Editing DNA Methylation in Mammalian Embryos
Abstract
1. Introduction
2. Editing DNA Methylation
2.1. Induction of DNA Methylation
2.2. Erasure of DNA Methylation
3. Evaluation of DNA Methylation
4. Editing DNA Methylation in Mammalian Embryos
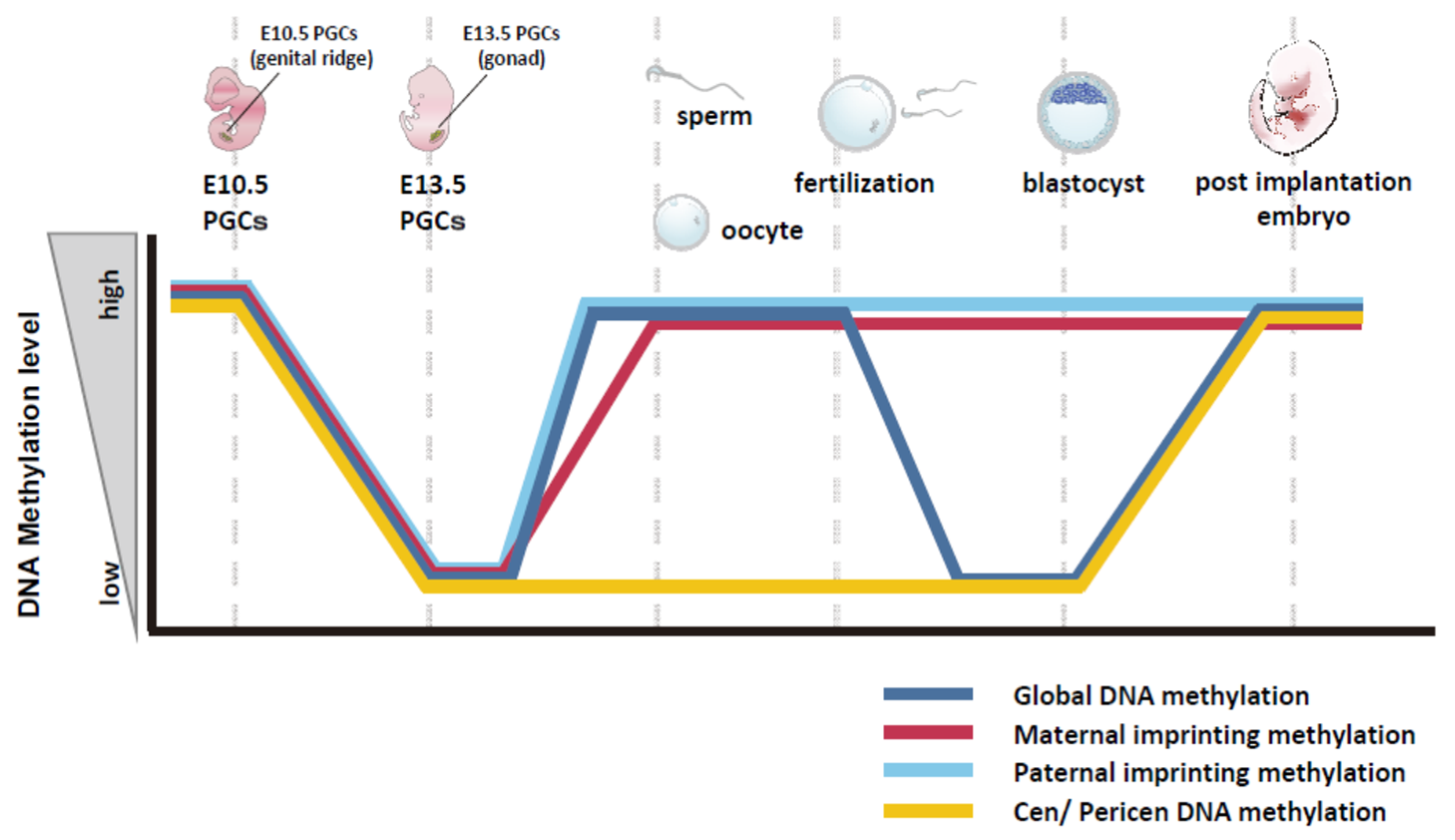
5. Features of DNA Methylation in Mouse Germline Cells
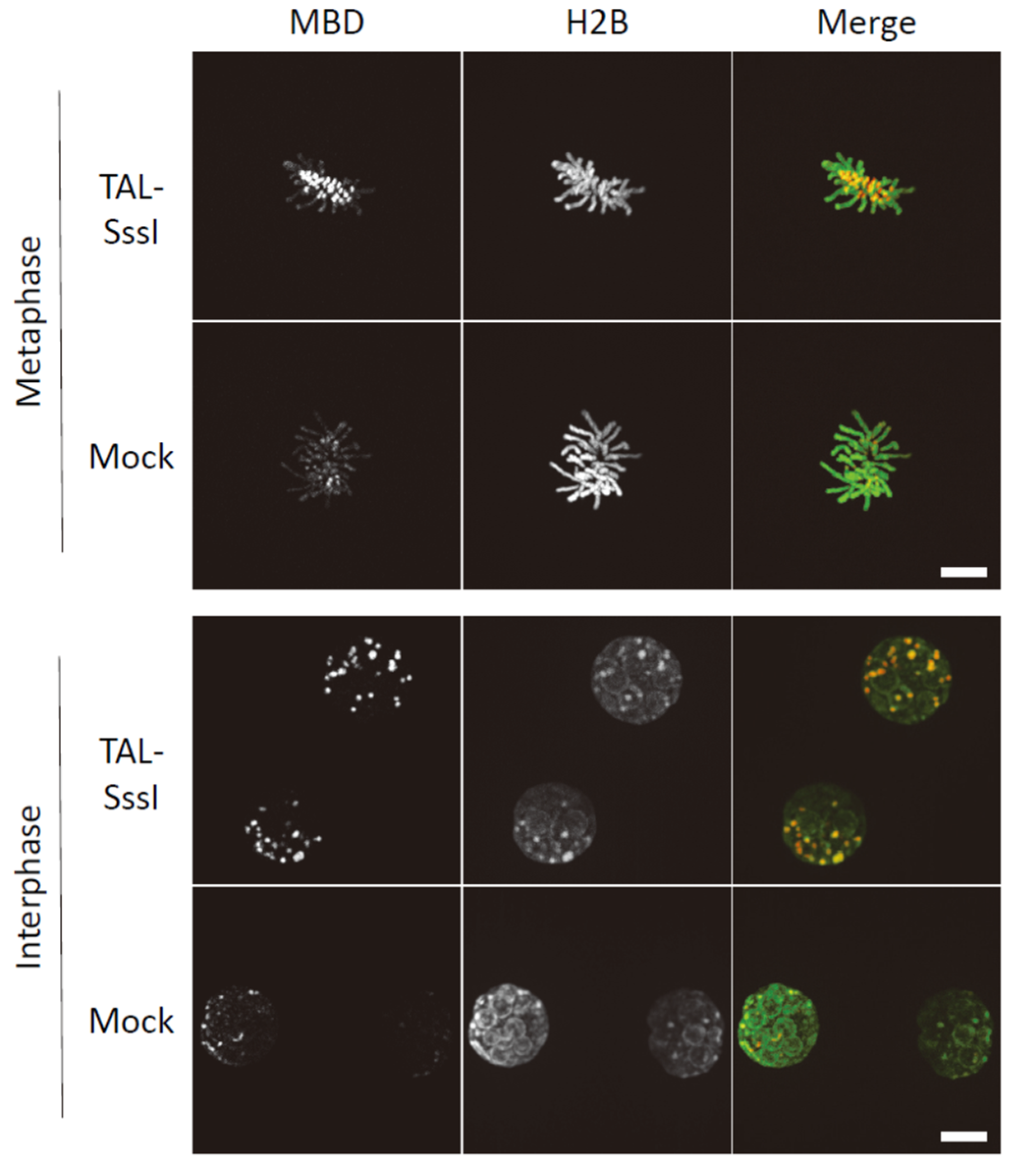
6. Artificial Introduction of DNA Methylation into the Pericentromeres of Mouse Embryos
7. Conclusions and Perspectives
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Smith, Z.D.; Meissner, A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Bergman, Y.; Cedar, H. DNA methylation dynamics in health and disease. Nat. Struct. Mol. Biol. 2013, 20, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Bestor, T.H.; Jaenisch, R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 1992, 69, 915–926. [Google Scholar] [CrossRef]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Hong, K.; Liu, R.; Shen, L.; Inoue, A.; Diep, D.; Zhang, K.; Zhang, Y. Tet1 controls meiosis by regulating meiotic gene expression. Nature 2012, 492, 443–447. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Shen, L.; Liu, Y.; Sendler, D.; Zhang, Y. Role of Tet1 in erasure of genomic imprinting. Nature 2013, 504, 460–464. [Google Scholar] [CrossRef]
- Hata, K.; Okano, M.; Lei, H.; Li, E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 2002, 129, 1983–1993. [Google Scholar]
- Gu, T.P.; Guo, F.; Yang, H.; Wu, H.P.; Xu, G.F.; Liu, W.; Xie, Z.G.; Shi, L.; He, X.; Jin, S.G.; et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 2011, 477, 606–610. [Google Scholar] [CrossRef]
- Liu, X.S.; Wu, H.; Krzisch, M.; Wu, X.; Graef, J.; Muffat, J.; Hnisz, D.; Li, C.H.; Yuan, B.; Xu, C.; et al. Rescue of Fragile X Syndrome Neurons by DNA Methylation Editing of the FMR1 Gene. Cell 2018, 172, 979–992. [Google Scholar] [CrossRef]
- Bustin, M.; Misteli, T. Nongenetic functions of the genome. Science 2016, 352, 6933. [Google Scholar] [CrossRef]
- Pabo, C.O.; Peisach, E.; Grant, R.A. Design and selection of novel Cys2His2 zinc finger proteins. Ann. Rev. Biochem. 2001, 70, 313–340. [Google Scholar] [CrossRef] [PubMed]
- Bogdanove, A.J.; Voytas, D.F. TAL effectors: Customizable proteins for DNA targeting. Science 2011, 333, 1843–1846. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A.; Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Gilbert, L.A.; Cimini, B.A.; Schnitzbauer, J.; Zhang, W.; Li, G.W.; Park, J.; Blackburn, E.H.; Weissman, J.S.; Qi, L.S.; et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 2013, 155, 1479–1491. [Google Scholar] [CrossRef]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef]
