Genome-Wide Investigation of Heat Shock Transcription Factor Family in Wheat (Triticum aestivum L.) and Possible Roles in Anther Development
Abstract
1. Introduction
2. Results
2.1. Identification and Characteristics of TaHsfs
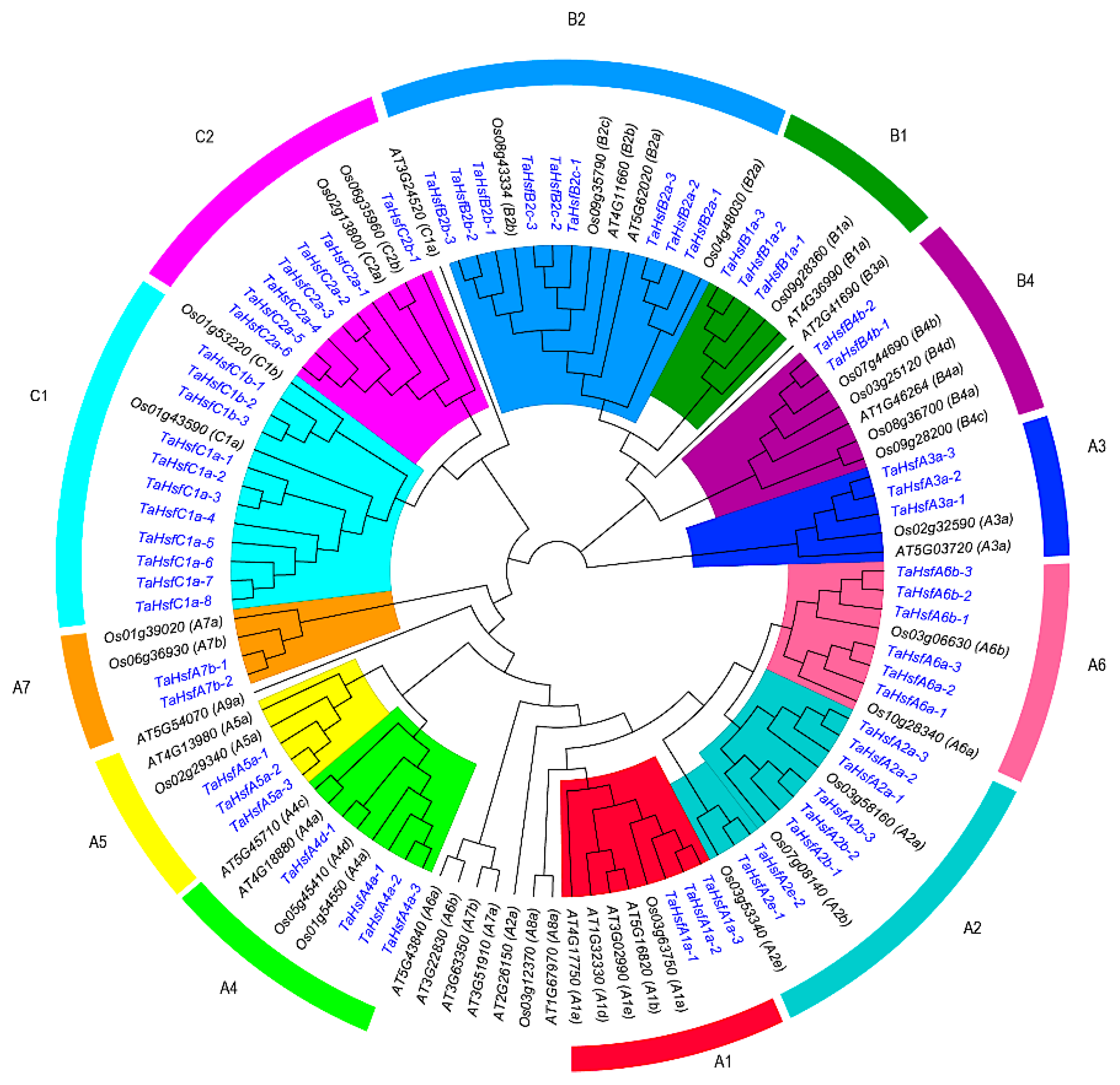
2.2. Evolution and Phylogenetic Analysis of TaHsfs
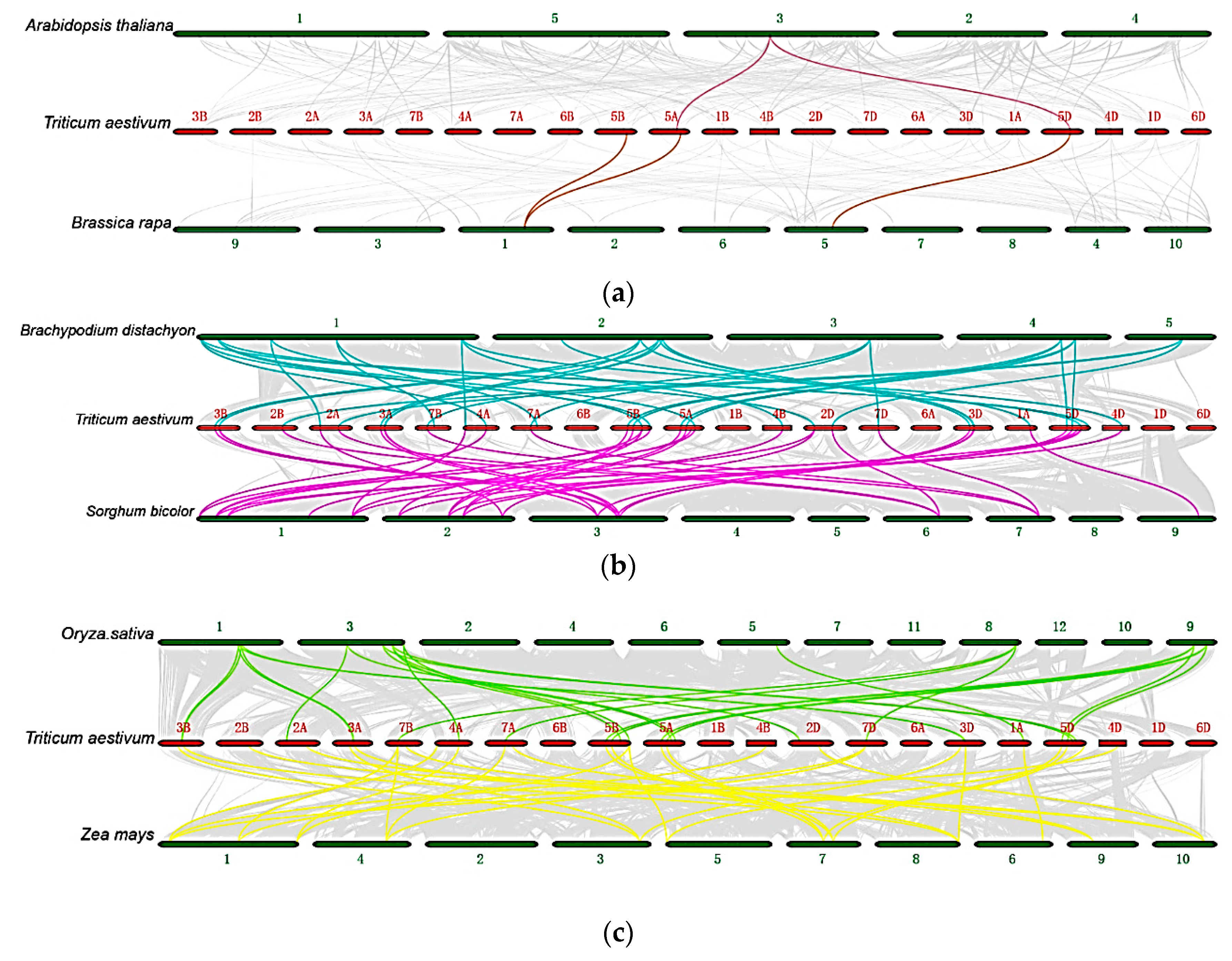
2.3. Duplication Events in Wheat HSF Family
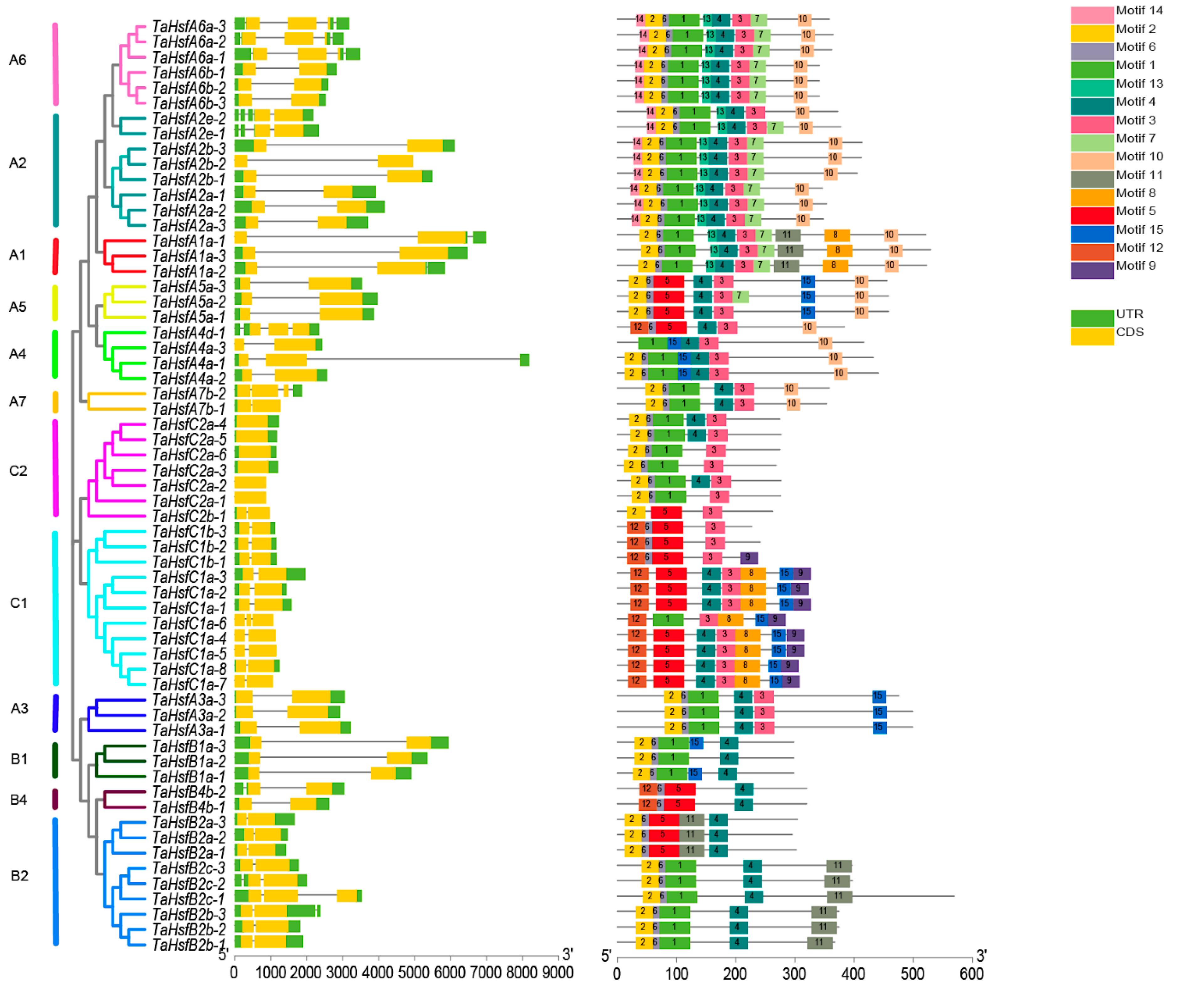
2.4. Gene Structure and Motif Composition in TaHsfs
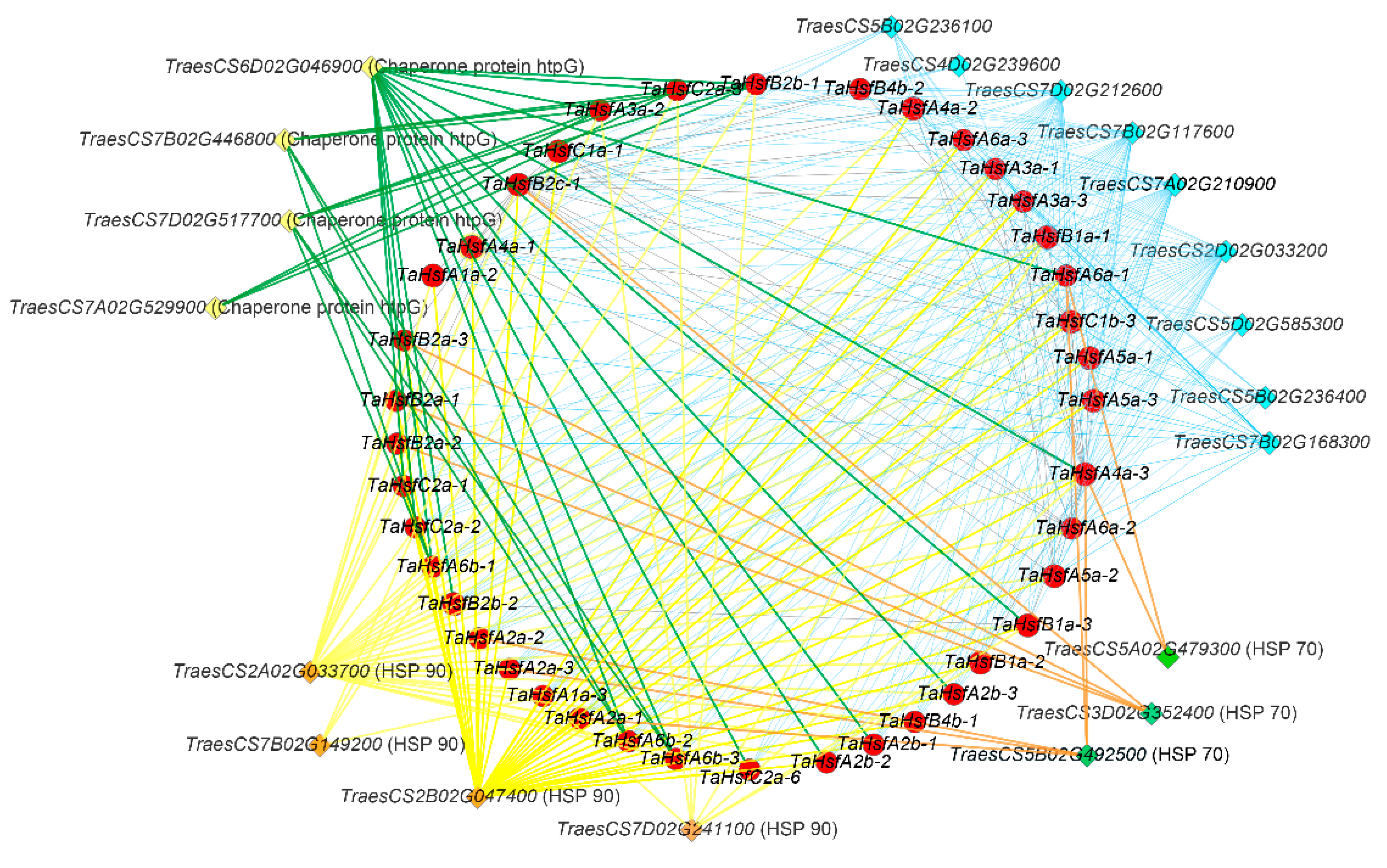
2.5. Interaction Network Analysis
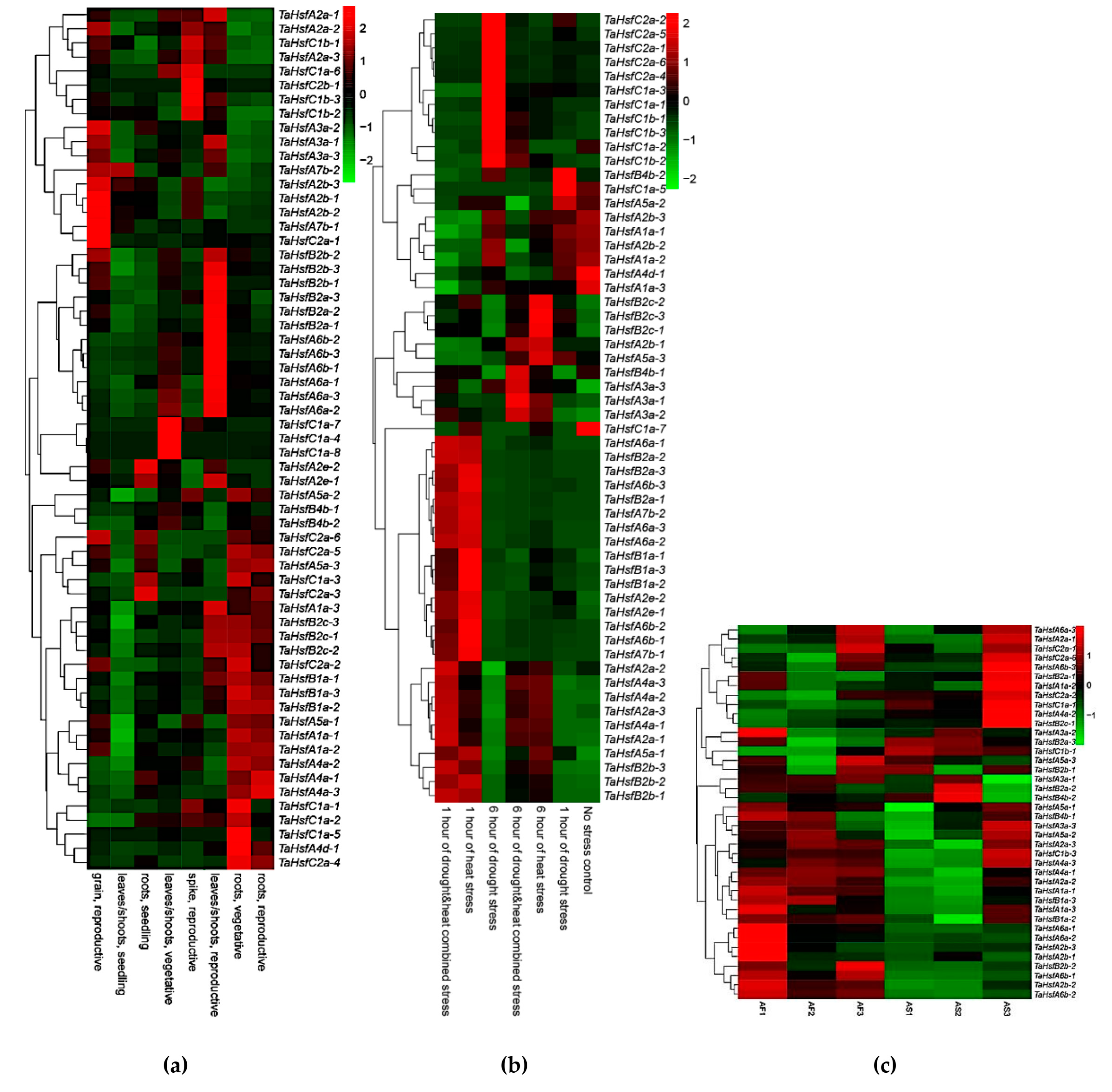
2.6. Expression Profiles of TaHsfs
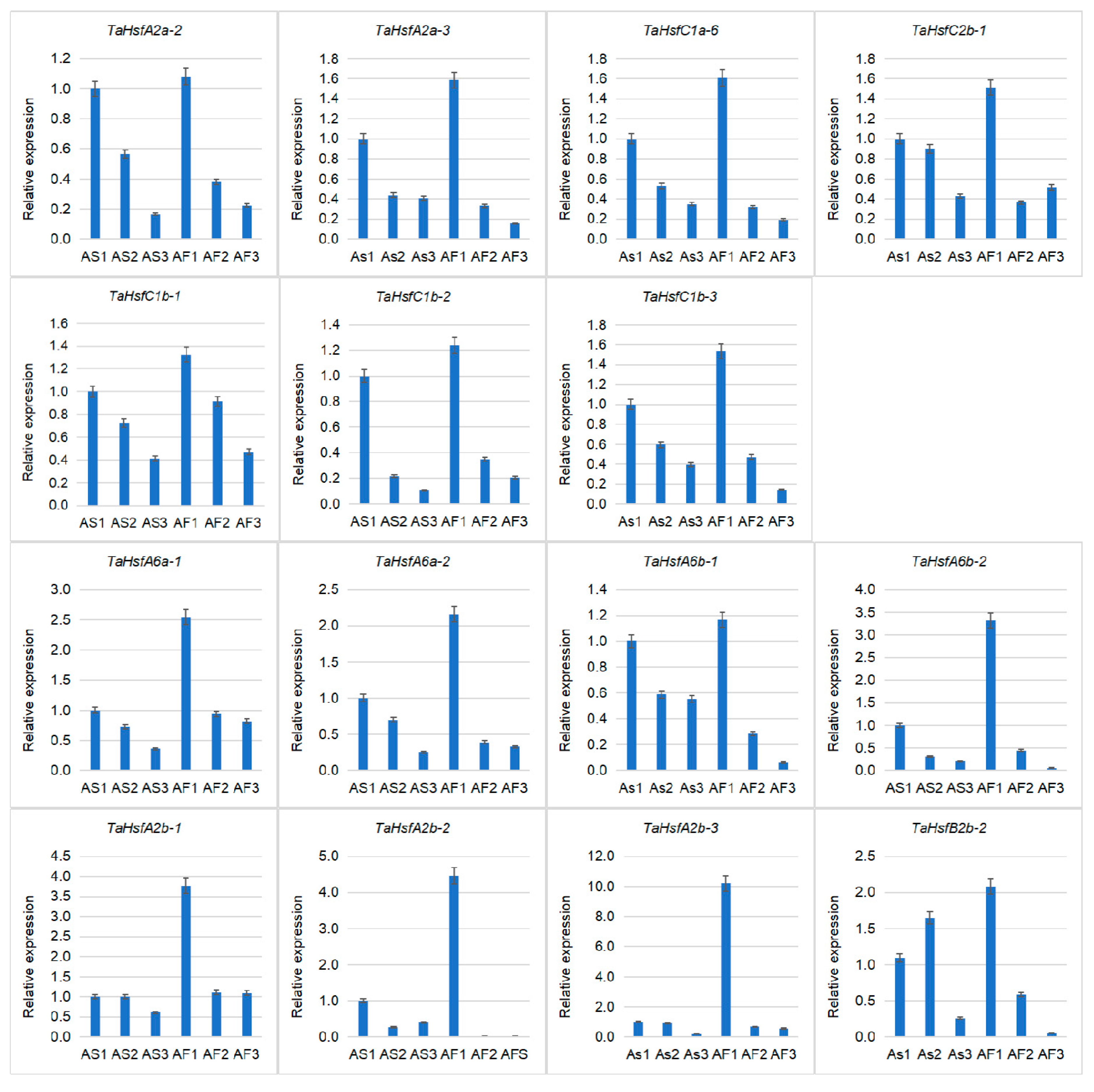
2.7. Validation of TaHsfs Involved in Anther Protection Mechanisms
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Genome-Wide Identification of HSF Family Members in Wheat
4.3. Multiple Sequence Alignment, Phylogenetic Analysis, and Classification of TaHsfs
4.4. Analysis of Gene Duplication and Synteny Relationships of TaHsfs
4.5. Structural Characterization, Conserved Motifs, and Construction of the Interaction Network for TaHsfs
4.6. RNA-seq Data Analysis
4.7. qRT-PCR Verification
