The Human Digestive Tract Is Capable of Degrading Gluten from Birth
Abstract
1. Introduction
2. Results
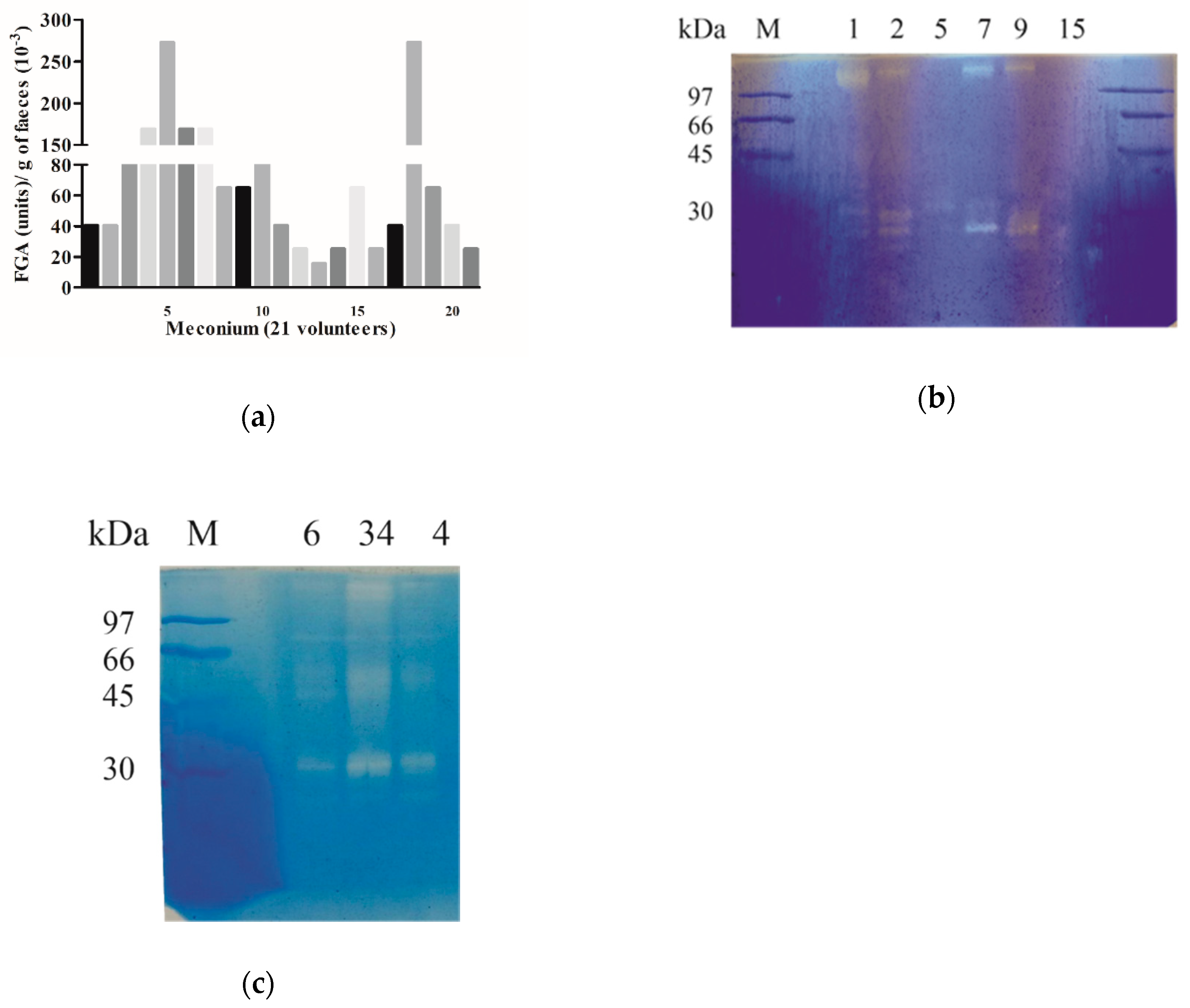
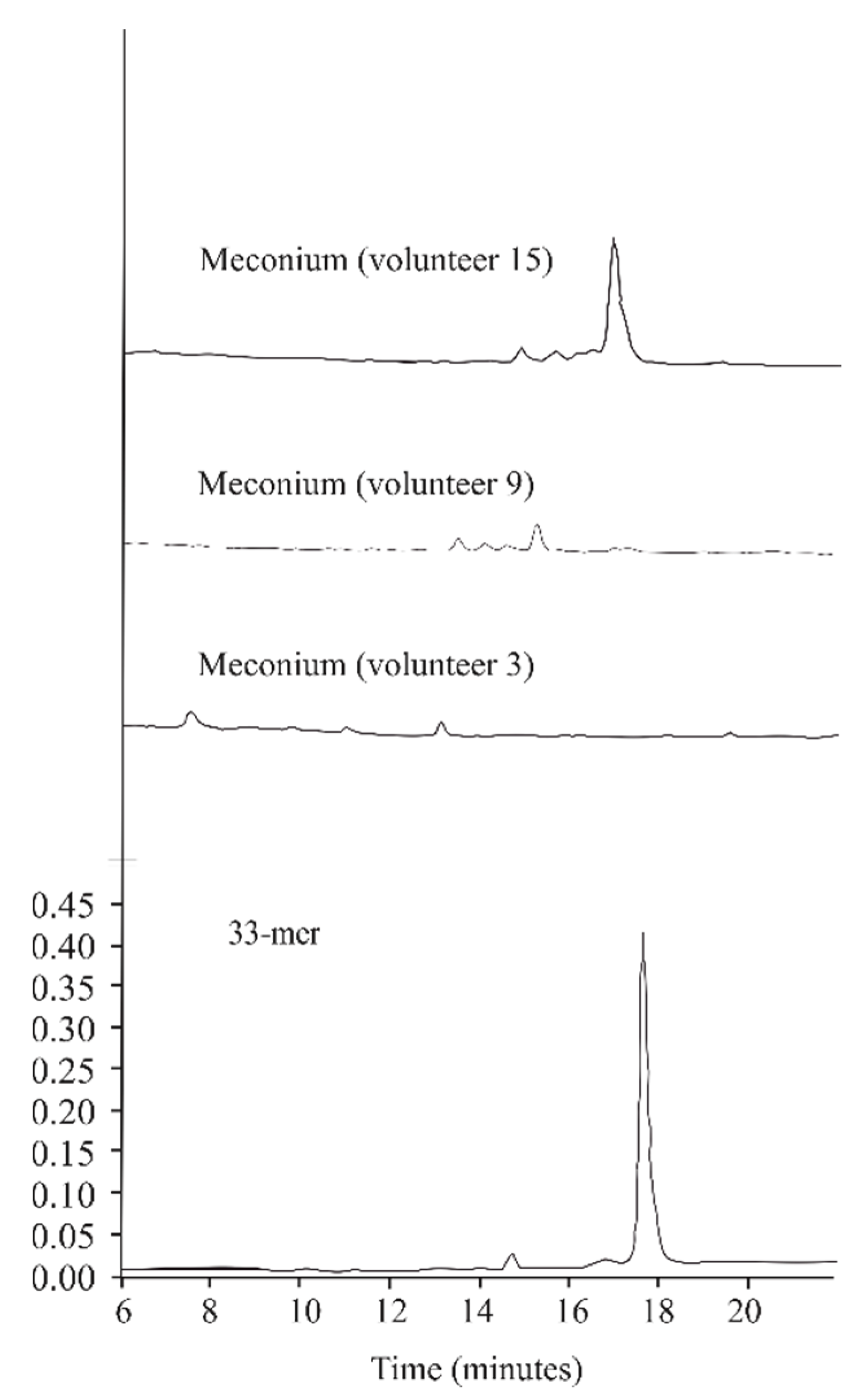
2.1. The Capacity to Metabolize Gluten Is Present in Humans from Birth
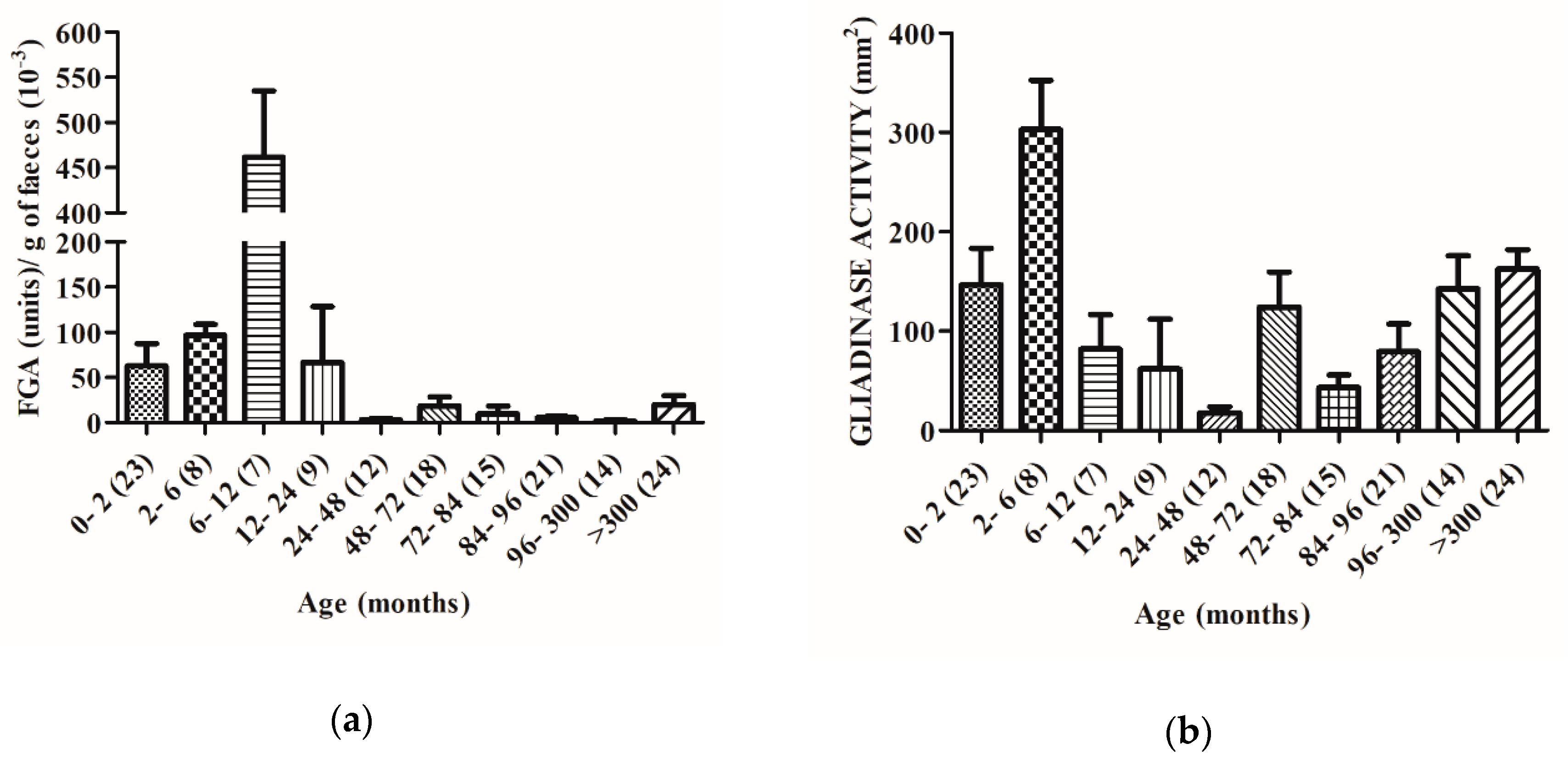
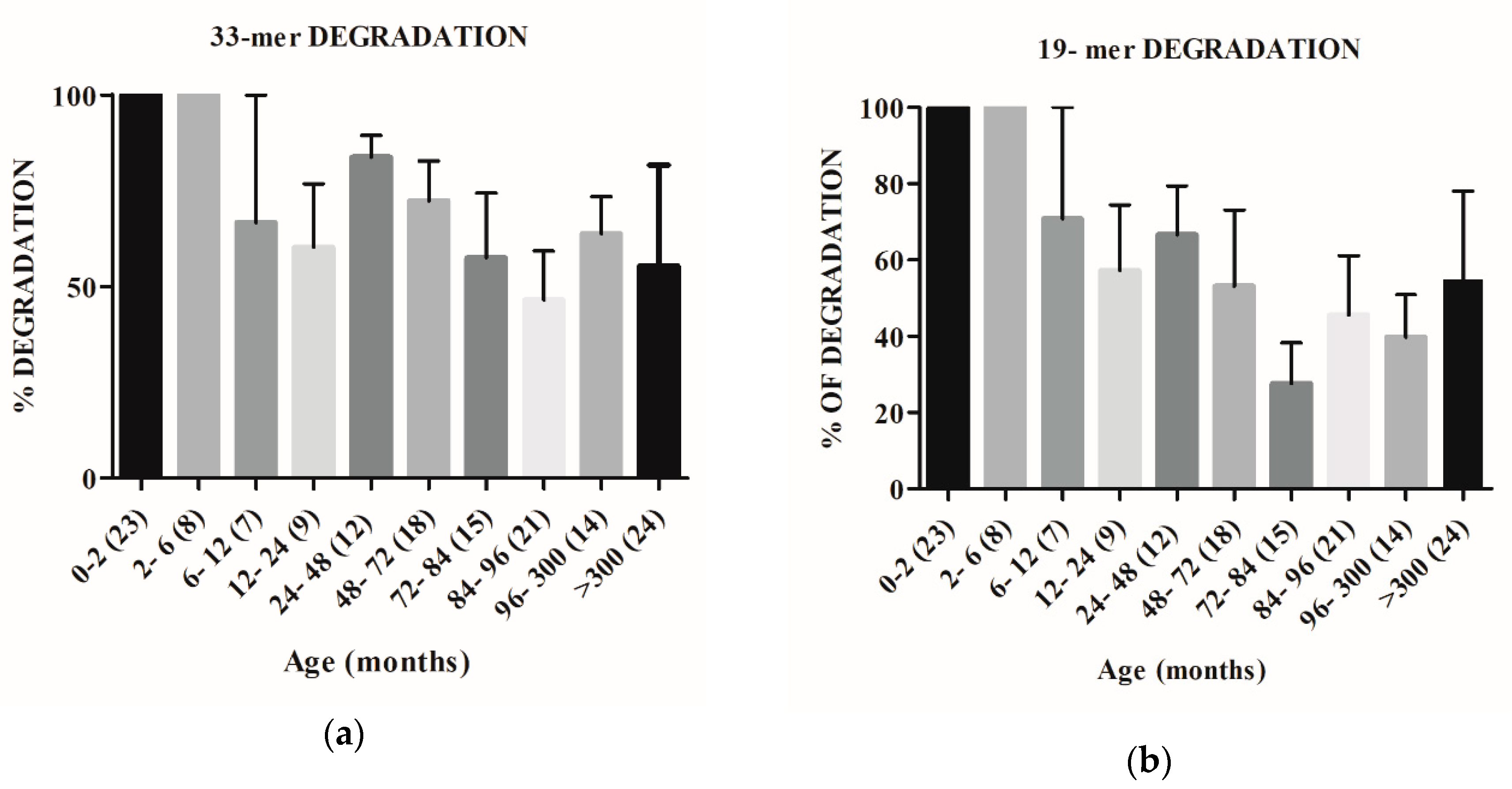
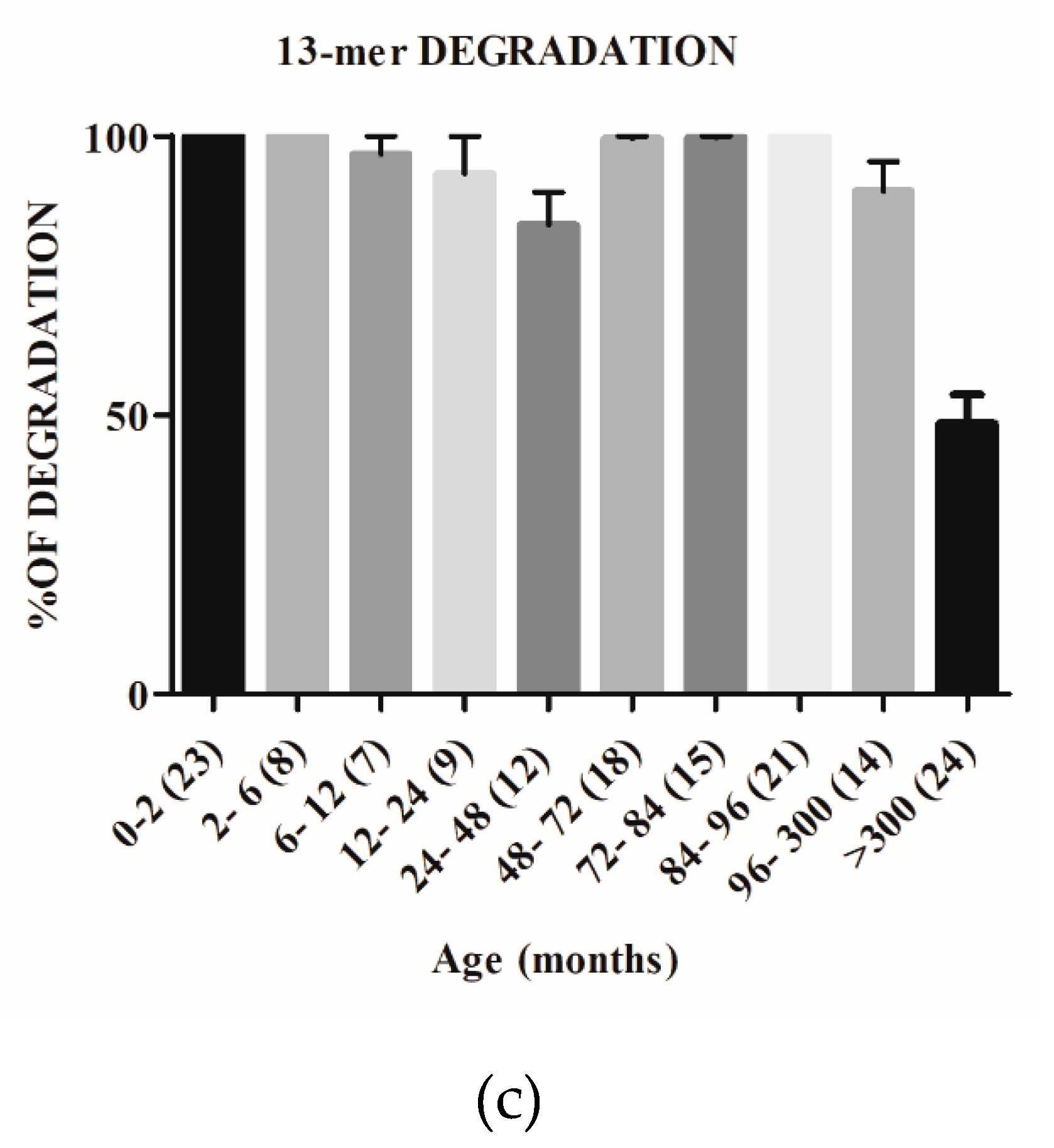
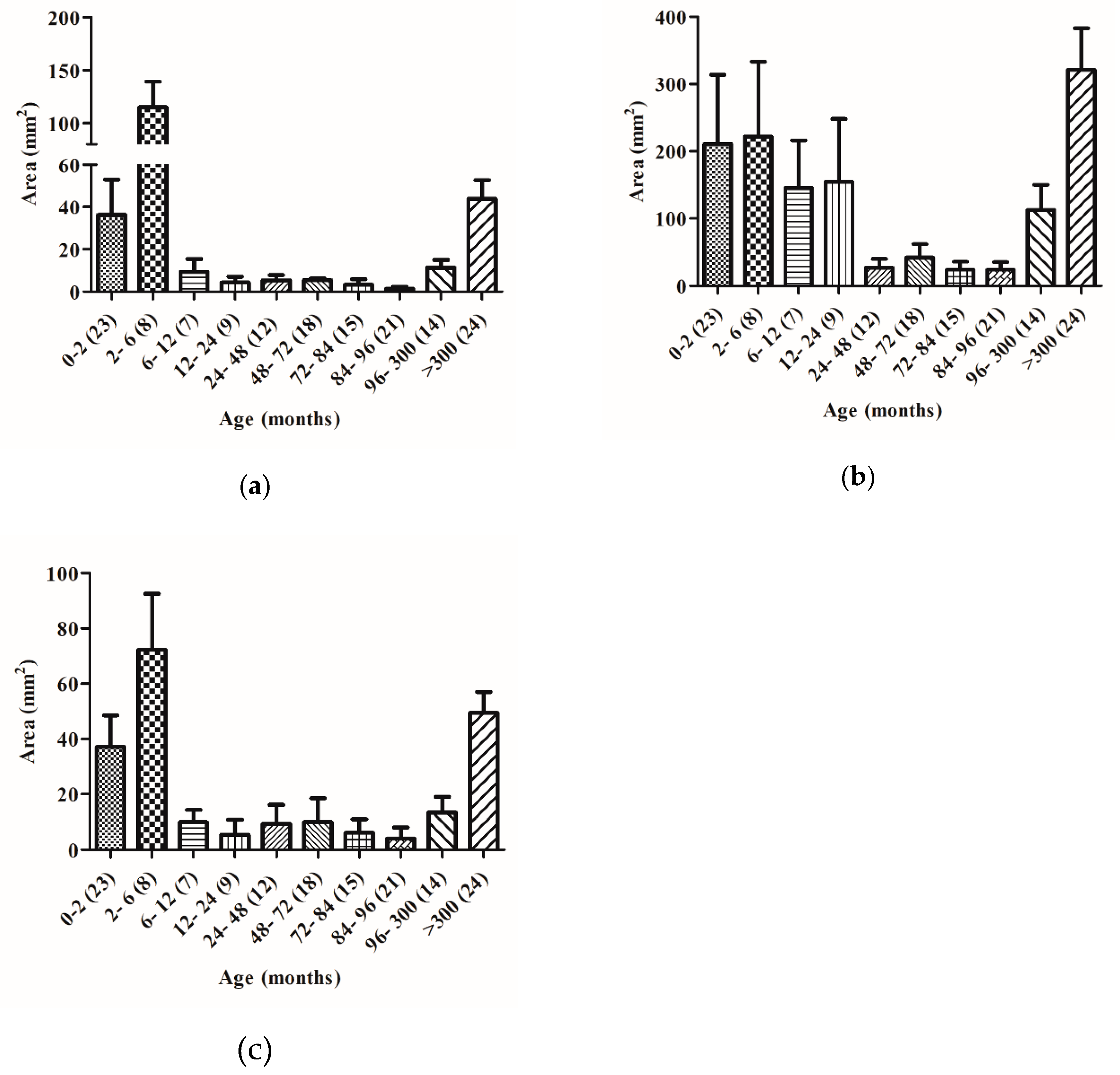
2.2. The Capacity to Degrade Gluten Changes with Age
2.3. The Gastrointestinal Proteases Involved in Gluten Human Digestion Are Present from Birth
3. Discussion
4. Materials and Methods
4.1. Fecal Sampling
