Isolation and Identification of a TaTDR-Like Wheat Gene Encoding a bHLH Domain Protein, Which Negatively Regulates Chlorophyll Biosynthesis in Arabidopsis
Abstract
1. Introduction
2. Results
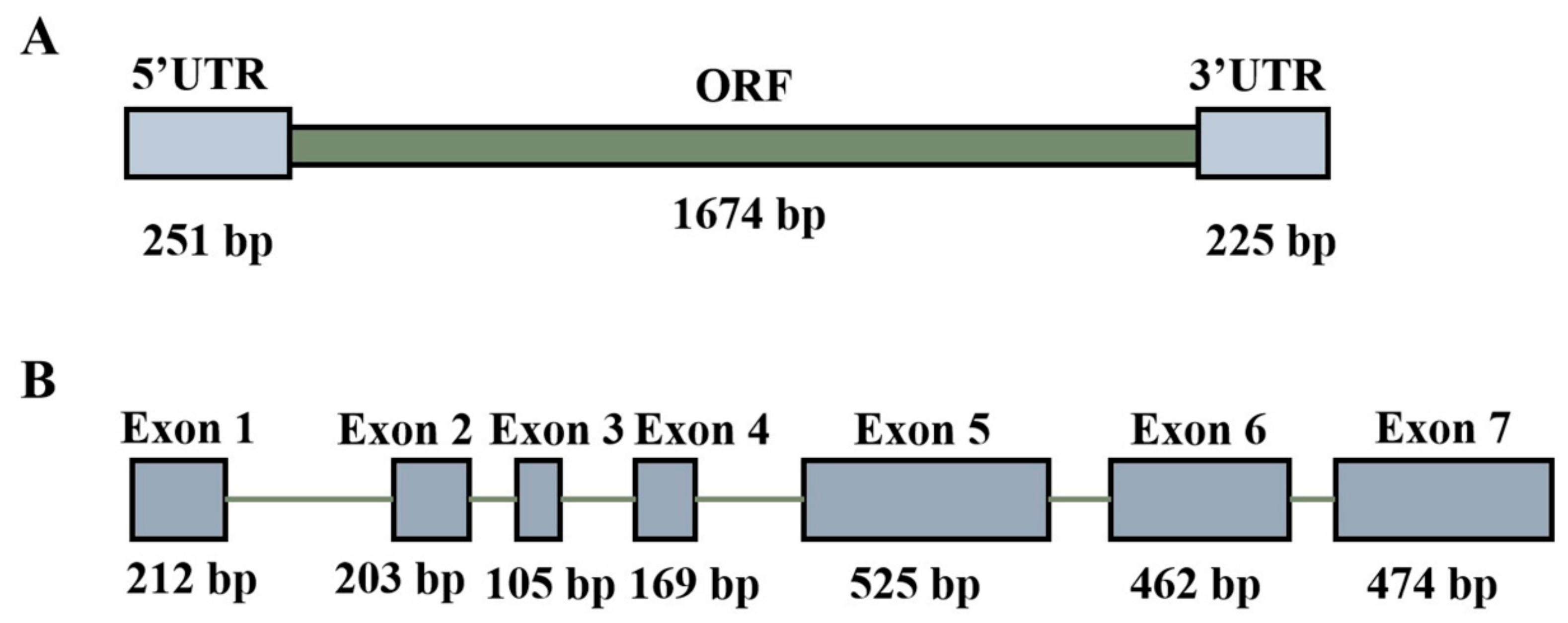
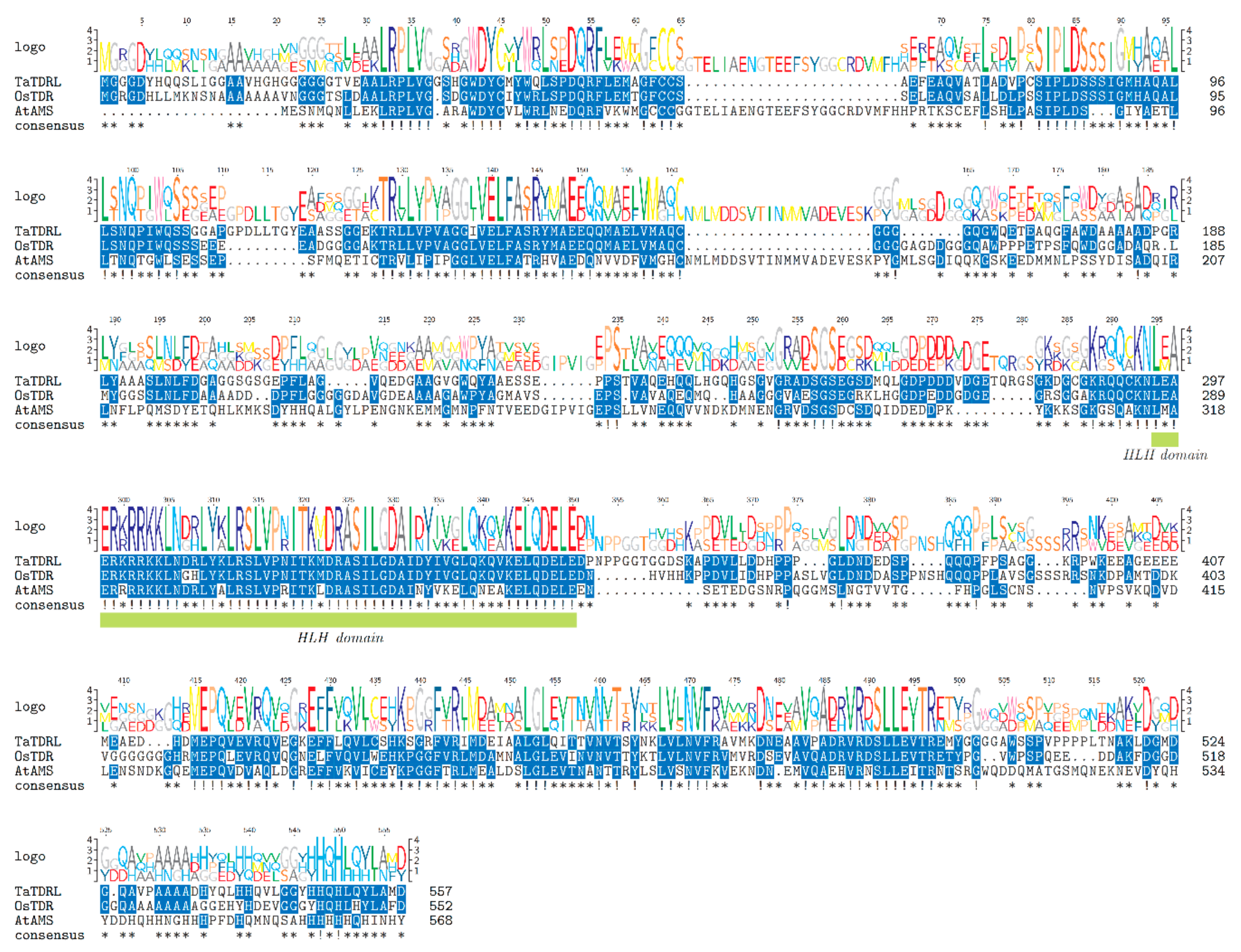
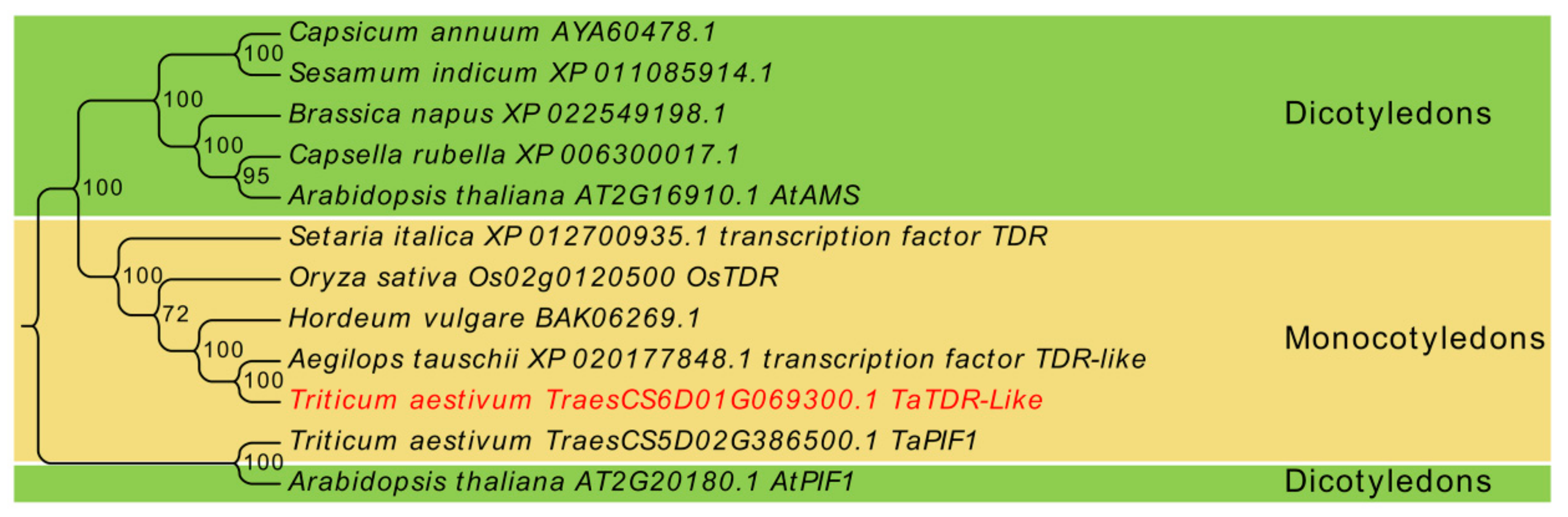
2.1. Identification and Sequence Analysis of TaTDRL
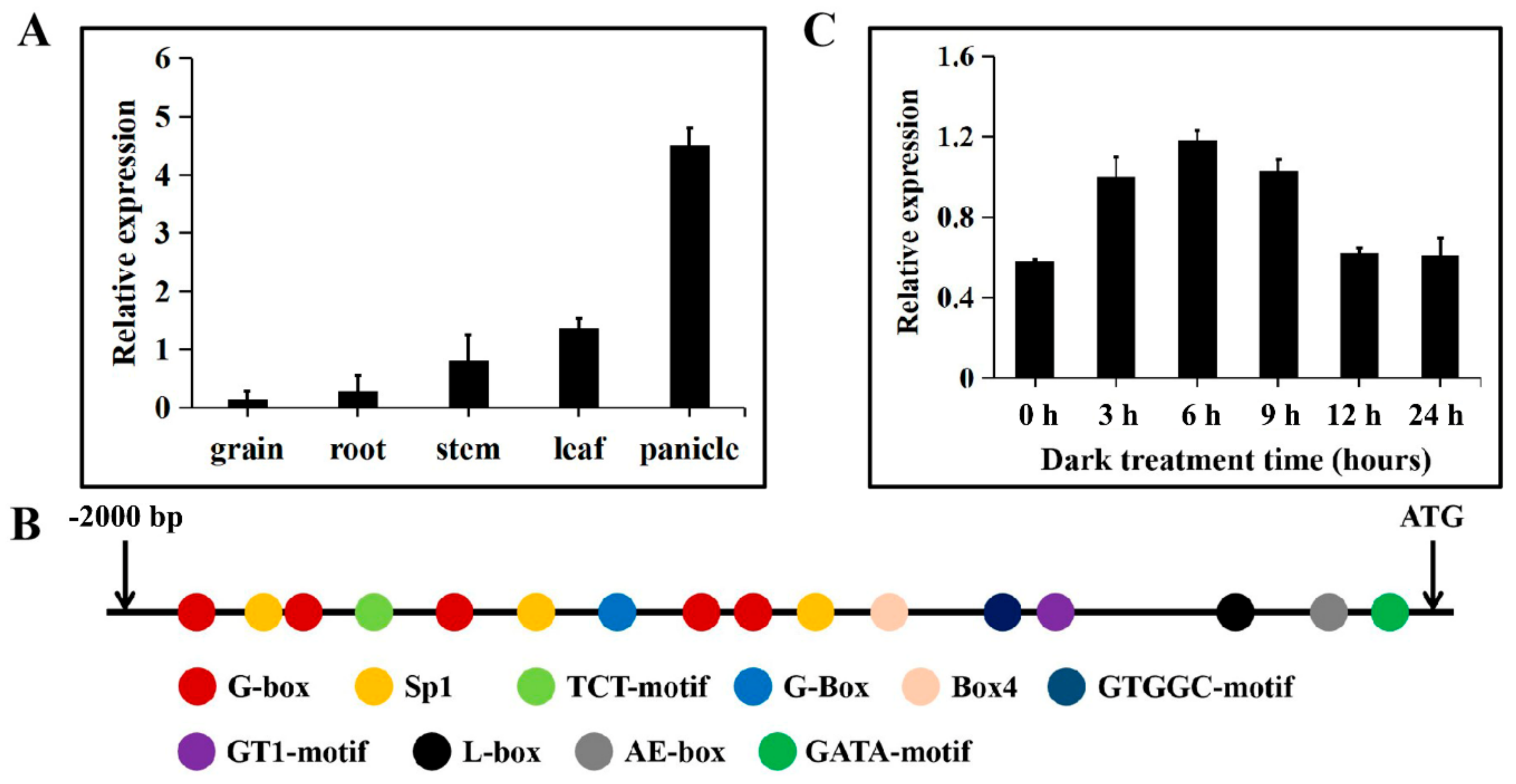
2.2. Expression Patterns and Promoter Analysis of TaTDRL
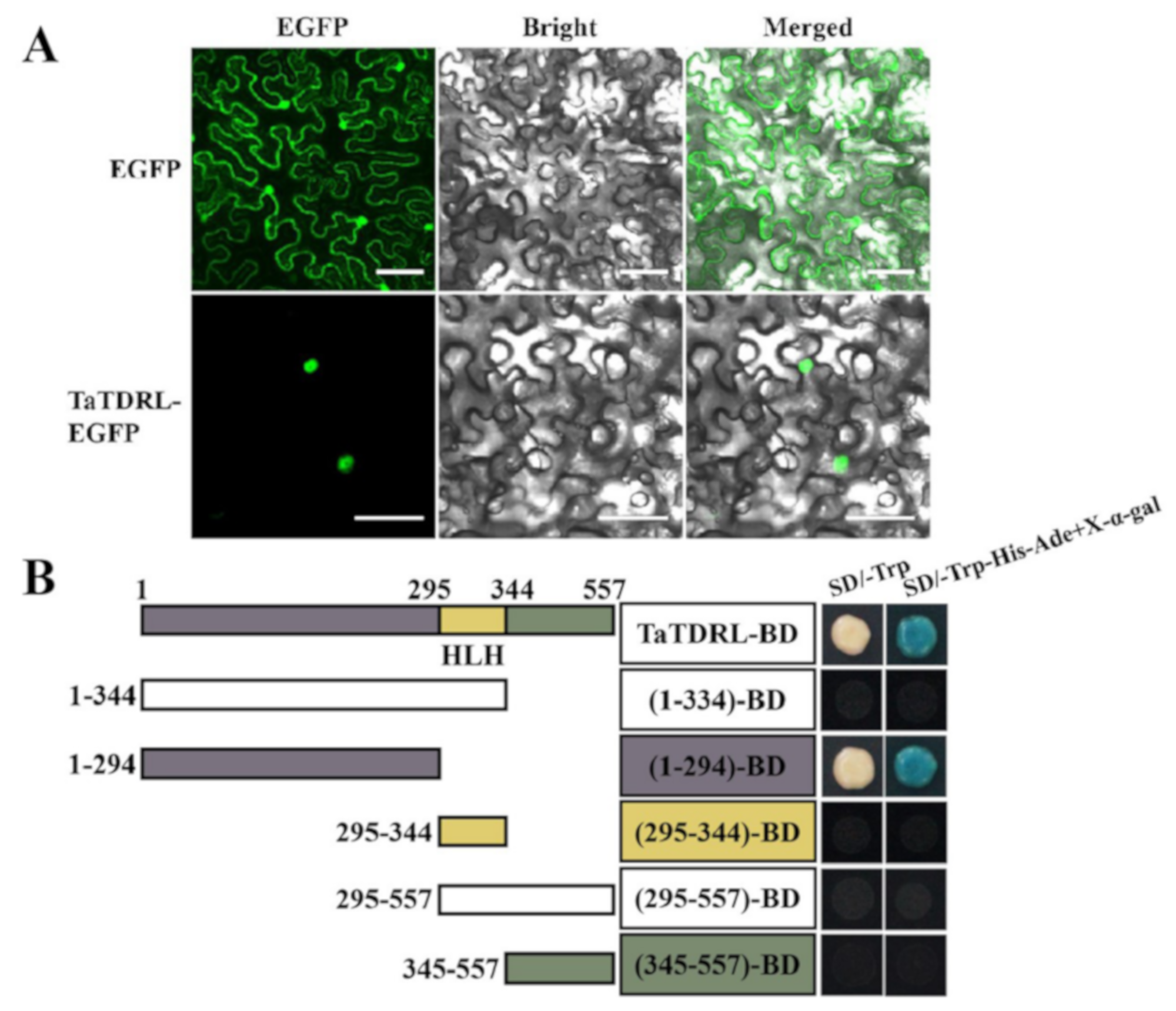
2.3. Subcellular Localization and Transactivation Activity of TaTDRL
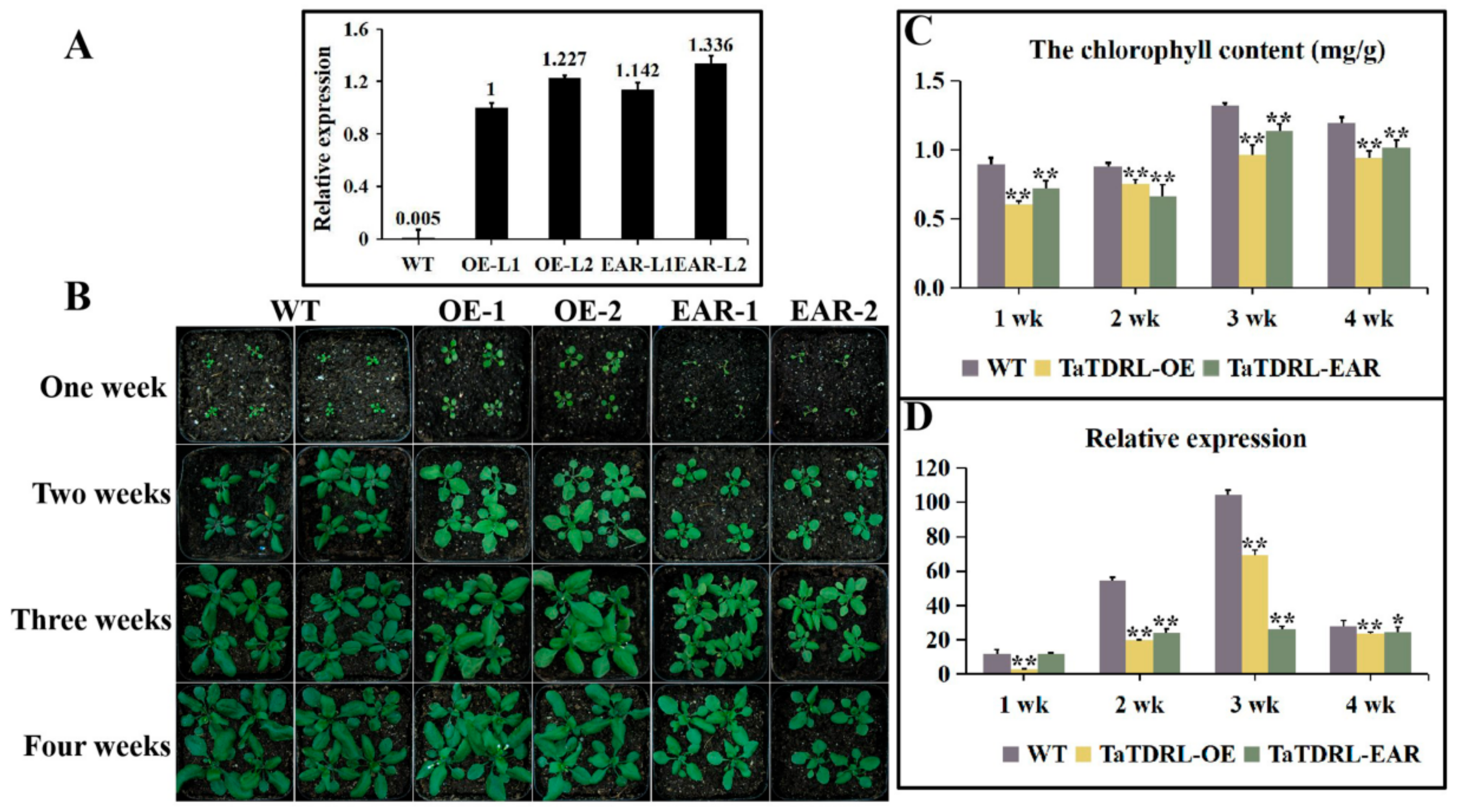
2.4. Effect of TaTDRL Overexpression in Arabidopsis Decreased the Content of Chlorophyll
2.5. Transgenic Plants Expressing the Chimeric TaTDRL Repressor
2.6. TaTDRL-OE and TaTDRL-EAR Affect the Expression of a Chlorophyll-Related Gene
3. Discussion
3.1. Structure and Localization of the TaTDR-Like Gene
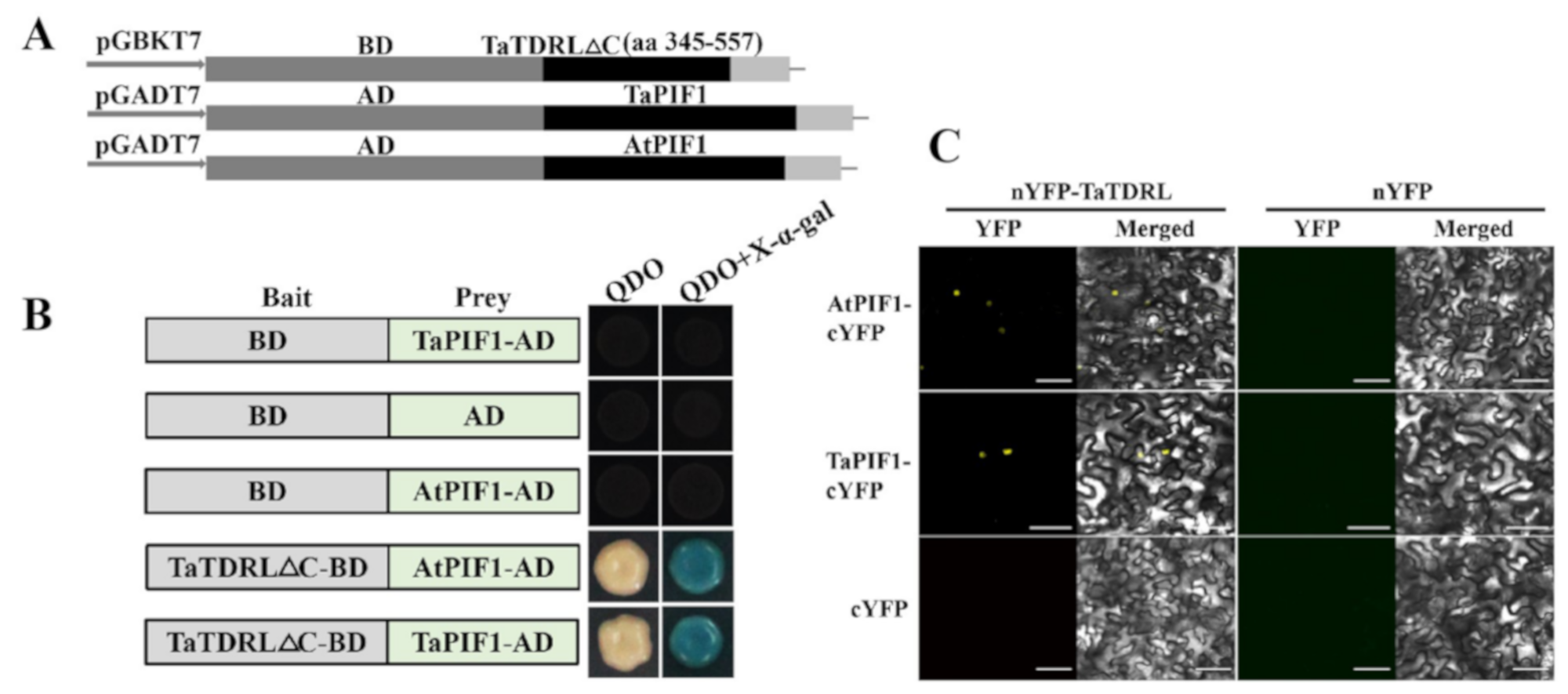
3.2. TaTDRL May Be a Direct Negative Regulator of Chlorophyll Biosynthesis
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Cloning and Sequence Analysis of TaTDRL Gene
4.3. Bioinformatics Analysis of TaTDRL
4.4. RNA Extraction, cDNA Synthesis, and Gene Expression Analysis
4.5. Promoter Sequence Analysis
4.6. Subcellular Localization and Transcription Activation Activity Assay of TaTDRL
4.7. Plant Transformation
4.8. Determination of Chlorophyll Content