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| HSF | Heat shock transcription factor |
| Hsf | HSF gene |
| TaHsf | Wheat HSF gene |
| DBD | DNA binding domain |
| OD | Oligomeric domain |
References
- Kumar, S.V.; Wigge, P.A. H2A.Z-Containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 2010, 140, 136–147. [Google Scholar] [CrossRef]
- Sung, D.; Kaplan, F.; Lee, K.; Guy, C.L. Acquired tolerance to temperature extremes. Trends Plant Sci. 2003, 8, 179–187. [Google Scholar] [CrossRef]
- Parsell, D.A.; Lindquist, S. The function of heat-shock proteins in stress tolerance: Degradation and reactivation of damaged proteins. Annu. Rev. Genet. 1993, 27, 437–496. [Google Scholar] [CrossRef] [PubMed]
- Hartl, F.U.; Hayerhartl, M. Molecular chaperones in the cytosol: From nascent chain to folded protein. Science 2002, 295, 1852–1858. [Google Scholar] [CrossRef] [PubMed]
- Nover, L.; Bharti, K.; Doring, P.; Mishra, S.K.; Ganguli, A.; Scharf, K. Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperones 2001, 6, 177–189. [Google Scholar] [CrossRef]
- Damberger, F.F.; Pelton, J.G.; Harrison, C.J.; Nelson, H.C.M.; Wemmer, D.E. Solution structure of the DNA-binding domain of the heat shock transcription factor determined by multidimensional heteronuclear magnetic resonance spectroscopy. Protein Sci. 1994, 3, 1806–1821. [Google Scholar] [CrossRef]