- Fujita, T.; Fujii, H. Efficient isolation of specific genomic regions and identification of associated proteins by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) using CRISPR. Biochem. Biophys. Res. Commun. 2013, 439, 132–136. [Google Scholar] [CrossRef]
- Stricker, S.H.; Koferle, A.; Beck, S. From profiles to function in epigenomics. Nat Rev Genet 2017, 18, 51–66. [Google Scholar] [CrossRef]
- Rivenbark, A.G.; Stolzenburg, S.; Beltran, A.S.; Yuan, X.; Rots, M.G.; Strahl, B.D.; Blancafort, P. Epigenetic reprogramming of cancer cells via targeted DNA methylation. Epigenetics 2012, 7, 350–360. [Google Scholar] [CrossRef]
- Siddique, A.N.; Nunna, S.; Rajavelu, A.; Zhang, Y.; Jurkowska, R.Z.; Reinhardt, R.; Rots, M.G.; Ragozin, S.; Jurkowski, T.P.; Jeltsch, A. Targeted methylation and gene silencing of VEGF-A in human cells by using a designed Dnmt3a-Dnmt3L single-chain fusion protein with increased DNA methylation activity. J. Mol. Biol. 2013, 425, 479–491. [Google Scholar] [CrossRef]
- Xirong, L.; Rui, L.; Xiaoli, Y.; Qiuyan, H.; Bikui, T.; Sibo, Z.; Naishuo, Z. Hepatitis B virus can be inhibited by DNA methyltransferase 3a via specific zinc-finger-induced methylation of the X promoter. Biochemistry 2014, 79, 111–123. [Google Scholar] [CrossRef]
- Kungulovski, G.; Nunna, S.; Thomas, M.; Zanger, U.M.; Reinhardt, R.; Jeltsch, A. Targeted epigenome editing of an endogenous locus with chromatin modifiers is not stably maintained. Epigenetics. Chromatin. 2015, 8, 12. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Gan, Y.; Gu, L.; Wilson, J.; Liu, Z.; Zhang, B.; Deng, D. P16-specific DNA methylation by engineered zinc finger methyltransferase inactivates gene transcription and promotes cancer metastasis. Genome. Biol. 2015, 16, 252. [Google Scholar] [CrossRef] [PubMed]
- Kojima-Kita, K.; Kuramochi-Miyagawa, S.; Nagamori, I.; Ogonuki, N.; Ogura, A.; Hasuwa, H.; Akazawa, T.; Inoue, N.; Nakano, T. MIWI2 as an Effector of DNA Methylation and Gene Silencing in Embryonic Male Germ Cells. Cell Rep. 2016, 16, 2819–2828. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, D.L.; Le Lay, J.E.; Ruano, E.G.; Kaestner, K.H. TALE-mediated epigenetic suppression of CDKN2A increases replication in human fibroblasts. J. Clin. Invest. 2015, 125, 1998–2006. [Google Scholar] [CrossRef]
