Genome and Karyotype Reorganization after Whole Genome Duplication in Free-Living Flatworms of the Genus Macrostomum
Abstract
1. Introduction
2. Results
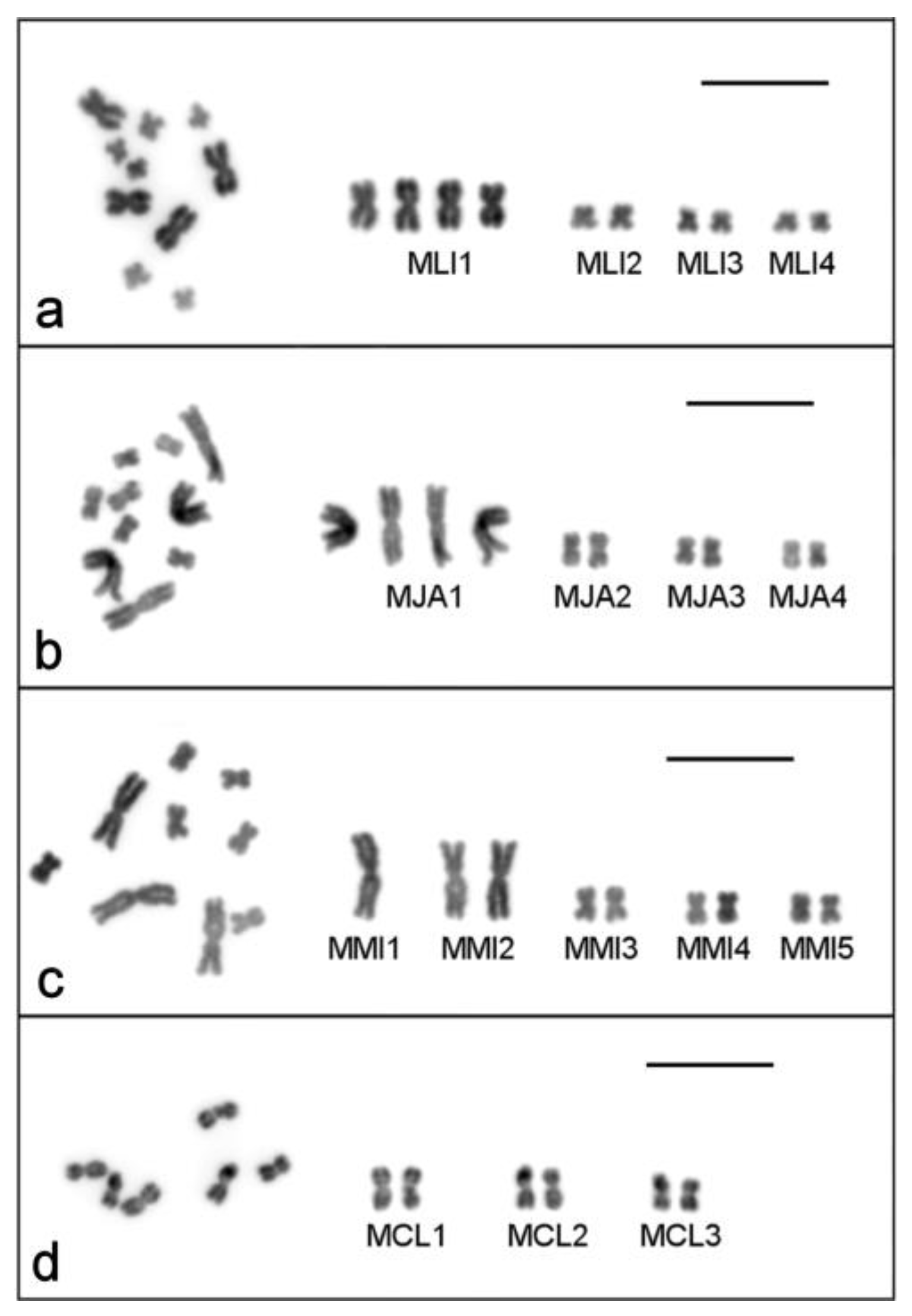
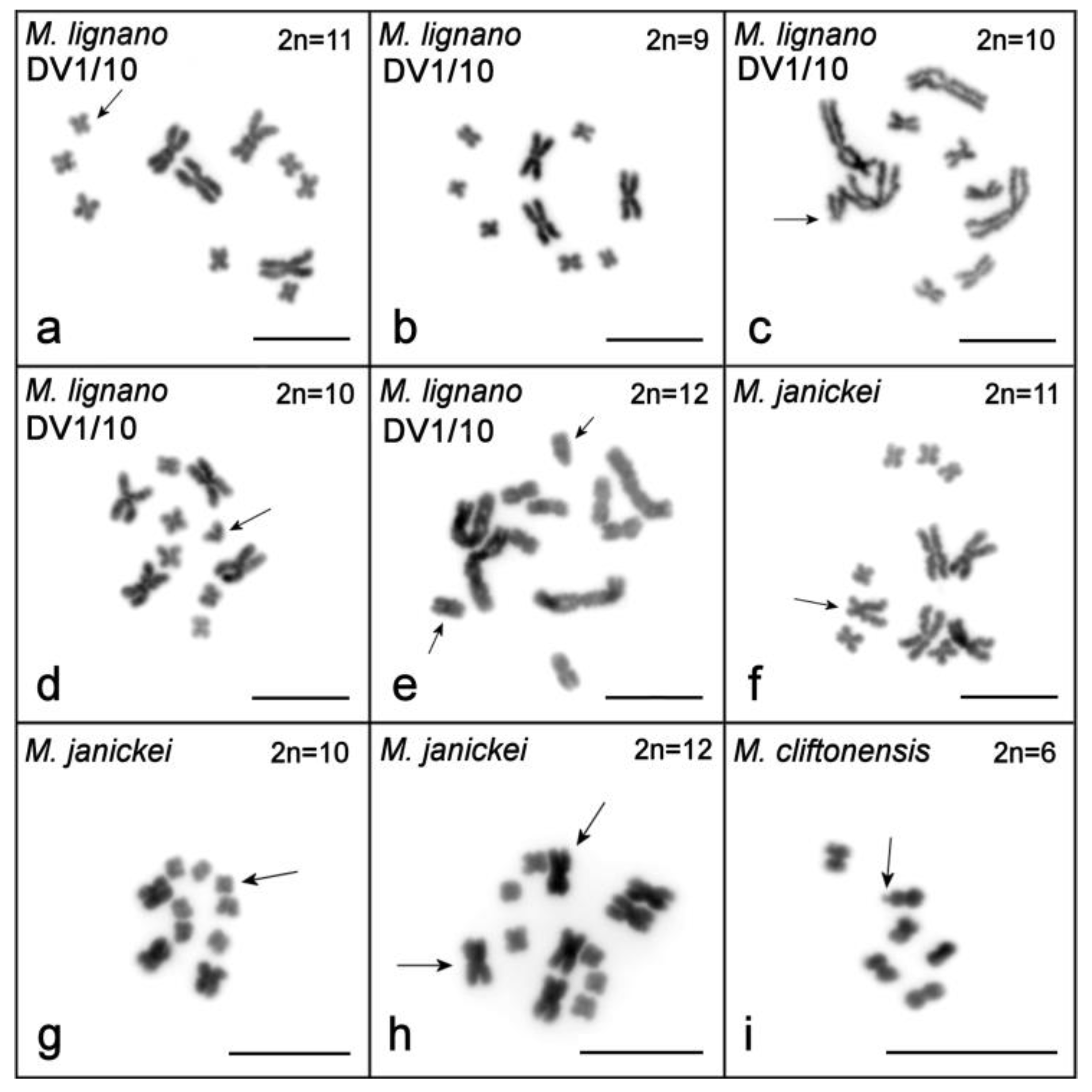
2.1. Single-Worm Karyotyping of Four Macrostomum Species
2.1.1. Macrostomum lignano
2.1.2. Macrostomum janickei
2.1.3. Macrostomum mirumnovem
2.1.4. Macrostomum cliftonensis
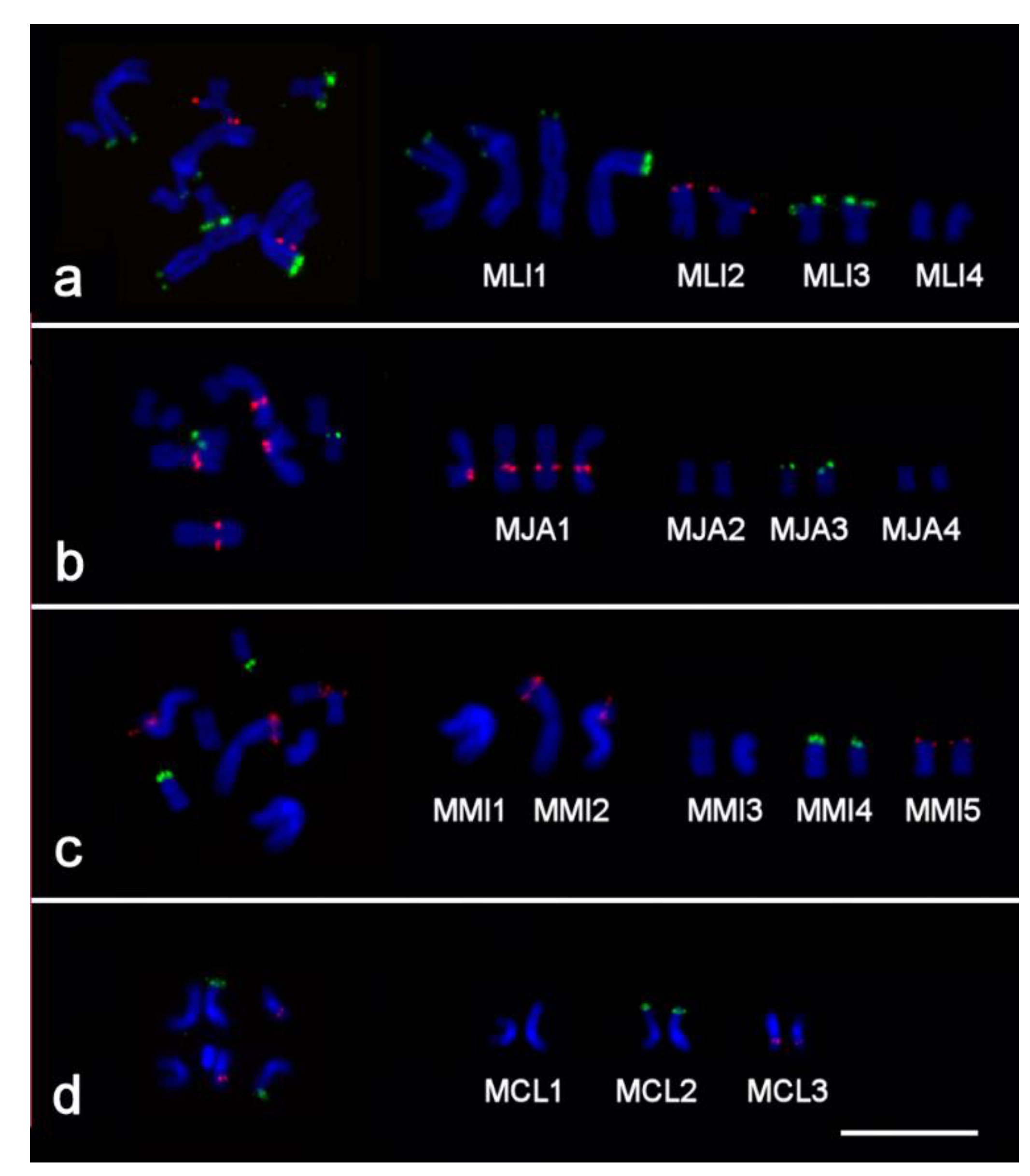
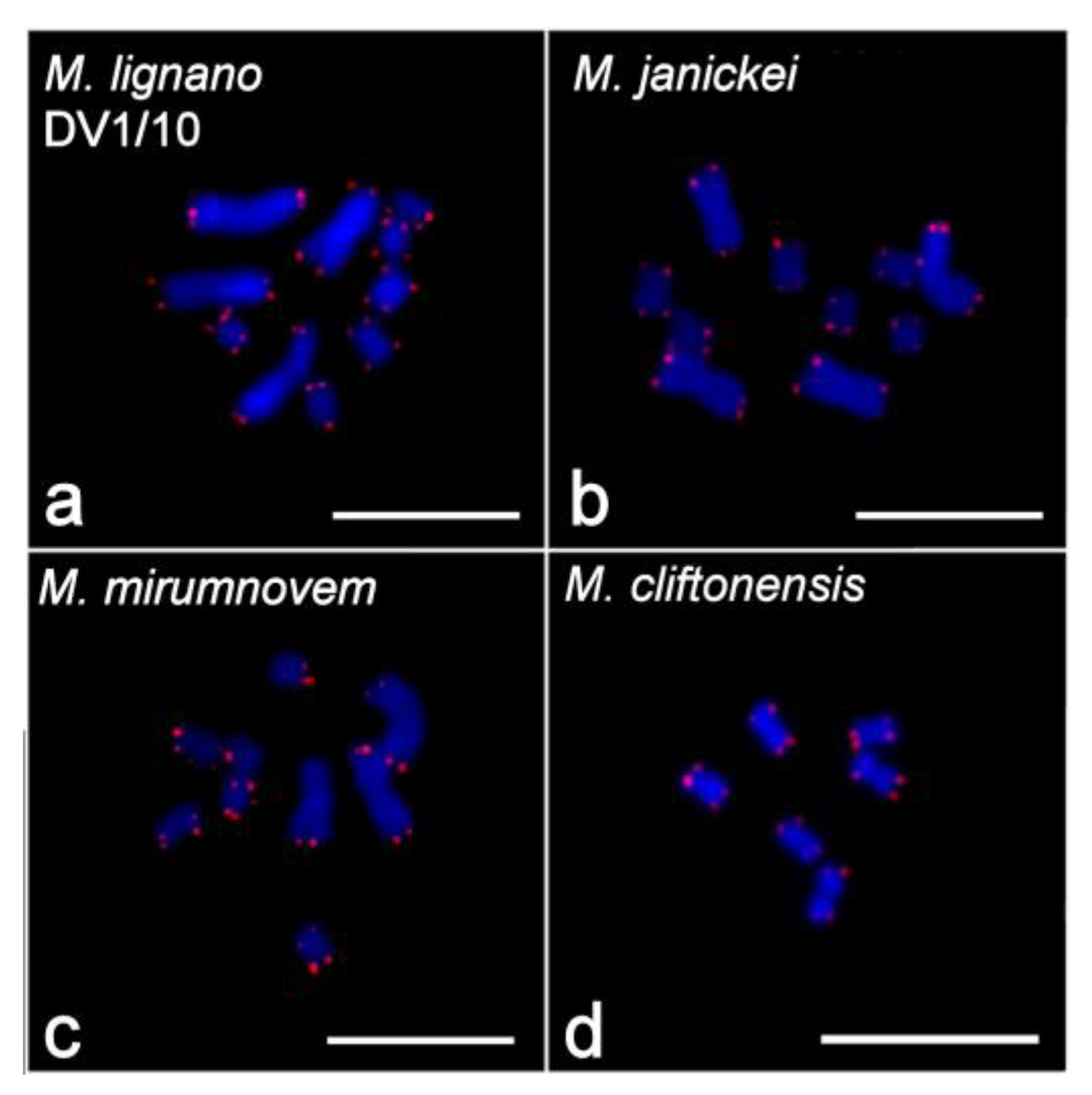
2.2. Clusters of 5S rDNA, 28S rDNA, and Telomeric Repeats in Chromosomes of Studied Species
2.3. FISH with Microdissected DNA Probes on Macrostomum Chromosomes
2.3.1. Cross-Species In Situ Hybridization
2.3.2. Microdissected Region-Specific DNA Probes Obtained from Large Chromosomes of M. mirumnovem
2.3.3. FISH with Single-Copy Microdissected DNA Probes Derived from Chromosomes of M. mirumnovem
2.3.4. FISH Analysis of Copy Number Variation of Large Chromosomes and Presence of B Chromosomes in M. mirumnovem
3. Discussion
3.1. Differentiation of Large Chromosomes and Origin of B Chromosomes in M. mirumnovem
3.2. Possible Reasons for Karyotypic Instability in Post-WGD Macrostomum Species
3.3. Comparative Cytogenetic Analysis of Chromosomes of Macrostomum Species
3.4. Hypothetical Scenarios of WGD in the M. lignano/M. janickei and M. mirumnovem Lineages
4. Materials and Methods
4.1. Study Organisms
4.2. Metaphase Chromosome Preparation
4.3. Chromosome Staining
4.4. Microscopy Analysis
4.5. Morphometric Analysis
4.6. Chromosome Microdissection and Microdissected DNA Probe Generation
4.7. Fluorescent In situ Hybridization
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| WGD | Whole genome duplication |
| TE | Transposable element |
References
- Kimura, M. The neutral theory of molecular evolution: A review of recent evidence. Jpn. J. Genet. 1991, 66, 367–386. [Google Scholar] [CrossRef]
- Ohta, T. Nearly neutral theory of molecular evolution. Annu. Rev. Ecol. Syst. 1992, 23, 263–286. [Google Scholar] [CrossRef]
- Loewe, L.; Hill, W.G. The population genetics of mutations: Good, bad and indifferent. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010, 365, 1153–1167. [Google Scholar] [CrossRef] [PubMed]
- Conrad, B.; Antonorakis, S.E. Gene duplication: A drive for phenotypic diversity and cause of human disease. Annu. Rev. Genom. Hum. Genet. 2007, 8, 17–35. [Google Scholar] [CrossRef] [PubMed]