- Cicero, M.P.; Hubl, S.T.; Harrison, C.J.; Littlefield, O.; Hardy, J.A.; Nelson, H.C. The wing in yeast heat shock transcription factor (HSF) DNA-binding domain is required for full activity. Nucleic Acids Res. 2001, 29, 1715–1723. [Google Scholar] [CrossRef][Green Version]
- Yura, T.; Nakahigashi, K. Regulation of the heat-shock response. Curr. Opin. Microbiol. 1999, 2, 153–158. [Google Scholar] [CrossRef]
- Scharf, K.; Berberich, T.; Ebersberger, I.; Nover, L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochim. Biophys. Acta 2012, 1819, 104–119. [Google Scholar] [CrossRef]
- Heerklotz, D.; Döring, P.; Bonzelius, F.; Winkelhaus, S.; Nover, L. The balance of nuclear import and export determines the intracellular distribution and function of tomato heat stress transcription factor HsfA2. Mol. Cell. Biol. 2001, 21, 1759–1768. [Google Scholar] [CrossRef]
- Kotak, S.; Port, M.; Ganguli, A.; Bicker, F.; Von Koskulldoring, P. Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J. 2004, 39, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.K.; Tripp, J.; Winkelhaus, S.; Tschiersch, B.; Theres, K.; Nover, L.; Scharf, K. In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev. 2002, 16, 1555–1567. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wu, J.; Ji, Q.; Wang, C.; Luo, L.; Yuan, Y.; Wang, Y.; Wang, J. Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. J. Genet. Genom. 2008, 35, 105–118. [Google Scholar] [CrossRef]
- Lin, Y.X.; Jiang, H.Y.; Chu, Z.X.; Tang, X.L.; Zhu, S.W.; Cheng, B.J. Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genom. 2011, 12, 76. [Google Scholar] [CrossRef]
- Li, P.S.; Yu, T.F.; He, G.H.; Chen, M.; Zhou, Y.B.; Chai, S.C.; Xu, Z.S.; Ma, Y.Z. Genome-wide analysis of the Hsf family in soybean and functional identification of GmHsf-34 involvement in drought and heat stresses. BMC Genom. 2014, 15, 1009. [Google Scholar] [CrossRef]
- Chidambaranathan, P.; Jagannadham, P.T.K.; Satheesh, V.; Kohli, D.; Basavarajappa, S.H.; Chellapilla, B.; Kumar, J.; Jain, P.K.; Srinivasan, R. Genome-wide analysis identifies chickpea (Cicer arietinum) heat stress transcription factors (Hsfs) responsive to heat stress at the pod development stage. J. Plant Res. 2018, 131, 525–542. [Google Scholar] [CrossRef]