- Yamazaki, T.; Hatano, Y.; Handa, T.; Kato, S.; Hoida, K.; Yamamura, R.; Fukuyama, T.; Uematsu, T.; Kobayashi, N.; Kimura, H.; et al. Targeted DNA methylation in pericentromeres with genome editing-based artificial DNA methyltransferase. PLoS ONE 2017, 12, e0177764. [Google Scholar] [CrossRef]
- Lo, C.L.; Choudhury, S.R.; Irudayaraj, J.; Zhou, F.C. Epigenetic Editing of Ascl1 Gene in Neural Stem Cells by Optogenetics. Sci. Rep. 2017, 7, 42047. [Google Scholar] [CrossRef]
- Vojta, A.; Dobrinic, P.; Tadic, V.; Bockor, L.; Korac, P.; Julg, B.; Klasic, M.; Zoldos, V. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 2016, 44, 5615–5628. [Google Scholar] [CrossRef]
- Liu, X.S.; Wu, H.; Ji, X.; Stelzer, Y.; Wu, X.; Czauderna, S.; Shu, J.; Dadon, D.; Young, R.A.; Jaenisch, R. Editing DNA Methylation in the Mammalian Genome. Cell 2016, 167, 233–247. [Google Scholar] [CrossRef]
- Lei, Y.; Zhang, X.; Su, J.; Jeong, M.; Gundry, M.C.; Huang, Y.H.; Zhou, Y.; Li, W.; Goodell, M.A. Targeted DNA methylation in vivo using an engineered dCas9-MQ1 fusion protein. Nat. Commun. 2017, 8, 16026. [Google Scholar] [CrossRef]
- Xiong, T.; Meister, G.E.; Workman, R.E.; Kato, N.C.; Spellberg, M.J.; Turker, F.; Timp, W.; Ostermeier, M.; Novina, C.D. Targeted DNA methylation in human cells using engineered dCas9-methyltransferases. Sci. Rep. 2017, 7, 6732. [Google Scholar] [CrossRef]
- Huang, Y.H.; Su, J.; Lei, Y.; Brunetti, L.; Gundry, M.C.; Zhang, X.; Jeong, M.; Li, W.; Goodell, M.A. DNA epigenome editing using CRISPR-Cas SunTag-directed DNMT3A. Genome Biol. 2017, 18, 176. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Lang, J.; Zhang, Q.; Yang, C.R.; Zhao, Z.A.; Zhang, Y.; Du, Y.; Sun, Y. DNA methylation analysis and editing in single mammalian oocytes. Proc. Natl. Acad. Sci. USA 2019, 116, 9883–9892. [Google Scholar] [CrossRef] [PubMed]
- Chedin, F.; Lieber, M.R.; Hsieh, C.L. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc. Natl. Acad. Sci. USA 2002, 99, 16916–16921. [Google Scholar] [CrossRef] [PubMed]
- Gowher, H.; Liebert, K.; Hermann, A.; Xu, G.; Jeltsch, A. Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J. Biol. Chem. 2005, 280, 13341–13348. [Google Scholar] [CrossRef] [PubMed]
- Jia, D.; Jurkowska, R.Z.; Zhang, X.; Jeltsch, A.; Cheng, X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature 2007, 449, 248–251. [Google Scholar] [CrossRef] [PubMed]