4.2. Extraction and Preparation of Fecal Proteins
4.3. Evaluation of Fecal Glutenase Activity
4.4. Gel Electrophoresis
4.5. Detection of Gliadinase Activity by Zymography
4.6. Densitometric Analyses of the Electrophoretic Results
4.7. Peptidase Activity against 33-Mer, 19-Mer, and 13-Mer
4.8. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CD | Celiac disease |
| FGA | Fecal glutenase activity |
| GI | Gastrointestinal |
| LPLC | Low-performance liquid chromatography |
| NCD | Non-celiac disease |
References
- Caminero, A.; Nistal, E.; Arias, L.; Vivas, S.; Comino, I.; Real, A.; Sousa, C.; De Morales, J.M.; Ferrero, M.A.; Rodríguez-Aparicio, L.B.; et al. A gluten metabolism study in healthy individuals shows the presence of faecal glutenasic activity. Eur. J. Nutr. 2012, 51, 293–299. [Google Scholar] [CrossRef]
- Wieser, H. Chemistry of gluten proteins. Food Microbiol. 2007, 24, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Schuppan, D.; Junker, Y.; Barisani, D. Celiac Disease: From Pathogenesis to Novel Therapies. Gastroenterology 2009, 137, 1912–1933. [Google Scholar] [CrossRef] [PubMed]
- Vergnolle, N. Protease inhibition as new therapeutic strategy for GI diseases. Gut 2016, 65, 1215–1224. [Google Scholar] [CrossRef]
- Sollid, L.M. Molecular Basis of Celiac Disease. Annu. Rev. Immunol. 2000, 18, 53–81. [Google Scholar] [CrossRef] [PubMed]