- De Peer, Y.; Maere, S.; Meyer, A. The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 2009, 10, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Bomblies, K.; Jones, G.; Franklin, C.; Zickler, D.; Kleckner, N. The challenge of evolving stable polyploidy: Could an increase in “crossover interference distance” play a central role? Chromosoma 2016, 125, 287–300. [Google Scholar] [CrossRef]
- Lenormand, T.; Engelstädter, J.; Johnston, S.E.; Wijnker, E.; Haag, C.R. Evolutionary mysteries in meiosis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016, 371, 20160001. [Google Scholar] [CrossRef]
- Wolfe, K.H.; Shields, D.C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 1997, 387, 708–713. [Google Scholar] [CrossRef]
- Dehal, P.; Boore, J.L. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005, 3, e314. [Google Scholar] [CrossRef]
- Aury, J.; Jaillon, O.; Duret, L.; Noel, B.; Jubin, C.; Porcel, B.M.; Ségurens, B.; Daubin, V.; Anthouard, V.; Aiach, N.; et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 2006, 444, 171–178. [Google Scholar] [CrossRef]
- Jiao, Y.; Wickett, N.J.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.; Soltis, P.S.; et al. Ancestral polyploidy in seed plants and angiosperms. Nature 2011, 473, 97–100. [Google Scholar] [CrossRef] [PubMed]
- Schwager, E.E.; Sharma, P.P.; Clarke, T.; Schwager, E.E.; Sharma, P.P.; Clarke, T.; Leite, D.J.; Wierschin, T.; Pechmann, M.; Akiyama-Oda, Y.; et al. The house spider genome reveals an ancient whole-genome duplication during arachnid evolution. BMC Biol. 2017, 15, 62. [Google Scholar] [CrossRef] [PubMed]
- Flot, J.F.; Hespeels, B.; Li, X.; Noel, B.; Arkhipova, I.; Danchin, E.G.; Hejnol, A.; Henrissat, B.; Koszul, R.; Aury, J.M.; et al. Genomic evidence for ameiotic evolution in the bdelloid rotifer Adineta vaga. Nature 2013, 500, 453–457. [Google Scholar] [CrossRef]
- Kenny, N.J.; Chan, K.W.; Nong, W.; Qu, Z.; Maeso, I.; Yip, H.Y.; Chan, T.F.; Kwan, H.S.; Holland, P.W.H.; Chu, K.H.; et al. Ancestral whole-genome duplication in the marine chelicerate horseshoe crabs. Heredity 2016, 116, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Glasauer, S.M.; Neuhauss, S.C. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol. Genet. Genom. 2014, 289, 1045–1060. [Google Scholar] [CrossRef] [PubMed]
- Mable, B.K.; Alexandrou, M.A.; Taylor, M.I. Genome duplication in amphibians and fish: An extended synthesis. J. Zool. 2011, 284, 151–182. [Google Scholar] [CrossRef]