- Carmell, M.A.; Girard, A.; van de Kant, H.J.; Bourc’his, D.; Bestor, T.H.; de Rooij, D.G.; Hannon, G.J. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell 2007, 12, 503–514. [Google Scholar] [CrossRef]
- Renbaum, P.; Abrahamove, D.; Fainsod, A.; Wilson, G.G.; Rottem, S.; Razin, A. Cloning, characterization, and expression in Escherichia coli of the gene coding for the CpG DNA methylase from Spiroplasma sp. strain MQ1(M.SssI). Nucleic Acids Res. 1990, 18, 1145–1152. [Google Scholar] [CrossRef]
- Darii, M.V.; Cherepanova, N.A.; Subach, O.M.; Kirsanova, O.V.; Rasko, T.; Slaska-Kiss, K.; Kiss, A.; Deville-Bonne, D.; Reboud-Ravaux, M.; Gromova, E.S. Mutational analysis of the CG recognizing DNA methyltransferase SssI: Insight into enzyme-DNA interactions. Biochim. Biophys. Acta 2009, 1794, 1654–1662. [Google Scholar] [CrossRef]
- Chaikind, B.; Ostermeier, M. Directed evolution of improved zinc finger methyltransferases. PLoS ONE 2014, 9, e96931. [Google Scholar] [CrossRef]
- Takashima, S.; Takehashi, M.; Lee, J.; Chuma, S.; Okano, M.; Hata, K.; Suetake, I.; Nakatsuji, N.; Miyoshi, H.; Tajima, S.; et al. Abnormal DNA methyltransferase expression in mouse germline stem cells results in spermatogenic defects. Biol. Reprod 2009, 81, 155–164. [Google Scholar] [CrossRef]
- Morita, S.; Noguchi, H.; Horii, T.; Nakabayashi, K.; Kimura, M.; Okamura, K.; Sakai, A.; Nakashima, H.; Hata, K.; Nakashima, K.; et al. Targeted DNA demethylation in vivo using dCas9-peptide repeat and scFv-TET1 catalytic domain fusions. Nat. Biotechnol. 2016, 34, 1060–1065. [Google Scholar] [CrossRef]
- Taghbalout, A.; Du, M.; Jillette, N.; Rosikiewicz, W.; Rath, A.; Heinen, C.D.; Li, S.; Cheng, A.W. Enhanced CRISPR-based DNA demethylation by Casilio-ME-mediated RNA-guided coupling of methylcytosine oxidation and DNA repair pathways. Nat. Commun. 2019, 10, 4296. [Google Scholar] [CrossRef]
- Kohli, R.M.; Zhang, Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 2013, 502, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Maeder, M.L.; Angstman, J.F.; Richardson, M.E.; Linder, S.J.; Cascio, V.M.; Tsai, S.Q.; Ho, Q.H.; Sander, J.D.; Reyon, D.; Bernstein, B.E.; et al. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat. Biotechnol. 2013, 31, 1137–1142. [Google Scholar] [CrossRef]
- Choudhury, S.R.; Cui, Y.; Lubecka, K.; Stefanska, B.; Irudayaraj, J. CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter. Oncotarget 2016, 7, 46545–46556. [Google Scholar] [CrossRef]