- Nagaraju, M.; Reddy, P.S.; Kumar, S.A.; Srivastava, R.K.; Kishor, P.B.K.; Rao, D.M. Genome-wide Scanning and Characterization of Sorghum bicolor L. Heat shock transcription factors. Curr. Genom. 2015, 16, 279–291. [Google Scholar] [CrossRef]
- Ma, J.; Xu, Z.S.; Wang, F.; Tan, G.F.; Li, M.Y.; Xiong, A.S. Genome-wide analysis of HSF family transcription factors and their responses to abiotic stresses in two Chinese cabbage varieties. Acta Physiol. Plant. 2014, 36, 513–523. [Google Scholar] [CrossRef]
- Guo, M.; Lu, J.P.; Zhai, Y.F.; Chai, W.G.; Gong, Z.H.; Lu, M.H. Genome-wide analysis, expression profile of heat shock factor gene family (CaHsfs) and characterisation of CaHsfA2 in pepper (Capsicum annuum L.). BMC Plant Biol. 2015, 15, 151. [Google Scholar] [CrossRef]
- Giorno, F.; Guerriero, G.; Baric, S.; Mariani, C. Heat shock transcriptional factors in Malus domestica: Identification, classification and expression analysis. BMC Genom. 2012, 13, 639. [Google Scholar] [CrossRef]
- Qiao, X.; Li, M.; Li, L.; Yin, H.; Wu, J.; Zhang, S. Genome-wide identification and comparative analysis of the heat shock transcription factor family in Chinese white pear (Pyrus bretschneideri) and five other Rosaceae species. BMC Plant Biol. 2015, 15, 12. [Google Scholar] [CrossRef] [PubMed]