4.9. Yeast Two-Hybrid Assay
4.10. Bimolecular Fuorescence Complementation (BiFC) Assays
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Eckhardt, U.; Grimm, B.; Hörtensteiner, S. Recent advances in chlorophyll biosynthesis and breakdown in higher plants. Plant Mol. Biol. 2004, 56, 1–14. [Google Scholar] [CrossRef]
- Hortensteiner, S. Update on the biochemistry of chlorophyll breakdown. Plant Mol. Biol. 2013, 82, 505–517. [Google Scholar] [CrossRef] [PubMed]
- Bae, G.; Choi, G. Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 2008, 59, 281–311. [Google Scholar] [CrossRef] [PubMed]
- Rizzini, L.; Favory, J.-J.; Cloix, C.; Faggionato, D.; O’Hara, A.; Kaiserli, E.; Baumeister, R.; Schäfer, E.; Nagy, F.; Jenkins, G.I.; et al. Perception of UV-B by the Arabidopsis UVR8 Protein. Science 2011, 332, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, N.C.; Su, Y.-S.; Lagarias, J.C. Phytochrome Structure and Signaling Mechanisms. Annu. Rev. Plant Biol. 2006, 57, 837–858. [Google Scholar] [CrossRef] [PubMed]
- Clack, T.; Mathews, S.; Sharrock, R.A. The phytochrome apoprotein family in Arabidopsis is encoded by five genes: The sequences and expression of PHYD and PHYE. Plant Mol. Biol. 1994, 25, 413–427. [Google Scholar] [CrossRef] [PubMed]
- Castillon, A.; Shen, H.; Huq, E. Phytochrome Interacting Factors: Central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 2007, 12, 514–521. [Google Scholar] [CrossRef]
