Constructing 3-Dimensional Atomic-Resolution Models of Nonsulfated Glycosaminoglycans with Arbitrary Lengths Using Conformations from Molecular Dynamics
Abstract
1. Introduction
2. Materials and Methods
2.1. Molecular Dynamics: System Construction, Energy Minimization, Heating, and Production Simulations
2.2. Conformational Analysis
2.3. Construction Algorithm to Generate GAG Conformational Ensembles
3. Results and Discussion
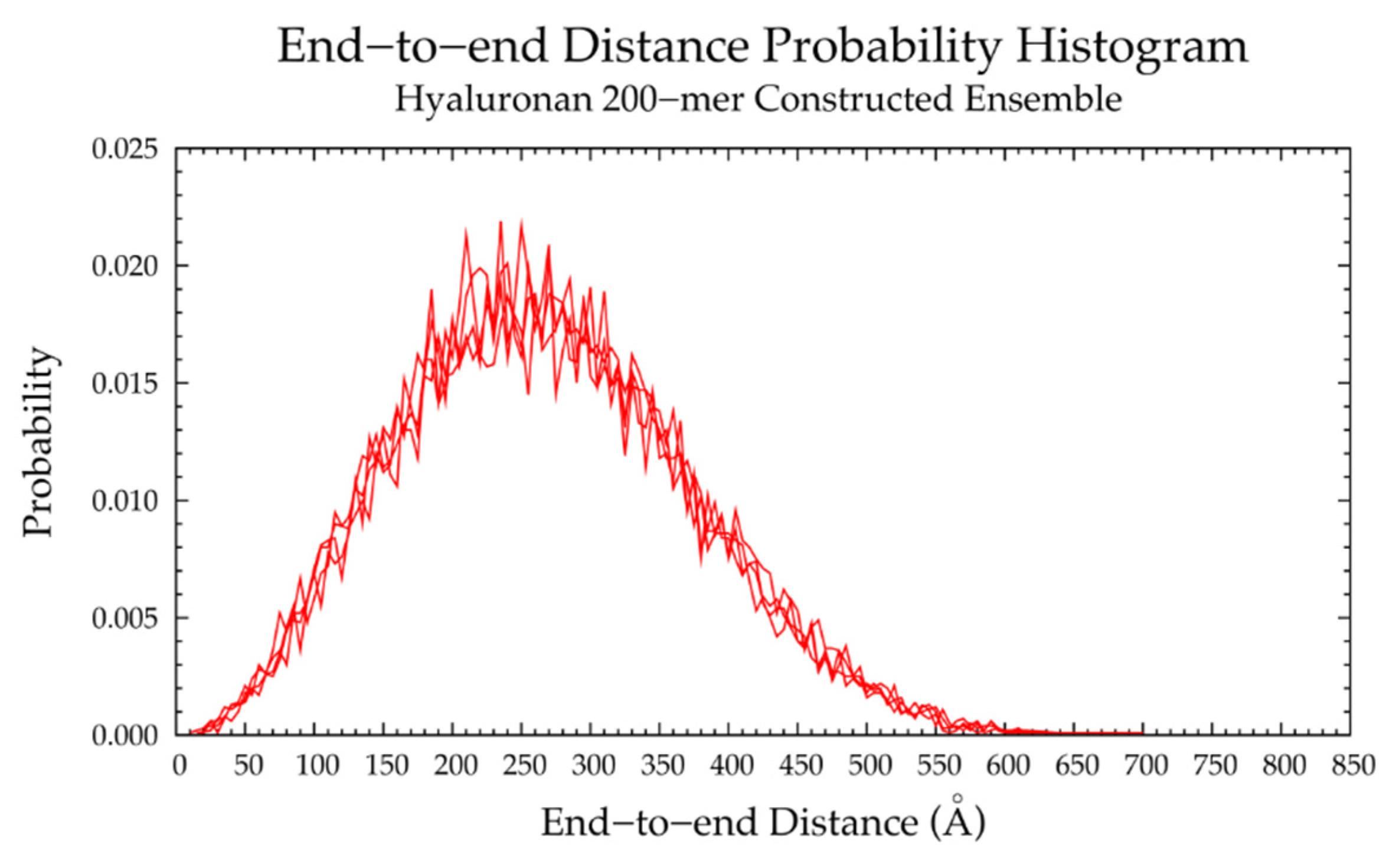
3.1. Hyaluronan
3.1.1. Molecular Dynamics Simulations: Glycosidic Linkage and Monosaccharide Ring Geometry Effects on Polymer Backbone Flexibility
3.1.2. Construction Algorithm
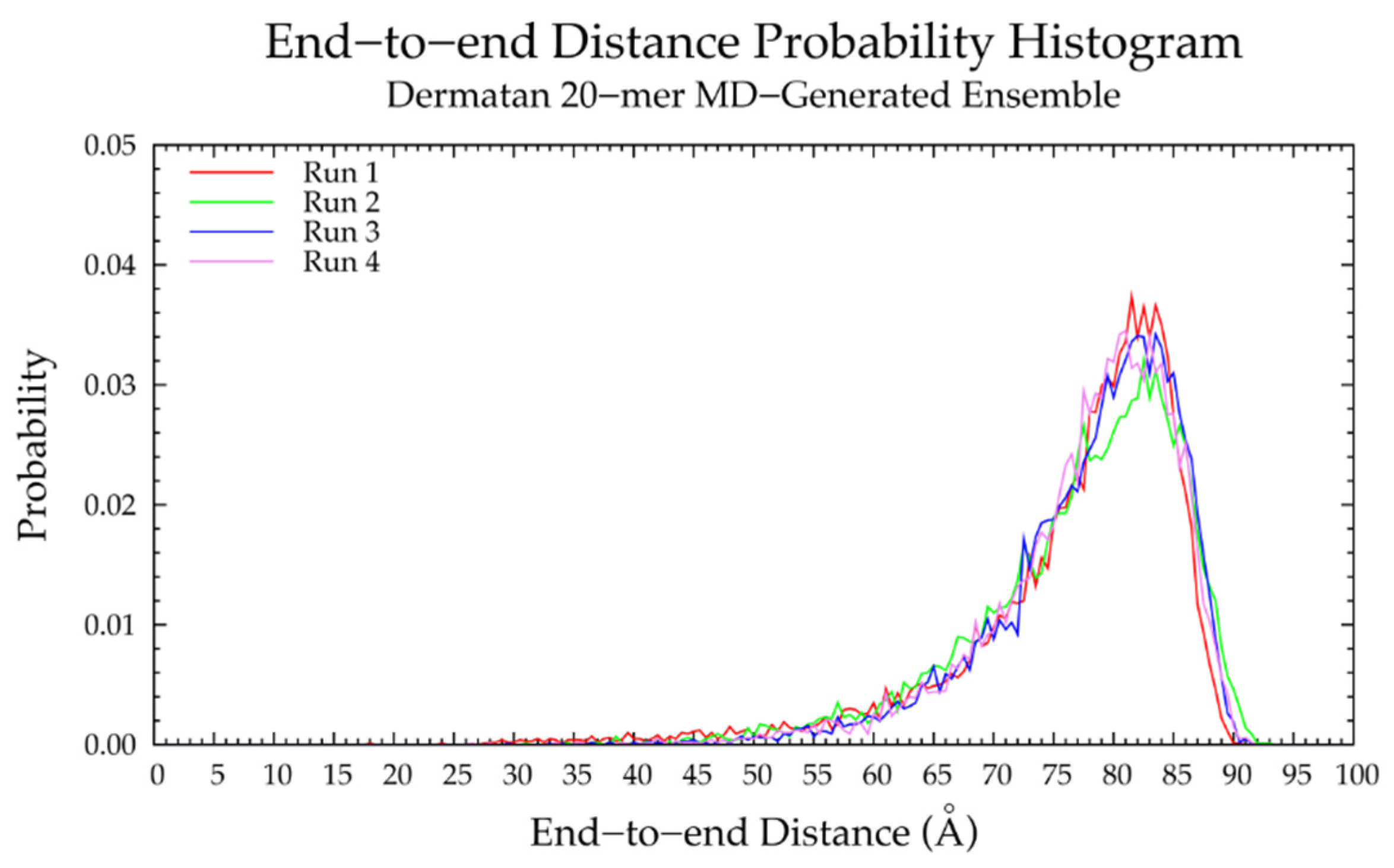
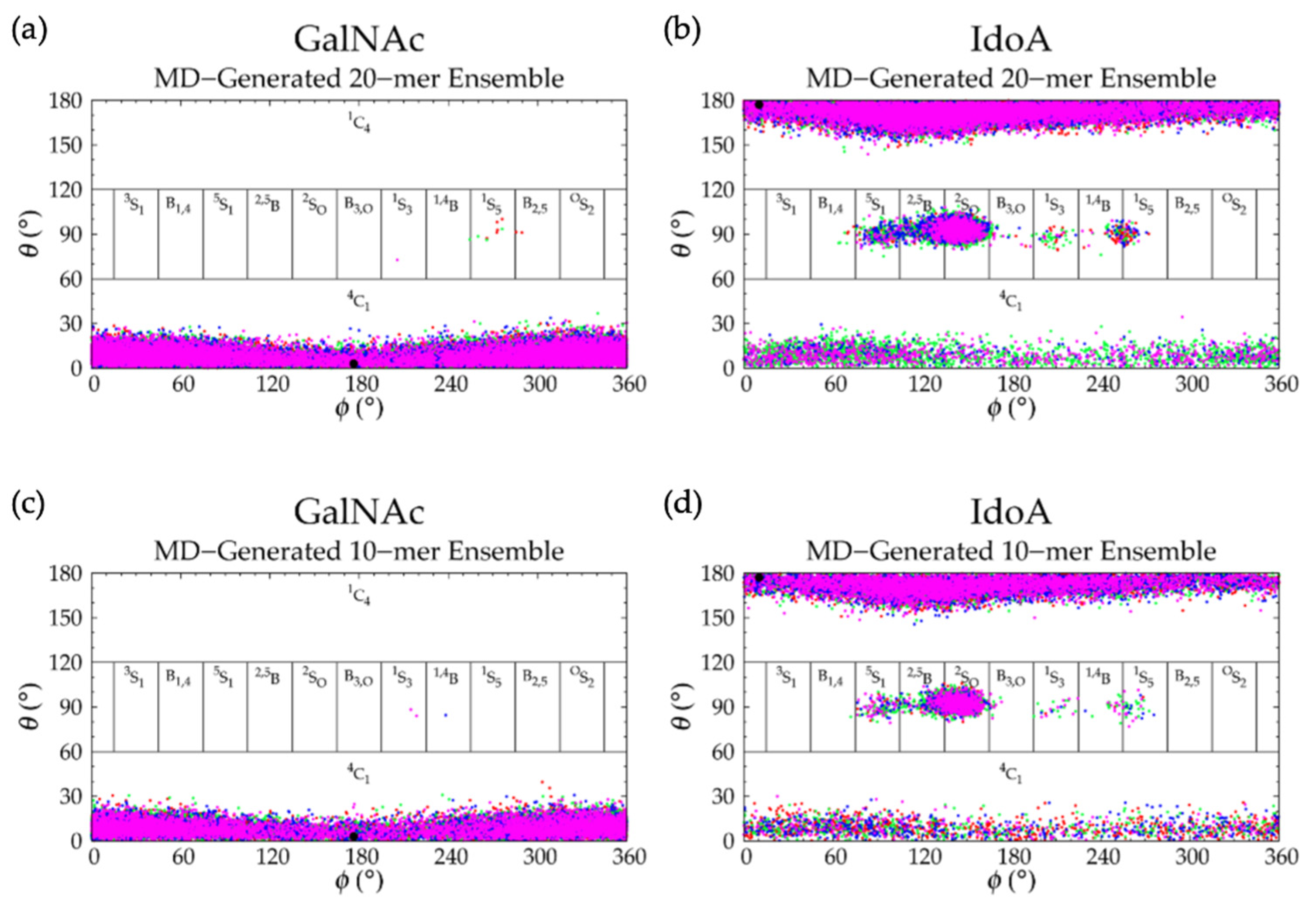
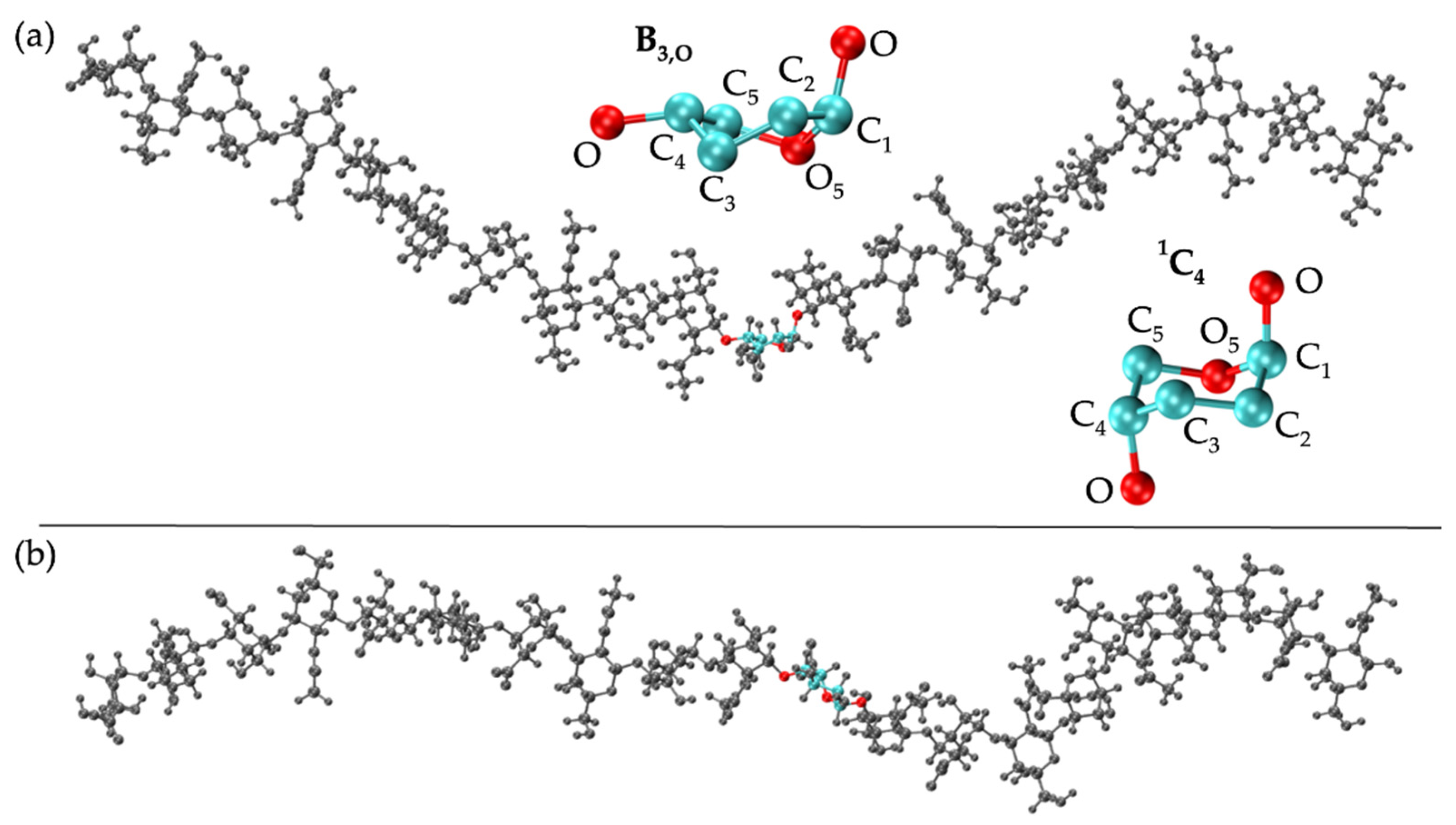
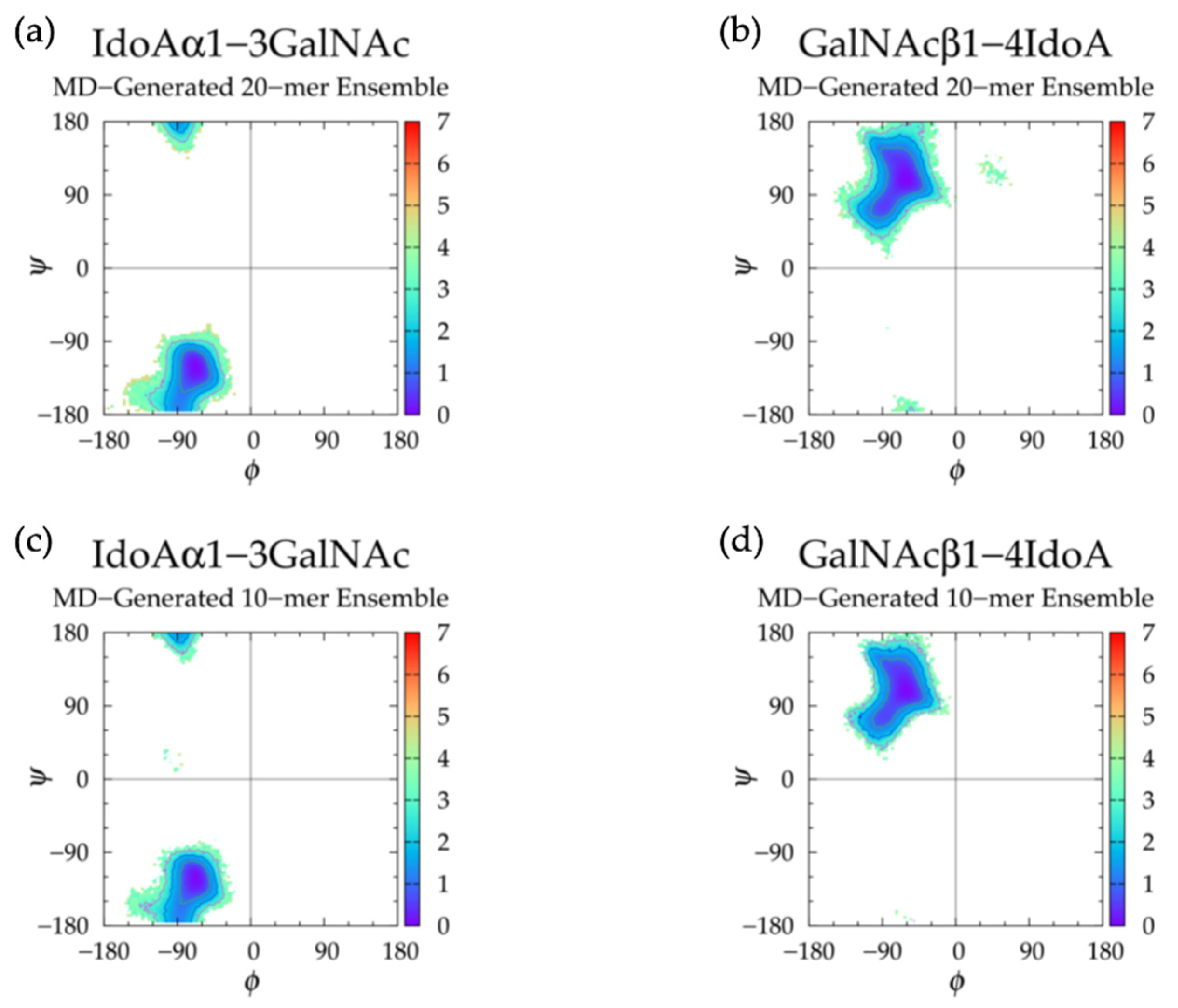
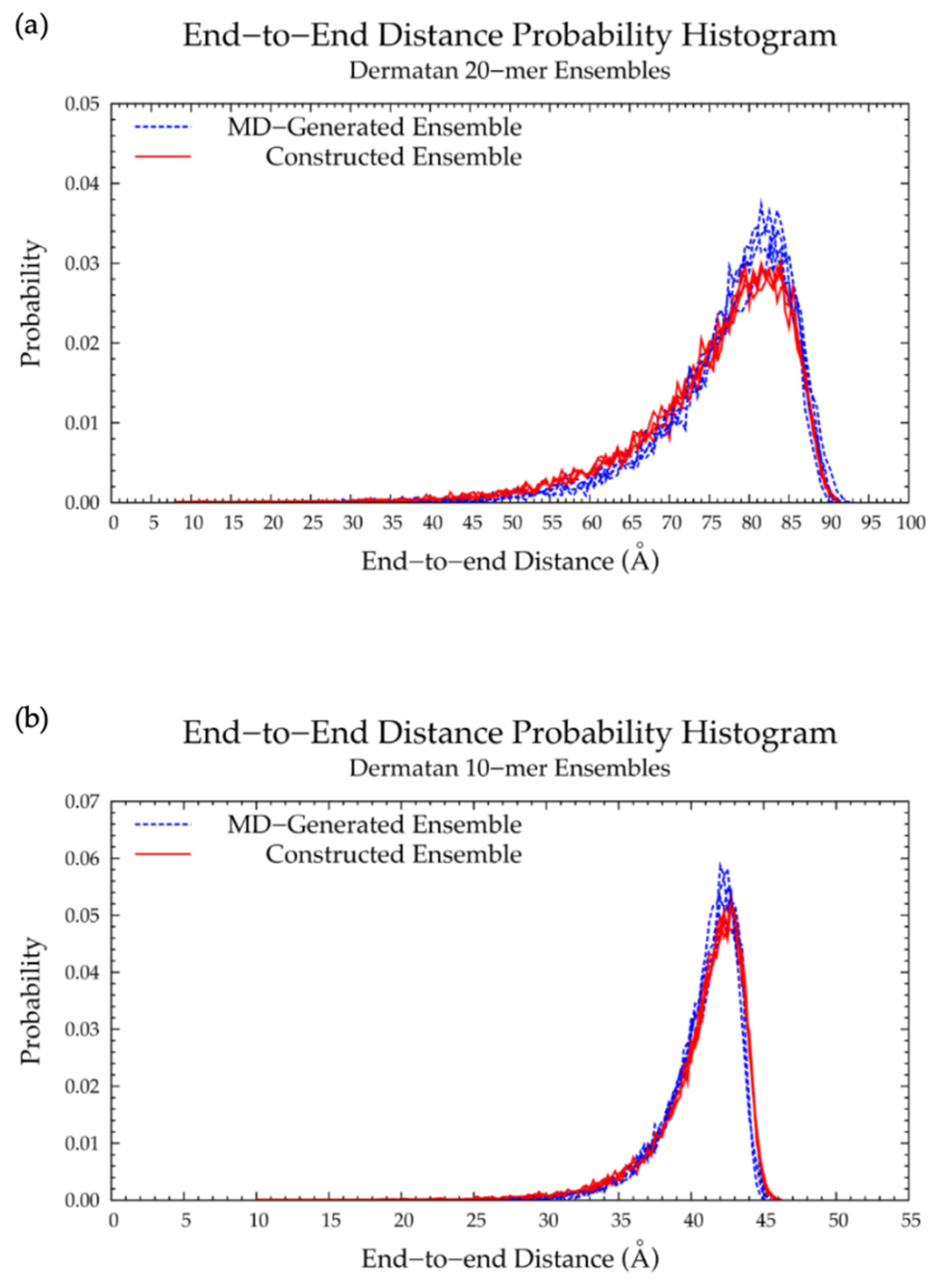
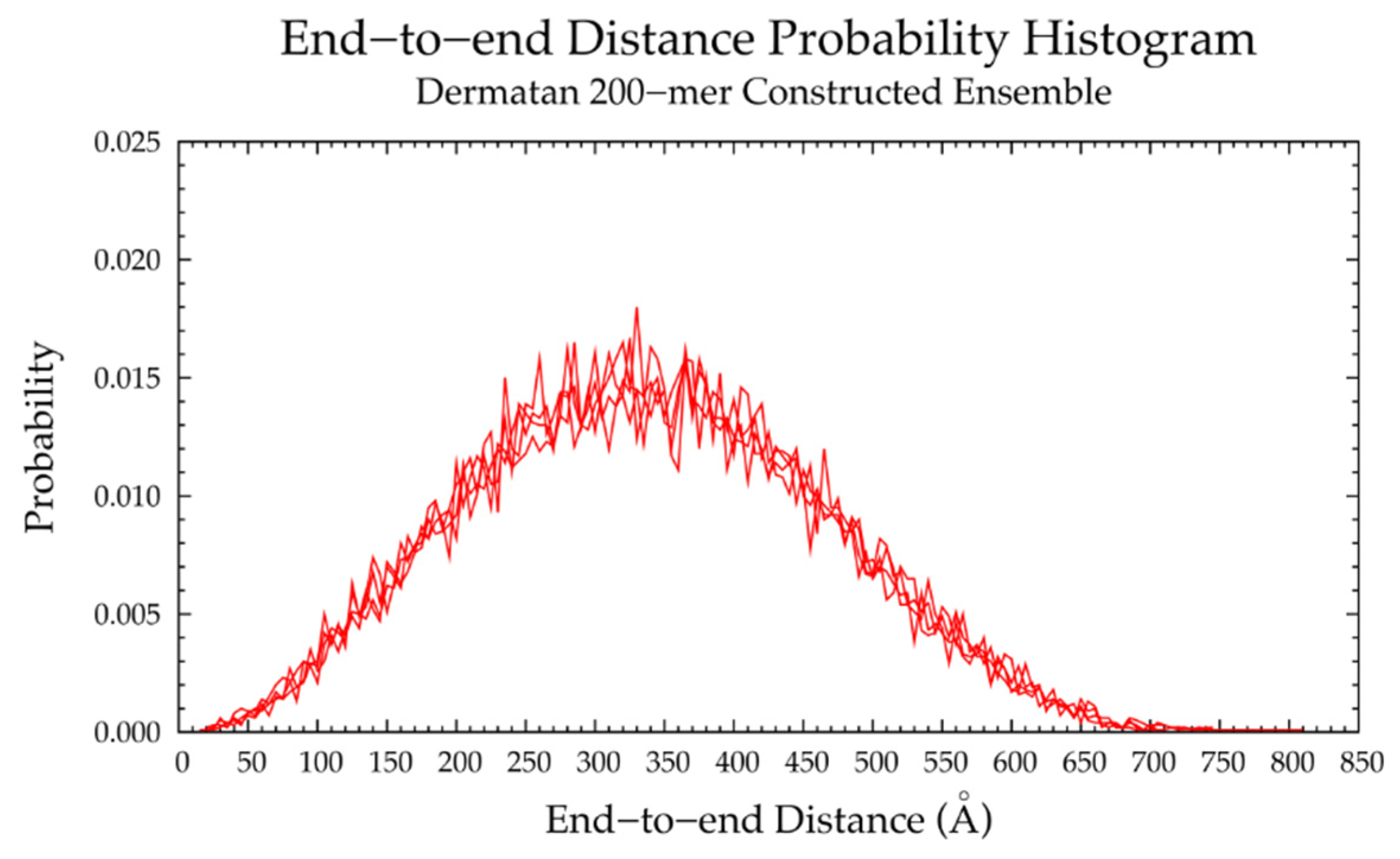
3.2. Nonsulfated Dermatan
3.2.1. Molecular Dynamics Simulations: Glycosidic Linkage and Monosaccharide Ring Geometry Effects on Polymer Backbone Flexibility
3.2.2. Construction Algorithm
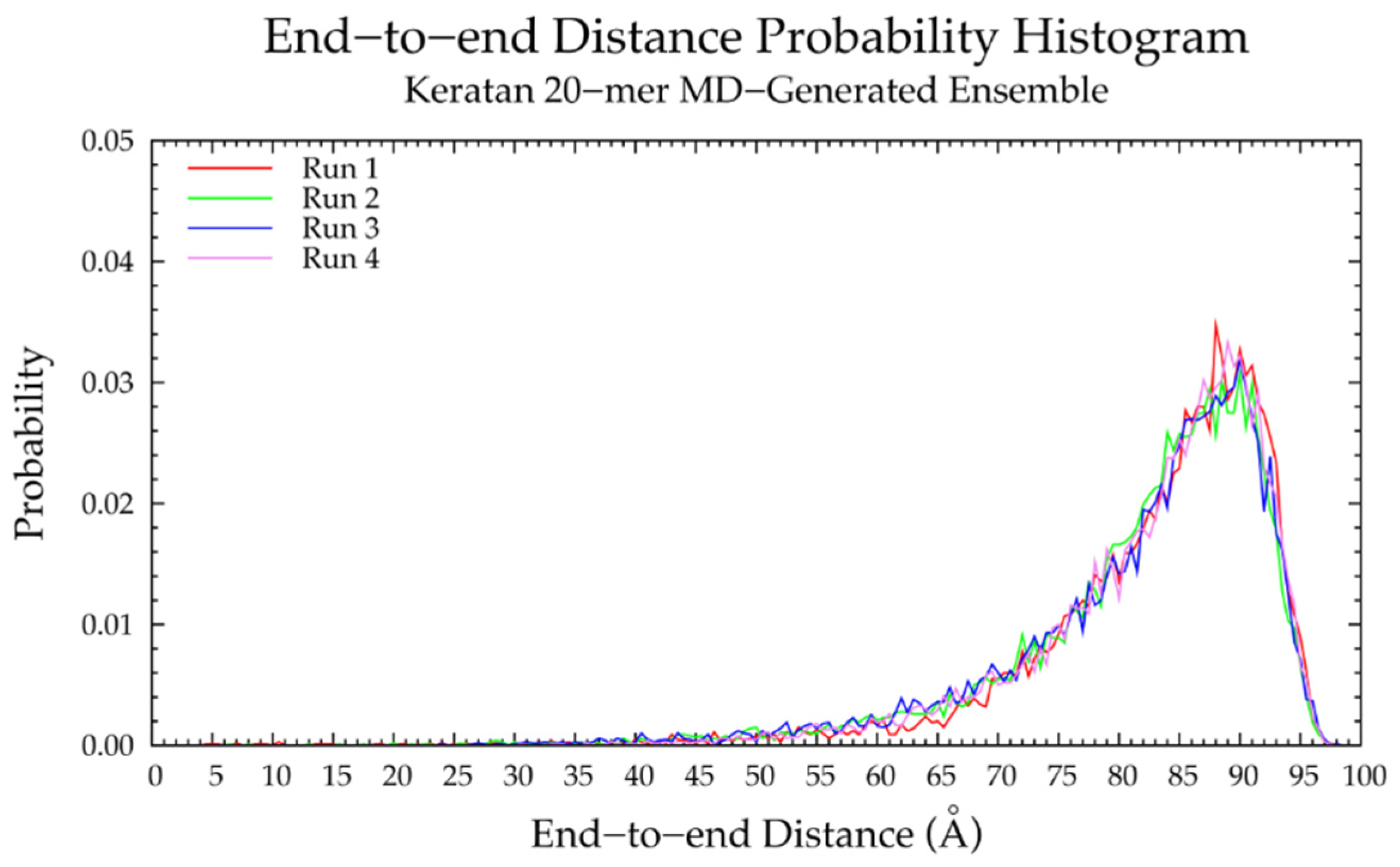
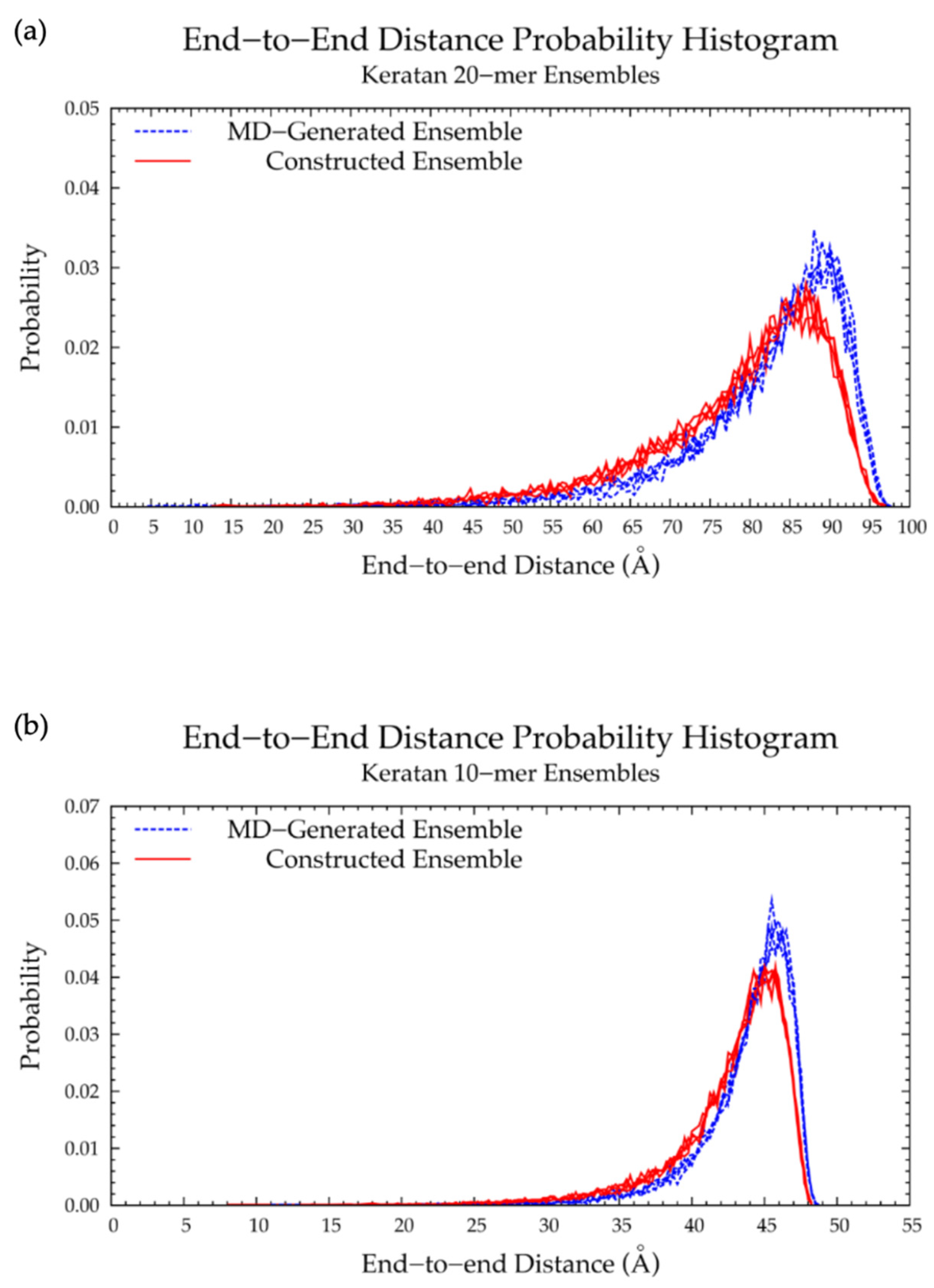
3.3. Nonsulfated Keratan
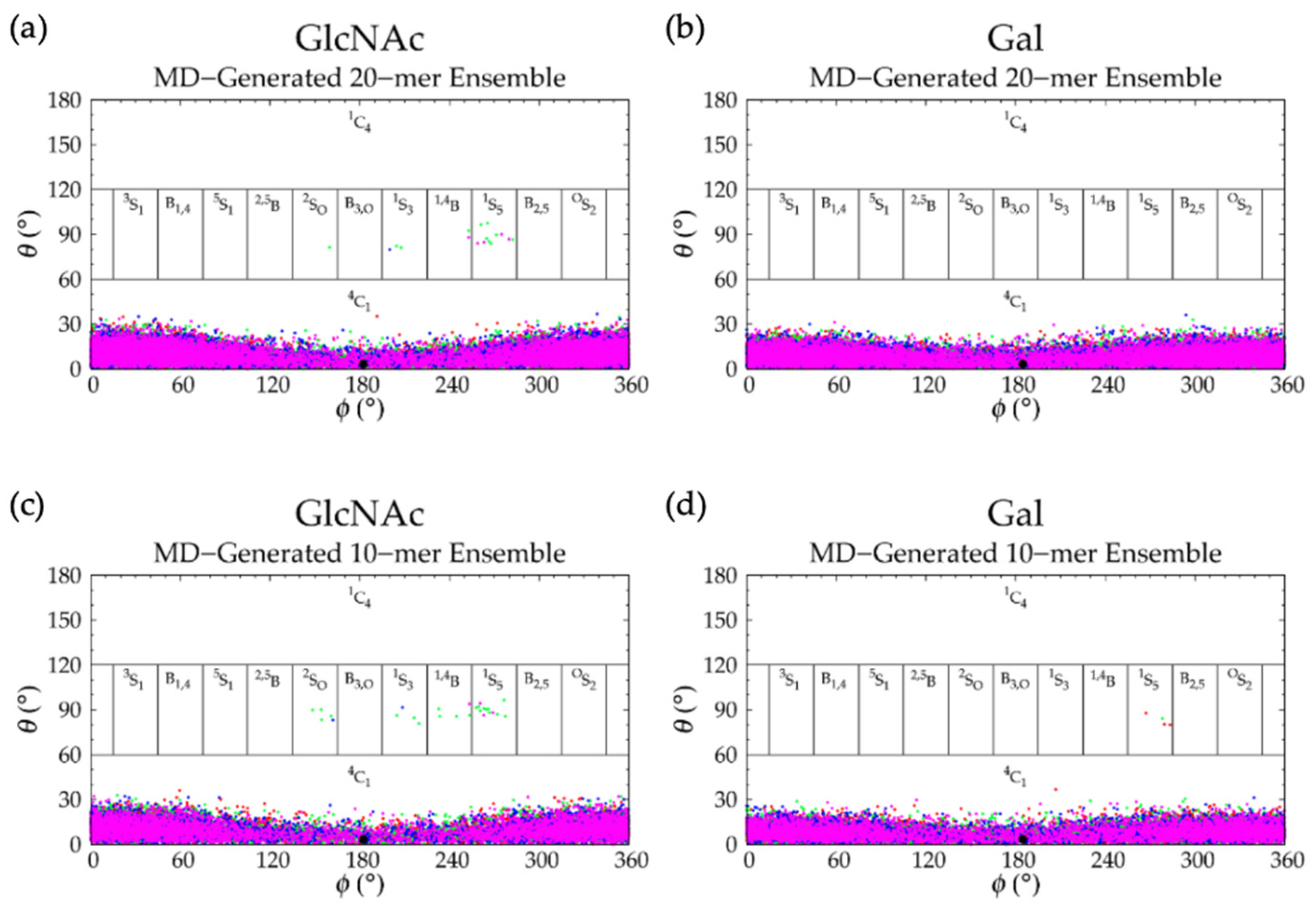
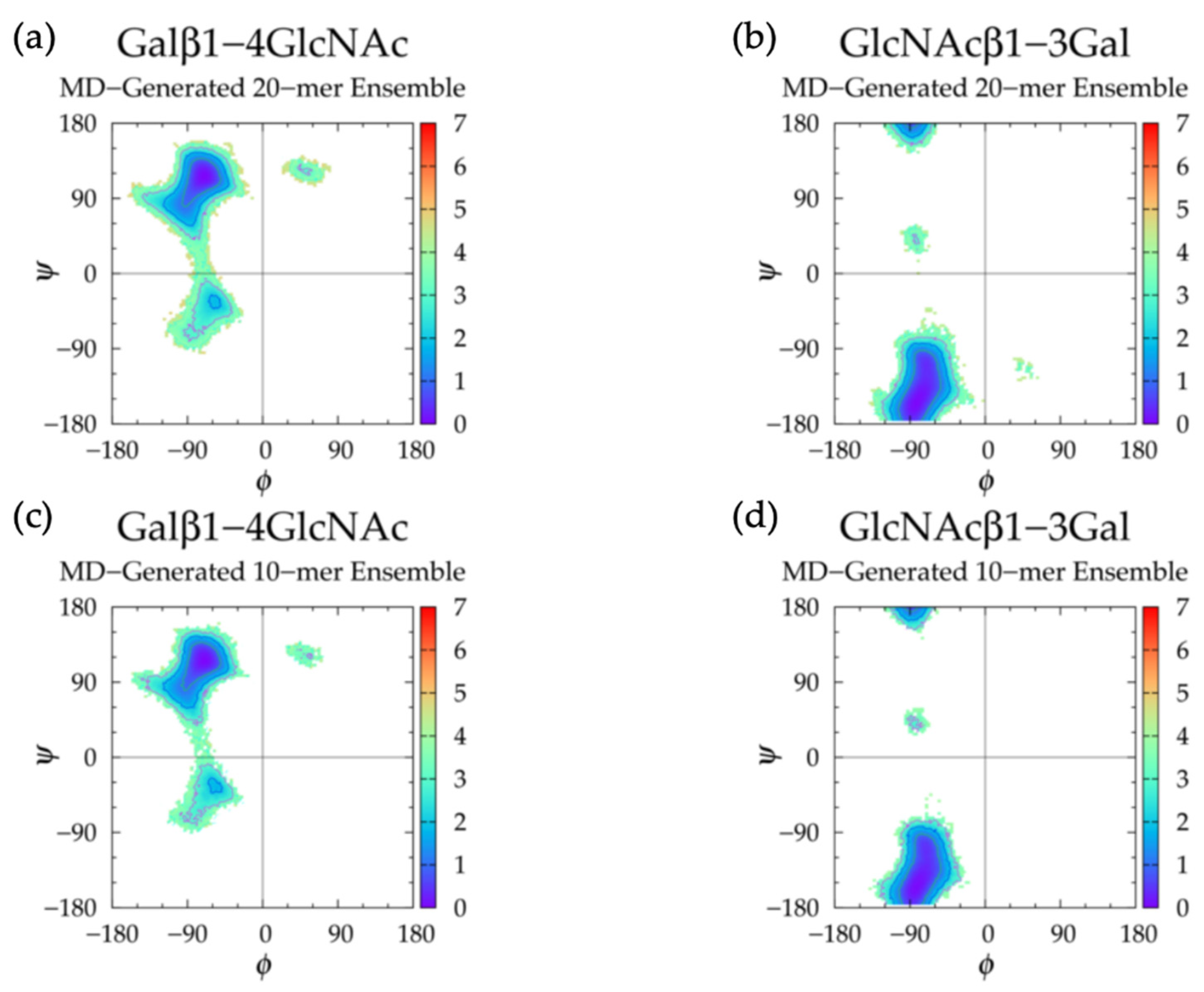
3.3.1. Molecular Dynamics Simulations: Glycosidic Linkage and Monosaccharide Ring Geometry Effects on Polymer Backbone Flexibility
3.3.2. Construction Algorithm
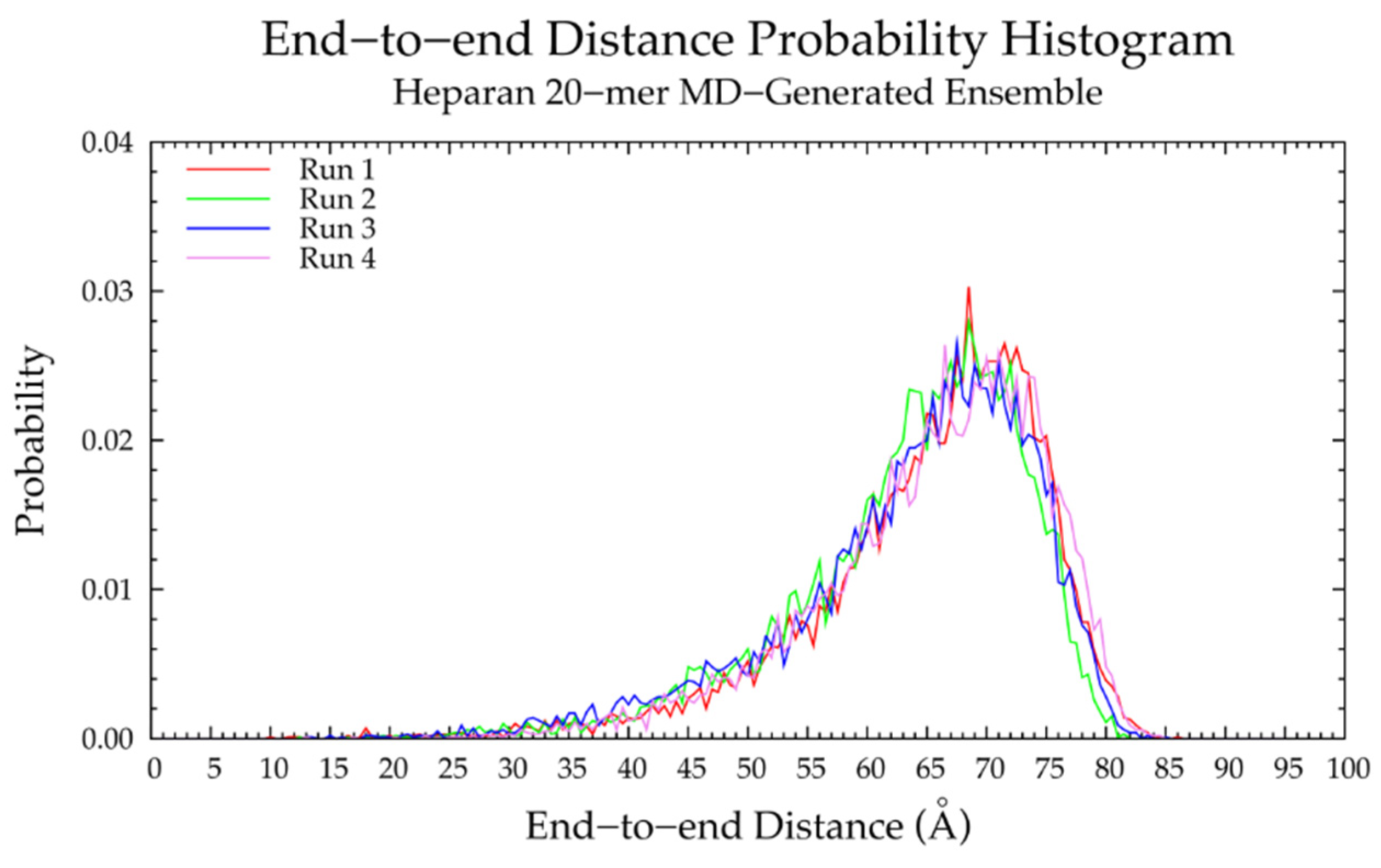
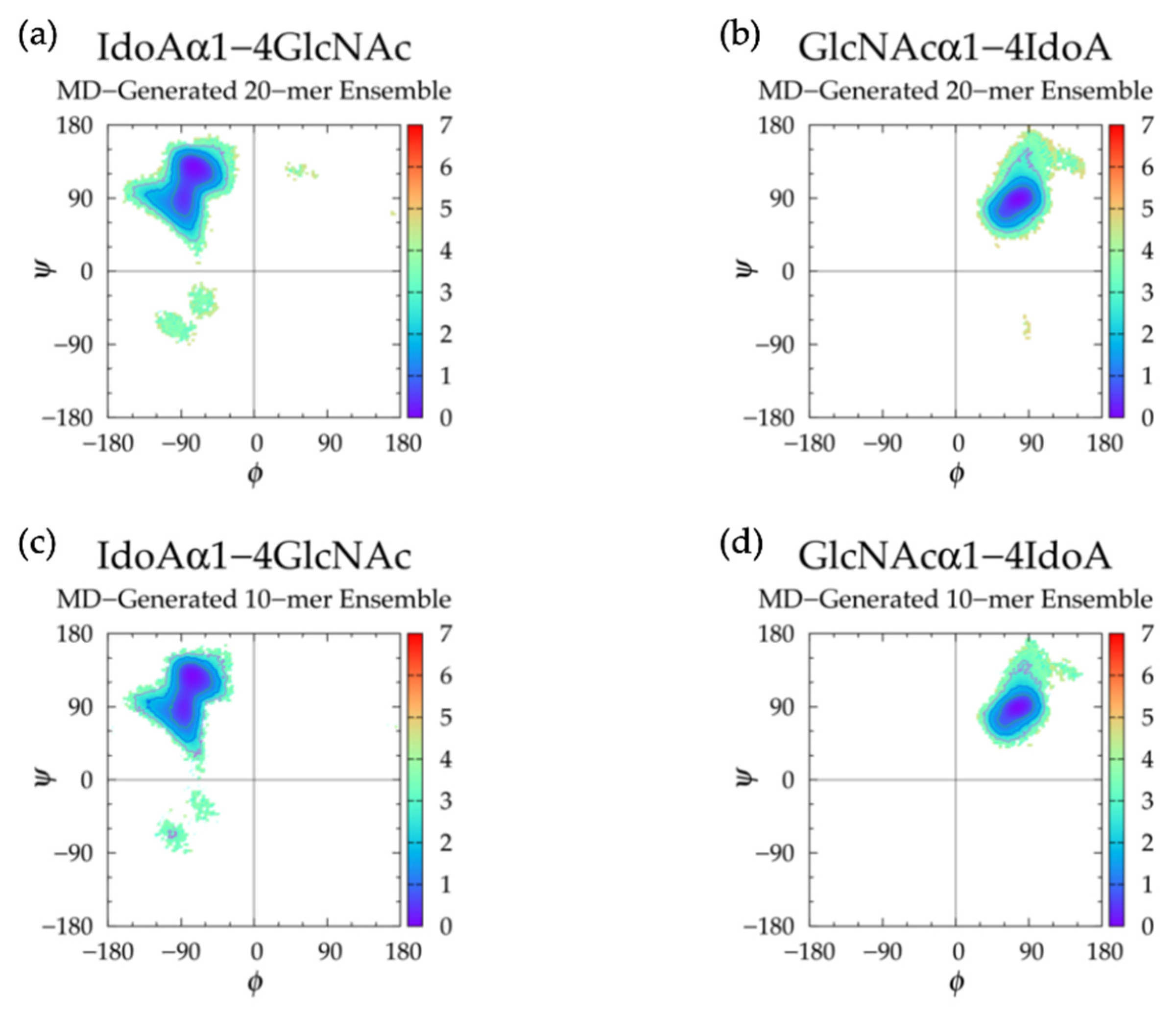
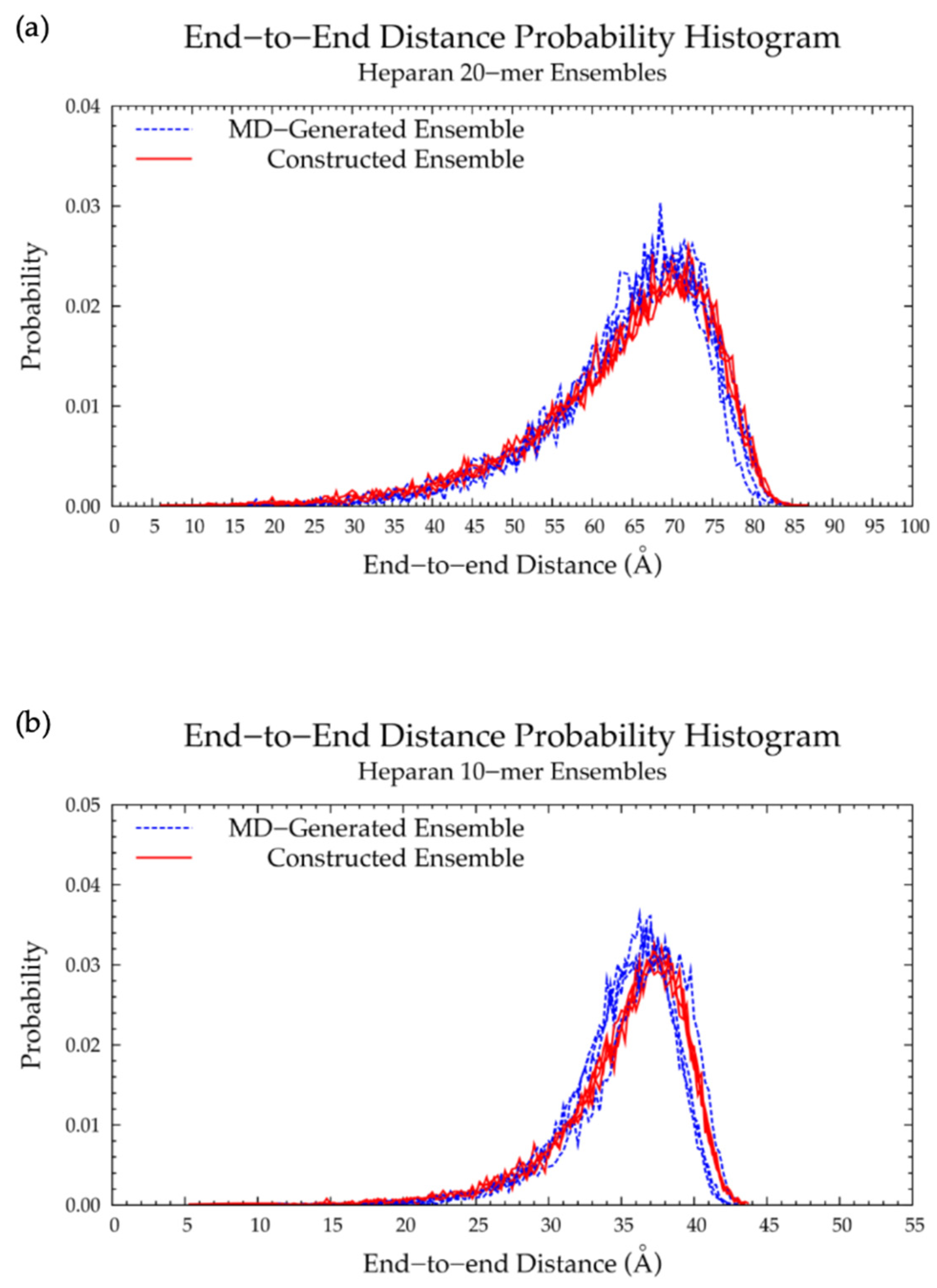
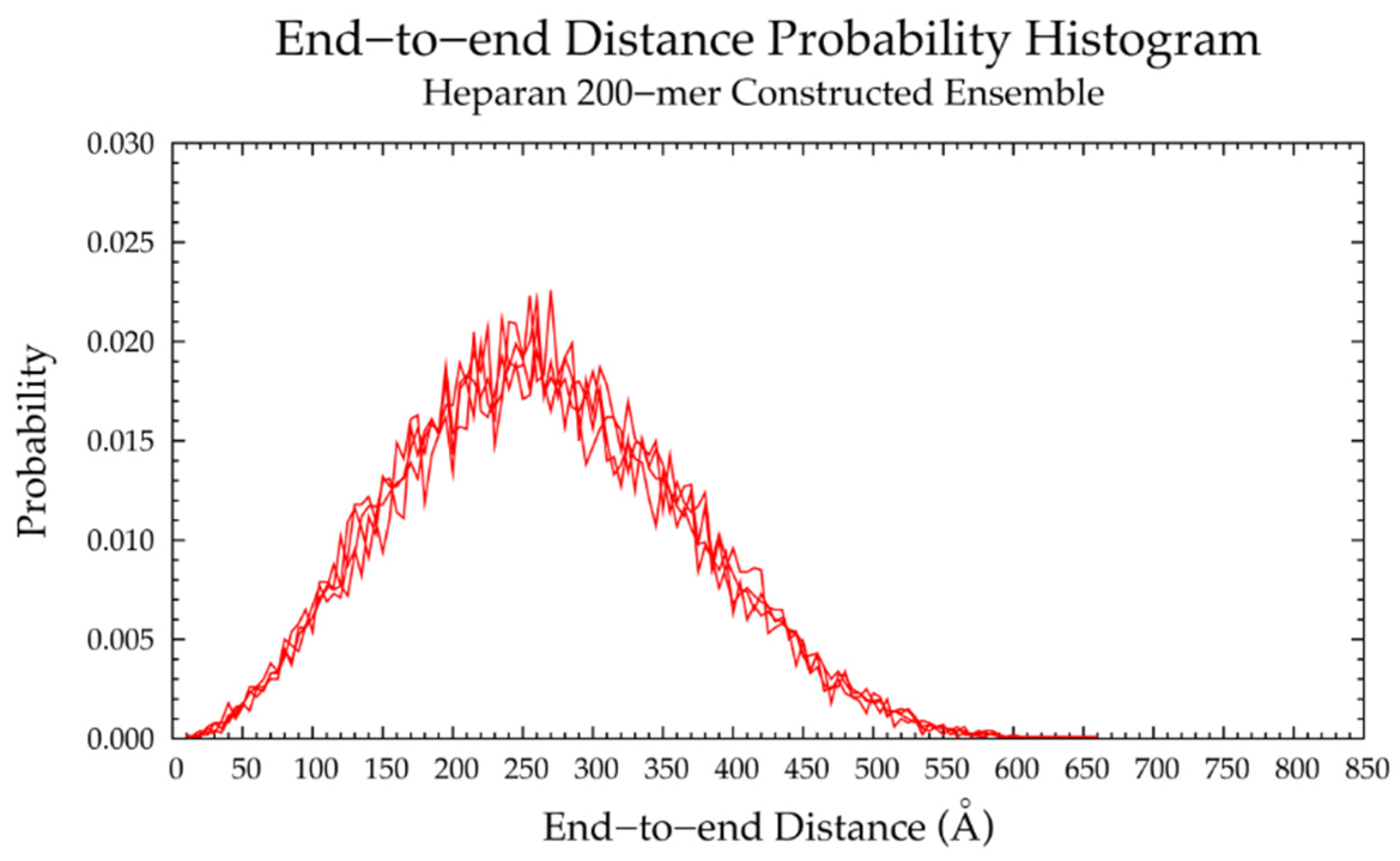
3.4. Nonsulfated Heparan
3.4.1. Molecular Dynamics Simulations: Glycosidic Linkage and Monosaccharide Ring Geometry Effects on Polymer Backbone Flexibility
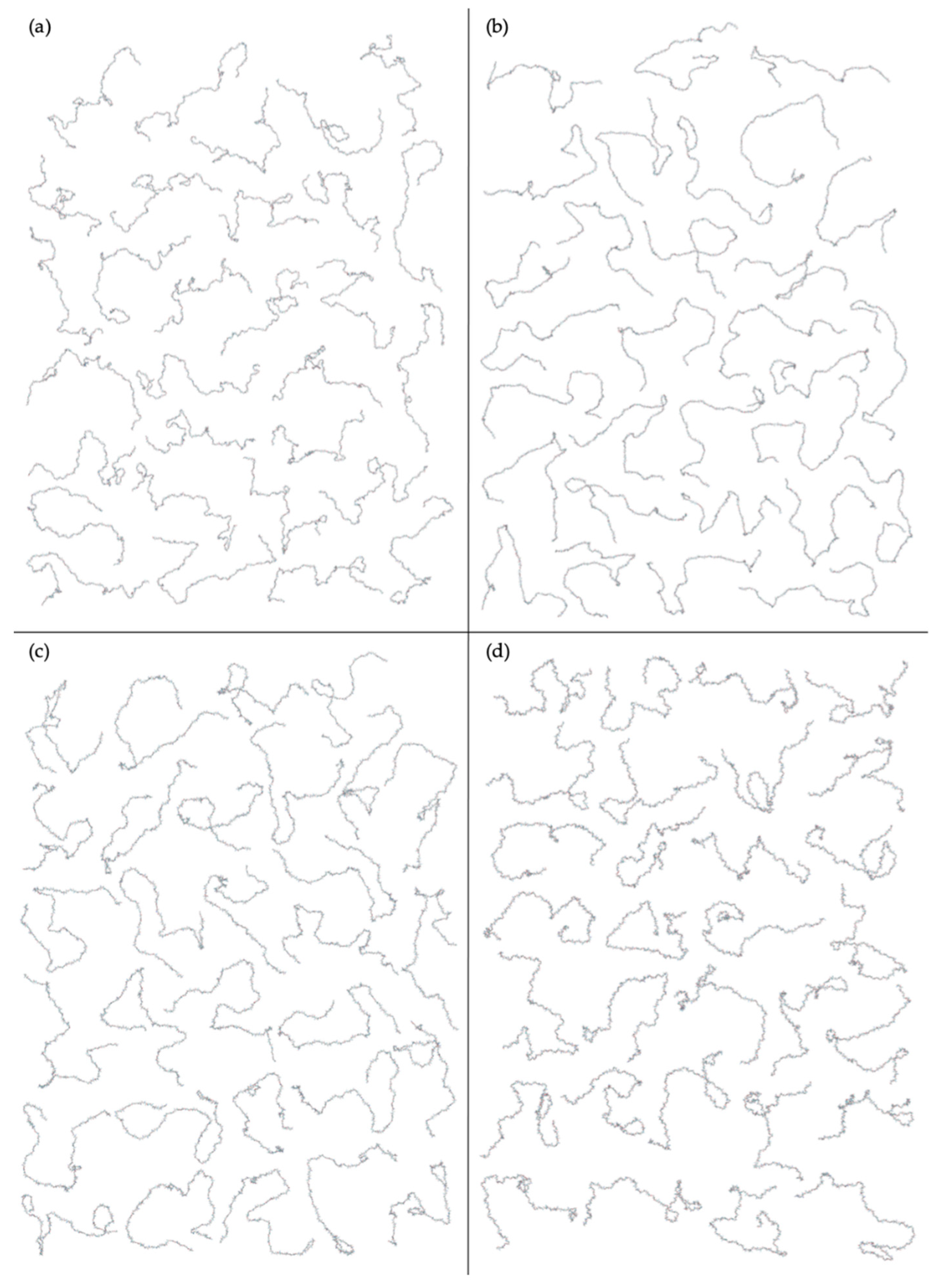
3.4.2. Construction Algorithm
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| PG | Proteoglycan |
| GAG | Glycosaminoglycan |
| MD | Molecular dynamics |
| GlcA | Glucuronate |
| IdoA | Iduronate |
| Gal | Galactose |
| GlcNAc | N-acetylglucosamine |
| GalNAc | N-acetylgalactosamine |
References
- Yoneda, A.; Couchman, J.R. Regulation of cytoskeletal organization by syndecan transmembrane proteoglycans. Matrix Biol. 2003, 22, 25–33. [Google Scholar] [CrossRef]
- Kolset, S.O.; Prydz, K.; Pejler, G. Intracellular proteoglycans. Biochem. J. 2004, 379, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Mizumoto, S.; Fongmoon, D.; Sugahara, K. Interaction of chondroitin sulfate and dermatan sulfate from various biological sources with heparin-binding growth factors and cytokines. Glycoconj. J. 2012, 30, 619–632. [Google Scholar] [CrossRef] [PubMed]
- Sugahara, K.; Mikami, T.; Uyama, T.; Mizuguchi, S.; Nomura, K.; Kitagawa, H. Recent advances in the structural biology of chondroitin sulfate and dermatan sulfate. Curr. Opin. Struct. Biol. 2003, 13, 612–620. [Google Scholar] [CrossRef] [PubMed]
- Sugahara, K.; Mikami, T. Chondroitin/dermatan sulfate in the central nervous system. Curr. Opin. Struct. Biol. 2007, 17, 536–545. [Google Scholar] [CrossRef]
- Kawashima, H.; Atarashi, K.; Hirose, M.; Hirose, J.; Yamada, S.; Sugahara, K.; Miyasaka, M. Oversulfated chondroitin/dermatan sulfates containing GlcAβ1/IdoAα1-3GalNAc(4,6-O-disulfate) interact with L- and P-selectin and chemokines. J. Biol. Chem. 2002, 277, 12921–12930. [Google Scholar] [CrossRef]
- Lander, A.D.; Selleck, S.B. The elusive functions of proteoglycans: In vivo veritas. J. Cell Biol. 2000, 148, 227–232. [Google Scholar] [CrossRef]
- Stallcup, W.B.; Dahlin, K.; Healy, P. Interaction of the NG2 chondroitin sulfate proteoglycan with type VI collagen. J. Cell Biol. 1990, 111, 3177–3188. [Google Scholar] [CrossRef]
- Van Susante, J.L.C.; Pieper, J.; Buma, P.; van Kuppevelt, T.H.; van Beuningen, H.; van Der Kraan, P.M.; Veerkamp, J.H.; van den Berg, W.B.; Veth, R.P.H. Linkage of chondroitin-sulfate to type I collagen scaffolds stimulates the bioactivity of seeded chondrocytes in vitro. Biomaterials 2001, 22, 2359–2369. [Google Scholar] [CrossRef]
- Streit, A.; Nolte, C.; Rásony, T.; Schachner, M. Interaction of astrochondrin with extracellular matrix components and its involvement in astrocyte process formation and cerebellar granule cell migration. J. Cell Biol. 1993, 120, 799–814. [Google Scholar] [CrossRef]
- Djerbal, L.; Lortat-Jacob, H.; Kwok, J. Chondroitin sulfates and their binding molecules in the central nervous system. Glycoconj. J. 2017, 34, 363–376. [Google Scholar] [CrossRef] [PubMed]
- Forsten-Williams, K.; Chu, C.L.; Fannon, M.; Buczek-Thomas, J.A.; Nugent, M.A. Control of growth factor networks by heparan sulfate proteoglycans. Ann. Biomed. Eng. 2008, 36, 2134–2148. [Google Scholar] [CrossRef] [PubMed]