- Session, A.M.; Uno, Y.; Kwon, T.; Chapman, J.A.; Toyoda, A.; Takahashi, S.; Fukui, A.; Hikosaka, A.; Suzuki, A.; Kondo, M.; et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature 2016, 538, 336–343. [Google Scholar] [CrossRef]
- Chen, Z.; Omori, Y.; Koren, S.; Shirokiya, T.; Kuroda, T.; Miyamoto, A.; Wada, H. De novo assembly of the goldfish (Carassius auratus) genome and the evolution of genes after whole-genome duplicaton. Sci. Adv. 2019, 5, aav0547. [Google Scholar] [CrossRef]
- McElroy, K.E.; Denton, R.D.; Sharbrough, J.; Bankers, L.; Neiman, M.; Lisle Gibbs, H. Genome expression balance in a triploid trihybrid vertebrate. Genome Biol. Evol. 2017, 9, 968–980. [Google Scholar] [CrossRef]
- Mason, A.S.; Chris Pires, J. Unreduced gametes: Meiotic mishap or evolutionary mechanism? Trends Genet. 2015, 31, 5–10. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Q.; Luo, K.; Chen, X.; Xiao, J.; Zhang, C.; Tao, M.; Zhao, R.; Liu, S. Cell fusion as the formation mechanism of unreduced gametes in the gynogenetic diploid hybrid fish. Sci. Rep. 2016, 31658, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Alix, K.; Gerard, P.R.; Schwarzacher, T.; Heslop-Harrison, J.S. Polyploidy and interspecific hybridization: Partners for adaptation, speciation and evolution in plants. Ann. Bot. 2017, 120, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Knytl, M.; Smolík, O.; Kubíčková, S.; Tlapáková, T.; Evans, B.J.; Krylov, V. Chromosome divergence during evolution of the tetraploid clawed frogs, Xenopus mellotropicalis and Xenopus epitropicalis as revealed by Zoo-FISH. PLoS ONE 2017, 12, e0177087. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef]
- Lien, S.; Koop, B.F.; Sandve, S.R.; Miller, J.R.; Kent, M.P.; Nome, T.; Hvidsten, T.R.; Leong, J.S.; Minkley, D.R.; Zimin, A.; et al. The Atlantic salmon genome provides insights into rediploidization. Nature 2016, 533, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ye, L.H.; Liu, Q.Z.; Peng, L.Y.; Liu, W.; Yi, X.G.; Wang, Y.D.; Xiao, J.; Xu, K.; Hu, F.Z.; et al. Rapid genomic DNA changes in allotetraploid fish hybrids. Heredity 2015, 114, 601–609. [Google Scholar] [CrossRef]
- Otto, S.P.; Yong, P. The evolution of gene duplicates. Adv. Genet. 2002, 46, 451–483. [Google Scholar]