- Lohmann, C.; Eggersschumacher, G.; Wunderlich, M.; Schoffl, F. Two different heat shock transcription factors regulate immediate early expression of stress genes in Arabidopsis. Mol. Genet. Genom. 2004, 271, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Busch, W.; Wunderlich, M.; Schöffl, F. Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana. Plant J. 2004, 41, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, D.; Yamaguchi, K.; Nishiuchi, T. High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased themotolerance but also salt/osmotic stress tolerance and enhanced callus growth. J. Exp. Bot. 2007, 58, 3373–3383. [Google Scholar] [CrossRef]
- Banti, V.; Mafessoni, F.; Loreti, E.; Alpi, A.; Perata, P. The heat-inducible transcriptionfactor HsfA2 enhances anoxia tolerance in Arabidopsis. Plant Physiol. 2010, 152, 1471–1483. [Google Scholar] [CrossRef]
- Zhang, L.; Li, Y.; Xing, D.; Gao, C. Characterization of mitochondrial dynamics and subcellular localization of ROS reveal that HsfA2 alleviates oxidative damage caused by heat stress in Arabidopsis. J. Exp. Bot. 2009, 60, 2073–2091. [Google Scholar] [CrossRef]
- Giorno, F.; Woltersarts, M.; Grillo, S.; Scharf, K.; Vriezen, W.H.; Mariani, C. Developmental and heat stress-regulated expression of HsfA2 and small heat shock proteins in tomato anthers. J. Exp. Bot. 2010, 61, 453–462. [Google Scholar] [CrossRef]
- Shim, D.; Hwang, J.; Lee, J.; Lee, S.; Choi, Y.; An, G.; Martinoia, E.; Lee, Y. Orthologs of the class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice. Plant Cell 2009, 21, 4031–4043. [Google Scholar] [CrossRef]
- Almoguera, C.; Prietodapena, P.; Diazmartin, J.; Espinosa, J.M.R.; Carranco, R.; Jordano, J. The HaDREB2 transcription factor enhances basal thermotolerance and longevity of seeds through functional interaction with HaHSFA9. BMC Plant Biol. 2009, 9, 75. [Google Scholar] [CrossRef]
- Bharti, K.; Von Koskulldoring, P.; Bharti, S.; Kumar, P.; Tintschlkorbitzer, A.; Treuter, E.; Nover, L. Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein ortholog HAC1. Plant Cell 2004, 16, 1521–1535. [Google Scholar] [CrossRef]
- Prasad, P.V.V.; Djanaguiraman, M. Response of floret fertility and individual grain weight of wheat to high temperature stress: Sensitive stages and thresholds for temperature and duration. Funct. Plant Biol. 2014, 41, 1261–1269. [Google Scholar] [CrossRef]