- Bhavanandan, V.P.; Davidson, E.A. Mucopolysaccharides associated with nuclei of cultured mammalian cells. Proc. Natl. Acad. Sci. USA 1975, 72, 2032–2036. [Google Scholar] [CrossRef]
- Fedarko, N.S.; Conrad, H.E. A unique heparan sulfate in the nuclei of hepatocytes: Strucural changes with the growth state of the cells. J. Cell Biol. 1986, 102, 587–599. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, M.; Fedarko, N.S.; Conrad, H.E. Transport of heparan sulfate into the nuclei of hepatocytes. J. Biol. Chem. 1986, 261, 13575–13580. [Google Scholar]
- Tumova, S.; Hatch, B.A.; Law, D.J.; Bame, K.J. Basic fibroblast growth factor does not prevent heparan sulphate proteoglycan catabolism in intact cells, but it alters the distribution of the glycosaminoglycan degradation products. Biochem. J. 1999, 337, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Kjellén, L.; Lindahl, U. Specificity of glycosaminoglycan-protein interactions. Curr. Opin. Struct. Biol. 2018, 50, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Sharif, M.; George, E.; Shepstone, L.; Knudson, W.; Thonar, E.J.M.; Cushnaghan, J.; Dieppe, P. Serum hyaluronic acid level as a predictor of disease progression in osteoarthritis of the knee. Arthritis Rheum. 1995, 38, 760–767. [Google Scholar] [CrossRef]
- Ghosh, P. The role of hyaluronic acid (hyaluronan) in health and disease: Interactions with cells, cartilage and components of synovial fluid. Clin. Exp. Rheum. 1994, 12, 75–82. [Google Scholar]
- Mizumoto, S.; Kosho, T.; Hatamochi, A.; Honda, T.; Yamaguchi, T.; Okamoto, N.; Miyake, N.; Yamada, S.; Sugahara, K. Defect in dermatan sulfate in urine of patients with Ehlers-Danlos syndrome caused by a CHST14/D4ST1 deficiency. Clin. Biochem. 2017, 50, 670–677. [Google Scholar] [CrossRef]
- Kosho, T. CHST14/D4ST1 deficiency: New form of Ehlers–Danlos syndrome. Pediatr. Int. 2016, 58, 88–99. [Google Scholar] [CrossRef]
- Lindahl, B.; Eriksson, L.; Spillmann, D.; Caterson, B.; Lindahl, U. Selective loss of cerebral keratan sulfate in Alzheimer’s disease. J. Biol. Chem. 1996, 271, 16991–16994. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.; Schmitz, S.; De Santis, R.; Doemming, S.; Haase, H.; Hoeger, J.; Heinbockel, L.; Brandenburg, K.; Marx, G.; Schuerholz, T. Peptide 19-2.5 inhibits heparan sulfate-triggered inflammation in murine cardiomyocytes stimulated with human sepsis serum. PLoS ONE 2015, 10, e0127584. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, E.P.; Overdier, K.H.; Sun, X.; Lin, L.; Liu, X.; Yang, Y.; Ammons, L.A.; Hiller, T.D.; Suflita, M.A.; Yu, Y. Urinary glycosaminoglycans predict outcomes in septic shock and acute respiratory distress syndrome. Am. J. Resp. Crit. Care 2016, 194, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, J. Fell-Muir Lecture: Heparan sulphate and the art of cell regulation: A polymer chain conducts the protein orchestra. Int. J. Exp. Pathol. 2015, 96, 203–231. [Google Scholar] [CrossRef] [PubMed]
- Shriver, Z.; Raguram, S.; Sasisekharan, R. Glycomics: A pathway to a class of new and improved therapeutics. Nat. Rev. Drug Discov. 2004, 3, 863–873. [Google Scholar] [CrossRef] [PubMed]
- Trottein, F.; Schaffer, L.; Ivanov, S.; Paget, C.; Vendeville, C.; Cazet, A.; Groux-Degroote, S.; Lee, S.; Krzewinski-Recchi, M.-A.; Faveeuw, C.; et al. Glycosyltransferase and sulfotransferase gene expression profiles in human monocytes, dendritic cells and macrophages. Glycoconj. J. 2009, 26, 1259–1274. [Google Scholar] [CrossRef]
- Gandhi, N.S.; Mancera, R.L. The Structure of glycosaminoglycans and their interactions with proteins. Chem. Biol. Drug Des. 2008, 72, 455–482. [Google Scholar] [CrossRef]
- Casu, B.; Petitou, M.; Provasoli, M.; Sinay, P. Conformational flexibility: A new concept for explaining binding and biological properties of iduronic acid-containing glycosaminoglycans. Trends Biochem. Sci. 1988, 13, 221–225. [Google Scholar] [CrossRef]
- Mulloy, B.; Forster, M.J.; Jones, C.; Drake, A.F.; Johnson, E.A.; Davies, D.B. The effect of variation of substitution on the solution conformation of heparin: A spectroscopic and molecular modelling study. Carbohydr. Res. 1994, 255, 1–26. [Google Scholar] [CrossRef]
- Zamparo, O.; Comper, W.D. The hydrodynamic frictional coefficient of polysaccharides: The role of the glycosidic linkage. Carbohydr. Res. 1991, 212, 193–200. [Google Scholar] [CrossRef]
- Samsonov, S.A.; Gehrcke, J.-P.; Pisabarro, M.T. Flexibility and explicit solvent in molecular-dynamics-based docking of protein–glycosaminoglycan systems. J. Chem. Inf. Model. 2014, 54, 582–592. [Google Scholar] [CrossRef] [PubMed]