- Liu, E.; Lee, H.-S.; Aronsson, C.A.; Hagopian, W.A.; Koletzko, S.; Rewers, M.J.; Eisenbarth, G.S.; Bingley, P.J.; Bonifacio, E.; Simell, V.; et al. Risk of Pediatric Celiac Disease According to HLA Haplotype and Country. N. Engl. J. Med. 2014, 371, 42–49. [Google Scholar] [CrossRef]
- Vriezinga, S.L.; Schweizer, J.J.; Koning, F.; Mearin, M.L. Coeliac disease and gluten-related disorders in childhood. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 527–536. [Google Scholar] [CrossRef]
- Vivas, S.; Vaquero, L.; Rodríguez-Martín, L.; Caminero, A. Age-related differences in celiac disease: Specific characteristics of adult presentation. World J. Gastrointest. Pharmacol. Ther. 2015, 6, 207–212. [Google Scholar] [CrossRef]
- Vivas, S.; De Morales, J.R.; Fernandez, M.; Hernando, M.; Herrero, B.; Casqueiro, J.; Gutierrez, S. Age-Related Clinical, Serological, and Histopathological Features of Celiac Disease. Am. J. Gastroenterol. 2008, 103, 2360–2365. [Google Scholar] [CrossRef]
- Guandalini, S.; Assiri, A. Celiac Disease: A Review. JAMA Pediatr. 2014, 168, 272–278. [Google Scholar] [CrossRef]
- Caminero, A.; Nistal, E.; Herrán, A.R.; Pérez-Andrés, J.; Ferrero, M.A.; Ayala, L.V.; Vivas, S.; Ruiz de Morales, J.M.; Albillos, S.M.; Casqueiro, F.J. Differences