- Mikulíček, P.; Kautman, M.; Kautman, J.; Pruvost, N.B.M. Mode of hybridogenesis and habitat preferences influence population composition of water frogs (Pelophylax esculentus complex, Anura: Ranidae) in a region of sympatric occurrence (western Slovakia). J. Zool. Syst. Evol. Res. 2014, 53, 124–132. [Google Scholar] [CrossRef]
- Ma, W.; Zhu, Z.H.; Bi, X.Y.; Murphy, R.W.; Wang, S.Y.; Gao, Y.; Xiao, H.; Luo, J. Allopolyploidization is not so simple: Evidence from the origin of the tribe Cyprinini (Teleostei: Cypriniformes). Curr. Mol. Med. 2014, 14, 1331–1338. [Google Scholar] [CrossRef]
- Meier, J.; Marques, D.A.; Mwaiko, S.; Wagner, C.E.; Excoffier, L.; Seehausen, O. Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nat. Commun. 2017, 8, 14363. [Google Scholar] [CrossRef]
- Gray, Y.H.M. It takes two transposons to tango: Transposable-element-mediated chromosomal rearrangements. Trends Genet. 2000, 16, 461–468. [Google Scholar] [CrossRef]
- Metcalfe, C.J.; Bulazel, K.V.; Ferreri, G.C.; Schroeder-Reiter, E.; Wanner, G.; Rens, W.; Obergfell, C.; Eldridge, M.D.; O’Neill, R.J. Genomic instability within centromeres of interspecific marsupial hybrids. Genetics 2007, 177, 2507–2517. [Google Scholar] [CrossRef]
- Zadesenets, K.S.; Rubtsov, N.B. Genome duplication in animal evolution. Russ. J. Genet. 2018, 54, 1125–1136. [Google Scholar] [CrossRef]
- Schärer, L.; Brand, J.N.; Singh, P.; Zadesenets, K.S.; Stelzer, C.-P.; Viktorin, G. A phylogenetically informed search for an alternative Macrostomum model species, with notes on taxonomy, mating behavior, karyology and genome size. J. Zool. Syst. Evol. Res. 2019. [Google Scholar] [CrossRef]
- Egger, B.; Ishida, S. Chromosome fission or duplication in Macrostomum lignano (Macrostomorpha, Plathelminthes)—Remarks on chromosome numbers in ‘archoophoran turbellarians’. J. Zool. Syst. Evol. Res. 2005, 43, 127–132. [Google Scholar] [CrossRef]
- Zadesenets, K.S.; Vizoso, D.B.; Schlatter, A.; Konopatskaia, I.D.; Berezikov, E.; Schärer, L.; Rubtsov, N.B. Evidence for karyotype polymorphism in the free-living flatworm, Macrostomum lignano, a model organism for evolutionary and developmental biology. PLoS ONE 2016, 11, e0164915. [Google Scholar] [CrossRef]
- Wasik, K.; Gurtowski, J.; Zhou, X.; Ramos, O.M.; Delás, M.J.; Battistoni, G.; El Demerdash, O.; Falciatori, I.; Vizoso, D.B.; Smith, A.D.; et al. Genome and transcriptome of the regeneration-complement flatworm, Macrostomum lignano. Proc. Natl. Acad. Sci. USA 2015, 112, 12462–12467. [Google Scholar] [CrossRef]