- Whitmore, E.K.; Vesenka, G.; Sihler, H.; Guvench, O. Efficient Construction of Atomic-Resolution Models of Non-Sulfated Chondroitin Glycosaminoglycan Using Molecular Dynamics Data. Biomolecules 2020, 10, 537. [Google Scholar] [CrossRef] [PubMed]
- Toyoda, H.; Kinoshita-Toyoda, A.; Selleck, S.B. Structural analysis of glycosaminoglycans in Drosophila and Caenorhabditis elegans and demonstration that tout-velu, a drosophila gene related to EXT tumor suppressors, affects heparan sulfate in vivo. J. Biol. Chem. 2000, 275, 2269–2275. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.O.; Gunay, N.S.; Toida, T.; Kuberan, B.; Yu, G.; Kim, Y.S.; Linhardt, R.J. Preparation and structural determination of dermatan sulfate-derived oligosaccharides. Glycobiology 2000, 10, 1033–1039. [Google Scholar] [CrossRef] [PubMed]
- Zaia, J.; Costello, C.E. Compositional analysis of glycosaminoglycans by electrospray mass spectrometry. Anal. Chem. 2001, 73, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Shao, C.; Zhang, F.; Kemp, M.M.; Linhardt, R.J.; Waisman, D.M.; Head, J.F.; Seaton, B.A. Crystallographic analysis of calcium-dependent heparin binding to annexin A2. J. Biol. Chem. 2006, 281, 31689–31695. [Google Scholar] [CrossRef]
- Capila, I.; Hernáiz, M.a.J.; Mo, Y.D.; Mealy, T.R.; Campos, B.; Dedman, J.R.; Linhardt, R.J.; Seaton, B.A. Annexin V–heparin oligosaccharide complex suggests heparan sulfate–mediated assembly on cell surfaces. Structure 2001, 9, 57–64. [Google Scholar] [CrossRef]
- Dementiev, A.; Petitou, M.; Herbert, J.-M.; Gettins, P.G.W. The ternary complex of antithrombin–anhydrothrombin–heparin reveals the basis of inhibitor specificity. Nat. Struct. Mol. Biol. 2004, 11, 863–867. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Johnson, D.J.D.; Esmon, C.T.; Huntington, J.A. Structure of the antithrombin–thrombin–heparin ternary complex reveals the antithrombotic mechanism of heparin. Nat. Struct. Mol. Biol. 2004, 11, 857–862. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Kienetz, M.; Cherney, M.M.; James, M.N.G.; Brömme, D. The crystal and molecular structures of a cathepsin K:chondroitin sulfate complex. J. Mol. Biol. 2008, 383, 78–91. [Google Scholar] [CrossRef]
- Imberty, A.; Lortat-Jacob, H.; Pérez, S. Structural view of glycosaminoglycan–protein interactions. Carbohydr. Res. 2007, 342, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Ferro, D.R.; Provasoli, A.; Ragazzi, M.; Torri, G.; Casu, B.; Gatti, G.; Jacquinet, J.C.; Sinay, P.; Petitou, M.; Choay, J. Evidence for conformational equilibrium of the sulfated L-iduronate residue in heparin and in synthetic heparin mono- and oligo-saccharides: NMR and force-field studies. J. Am. Chem. Soc. 1986, 108, 6773–6778. [Google Scholar] [CrossRef]
- Bossennec, V.; Petitou, M.; Perly, B. 1H-n.m.r. investigation of naturally occurring and chemically oversulphated dermatan sulphates. Identification of minor monosaccharide residues. Biochem. J. 1990, 267, 625–630. [Google Scholar] [CrossRef] [PubMed]
- Ferro, D.R.; Provasoli, A.; Ragazzi, M.; Casu, B.; Torri, G.; Bossennec, V.; Perly, B.; Sinaÿ, P.; Petitou, M.; Choay, J. Conformer populations of l-iduronic acid residues in glycosaminoglycan sequences. Carbohydr. Res. 1990, 195, 157–167. [Google Scholar] [CrossRef]
- Yamada, S.; Yamane, Y.; Sakamoto, K.; Tsuda, H.; Sugahara, K. Structural determination of sulfated tetrasaccharides and hexasaccharides containing a rare disaccharide sequence, -3GalNAc(4,6-disulfate)beta1-4IdoAalpha1-, isolated from porcine intestinal dermatan sulfate. Eur. J. Biochem. 1998, 258, 775–783. [Google Scholar] [CrossRef]
- Silipo, A.; Zhang, Z.; Cañada, F.J.; Molinaro, A.; Linhardt, R.J.; Jiménez-Barbero, J. Conformational analysis of a dermatan sulfate-derived tetrasaccharide by NMR, molecular modeling, and residual dipolar couplings. Chembiochem 2008, 9, 240–252. [Google Scholar] [CrossRef]
- Sattelle, B.M.; Shakeri, J.; Cliff, M.J.; Almond, A. Proteoglycans and their heterogeneous glycosaminoglycans at the atomic scale. Biomacromolecules 2015, 16, 951–961. [Google Scholar] [CrossRef]
- Almond, A.; Sheehan, J.K. Glycosaminoglycan conformation: Do aqueous molecular dynamics simulations agree with x-ray fiber diffraction? Glycobiology 2000, 10, 329–338. [Google Scholar] [CrossRef]
- Gandhi, N.S.; Mancera, R.L. Can current force fields reproduce ring puckering in 2-O-sulfo-α-l-iduronic acid? A molecular dynamics simulation study. Carbohydr. Res. 2010, 345, 689–695. [Google Scholar] [CrossRef]
- Sattelle, B.M.; Hansen, S.U.; Gardiner, J.; Almond, A. Free energy landscapes of iduronic acid and related monosaccharides. J. Am. Chem. Soc. 2010, 132, 13132–13134. [Google Scholar] [CrossRef]