- Leivar, P.; Quail, P.H. PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 2011, 16, 19–28. [Google Scholar] [CrossRef]
- Toledo-Ortiz, G.; Hyq, E.; Rodríguez-Concepción, M. Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 2010, 107, 11626–11631. [Google Scholar] [CrossRef]
- Moon, J.; Zhu, L.; Shen, H.; Huq, E. PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc. Natl. Acad. Sci. USA 2008, 105, 9433–9438. [Google Scholar] [CrossRef]
- Huq, E.; Al-Sady, B.; Hudson, M.; Kim, C.; Apel, K.; Quail, P.H. Phytochrome-Interacting factor 1 Is a Critical bHLH Regulator of Chlorophyll Biosynthesis. Science 2004, 305, 1937–1941. [Google Scholar] [CrossRef] [PubMed]
- Bu, Q.; Castillon, A.; Chen, F.; Zhu, L.; Huq, E. Dimerization and blue light regulation of PIF1 interacting bHLH proteins in Arabidopsis. Plant Mol. Biol. 2011, 77, 501–511. [Google Scholar] [CrossRef] [PubMed]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis basic/helix-Loop-Helix transcription factor family. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef] [PubMed]
- Fairchild, C.; Schumaker, M.A.; Quail, P.H. HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 2000, 14, 2377–2391. [Google Scholar]
- Huq, E.; Quail, P.H. PIF4, a phytochrome-Interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 2002, 21, 2441–2450. [Google Scholar] [CrossRef]