- Chen, H.; Kazemier, H.G.; de Groote, M.L.; Ruiters, M.H.; Xu, G.L.; Rots, M.G. Induced DNA demethylation by targeting Ten-Eleven Translocation 2 to the human ICAM-1 promoter. Nucleic Acids Res. 2014, 42, 1563–1574. [Google Scholar] [CrossRef] [PubMed]
- Baumann, V.; Wiesbeck, M.; Breunig, C.T.; Braun, J.M.; Koferle, A.; Ninkovic, J.; Gotz, M.; Stricker, S.H. Targeted removal of epigenetic barriers during transcriptional reprogramming. Nat. Commun. 2019, 10, 2119. [Google Scholar] [CrossRef]
- Cheng, A.W.; Jillette, N.; Lee, P.; Plaskon, D.; Fujiwara, Y.; Wang, W.; Taghbalout, A.; Wang, H. Casilio: A versatile CRISPR-Cas9-Pumilio hybrid for gene regulation and genomic labeling. Cell Res. 2016, 26, 254–257. [Google Scholar] [CrossRef] [PubMed]
- Harrison, A.; Parle-McDermott, A. DNA methylation: A timeline of methods and applications. Front. Genet. 2011, 2, 74. [Google Scholar] [CrossRef]
- Yong, W.S.; Hsu, F.M.; Chen, P.Y. Profiling genome-wide DNA methylation. Epigenet. Chrom. 2016, 9, 26. [Google Scholar] [CrossRef]
- Ueda, J.; Maehara, K.; Mashiko, D.; Ichinose, T.; Yao, T.; Hori, M.; Sato, Y.; Kimura, H.; Ohkawa, Y.; Yamagata, K. Heterochromatin dynamics during the differentiation process revealed by the DNA methylation reporter mouse, MethylRO. Stem Cell Rep. 2014, 2, 910–924. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, T.; Yamagata, K.; Baba, T. Time-lapse and retrospective analysis of DNA methylation in mouse preimplantation embryos by live cell imaging. Dev. Biol. 2007, 304, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Yamagata, K.; Yamazaki, T.; Miki, H.; Ogonuki, N.; Inoue, K.; Ogura, A.; Baba, T. Centromeric DNA hypomethylation as an epigenetic signature discriminates between germ and somatic cell lineages. Dev. Biol. 2007, 312, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Morgan, H.D.; Sutherland, H.G.; Martin, D.I.; Whitelaw, E. Epigenetic inheritance at the agouti locus in the mouse. Nat. Genet. 1999, 23, 314–318. [Google Scholar] [CrossRef] [PubMed]
- Kono, T.; Obata, Y.; Wu, Q.; Niwa, K.; Ono, Y.; Yamamoto, Y.; Park, E.S.; Seo, J.S.; Ogawa, H. Birth of parthenogenetic mice that can develop to adulthood. Nature 2004, 428, 860–864. [Google Scholar] [CrossRef] [PubMed]
- Reik, W.; Dean, W.; Walter, J. Epigenetic reprogramming in mammalian development. Science 2001, 293, 1089–1093. [Google Scholar] [CrossRef] [PubMed]