in gluten metabolism among healthy volunteers, coeliac disease patients and first-degree relatives. Br. J. Nutr. 2015, 114, 1157–1167. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, S.; Pérez-Andrés, J.; Martínez-Blanco, H.; Ferrero, M.A.; Vaquero, L.; Vivas, S.; Casqueiro, J.; Rodríguez-Aparicio, L.B. The human digestive tract has proteases capable of gluten hydrolysis. Mol. Metab. 2017, 6, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Wilczyńska, P.; Skarżyńska, E.; Lisowska-Myjak, B. Meconium microbiome as a new source of information about long-term health and disease: Questions and answers. J. Matern Fetal Neonatal Med. 2019, 32, 681–686. [Google Scholar] [CrossRef] [PubMed]
- Rescigno, M.; Urbano, M.; Valzasina, B.; Francolini, M.; Rotta, G.; Bonasio, R.; Granucci, F.; Kraehenbuhl, J.P.; Ricciardi-Castagnoli, P. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2001, 2, 361–367. [Google Scholar] [CrossRef]
- Pérez-Andrés, J. Aislamiento, Identificación Y Caracterización DE Microorganismos Involucrados en El Metabolismo Del Gluten: Implicaciones Para La Enfermedad Celiaca Y La Salud Humana. Ph.D. Thesis, Universidad de León, León, Spain, 2017. Available online: https://buleria.unileon.es/handle/10612/8862?show=full (accessed on 30 October 2018).