- Li, X.; Duan, X.; Jiang, H.; Sun, Y.; Tang, Y.; Yuan, Z.; Guo, J.; Liang, W.; Chen, L.; Yin, J.; et al. Genome-Wide analysis of basic/helix-Loop-Helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006, 141, 1167–1184. [Google Scholar] [CrossRef]
- Li, N.; Zhang, D.S.; Liu, H.S.; Yin, C.S.; Li, X.X.; Liang, W.Q.; Yuan, Z.; Xu, B.; Chu, H.W.; Wang, J.; et al. The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 2006, 18, 2999–3014. [Google Scholar] [CrossRef]
- Hiratsu, K.; Matsui, K.; Oyama, T.; Ohme-Takagi, M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003, 34, 733–739. [Google Scholar] [CrossRef]
- Coller, H.A.; Grandori, C.; Tamayo, P.; Colbert, T.; Lander, E.S.; Eisenman, R.N.; Golub, T.R. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc. Natl. Acad. Sci. USA 2000, 97, 3260–3265. [Google Scholar] [CrossRef]
- Elbadawy, M.; Usui, T.; Yamawaki, H.; Sasaki, K. Emerging Roles of C-Myc in Cancer Stem Cell-Related Signaling and Resistance to Cancer Chemotherapy: A Potential Therapeutic Target Against Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 2340. [Google Scholar] [CrossRef]
- Blackwood, E.; Eisenman, R.N. Max: A helix-Loop-Helix zipper protein that forms a sequence-Specific DNA-binding complex with Myc. Science 1991, 251, 1211–1217. [Google Scholar] [CrossRef] [PubMed]