- Zadesenets, K.S.; Schärer, L.; Rubtsov, N.B. New insights into the karyotype evolution of the free-living flatworm Macrostomum lignano (Platyhelminthes, Turbellaria). Sci. Rep. 2017, 7, 6066. [Google Scholar] [CrossRef]
- Zadesenets, K.S.; Ershov, N.I.; Berezikov, E.; Rubtsov, N.B. Chromosome evolution in the free-living flatworms: First evidence of intrachromosomal rearrangements in karyotype evolution of Macrostomum lignano (Platyhelminthes, Macrostomida). Genes 2017, 8, 298. [Google Scholar] [CrossRef]
- Rebollo, R.; Horard, B.; Hubert, B.; Vieira, C. Jumping genes and epigenetics: Towards new species. Gene 2010, 454, 1–7. [Google Scholar] [CrossRef]
- Serrato-Capuchina, A.; Matute, D.R. The role of transposable elements in speciation. Genes 2018, 9, 254. [Google Scholar] [CrossRef] [PubMed]
- Jetybayev, I.E.; Bugrov, A.G.; Buleu, O.G.; Bogomolov, A.G.; Rubtsov, N.B. Origin and evolution of the neo-sex chromosomes in Pamphagidae grasshoppers through chromosome fusion and following heteromorphization. Genes 2017, 8, 323. [Google Scholar] [CrossRef] [PubMed]
- Jetybayev, I.Y.; Bugrov, A.G.; Dzyubenko, V.V.; Rubtsov, N.B. B Chromosomes in grasshoppers: Different origins and pathways to the modern Bs. Genes 2018, 9, 509. [Google Scholar] [CrossRef] [PubMed]
- Valente, G.T.; Conte, M.A.; Fantinatti, B.E.A.; Cabral-De-Mello, D.C.; Carvalho, R.F.; Vicari, M.R.; Kocher, T.D.; Martins, C. Origin and evolution of B chromosomes in the cichlid fish Astatotilapia latifasciata based on integrated genomic analyses. Mol. Biol. Evol. 2014, 31, 2061–2072. [Google Scholar] [CrossRef]
- Kichigin, I.G.; Lisachov, A.P.; Giovannotti, M.; Makunin, A.I.; Kabilov, M.R.; O’Brien, P.C.M.; Ferguson-Smith, M.A.; Graphodatsky, A.S.; Trifonov, V.A. First report on B chromosome content in a reptilian species: The case of Anolis carolinensis. Mol. Genet. Genom. 2018, 294, 13–21. [Google Scholar] [CrossRef]
- Makunin, A.I.; Romanenko, S.A.; Beklemisheva, V.R.; Perelman, P.L.; Druzhkova, A.S.; Petrova, K.O.; Prokopov, D.Y.; Chernyaeva, E.N.; Johnson, J.L.; Kukekova, A.V.; et al. Sequencing of supernumerary chromosomes of red fox and raccoon dog confirms a non-random gene acquisition by B chromosomes. Genes 2018, 9, 405. [Google Scholar] [CrossRef]
- Singh, P.; Ballmer, D.; Laubscher, M.; Schärer, L. Hybridisation between two closely related flatworm species despite differences in reproductive morphology and behavior. bioRxiv 2019, 851972. [Google Scholar] [CrossRef]