- Balogh, G.; Gyöngyö;si, T.S.; Timári, I.; Herczeg, M.; Borbás, A.; Fehér, K.; Kövér, K.E. Comparison of carbohydrate force fields using Gaussian Accelerated Molecular Dynamics simulations and development of force field parameters for heparin-analogue pentasaccharides. J. Chem. Inf. Model. 2019, 59, 4855–4867. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.; Nandha Premnath, P.; Guvench, O. Rigidity and flexibility in the tetrasaccharide linker of proteoglycans from atomic-resolution molecular simulation. J. Comput. Chem. 2017, 38, 1438–1446. [Google Scholar] [CrossRef] [PubMed]
- Guvench, O. Revealing the mechanisms of protein disorder and N-glycosylation in CD44-hyaluronan binding using molecular simulation. Front. Immunol. 2015, 6, 305. [Google Scholar] [CrossRef] [PubMed]
- Faller, C.E.; Guvench, O. Sulfation and cation effects on the conformational properties of the glycan backbone of chondroitin sulfate disaccharides. J. Phys. Chem. B 2015, 119, 6063–6073. [Google Scholar] [CrossRef] [PubMed]
- Favreau, A.J.; Faller, C.E.; Guvench, O. CD44 receptor unfolding enhances binding by freeing basic amino acids to contact carbohydrate ligand. Biophys. J. 2013, 105, 1217–1226. [Google Scholar] [CrossRef] [PubMed]
- Guvench, O.; Mallajosyula, S.S.; Raman, E.P.; Hatcher, E.; Vanommeslaeghe, K.; Foster, T.J.; Jamison, F.W., II; Mackerell, A.D., Jr. CHARMM additive all-atom force field for carbohydrate derivatives and its utility in polysaccharide and carbohydrate-protein modeling. J. Chem. Theory Comput. 2011, 7, 3162–3180. [Google Scholar] [CrossRef]
- Brannigan, G.; Lin, L.C.; Brown, F.L. Implicit solvent simulation models for biomembranes. Eur. Biophys. J. 2006, 35, 104–124. [Google Scholar] [CrossRef]
- Srinivas, G.; Cheng, X.; Smith, J.C. A solvent-free coarse grain model for crystalline and amorphous cellulose fibrils. J. Chem. Theory Comput. 2011, 7, 2539–2548. [Google Scholar] [CrossRef]
- Ingólfsson, H.I.; Lopez, C.A.; Uusitalo, J.J.; de Jong, D.H.; Gopal, S.M.; Periole, X.; Marrink, S.J. The power of coarse graining in biomolecular simulations. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2014, 4, 225–248. [Google Scholar] [CrossRef]
- Guvench, O.; Greene, S.N.; Kamath, G.; Brady, J.W.; Venable, R.M.; Pastor, R.W.; Mackerell, A.D., Jr. Additive empirical force field for hexopyranose monosaccharides. J. Comput. Chem. 2008, 29, 2543–2564. [Google Scholar] [CrossRef] [PubMed]
- Guvench, O.; Hatcher, E.R.; Venable, R.M.; Pastor, R.W.; Mackerell, A.D. CHARMM additive all-atom force field for glycosidic linkages between hexopyranoses. J. Chem. Theory Comput. 2009, 5, 2353–2370. [Google Scholar] [CrossRef] [PubMed]
- Mallajosyula, S.S.; Guvench, O.; Hatcher, E.; Mackerell, A.D., Jr. CHARMM additive all-atom force field for phosphate and sulfate linked to carbohydrates. J. Chem. Theory Comput. 2012, 8, 759–776. [Google Scholar] [CrossRef] [PubMed]
- Hascall, V.C.; Heinegård, D. Aggregation of cartilage proteoglycans II. Oligosaccharide competitors of the proteoglycan-hyaluronic acid interaction. J. Biol. Chem. 1974, 249, 4242–4249. [Google Scholar] [PubMed]
- Winter, W.; Smith, P.; Arnott, S. Hyaluronic acid: Structure of a fully extended 3-fold helical sodium salt and comparison with the less extended 4-fold helical forms. J. Mol. Biol. 1975, 99, 219–235. [Google Scholar] [CrossRef]
- Sheehan, J.; Gardner, K.; Atkins, E. Hyaluronic acid: A double-helical structure in the presence of potassium at low pH and found also with the cations ammonium, rubidium and caesium. J. Mol. Biol. 1977, 117, 113–135. [Google Scholar] [CrossRef]
- Almond, A.; Brass, A.; Sheehan, J.K. Dynamic exchange between stabilized conformations predicted for hyaluronan tetrasaccharides: Comparison of molecular dynamics simulations with available NMR data. Glycobiology 1998, 8, 973–980. [Google Scholar] [CrossRef]
- Almond, A.; Sheehan, J.K.; Brass, A. Molecular dynamics simulations of the two disaccharides of hyaluronan in aqueous solution. Glycobiology 1997, 7, 597–604. [Google Scholar] [CrossRef]
- Gribbon, P.; Heng, B.C.; Hardingham, T.E. The molecular basis of the solution properties of hyaluronan investigated by confocal fluorescence recovery after photobleaching. Biophys. J. 1999, 77, 2210–2216. [Google Scholar] [CrossRef][Green Version]
- Atkins, E.; Meader, D.; Scott, J. Model for hyaluronic acid incorporating four intramolecular hydrogen bonds. Int. J. Biol. Macromol. 1980, 2, 318–319. [Google Scholar] [CrossRef]
- Scott, J.E. Supramolecular organization of extracellular matrix glycosaminoglycans, in vitro and in the tissues. FASEB J. 1992, 6, 2639–2645. [Google Scholar] [CrossRef]
- Scott, J.E.; Cummings, C.; Brass, A.; Chen, Y. Secondary and tertiary structures of hyaluronan in aqueous solution, investigated by rotary shadowing-electron microscopy and computer simulation. Biochem. J. 1991, 274, 699–705. [Google Scholar] [CrossRef] [PubMed]