- Serena, G.; Fasano, A. Use of Probiotics to Prevent Celiac Disease and IBD in Pediatrics. In Advances in Microbiology, Infectious Diseases and Public Health; Guandalini, S., Indrio, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2018; Volume 1125, pp. 69–81. [Google Scholar] [CrossRef]
- Quero Acosta, L.; Coronel Rodríguez, C.; Argüelles Martín, F. Influence of infant feeding on the excretion of gluten immunopeptides in feces. Rev. Esp. Enferm. Dig. 2019, 111, 134–139. [Google Scholar] [CrossRef]
- Cuccioloni, M.; Mozzicafreddo, M.; Ali, I.; Bonfili, L.; Cecarini, V.; Eleuteri, A.M.; Angeletti, M. Interaction between wheat alpha-amylase/trypsin bi-functional inhibitor and mammalian digestive enzymes: Kinetic, equilibrium and structural characterization of binding. Food Chem. 2016, 213, 571–578. [Google Scholar] [CrossRef]
- Gamage, H.K.A.H.; Tetu, S.G.; Chong, R.W.W.; Ashton, J.; Packer, N.H.; Paulsen, I.T. Cereal products derived from wheat, sorghum, rice and oats alter the infant gut microbiota in vitro. Sci. Rep. 2017, 7, 14312. [Google Scholar] [CrossRef]
- Arrieta, M.-C.; Stiemsma, L.T.; Amenyogbe, N.; Brown, E.M. The Intestinal Microbiome in Early Life: Health and Disease. Front. Immunol. 2014, 5, 427. [Google Scholar] [CrossRef]
- Vriezinga, S.L.; Auricchio, R.; Bravi, E.; Castillejo, G.; Chmielewska, A.; Crespo Escobar, P.; Kolaček, S.; Koletzko, S.; Korponay-Szabo, I.R.; Mummert, E.; et al. Randomized feeding intervention in infants at high risk for celiac disease. N. Engl. J. Med. 2014, 371, 1304–1315. [Google Scholar] [CrossRef]
- Szajewska, H.; Shamir, R.; Mearin, L.; Ribes-Koninckx, C.; Catassi, C.; Domellöf, M.; Fewtrell, M.S.; Husby, S.; Papadopoulou, A.; Vandenplas, Y.; et al. Gluten Introduction and the Risk of Coeliac Disease: A Position Paper by the European Society for Pediatric Gastroenterology, Hepatology, and Nutrition. J. Pediatr. Gastroenterol. Nutr. 2016, 62, 507–513. [Google Scholar] [CrossRef]
- Junker, Y.; Zeissig, S.; Kim, S.-J.; Barisani, D.; Wieser, H.; Leffler, D.A.; Zevallos, V.; Libermann, T.A.; Dillon, S.; Freitag, T.L.; et al. Wheat amylase trypsin inhibitors drive intestinal inflammation via activation of toll-like receptor 4. J. Exp. Med. 2012, 209, 2395–2408. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680. [Google Scholar] [CrossRef] [PubMed]
- Helmerhorst, E.J.; Wei, G. Experimental strategy to discover microbes with gluten-degrading enzyme activities. Proc. SPIE Int. Soc. Opt. Eng. 2014, 9112. [Google Scholar] [CrossRef]
- Caminero, A.; Herrán, A.R.; Nistal, E.; Pérez-Andrés, J.; Vaquero, L.; Vivas, S.; Ruiz de Morales, J.M.; Albillos, S.M.; Casqueiro, J. Diversity of the cultivable human gut microbiome involved in gluten metabolism: Isolation of microorganisms with potential interest for coeliac disease. FEMS Microbiol. Ecol. 2014, 88, 309–319. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández-Pérez, S.; Pérez-Andrés, J.; Gutiérrez, S.; Navasa, N.; Martínez-Blanco, H.; Ferrero, M.Á.; Vivas, S.; Vaquero, L.; Iglesias, C.; Casqueiro, J.; et al. The Human Digestive Tract Is Capable of Degrading Gluten from Birth. Int. J. Mol. Sci. 2020, 21, 7696. https://doi.org/10.3390/ijms21207696
Fernández-Pérez S, Pérez-Andrés J, Gutiérrez S, Navasa N, Martínez-Blanco H, Ferrero MÁ, Vivas S, Vaquero L, Iglesias C, Casqueiro J, et al. The Human Digestive Tract Is Capable of Degrading Gluten from Birth. International Journal of Molecular Sciences. 2020; 21(20):7696. https://doi.org/10.3390/ijms21207696
Chicago/Turabian StyleFernández-Pérez, Silvia, Jenifer Pérez-Andrés, Sergio Gutiérrez, Nicolás Navasa, Honorina Martínez-Blanco, Miguel Ángel Ferrero, Santiago Vivas, Luis Vaquero, Cristina Iglesias, Javier Casqueiro, and et al. 2020. "The Human Digestive Tract Is Capable of Degrading Gluten from Birth" International Journal of Molecular Sciences 21, no. 20: 7696. https://doi.org/10.3390/ijms21207696
APA StyleFernández-Pérez, S., Pérez-Andrés, J., Gutiérrez, S., Navasa, N., Martínez-Blanco, H., Ferrero, M. Á., Vivas, S., Vaquero, L., Iglesias, C., Casqueiro, J., & Rodríguez-Aparicio, L. B. (2020). The Human Digestive Tract Is Capable of Degrading Gluten from Birth. International Journal of Molecular Sciences, 21(20), 7696. https://doi.org/10.3390/ijms21207696