- Hackett, J.A.; Surani, M.A. DNA methylation dynamics during the mammalian life cycle. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2013, 368, 20110328. [Google Scholar] [CrossRef]
- Shiota, K.; Kogo, Y.; Ohgane, J.; Imamura, T.; Urano, A.; Nishino, K.; Tanaka, S.; Hattori, N. Epigenetic marks by DNA methylation specific to stem, germ and somatic cells in mice. Genes Cells 2002, 7, 961–969. [Google Scholar] [CrossRef]
- Smith, Z.D.; Chan, M.M.; Humm, K.C.; Karnik, R.; Mekhoubad, S.; Regev, A.; Eggan, K.; Meissner, A. DNA methylation dynamics of the human preimplantation embryo. Nature 2014, 511, 611–615. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, J.; Duan, J.; Gao, X.; Zhu, W.; Lu, X.; Yang, L.; Zhang, J.; Li, G.; Ci, W.; et al. Programming and inheritance of parental DNA methylomes in mammals. Cell 2014, 157, 979–991. [Google Scholar] [CrossRef]
- Shen, L.; Inoue, A.; He, J.; Liu, Y.; Lu, F.; Zhang, Y. Tet3 and DNA replication mediate demethylation of both the maternal and paternal genomes in mouse zygotes. Cell Stem Cell 2014, 15, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Zhu, P.; Yan, L.; Li, R.; Hu, B.; Lian, Y.; Yan, J.; Ren, X.; Lin, S.; Li, J.; et al. The DNA methylation landscape of human early embryos. Nature 2014, 511, 606–610. [Google Scholar] [CrossRef] [PubMed]
- Howell, C.Y.; Bestor, T.H.; Ding, F.; Latham, K.E.; Mertineit, C.; Trasler, J.M.; Chaillet, J.R. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell 2001, 104, 829–838. [Google Scholar] [CrossRef]
- Nakamura, T.; Arai, Y.; Umehara, H.; Masuhara, M.; Kimura, T.; Taniguchi, H.; Sekimoto, T.; Ikawa, M.; Yoneda, Y.; Okabe, M.; et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat. Cell Biol. 2007, 9, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Hirasawa, R.; Chiba, H.; Kaneda, M.; Tajima, S.; Li, E.; Jaenisch, R.; Sasaki, H. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 2008, 22, 1607–1616. [Google Scholar] [CrossRef] [PubMed]
- Lawson, K.A.; Dunn, N.R.; Roelen, B.A.; Zeinstra, L.M.; Davis, A.M.; Wright, C.V.; Korving, J.P.; Hogan, B.L. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 1999, 13, 424–436. [Google Scholar] [CrossRef]
- Seisenberger, S.; Andrews, S.; Krueger, F.; Arand, J.; Walter, J.; Santos, F.; Popp, C.; Thienpont, B.; Dean, W.; Reik, W. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell 2012, 48, 849–862. [Google Scholar] [CrossRef]