- Scott, J.; Heatley, F.; Moorcroft, D.; Olavesen, A. Secondary structures of hyaluronate and chondroitin sulphates. A 1H nmr study of NH signals in dimethyl sulphoxide solution. Biochem. J. 1981, 199, 829. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Montgomery, D.; Xue, X.; Foley, B.L.; Woods, R.J. GAG Builder: A web-tool for modeling 3D structures of glycosaminoglycans. Glycobiology 2019, 29, 515–518. [Google Scholar] [CrossRef] [PubMed]
- Engelsen, S.B.; Hansen, P.I.; Perez, S. POLYS 2.0: An open source software package for building three-dimensional structures of polysaccharides. Biopolymers 2014, 101, 733–743. [Google Scholar] [CrossRef]
- Kuttel, M.M.; Ståhle, J.; Widmalm, G. CarbBuilder: Software for building molecular models of complex oligo-and polysaccharide structures. J. Comput. Chem. 2016, 37, 2098–2105. [Google Scholar] [CrossRef]
- Clerc, O.; Deniaud, M.; Vallet, S.D.; Naba, A.; Rivet, A.; Perez, S.; Thierry-Mieg, N.; Ricard-Blum, S. MatrixDB: Integration of new data with a focus on glycosaminoglycan interactions. Nucleic Acids Res. 2018, 47, D376–D381. [Google Scholar] [CrossRef] [PubMed]
- Clerc, O.; Mariethoz, J.; Rivet, A.; Lisacek, F.; Pérez, S.; Ricard-Blum, S. A pipeline to translate glycosaminoglycan sequences into 3D models. Application to the exploration of glycosaminoglycan conformational space. Glycobiology 2018, 29, 36–44. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Song, K.C.; Desaire, H.; MacKerell, A.D., Jr.; Im, W. Glycan Reader: Automated sugar identification and simulation preparation for carbohydrates and glycoproteins. J. Comput. Chem. 2011, 32, 3135–3141. [Google Scholar] [CrossRef]
- Park, S.-J.; Lee, J.; Patel, D.S.; Ma, H.; Lee, H.S.; Jo, S.; Im, W. Glycan Reader is improved to recognize most sugar types and chemical modifications in the Protein Data Bank. Bioinformatics 2017, 33, 3051–3057. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-J.; Lee, J.; Qi, Y.; Kern, N.R.; Lee, H.S.; Jo, S.; Joung, I.; Joo, K.; Lee, J.; Im, W. CHARMM-GUI glycan modeler for modeling and simulation of carbohydrates and glycoconjugates. Glycobiology 2019, 29, 320–331. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.R.; Bruccoleri, R.E.; Olafson, B.D.; States, D.J.; Swaminathan, S.; Karplus, M. CHARMM: A Program for Macromolecular Energy, Minimization, and Dynamics Calculations. J. Comput. Chem. 1983, 4, 187–217. [Google Scholar] [CrossRef]
- Brooks, B.R.; Brooks, C.L., III; Mackerell, A.D., Jr.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef]
- MacKerell, A.D., Jr.; Brooks, B.; Brooks, C.L., III; Nilsson, L.; Roux, B.; Won, Y.; Karplus, M. CHARMM: The energy function and its parameterization with an overview of the program. In Encyclopedia of Computational Chemistry; John Wiley and Sons, Ltd.: New York, NY, USA, 1998; Volume 1, pp. 271–277. [Google Scholar]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Durell, S.R.; Brooks, B.R.; Ben-Naim, A. Solvent-induced forces between two hydrophilic groups. J. Phys. Chem. 1994, 98, 2198–2202. [Google Scholar] [CrossRef]
- Beglov, D.; Roux, B. Finite representation of an infinite bulk system: Solvent boundary potential for computer simulations. J. Chem. Phys. 1994, 100, 9050–9063. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graphics 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Cremer, D.; Pople, J. General definition of ring puckering coordinates. J. Am. Chem. Soc. 1975, 97, 1354–1358. [Google Scholar] [CrossRef]
- Banerji, S.; Wright, A.J.; Noble, M.; Mahoney, D.J.; Campbell, I.D.; Day, A.J.; Jackson, D.G. Structures of the CD44–hyaluronan complex provide insight into a fundamental carbohydrate-protein interaction. Nat. Struct. Mol. Biol. 2007, 14, 234. [Google Scholar] [CrossRef]
- Jedrzejas, M.J.; Mello, L.V.; de Groot, B.L.; Li, S. Mechanism of hyaluronan degradation by Streptococcus pneumoniae Hyaluronate Lyase. J. Biol. Chem. 2002, 277, 28287–28297. [Google Scholar] [CrossRef] [PubMed]
- Mello, L.V.; de Groot, B.L.; Li, S.; Jedrzejas, M.J. Structure and flexibility of Streptococcus agalactiae hyaluronate lyase complex with its substrate. J. Biol. Chem. 2002, 277, 36678–36688. [Google Scholar] [CrossRef] [PubMed]
- Marković-Housley, Z.; Miglierini, G.; Soldatova, L.; Rizkallah, P.J.; Müller, U.; Schirmer, T. Crystal structure of hyaluronidase, a major allergen of bee venom. Structure 2000, 8, 1025–1035. [Google Scholar] [CrossRef]
- Tashima, T.; Nagatoishi, S.; Caaveiro, J.M.; Nakakido, M.; Sagara, H.; Kusano-Arai, O.; Iwanari, H.; Mimuro, H.; Hamakubo, T.; Ohnuma, S.-i. Molecular basis for governing the morphology of type-I collagen fibrils by Osteomodulin. Commun. Biol. 2018, 1, 1–10. [Google Scholar] [CrossRef]