- Cheng, F.; Wu, J.; Cai, X.; Liang, J.; Freeling, M.; Wang, X. Gene retention, fractionation and subgenome differences in polyploidy plants. Nat. Plants 2018, 4, 258–268. [Google Scholar] [CrossRef]
- Bakloushinskaya, I.; Romanenko, S.A.; Serdukova, N.A.; Graphodatsky, A.S.; Lyapunova, E.A. A new form of the mole vole Ellobius tancrei Blasius, 1884 (Mammalia, Rodentia) with the lowest chromosome number. Comp. Cytogenet. 2013, 7, 163–169. [Google Scholar] [CrossRef]
- Zima, J.; Fedyk, S.; Fredga, K.; Hausser, J.; Mishta, A.; Searle, J.B.; Volobouev, V.T.; Wójcik, J.M. The list of the chromosome races of the common shrew (Sorex araneus). Hereditas 1996, 125, 97–107. [Google Scholar] [CrossRef]
- Mudd, A.B.; Bredeson, J.B.; Baum, R.; Hockemeyer, D.; Rokhsar, D.S. Muntijac chromosome evolution and architecture. bioRxiv 2019, 772343. [Google Scholar] [CrossRef]
- Ijdo, J.; Baldini, A.; Ward, D.C.; Reeders, S.T.; Wells, R.A. Origin of human chromosome 2: An ancestral telomere–telomere fusion. Proc. Natl. Acad. Sci. USA 1991, 88, 9051–9055. [Google Scholar] [CrossRef]
- Hartmann, N.; Scherthan, H. Characterization of ancestral chromosome fusion points in the Indian muntjac deer. Chromosoma 2004, 112, 213–220. [Google Scholar] [CrossRef]
- Chi, J.X.; Huang, L.; Nie, W.; Wang, J.; Su, B.; Yang, F. Defining the orientation of the tandem fusions that occurred during the evolution of Indian muntjac chromosomes by BAC mapping. Chromosoma 2005, 114, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Rieger, R.; Gehlen, M.; Haszprunar, G.; Holmlund, M.; Legniti, A.; Salvenmoser, W.; Tyler, R. Laboratory cultures of marine Macrostomida (Turbellaria). Forts Zool. 1988, 36, 525. [Google Scholar]
- Ladurner, P.; Schärer, L.; Salvenmoser, W.; Rieger, R.M. A new model organism among the lower Bilateria and the use of digital microscopy in taxonomy of meiobenthic Platyhelminthes: Macrostomum lignano, n.sp (Rhabditophora, Macrostomorpha). J. Zool. Syst. Evol. Res. 2005, 43, 114–126. [Google Scholar] [CrossRef]
- Janicke, T.; Marie-Orleach, L.; De Mulder, K.; Berezikov, E.; Ladurner, P.; Vizoso, D.B.; Schärer, L. Sex allocation adjustment to mating group size in a simultaneous hermaphrodite. Evolution 2013, 67, 3233–3242. [Google Scholar] [CrossRef]
- Vellnow, N.; Vizoso, D.B.; Viktorin, G.; Schärer, L. No evidence for strong cytonuclear conflict over sex allocation in a simultaneously hermaphroditic flatworm. BMC Evol. Biol. 2017, 17, 103. [Google Scholar] [CrossRef] [PubMed]