- Kobayashi, H.; Sakurai, T.; Miura, F.; Imai, M.; Mochiduki, K.; Yanagisawa, E.; Sakashita, A.; Wakai, T.; Suzuki, Y.; Ito, T.; et al. High-resolution DNA methylome analysis of primordial germ cells identifies gender-specific reprogramming in mice. Genome Res. 2013, 23, 616–627. [Google Scholar] [CrossRef]
- Kagiwada, S.; Kurimoto, K.; Hirota, T.; Yamaji, M.; Saitou, M. Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. EMBO J. 2013, 32, 340–353. [Google Scholar] [CrossRef]
- Kaneda, M.; Okano, M.; Hata, K.; Sado, T.; Tsujimoto, N.; Li, E.; Sasaki, H. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 2004, 429, 900–903. [Google Scholar] [CrossRef]
- Muller, S.; Almouzni, G. Chromatin dynamics during the cell cycle at centromeres. Nat. Rev. Genet. 2017, 18, 192–208. [Google Scholar] [CrossRef] [PubMed]
- Widschwendter, M.; Jiang, G.; Woods, C.; Muller, H.M.; Fiegl, H.; Goebel, G.; Marth, C.; Muller-Holzner, E.; Zeimet, A.G.; Laird, P.W.; et al. DNA hypomethylation and ovarian cancer biology. Cancer Res. 2004, 64, 4472–4480. [Google Scholar] [CrossRef] [PubMed]
- Molaro, A.; Hodges, E.; Fang, F.; Song, Q.; McCombie, W.R.; Hannon, G.J.; Smith, A.D. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell 2011, 146, 1029–1041. [Google Scholar] [CrossRef] [PubMed]
- Miyanari, Y.; Ziegler-Birling, C.; Torres-Padilla, M.E. Live visualization of chromatin dynamics with fluorescent TALEs. Nat. Struct. Mol. Biol. 2013, 20, 1321–1324. [Google Scholar] [CrossRef]
- Tsumura, A.; Hayakawa, T.; Kumaki, Y.; Takebayashi, S.; Sakaue, M.; Matsuoka, C.; Shimotohno, K.; Ishikawa, F.; Li, E.; Ueda, H.R.; et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 2006, 11, 805–814. [Google Scholar] [CrossRef]
- Fleming, T.P.; Watkins, A.J.; Velazquez, M.A.; Mathers, J.C.; Prentice, A.M.; Stephenson, J.; Barker, M.; Saffery, R.; Yajnik, C.S.; Eckert, J.J.; et al. Origins of lifetime health around the time of conception: Causes and consequences. Lancet 2018, 391, 1842–1852. [Google Scholar] [CrossRef]
- DiTroia, S.P.; Percharde, M.; Guerquin, M.J.; Wall, E.; Collignon, E.; Ebata, K.T.; Mesh, K.; Mahesula, S.; Agathocleous, M.; Laird, D.J.; et al. Maternal vitamin C regulates reprogramming of DNA methylation and germline development. Nature 2019, 573, 271–275. [Google Scholar] [CrossRef]
- Ryan, D.P.; Henzel, K.S.; Pearson, B.L.; Siwek, M.E.; Papazoglou, A.; Guo, L.; Paesler, K.; Yu, M.; Muller, R.; Xie, K.; et al. A paternal methyl donor-rich diet altered cognitive and neural functions in offspring mice. Mol. Psychiatry 2018, 23, 1345–1355. [Google Scholar] [CrossRef]