- Paracuellos, P.; Kalamajski, S.; Bonna, A.; Bihan, D.; Farndale, R.W.; Hohenester, E. Structural and functional analysis of two small leucine-rich repeat proteoglycans, fibromodulin and chondroadherin. Matrix Biol. 2017, 63, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Ohto, U.; Usui, K.; Ochi, T.; Yuki, K.; Satow, Y.; Shimizu, T. Crystal structure of human β-galactosidase structural basis of gm1 gangliosidosis and morquio b diseases. J. Biol. Chem. 2012, 287, 1801–1812. [Google Scholar] [CrossRef]
- Scally, S.W.; Petersen, J.; Law, S.C.; Dudek, N.L.; Nel, H.J.; Loh, K.L.; Wijeyewickrema, L.C.; Eckle, S.B.; van Heemst, J.; Pike, R.N. A molecular basis for the association of the HLA-DRB1 locus, citrullination, and rheumatoid arthritis. J. Exp. Med. 2013, 210, 2569–2582. [Google Scholar] [CrossRef] [PubMed]
- Sattelle, B.M.; Almond, A. Is N-acetyl-d-glucosamine a rigid 4C1 chair? Glycobiology 2011, 21, 1651–1662. [Google Scholar] [CrossRef]
- Siciňska, W.; Adams, B.; Lerner, L. A detailed 1H and 13C NMR study of a repeating disaccharide of hyaluronan: The effects of temperature and counterion type. Carbohydr. Res. 1993, 242, 29–51. [Google Scholar] [CrossRef]
- Gargiulo, V.; Morando, M.A.; Silipo, A.; Nurisso, A.; Pérez, S.; Imberty, A.; Cañada, F.J.; Parrilli, M.; Jiménez-Barbero, J.; De Castro, C. Insights on the conformational properties of hyaluronic acid by using NMR residual dipolar couplings and MD simulations. Glycobiology 2010, 20, 1208–1216. [Google Scholar] [CrossRef] [PubMed]
- Ikemizu, S.; Sparks, L.M.; van der Merwe, P.A.; Harlos, K.; Stuart, D.I.; Jones, E.Y.; Davis, S.J. Crystal structure of the CD2-binding domain of CD58 (lymphocyte function-associated antigen 3) at 1.8-Å resolution. Proc. Natl. Acad. Sci. USA 1999, 96, 4289–4294. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.; Magracheva, E.; Kozlov, S.; Fong, S.; Tobin, G.; Kotenko, S.; Wlodawer, A.; Zdanov, A. Crystal structure of interleukin-19 defines a new subfamily of helical cytokines. J. Biol. Chem. 2003, 278, 3308–3313. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; McBride, R.; Paulson, J.C.; Basler, C.F.; Wilson, I.A. Structure, receptor binding, and antigenicity of influenza virus hemagglutinins from the 1957 H2N2 pandemic. J. Virol. 2010, 84, 1715–1721. [Google Scholar] [CrossRef] [PubMed]
- Maveyraud, L.; Niwa, H.; Guillet, V.; Svergun, D.I.; Konarev, P.V.; Palmer, R.A.; Peumans, W.J.; Rougé, P.; Van Damme, E.J.; Reynolds, C.D. Structural basis for sugar recognition, including the Tn carcinoma antigen, by the lectin SNA-II from Sambucus nigra. Proteins Struct. Funct. Bioinf. 2009, 75, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Alibay, I.; Bryce, R.A. Ring puckering landscapes of glycosaminoglycan-related monosaccharides from molecular dynamics simulations. J. Chem. Inf. Model. 2019, 59, 4729–4741. [Google Scholar] [CrossRef]
- Alibay, I.; Burusco, K.K.; Bruce, N.J.; Bryce, R.A. Identification of rare Lewis oligosaccharide conformers in aqueous solution using enhanced sampling molecular dynamics. J. Phys. Chem. B 2018, 122, 2462–2474. [Google Scholar] [CrossRef]
- Nyerges, B.; Kovacs, A. Density functional study of the conformational space of 4C1 D-glucuronic acid. J. Phys. Chem. A 2005, 109, 892–897. [Google Scholar] [CrossRef]
- Blundell, C.D.; Roberts, I.S.; Sheehan, J.K.; Almond, A. Investigating the molecular basis for the virulence of Escherichia coli K5 by nuclear magnetic resonance analysis of the capsule polysaccharide. J. Mol. Microbiol. Biotechnol. 2009, 17, 71–82. [Google Scholar] [CrossRef]
- Furlan, S.; La Penna, G.; Perico, A.; Cesàro, A. Hyaluronan chain conformation and dynamics. Carbohydr. Res. 2005, 340, 959–970. [Google Scholar] [CrossRef]
- Almond, A.; DeAngelis, P.L.; Blundell, C.D. Hyaluronan: The local solution conformation determined by NMR and computer modeling is close to a contracted left-handed 4-fold helix. J. Mol. Biol. 2006, 358, 1256–1269. [Google Scholar] [CrossRef] [PubMed]
- Holmbeck, S.M.; Petillo, P.A.; Lerner, L.E. The solution conformation of hyaluronan: A combined NMR and molecular dynamics study. Biochemistry 1994, 33, 14246–14255. [Google Scholar] [CrossRef] [PubMed]
- Adrian-Scotto, M.; Abdallah, K.B.; Mallet, G.; Vasilescu, D. Quantum molecular modeling of free radical saccharides from hyaluronan. J. Molec. Struct. 2003, 636, 89–113. [Google Scholar] [CrossRef]
- Moulabbi, M.; Broch, H.; Robert, L.; Vasilescu, D. Quantum molecular modeling of hyaluronan. J. Mol. Struct. 1997, 395, 477–508. [Google Scholar] [CrossRef]