- Ferre-D’Amaré, A.R.; Pognonec, P.; Roeder, R.G.; Burley, S.K. Structure and function of the b/HLH/Z domain of USF. EMBO J. 1994, 13, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Thorstensen, T.; Grini, P.E.; Mercy, I.S.; Alm, V.; Erdal, S.; Aasland, R.; Aalen, R.B. The Arabidopsis SET-Domain protein ASHR3 is involved in stamen development and interacts with the bHLH transcription factor ABORTED MICROSPORES (AMS). Plant Mol. Biol. 2008, 66, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Ellenberger, T.; Fass, D.; Arnaud, M.; Harrison, S.C. Crystal structure of transcription factor E47: E-Box recognition by a basic region helix-Loop-Helix dimer. Genes Dev. 1994, 8, 970–980. [Google Scholar] [CrossRef]
- Marie Henriksson, B.L. Proteins of the Myc network: Essential regulators of cell growth and differentiation. Adv. Cancer Res. 1996, 68, 109–182. [Google Scholar]
- Badawi, M.; Reddy, Y.V.; Agharbaoui, Z.; Tominaga, Y.; Danyluk, J.; Sarhan, F.; Houde, M. Structure and functional analysis of wheat ICE (inducer of CBF expression) genes. Plant Cell Physiol. 2008, 49, 1237–1249. [Google Scholar] [CrossRef]
- Castilhos, G.; Lazzarotto, F.; Spagnolo-Fonini, L.; Bodanese-Zanettini, M.H.; Margis-Pinheiro, M. Possible roles of basic helix-Loop-Helix transcription factors in adaptation to drought. Plant Sci. Int. J. Exp. Plant Biol. 2014, 223, 1–7. [Google Scholar] [CrossRef]
- Duek, P.D.; Fankhauser, C. bHLH class transcription factors take centre stage in phytochrome signalling. Trends Plant Sci. 2005, 10, 51–54. [Google Scholar] [CrossRef]
