen, R.; Doležel, J. Molecular and cytogenetic characterization of wild Musa species. PLoS ONE 2015, 10, e0134096. [Google Scholar] [CrossRef]
- Roux, N.; Toloza, A.; Radecki, Z.; Zapata-Arias, F.J.; Doležel, J. Rapid detection of aneuploidy in Musa using flow cytometry. Plant. Cell Rep. 2003, 21, 483–490. [Google Scholar] [CrossRef]
- Sedlazeck, F.J.; Rescheneder, P.; Smolka, M.; Fang, H.; Nattestad, M.; von Haeseler, A.; Schatz, M.C. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods 2018, 15, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Liang, F.; Cheng, D.; Zhang, Z.; Yu, G.; Zha, J.; Wang, Y.; Xia, Q.; Yuan, D.; Tan, Y.; et al. Location of balanced chromosome-translocation breakpoints by long-read sequencing on the Oxford Nanopore platform. Front. Genet. 2020, 10, 1330. [Google Scholar] [CrossRef] [PubMed]
- Soto, D.C.; Shew, C.; Mastoras, M.; Schmidt, J.M.; Sahasrabudhe, R.; Kaya, G.; Andrés, A.M.; Dennis, M.Y. Identification of structural variations in chimpanzees using optical mapping and nanopore sequencing. Genes 2020, 11, 276. [Google Scholar] [CrossRef] [PubMed]
- Perrier, X.; Jacquemoud-Collet, J.P. DARwin Software. 2006. Available online: http://darwin.cirad.fr/ (accessed on 20 May 2020).
- FigTree v1.4.0. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 20 May 2020).
- D’Hont, A.; Denoeud, F.; Aury, J.M.; Baurens, F.C.; Carreel, F.; Garsmeur, O.; Noel, B.; Bocs, S.; Droc, G.; Rouard, M.; et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 2012, 488, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Doležel, J.; Doleželová, M.; Roux, N.; van den Houwe, I. A novel method to prepare slides for high resolution chromosome studies in Musa spp. Infomusa 1998, 7, 3–4. [Google Scholar]
| Species | Subspecies/Subgroup | Accession Name | ITC Code a | Genomic Constitution | Chromosome Number (2n) |
|---|---|---|---|---|---|
| M. acuminata | banksii | Banksii | 0806 | AA | 22 |
| microcarpa | Borneo | 0253 | AA | 22 | |
| zebrina | Maia Oa | 0728 | AA | 22 | |
| burmannica | Tavoy | 0072 | AA | 22 | |
| burmannicoides | Calcutta 4 | 0249 | AA | 22 | |
| siamea | Pa Rayong | 0672 | AA | 22 | |
| M. balbisiana | - | Pisang Klutuk Wulung | - | BB | 22 |
| Cultivars | unknown | Pisang Lilin | - | AA | 22 |
| Mchare | Huti White | - | AA | 22 | |
| Mchare | Huti (Shumba nyeelu) | 1452 | AA | 22 | |
| Mchare | Ndyali | 1552 | AA | 22 | |
| Cavendish | Poyo | 1482 | AAA | 33 | |
| Gros Michel | Gros Michel | 0484 | AAA | 33 | |
| Mutika/Lujugira | Imbogo | 0168 | AAA | 32* | |
| Mutika/Lujugira | Kagera | 0141 | AAA | 33 | |
| Plantain (Horn) | 3 Hands Planty | 1132 | AAB | 33 | |
| Plantain (French) | Amou | 0963 | AAB | 32 * | |
| Plantain (French) | Obino l’Ewai | 0109 | AAB | 33 | |
| Pelipita | Pelipita | 0472 | ABB | 33 | |
| Saba | Saba sa Hapon | 1777 | ABB | 33 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Šimoníková, D.; Němečková, A.; Čížková, J.; Brown, A.; Swennen, R.; Doležel, J.; Hřibová, E. Chromosome Painting in Cultivated Bananas and Their Wild Relatives (Musa spp.) Reveals Differences in Chromosome Structure. Int. J. Mol. Sci. 2020, 21, 7915. https://doi.org/10.3390/ijms21217915
Šimoníková D, Němečková A, Čížková J, Brown A, Swennen R, Doležel J, Hřibová E. Chromosome Painting in Cultivated Bananas and Their Wild Relatives (Musa spp.) Reveals Differences in Chromosome Structure. International Journal of Molecular Sciences. 2020; 21(21):7915. https://doi.org/10.3390/ijms21217915
Chicago/Turabian StyleŠimoníková, Denisa, Alžběta Němečková, Jana Čížková, Allan Brown, Rony Swennen, Jaroslav Doležel, and Eva Hřibová. 2020. "Chromosome Painting in Cultivated Bananas and Their Wild Relatives (Musa spp.) Reveals Differences in Chromosome Structure" International Journal of Molecular Sciences 21, no. 21: 7915. https://doi.org/10.3390/ijms21217915
APA StyleŠimoníková, D., Němečková, A., Čížková, J., Brown, A., Swennen, R., Doležel, J., & Hřibová, E. (2020). Chromosome Painting in Cultivated Bananas and Their Wild Relatives (Musa spp.) Reveals Differences in Chromosome Structure. International Journal of Molecular Sciences, 21(21), 7915. https://doi.org/10.3390/ijms21217915