- Guss, J.; Hukins, D.; Smith, P.; Winter, W.; Arnott, S.; Moorhouse, R.; Rees, D. Hyaluronic acid: Molecular conformations and interactions in two sodium salts. J. Mol. Biol. 1975, 95, 359–384. [Google Scholar] [CrossRef]
- Rees, D. Conformational analysis of polysaccharides. Part II. Alternating copolymers of the agar–carrageenan–chondroitin type by model building in the computer with calculation of helical parameters. J. Chem. Soc. B 1969, 217–226. [Google Scholar] [CrossRef]
- Mitra, A.; Arnott, S.; Atkins, E.; Isaac, D. Dermatan sulfate: Molecular conformations and interactions in the condensed state. J. Mol. Biol. 1983, 169, 873–901. [Google Scholar] [CrossRef]
- Virudachalam, R.; Rao, V.S. Theoretical investigations on 2-acetamido-2-deoxy aldohexopyranoses: Conformation and the anomeric effect. Carbohydr. Res. 1976, 51, 135–139. [Google Scholar] [CrossRef]
- Hsieh, P.-H.; Thieker, D.F.; Guerrini, M.; Woods, R.J.; Liu, J. Uncovering the relationship between sulphation patterns and conformation of iduronic acid in heparan sulphate. Nature 2016, 6, 29602. [Google Scholar] [CrossRef]
- Gatti, G.; Casu, B.; Torri, G.; Vercellotti, J.R. Resolution-enhanced 1H-nmr spectra of dermatan sulfate and chondroitin sulfates: Conformation of the uronic acid residues. Carbohydr. Res. 1979, 68, C3–C7. [Google Scholar] [CrossRef]
- Ernst, S.; Venkataraman, G.; Sasisekharan, V.; Langer, R.; Cooney, C.L.; Sasisekharan, R. Pyranose ring flexibility. Mapping of physical data for iduronate in continuous conformational space. J. Am. Chem. Soc. 1998, 120, 2099–2107. [Google Scholar] [CrossRef]
- Van Boeckel, C.; Van Aelst, S.; Wagenaars, G.; Mellema, J.R.; Paulsen, H.; Peters, T.; Pollex, A.; Sinnwell, V. Conformational analysis of synthetic heparin-like oligosaccharides containing α-L-idopyranosyluronic acid. Recl. Trav. Chim. Pays Bas. 1987, 106, 19–29. [Google Scholar] [CrossRef]
- Inoue, Y.; Inouye, Y.; Nagasawa, K. Conformational equilibria of the L-iduronate residue in non-sulphated di-, tetra-and hexa-saccharides and their alditols derived from dermatan sulphate. Biochem. J. 1990, 265, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Ragazzi, M.; Provasoli, A.; Ferro, D. Molecular mechanics and the structure of iduronate-containing carbohydrates. In Computer Modeling of Carbohydrate Molecules; French, A., Ed.; ACS Publications: Washington, DC, USA, 1990; pp. 332–344. [Google Scholar]
- Forster, M.J.; Mulloy, B. Molecular dynamics study of iduronate ring conformation. Biopolymers 1993, 33, 575–588. [Google Scholar] [CrossRef]
- Venkataraman, G.; Sasisekharan, V.; Cooney, C.L.; Langer, R.; Sasisekharan, R. A stereochemical approach to pyranose ring flexibility: Its implications for the conformation of dermatan sulfate. Proc. Natl. Acad. Sci. USA 1994, 91, 6171–6175. [Google Scholar] [CrossRef]
- Arnott, S.; Guss, J.; Hukins, D.; Dea, I.; Rees, D. Conformation of keratan sulphate. J. Mol. Biol. 1974, 88, 175–184. [Google Scholar] [CrossRef]
- Rudrum, M.; Shaw, D. 10. The structure and conformation of some monosaccharides in solution. J. Chem. Soc. 1965, 52–57. [Google Scholar] [CrossRef]
- Angyal, S.J. Conformational analysis in carbohydrate chemistry: I. Conformational free energies. The conformations and α: β ratios of aldopyranoses in aqueous solution. Aust. J. Chem. 1968, 21, 2737–2746. [Google Scholar] [CrossRef]
- Bush, C.A.; Feeney, R.E. Conformation of the glycotripeptide repeating unit of antifreeze glycoprotein of polar fish as determined from the fully assigned proton nmr spectrum. Int. J. Pept. Prot. Res. 1986, 28, 386–397. [Google Scholar] [CrossRef] [PubMed]
- Mulloy, B.; Forster, M.J. Conformation and dynamics of heparin and heparan sulfate. Glycobiology 2000, 10, 1147–1156. [Google Scholar] [CrossRef]
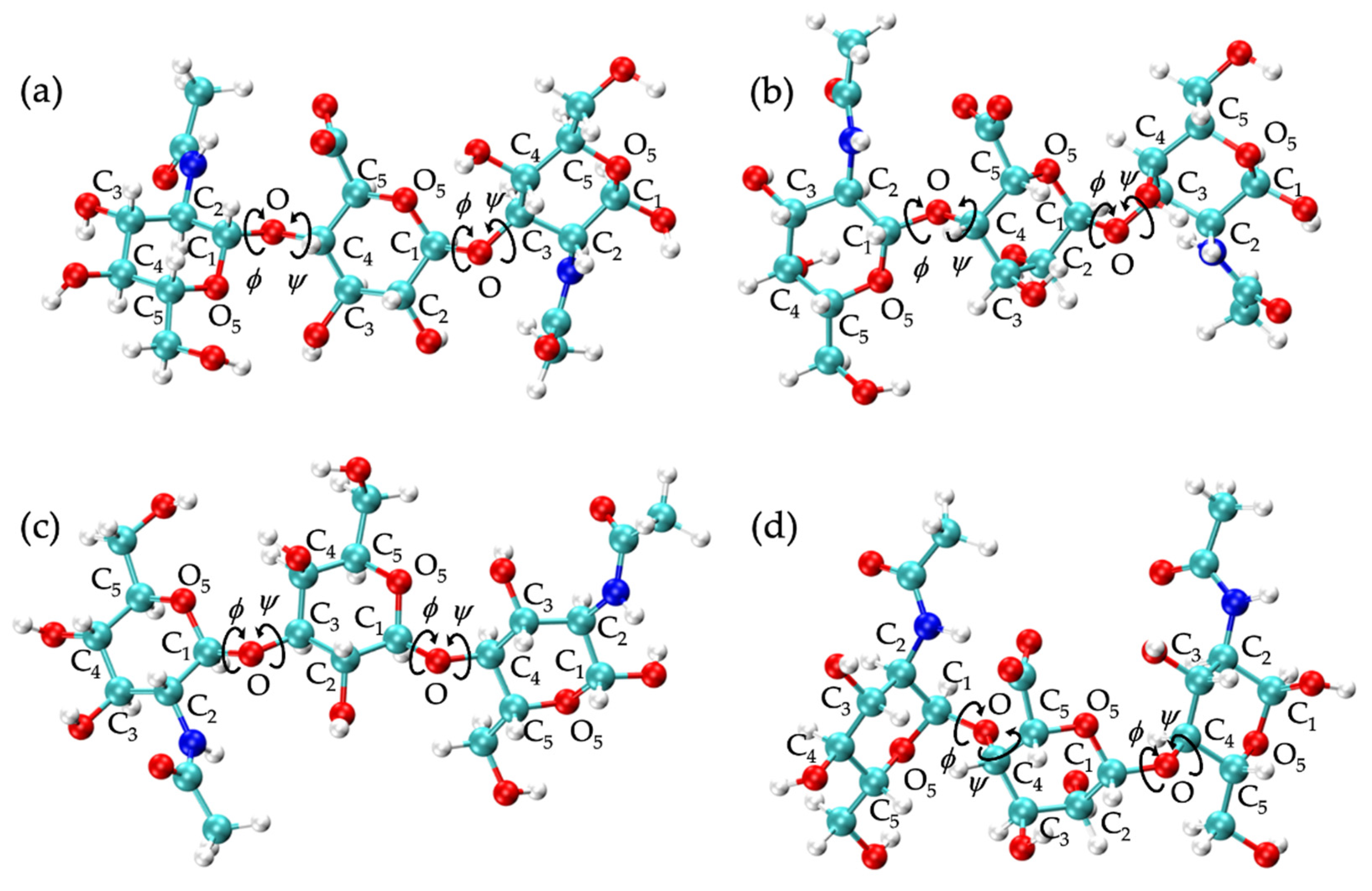
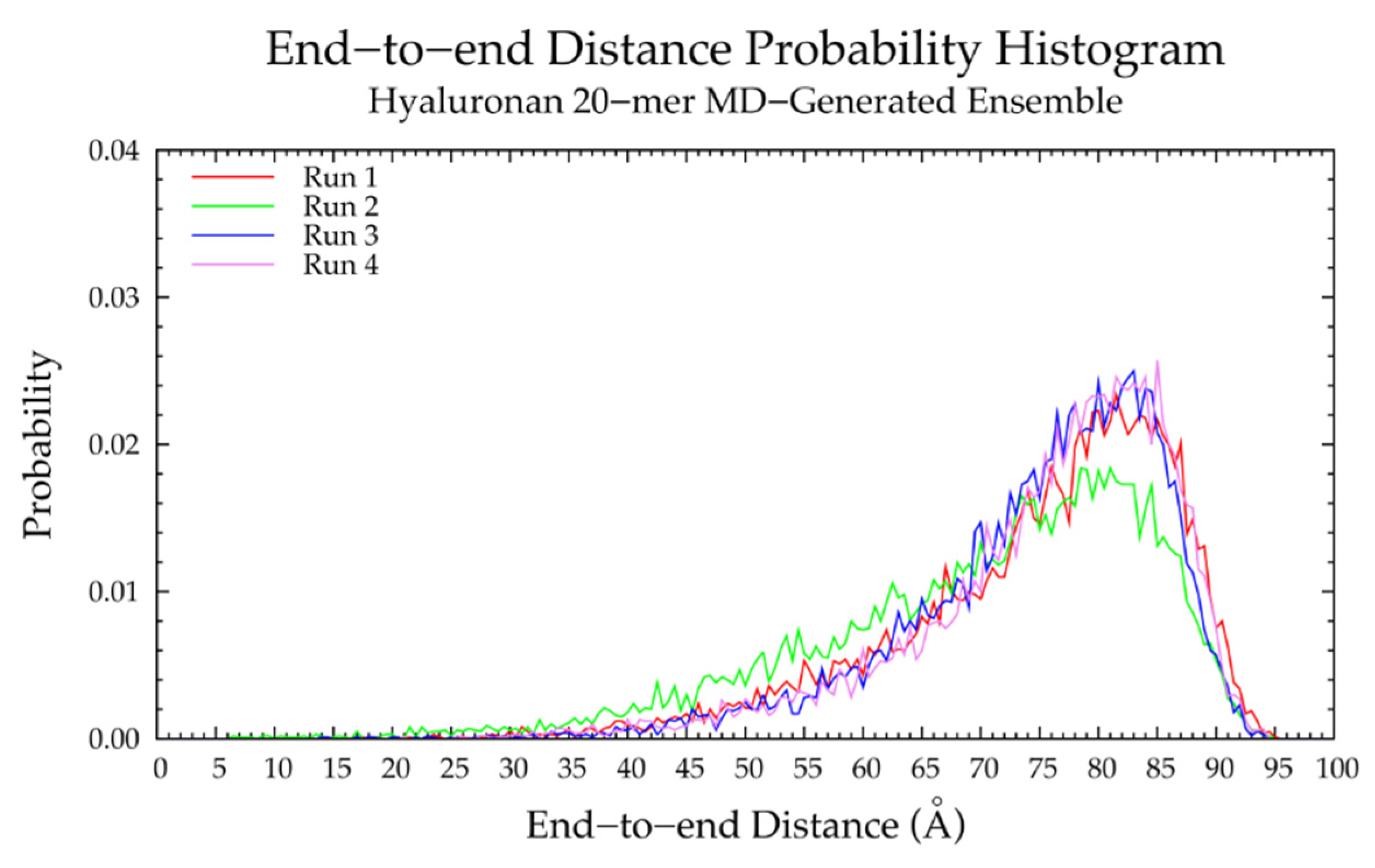
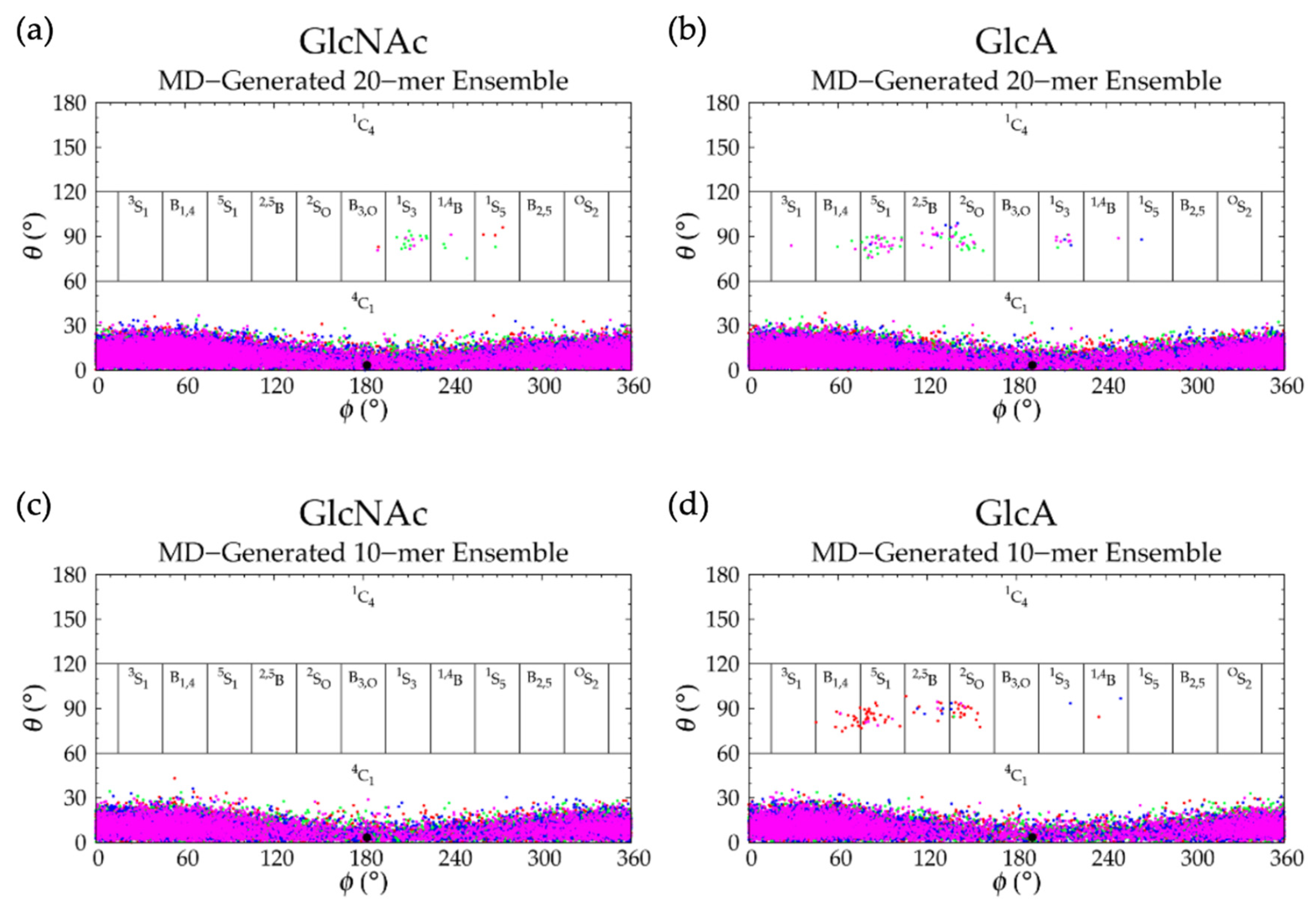
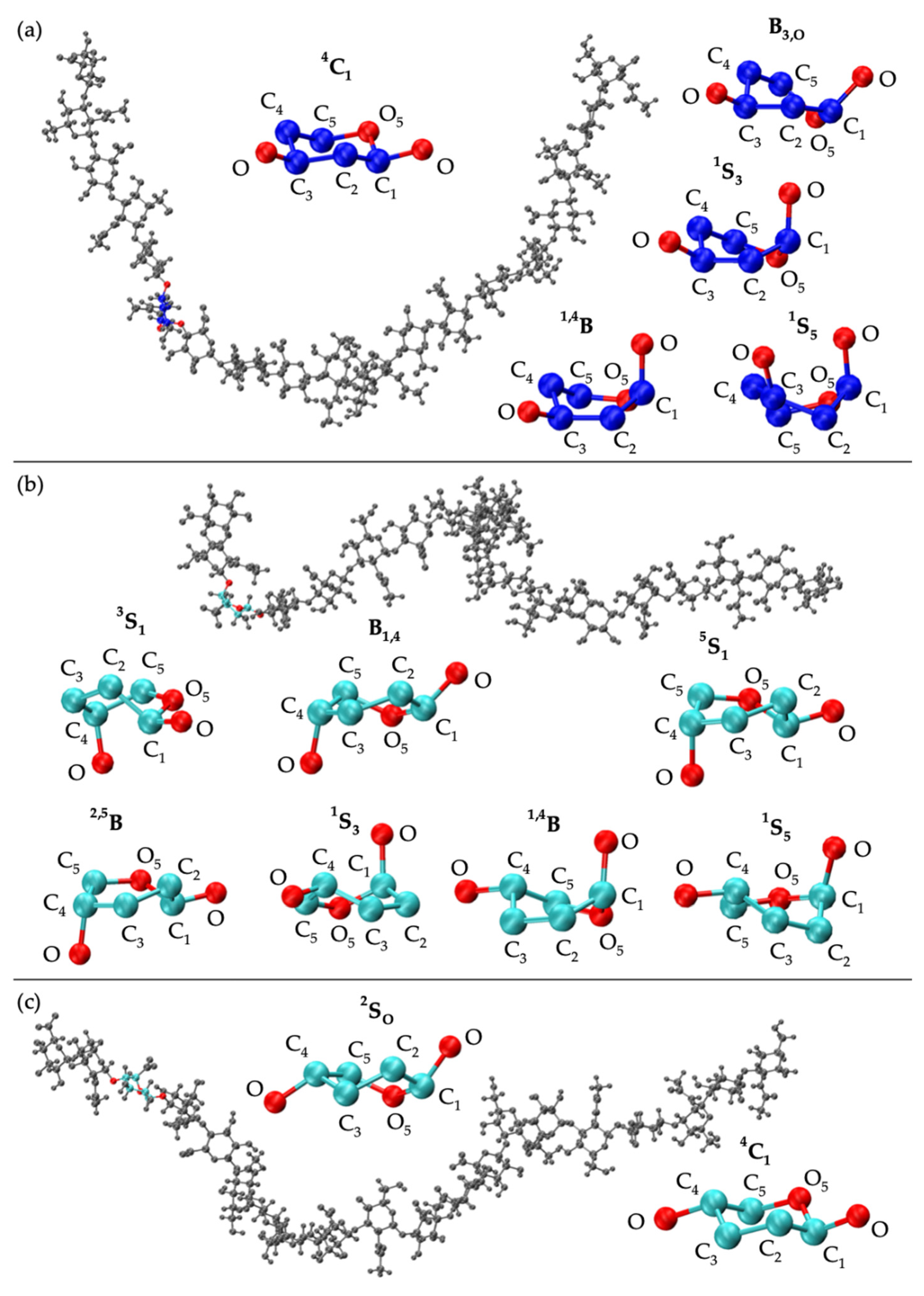
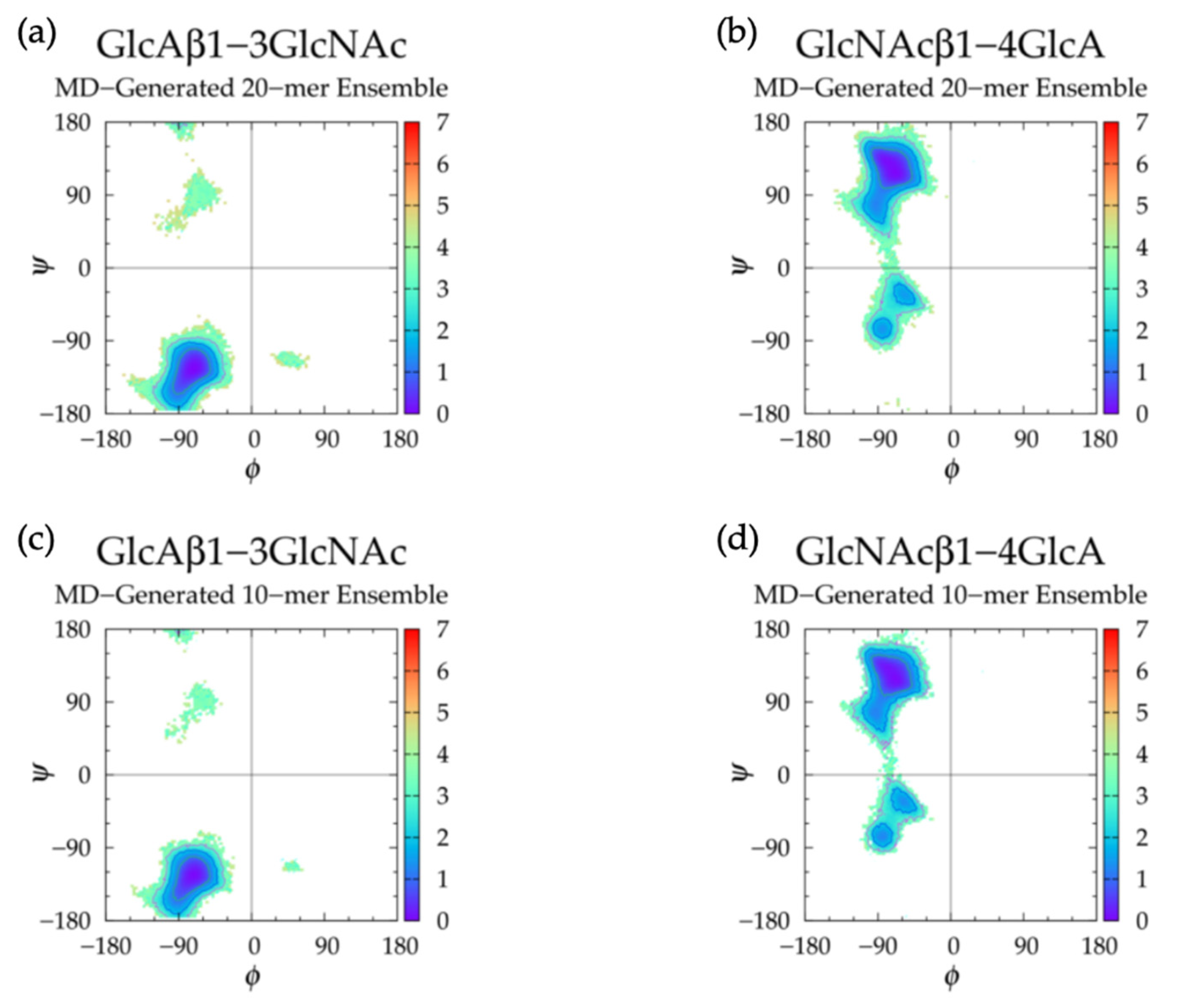
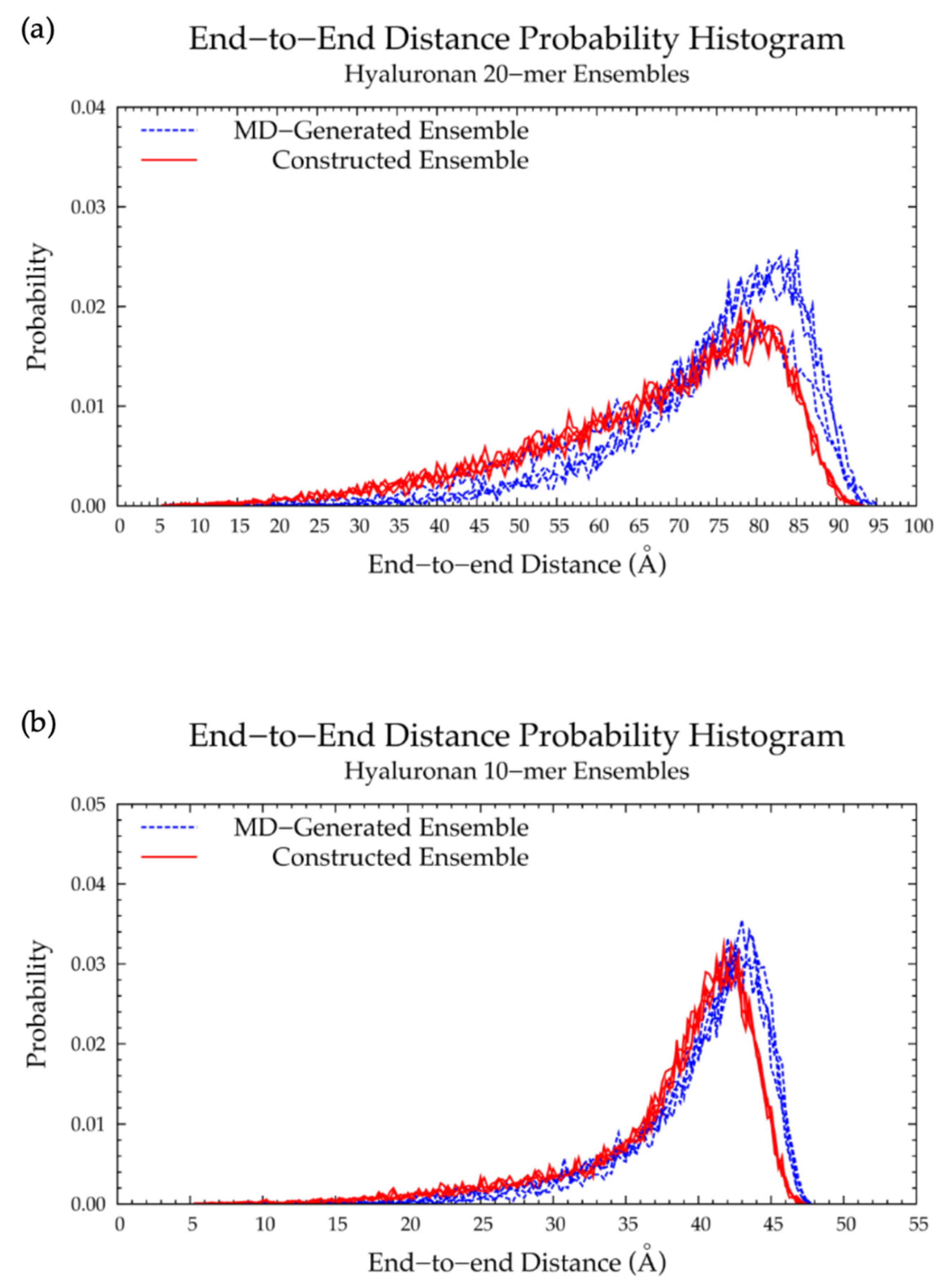


| 20-mer Ensembles | 10-mer Ensembles | |||||
|---|---|---|---|---|---|---|
| MD-Generated d (Å) | Constructed d (Å) | % Difference | MD-Generated d (Å) | Constructed d (Å) | % Difference | |
| Run 1 | 81.5 | 80.5 | 43.00 | 42.25 | ||
| Run 2 | 81.0 | 78.0 | 42.50 | 41.75 | ||
| Run 3 | 83.0 | 80.5 | 44.00 | 42.75 | ||
| Run 4 | 85.0 | 78.0 | 43.50 | 42.75 | ||
| All 2 | 80.0 | 78.0 | 2.53% | 43.50 | 41.75 | 4.11% |
| MD-Generated 20-mer Ensemble | MD-Generated 10-mer Ensemble | Constructed 20-mer Ensemble | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GlcAβ1-3 GlcNAc | GlcNAcβ1-4 GlcA | GlcAβ1-3 GlcNAc | GlcNAcβ1-4 GlcA | GlcAβ1-3 GlcNAc | GlcNAcβ1-4 GlcA | |||||||
| Min | ϕ, ψ | ΔG | ϕ, ψ | ΔG | ϕ, ψ | ΔG | ϕ, ψ | ΔG | ϕ, ψ | ΔG | ϕ, ψ | ΔG |