- Li, H.; Sun, J.; Xu, Y.; Jiang, H.; Wu, X.; Li, C. The bHLH-Type transcription factor AtAIB positively regulates ABA response in Arabidopsis. Plant Mol. Biol. 2007, 65, 655–665. [Google Scholar] [CrossRef]
- Schiefelbein, J. Cell-Fate specification in the epidermis: A common patterning mechanism in the root and shoot. Growth Dev. 2003, 6, 74–78. [Google Scholar] [CrossRef]
- Zhang, L.Y.; Bai, M.Y.; Wu, J.; Zhu, J.Y.; Wang, H.; Zhang, Z.; Wang, W.; Sun, Y.; Zhao, J.; Sun, X.; et al. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 2009, 21, 3767–3780. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, F.; Wang, J.L.; Ma, Y.; Chong, K.; Xu, Y.Y. Basic helix-Loop-Helix transcription factor from wild rice (OrbHLH2) improves tolerance to salt- and osmotic stress in Arabidopsis. J. Plant Physiol. 2009, 166, 1296–1306. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.-S.; Liang, W.-Q.; Yuan, Z.; Li, N.; Shi, J.; Wang, J.; Liu, Y.-M.; Yu, W.-J.; Zhang, D.-B. Tapetum Degeneration Retardation is Critical for Aliphatic Metabolism and Gene Regulation during Rice Pollen Development. Mol. Plant 2008, 1, 599–610. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Sotomayor, P.; Chavez Montes, R.A.; Silvestre-Vano, M.; Herrera-Ubaldo, H.; Greco, R.; Pablo-Villa, J.; Galliani, B.M.; Diaz-Ramirez, D.; Weemen, M.; Boutilier, K.; et al. Altered expression of the bZIP transcription factor DRINK ME affects growth and reproductive development in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 2016, 88, 437–451. [Google Scholar] [CrossRef] [PubMed]