- Schärer, L.; Littlewood, D.T.J.; Waeschenbach, A.; Yoshida, W.; Vizoso, D.B. Mating behavior and the evolution of sperm design. Proc. Natl. Acad. Sci. USA 2011, 108, 1490–1495. [Google Scholar] [CrossRef]


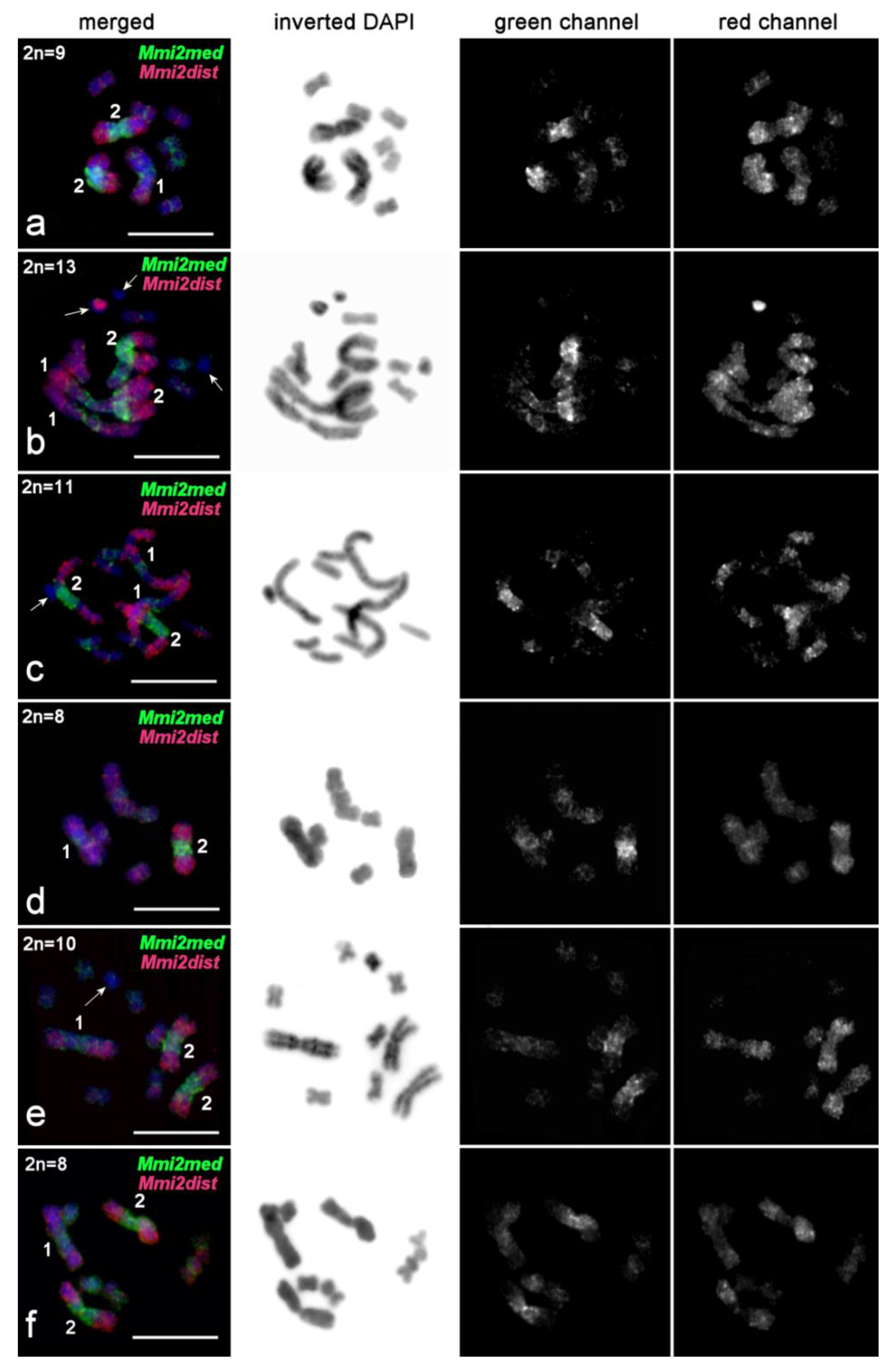
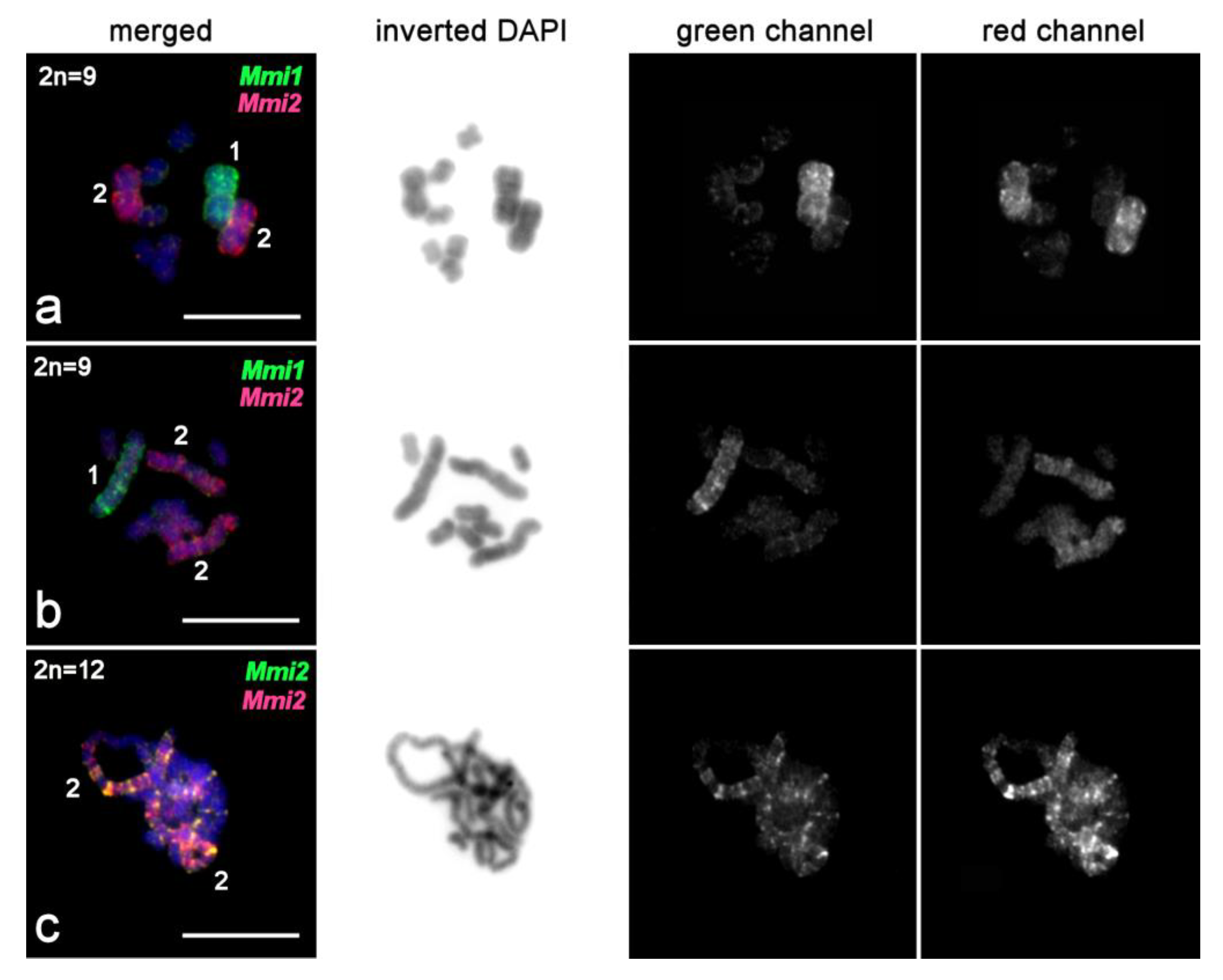

| Line, Culture or Field Collection | Year Analyzed | n | Diploid Karyotype, n (%) | Aneuploid Variants of Karyotype, n (%) | References | ||
|---|---|---|---|---|---|---|---|
| (A) M. lignano (2n = 8, 2n = 9, 2n = 10, 2n = 11, and 2n = 12 karyotypes carry 2, 3, 4, 5, or 6 large metacentrics, respectively) | |||||||
| Field collection | 2n = 8 | 2n = 9 | 2n = 10 | Other | |||
| 2014 | 122 | 120 (98.4%) | 1 (0.8%) | – | 1 (0.8%) | [36] | |
| DV1 (2003) | 2n = 8 | 2n = 9 | 2n = 10 | Other | |||
| 2014 | 134 | 16 (11.9%) | 77 (57.5%) | 36 (26.9%) | 5 (3.7%) | [36] | |
| 2015 | 78 | 3 (3.8%) | 23 (29.5%) | 51 (65.4%) | 1 (1.3%) | This study | |
| DV1/10 (2015) | 2n = 10 | 2n = 11 | 2n = 12 | Other | |||
| 2015 | 100 | 97 (97%) | 1 (1%) | – | 2 (2%) | [38] | |
| 2016 | 100 | 96 (96%) | 2(1%) | – | 3 (3%) | [38] | |
| 2017 | 94 | 89 (94.7%) | – | – | 5 (5.3%) | This study | |
| 2018 | 100 | 95 (95%) | 5 (5%) | – | – | This study | |
| (B) M. janickei (2n = 9, 2n = 10, and 2n = 11 karyotypes carry 3, 4, or 5 large metacentrics, respectively) | |||||||
| Field collection | 2n = 10 | 2n = 9 | 2n = 11 | Other | |||
| 2014 | 22 | 18 (81.8%) | 2 (9.09%) | 2 (9.09%) | – | [36] | |
| 2014 culture | 2018 | 100 | 48 (48%) | – | 39 (39%) | 13 (13%) | This study |
| (C) M. mirumnovem (2n = 10 and 2n = 11 karyotypes carry 1 or 2 additional large metacentrics, respectively) | |||||||
| 2017 Culture | 2n = 9 | 2n = 10 | 2n = 11 | Other | |||
| 2017 | 52 | 34 (65.4%) | 1 (1.9%) | – | 17 (32.7%) | This study | |
| 2018 | 100 | 20 (20%) | 26 (26%) | 8 (8%) | 46 (46%) | This study | |
| (D) M. cliftonensis (no other karyotypes have so far been observed) | |||||||
| 2017 Culture | 2n = 6 | Other | |||||
| 2017 | 10 | 10 (100%) | – | – | – | This study | |
| 2018 | 100 | 100 (100%) | - | - | - | This study | |
| AL (µm) | RL (%) | L (µm) | S (µm) | R | CI | |
|---|---|---|---|---|---|---|
| (A) Macrostomum mirumnovem | ||||||
| 1 | 9.63 ± 0.46 | 40.00 ± 0.35 | 5.02 ± 0.44 | 4.53 ± 0.45 | 1.09 ± 0.67 | 0.48 ± 0.01 (m) |
| 2 | 8.32 ± 0.59 | 34.87 ± 0.49 | 4.46 ± 0.47 | 3.93 ± 0.49 | 1.12 ± 0.07 | 0.46 ± 0.02 (m) |