| I | −71.25°, −123.75° | 0.00 | −68.75°, 116.25° | 0.00 | −71.25°, −123.75° | 0.00 | −68.75°, 118.75° | 0.00 | −71.25°, −123.75° | 0.00 | −71.25°, 116.25° | 0.00 |
| II | −53.75°, 91.25° | 3.06 | −83.75°, −73.75° | 1.31 | −58.75°, 86.25° | 3.08 | −81.25°, −76.25° | 1.16 | −56.25°, 91.25° | 3.01 | −83.75°, −73.75° | 1.29 |
| II’ | −58.75°, −33.75° | 1.42 | −53.75°, −36.25° | 1.27 | −56.25°, −36.25° | 1.40 | ||||||
| 20-mer Ensembles | 10-mer Ensembles | |||||
|---|---|---|---|---|---|---|
| MD-Generated d (Å) | Constructed d (Å) | % Difference | MD-Generated d (Å) | Constructed d (Å) | % Difference | |
| Run 1 | 81.5 | 81.5 | 42.25 | 42.75 | ||
| Run 2 | 82.5 | 83.5 | 42.50 | 42.00 | ||
| Run 3 | 83.5 | 84.0 | 42.00 | 42.75 | ||
| Run 4 | 81.0 | 81.5 | 42.25 | 42.75 | ||
| All 2 | 83.5 | 81.5 | 2.42% | 42.25 | 42.75 | 1.18% |
| MD-Generated 20-mer Ensemble | MD-Generated 10-mer Ensemble | Constructed 20-mer Ensemble | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IdoAβ1-3 GalNAc | GalNAcβ1-4 IdoA | IdoAβ1-3 GalNAc | GalNAcβ1-4 IdoA | IdoAβ1-3 GalNAc | GalNAcβ1-4 IdoA | |||||||
| Min | ϕ, ψ | ΔG | ϕ, ψ | ΔG | ϕ, ψ | ΔG | ϕ, ψ | ΔG | ϕ, ψ | ΔG | ϕ, ψ | ΔG |
| I | −66.25°, −123.75° | 0.00 | −61.25°, 106.25° | 0.00 | −68.75°, −121.25° | 0.00 | −58.75°, 108.75° | 0.00 | −68.75°, −121.25° | 0.00 | −61.25°, 108.75° | 0.00 |
| 20-mer Ensembles | 10-mer Ensembles | |||||
|---|---|---|---|---|---|---|
| MD-Generated d (Å) | Constructed d (Å) | % Difference | MD-Generated d (Å) | Constructed d (Å) | % Difference | |
| Run 1 | 88.0 | 86.0 | 45.50 | 45.00 | ||
| Run 2 | 90.0 | 87.5 | 46.00 | 45.75 | ||
| Run 3 | 90.0 | 87.0 | 45.75 | 45.00 | ||
| Run 4 | 89.0 | 86.0 | 45.50 | 45.00 | ||
| All 2 | 90.0 | 87.0 | 3.39% | 45.50 | 45.00 | 1.10% |
| MD-Generated 20-mer Ensemble | MD-Generated 10-mer Ensemble | Constructed 20-mer Ensemble | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Galβ1-4 GlcNAc | GlcNAcβ1-3 Gal | Galβ1-4 GlcNAc | GlcNAcβ1-3 Gal | Galβ1-4 GlcNAc | GlcNAcβ1-3 Gal | |||||||
| Min | ϕ, ψ | ΔG | ϕ, ψ | ΔG | ϕ, ψ | ΔG | ϕ, ψ | ΔG | ϕ, ψ | ΔG | ϕ, ψ | ΔG |
| I | −73.75°, 116.25° | 0.00 | −81.25°, −156.25° | 0.00 | −71.25°, 116.25° | 0.00 | −81.25°, −156.25° | 0.00 | −71.25°, 118.75° | 0.00 | −81.25°, −158.75° | 0.00 |
| II | −56.25°, −31.25° | 1.75 | −56.25°, −36.25° | 1.67 | −53.75°, −33.75° | 1.75 | ||||||
| 20-mer Ensembles | 10-mer Ensembles | |||||
|---|---|---|---|---|---|---|
| MD-Generated d (Å) | Constructed d (Å) | % Difference | MD-Generated d (Å) | Constructed d (Å) | % Difference | |
| Run 1 | 68.5 | 70.0 | 37.00 | 37.25 | ||
| Run 2 | 68.5 | 72.0 | 36.25 | 38.25 | ||
| Run 3 | 67.5 | 67.5 | 37.50 | 37.25 | ||
| Run 4 | 66.5 | 71.5 | 36.75 | 36.75 | ||
| All 2 | 68.5 | 72.0 | 4.98% | 37.00 | 37.25 | 0.67% |
| MD-Generated 20-mer Ensemble | MD-Generated 10-mer Ensemble | Constructed 20-mer Ensemble | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IdoAα1-4 GlcNAc | GlcNAcα1-4 IdoA | IdoAα1-4 GlcNAc | GlcNAcα1-4 IdoA | IdoAα1-4 GlcNAc | GlcNAcα1-4 IdoA | |||||||
| Min | ϕ, ψ | ΔG | ϕ, ψ | ΔG | ϕ, ψ | ΔG | ϕ, ψ | ΔG | ϕ, ψ | ΔG | ϕ, ψ | ΔG |
| I | −73.75°, 128.75° | 0.00 | 76.25°, 88.75° | 0.00 | −76.25°, 128.75° | 0.00 | 73.75°, 88.75° | 0.00 | −76.25°, 128.75° | 0.00 | −71.25°, 116.25° | 0.00 |
| II | −103.75°, −68.75° | 3.13 | −103.75°, −63.75° | 2.66 | −98.75°, −68.75° | 3.23 | ||||||
| II’ | −66.25°, −31.25° | 3.38 | −66.25°, −33.75° | 2.99 | −61.25°, −36.25° | 3.34 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Whitmore, E.K.; Martin, D.; Guvench, O. Constructing 3-Dimensional Atomic-Resolution Models of Nonsulfated Glycosaminoglycans with Arbitrary Lengths Using Conformations from Molecular Dynamics. Int. J. Mol. Sci. 2020, 21, 7699. https://doi.org/10.3390/ijms21207699
Whitmore EK, Martin D, Guvench O. Constructing 3-Dimensional Atomic-Resolution Models of Nonsulfated Glycosaminoglycans with Arbitrary Lengths Using Conformations from Molecular Dynamics. International Journal of Molecular Sciences. 2020; 21(20):7699. https://doi.org/10.3390/ijms21207699
Chicago/Turabian StyleWhitmore, Elizabeth K., Devon Martin, and Olgun Guvench. 2020. "Constructing 3-Dimensional Atomic-Resolution Models of Nonsulfated Glycosaminoglycans with Arbitrary Lengths Using Conformations from Molecular Dynamics" International Journal of Molecular Sciences 21, no. 20: 7699. https://doi.org/10.3390/ijms21207699
APA StyleWhitmore, E. K., Martin, D., & Guvench, O. (2020). Constructing 3-Dimensional Atomic-Resolution Models of Nonsulfated Glycosaminoglycans with Arbitrary Lengths Using Conformations from Molecular Dynamics. International Journal of Molecular Sciences, 21(20), 7699. https://doi.org/10.3390/ijms21207699