- Das, P.; Ito, T.; Wellmer, F.; Vernoux, T.; Dedieu, A.; Traas, J.; Meyerowitz, E.M. Floral stem cell termination involves the direct regulation of AGAMOUS by PERIANTHIA. Development 2009, 136, 1605–1611. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Li, Y.; Zhang, X.; Zha, P.; Lin, R. The SWI2/SNF2 Chromatin-Remodeling ATPase BRAHMA Regulates Chlorophyll Biosynthesis in Arabidopsis. Mol. Plant 2017, 10, 155–167. [Google Scholar] [CrossRef] [PubMed]
- Beitz, E. TEXshade: Shading and labeling of multiple sequence alignments using LATEX2 epsilon. Bioinformatics 2000, 16, 135–139. [Google Scholar] [CrossRef]
- Walter, M.; Chaban, C.; Schütze, K.; Batistic, O.; Weckermann, K.; Näke, C.; Blazevic, D.; Grefen, C.; Schumacher, K.; Oecking, C.; et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004, 40, 428–438. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper enzymes in isolated chloroplasts. polyphenoloxidase in beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef]
- Lu, X.-D.; Zhou, C.-M.; Xu, P.-B.; Luo, Q.; Lian, H.-L.; Yang, H.-Q. Red-Light-Dependent Interaction of phyB with SPA1 Promotes COP1–SPA1 Dissociation and Photomorphogenic Development in Arabidopsis. Mol. Plant 2015, 8, 467–478. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, Y.; Li, Z.; Wang, J.; Li, Y.; Ren, Y.; Du, J.; Song, Q.; Ma, S.; Song, Y.; Zhao, H.; et al. Isolation and Identification of a TaTDR-Like Wheat Gene Encoding a bHLH Domain Protein, Which Negatively Regulates Chlorophyll Biosynthesis in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 629. https://doi.org/10.3390/ijms21020629
Xia Y, Li Z, Wang J, Li Y, Ren Y, Du J, Song Q, Ma S, Song Y, Zhao H, et al. Isolation and Identification of a TaTDR-Like Wheat Gene Encoding a bHLH Domain Protein, Which Negatively Regulates Chlorophyll Biosynthesis in Arabidopsis. International Journal of Molecular Sciences. 2020; 21(2):629. https://doi.org/10.3390/ijms21020629
Chicago/Turabian StyleXia, Yu, Zheng Li, Junwei Wang, Yanhong Li, Yang Ren, Jingjing Du, Qilu Song, Shoucai Ma, Yulong Song, Huiyan Zhao, and et al. 2020. "Isolation and Identification of a TaTDR-Like Wheat Gene Encoding a bHLH Domain Protein, Which Negatively Regulates Chlorophyll Biosynthesis in Arabidopsis" International Journal of Molecular Sciences 21, no. 2: 629. https://doi.org/10.3390/ijms21020629
APA StyleXia, Y., Li, Z., Wang, J., Li, Y., Ren, Y., Du, J., Song, Q., Ma, S., Song, Y., Zhao, H., Yang, Z., Zhang, G., & Niu, N. (2020). Isolation and Identification of a TaTDR-Like Wheat Gene Encoding a bHLH Domain Protein, Which Negatively Regulates Chlorophyll Biosynthesis in Arabidopsis. International Journal of Molecular Sciences, 21(2), 629. https://doi.org/10.3390/ijms21020629