- Lambrot, R.; Xu, C.; Saint-Phar, S.; Chountalos, G.; Cohen, T.; Paquet, M.; Suderman, M.; Hallett, M.; Kimmins, S. Low paternal dietary folate alters the mouse sperm epigenome and is associated with negative pregnancy outcomes. Nat. Commun. 2013, 4, 2889. [Google Scholar] [CrossRef]

| Target | DNA-Binding Module | Effector | References |
|---|---|---|---|
| Maspin | Zinc Finger | DNMT3A CD | [18] |
| VEGF-A | Zinc Finger | DNMT3A CD-DNMT3L | [19] |
| HBV x promoter | Zinc Finger | DNMT3A C-term | [20] |
| Line1 | Zinc Finger | MIWI2 | [23] |
| P16 (CDKN2A) | TALE | DNMT3A-DNMT3L | [24] |
| Major satellite | TALE, dCas9 | SssI | [25] |
| AsclI | TALE-CIB1 | DNMT3A CD-CRY2 | [26] |
| BACH-2, IL6ST | dCas9 | DNMT3A CD | [27] |
| Snrpn, CTCF | dCas9 | DNMT3A | [28] |
| Hox genes, Runx1, H19 | dCas9 | SssI (Q147L) | [29] |
| SALL2, HBG | dCas9 | Split SssI | [30] |
| HoxA5, KLF4 | dCas9-SunTag | scFv-DNMT3A | [31] |
| IAP (Agouti), H19, IG-DMR, Snrpn DMR | dCas9 | DNMT3A | [32] |
| Target | DNA-Binding Module | Effector | References |
|---|---|---|---|
| KLF4, RHOX, HBB | TALE | TET1 CD | [44] |
| ICAM1 | Zinc Finger | TET2 CD | [46] |
| AscI | TALE-CIB1 | TET1 CD-CRY2 | [26] |
| Snrpn, BDNF, MyoD | dCas9 | TET1 CD | [28] |
| Gfap, H19 DMR | dCas9-SunTag | scFv-TET1 CD | [41] |
| BRCA1 | dCas9 | TET1 CD | [45] |
| FMR1 | dCas9 | TET1 CD | [9] |
| Sox1 | dCas9 | TET1 CD | [47] |
| IAP (Agouti) | dCas9 | TET1 CD | [32] |
| hMLH1 | dCas9 + gRNA with PUFa-binding site | PUFa-TET1 CD with GADD45A or NEIL2 | [42] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamazaki, T.; Hatano, Y.; Taniguchi, R.; Kobayashi, N.; Yamagata, K. Editing DNA Methylation in Mammalian Embryos. Int. J. Mol. Sci. 2020, 21, 637. https://doi.org/10.3390/ijms21020637
Yamazaki T, Hatano Y, Taniguchi R, Kobayashi N, Yamagata K. Editing DNA Methylation in Mammalian Embryos. International Journal of Molecular Sciences. 2020; 21(2):637. https://doi.org/10.3390/ijms21020637
Chicago/Turabian StyleYamazaki, Taiga, Yu Hatano, Ryoya Taniguchi, Noritada Kobayashi, and Kazuo Yamagata. 2020. "Editing DNA Methylation in Mammalian Embryos" International Journal of Molecular Sciences 21, no. 2: 637. https://doi.org/10.3390/ijms21020637
APA StyleYamazaki, T., Hatano, Y., Taniguchi, R., Kobayashi, N., & Yamagata, K. (2020). Editing DNA Methylation in Mammalian Embryos. International Journal of Molecular Sciences, 21(2), 637. https://doi.org/10.3390/ijms21020637