| 3 | 4.12 ± 0.45 | 16.80 ± 0.41 | 2.17 ± 0.25 | 1.94 ± 0.22 | 1.12 ± 0.09 | 0.47 ± 0.02 (m) |
| 4 | 3.68 ± 0.39 | 15.03 ± 0.32 | 1.96 ± 0.23 | 1.72 ± 0.19 | 1.13 ± 0.09 | 0.47 ± 0.02 (m) |
| 5 | 3.24 ± 0.38 | 13.29 ± 0.33 | 1.74 ± 0.24 | 1.49 ± 0.19 | 1.17 ± 0.15 | 0.46 ± 0.03 (m) |
| (B) Macrostomum cliftonensis | ||||||
| 1 | 4.27 ± 0.36 | 37.62 ± 0.57 | 2.3 ± 0.23 | 1.97 ± 0.16 | 1.18 ± 0.09 | 0.46 ± 0.02 (m) |
| 2 | 3.71 ± 0.33 | 32.71 ± 0.48 | 1.93 ± 0.22 | 1.7 ± 0.21 | 1.16 ± 0.08 | 0.47 ± 0.02 (m) |
| 3 | 3.31 ± 0.3 | 29.67 ± 0.38 | 1.76 ± 0.17 | 1.55 ± 0.16 | 1.15 ± 0.09 | 0.47 ± 0.02 (m) |
| DNA Probe | Species | Original Chromosome or Chromosomal Region(s) * | n ** | Generation of DNA Probe |
|---|---|---|---|---|
| Mli2 | M. lignano | MLI2 | 15 | [38] |
| Mli3_4 | M. lignano | MLI3 and MLI4 | 15 | [38] |
| Mmi2med | M. mirumnovem | Medial part of chromosome MMI2 | 15 | This study |
| Mmi2dist | M. mirumnovem | Distal parts of p and q-arms of chromosome MMI2 | 15 | This study |
| Mmi1 | M. mirumnovem | MMI1 | 1 | This study |
| Mmi2 | M. mirumnovem | MMI2 | 2 | This study |
| Mmi3 | M. mirumnovem | MMI3 | 2 | This study |
| Mmi4 | M. mirumnovem | MMI4 | 2 | This study |
| Mmi5 | M. mirumnovem | MMI5 | 2 | This study |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zadesenets, K.S.; Jetybayev, I.Y.; Schärer, L.; Rubtsov, N.B. Genome and Karyotype Reorganization after Whole Genome Duplication in Free-Living Flatworms of the Genus Macrostomum. Int. J. Mol. Sci. 2020, 21, 680. https://doi.org/10.3390/ijms21020680
Zadesenets KS, Jetybayev IY, Schärer L, Rubtsov NB. Genome and Karyotype Reorganization after Whole Genome Duplication in Free-Living Flatworms of the Genus Macrostomum. International Journal of Molecular Sciences. 2020; 21(2):680. https://doi.org/10.3390/ijms21020680
Chicago/Turabian StyleZadesenets, Kira S., Ilyas Y. Jetybayev, Lukas Schärer, and Nikolay B. Rubtsov. 2020. "Genome and Karyotype Reorganization after Whole Genome Duplication in Free-Living Flatworms of the Genus Macrostomum" International Journal of Molecular Sciences 21, no. 2: 680. https://doi.org/10.3390/ijms21020680
APA StyleZadesenets, K. S., Jetybayev, I. Y., Schärer, L., & Rubtsov, N. B. (2020). Genome and Karyotype Reorganization after Whole Genome Duplication in Free-Living Flatworms of the Genus Macrostomum. International Journal of Molecular Sciences, 21(2), 680. https://doi.org/10.3390/ijms21020680