- Xue, G.; Sadat, S.; Drenth, J.; McIntyre, C.L. The heat shock factor family from Triticum aestivum in response to heat and other major abiotic stresses and their role in regulation of heat shock protein genes. J. Exp. Bot. 2014, 65, 539–557. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Xu, Z.; Li, P.; Yang, L.; Wei, Y.; Chen, M.; Li, L.; Zhang, G.; Ma, Y. Overexpression of TaHSF3 in transgenic Arabidopsis enhances tolerance to extreme temperatures. Plant Mol. Biol. Rep. 2013, 31, 688–697. [Google Scholar] [CrossRef]
- Kumar, R.R.; Goswami, S.; Singh, K.; Dubey, K.; Rai, G.K.; Singh, B.; Singh, S.; Grover, M.; Mishra, D.; Kumar, S.; et al. Characterization of novel heat-responsive transcription factor (TaHSFA6e) gene involved in regulation of heat shock proteins (HSPs)—A key member of heat stress-tolerance network of wheat. J. Biotechnol. 2018, 279, 1–12. [Google Scholar] [CrossRef]
- Hu, X.J.; Chen, D.; Lynne Mclntyre, C.; Fernanda Dreccer, M.; Zhang, Z.B.; Drenth, J.; Kalaipandian, S.; Chang, H.; Xue, G.P. Heat shock factor C2a serves as a proactive mechanism for heat protection in developing grains in wheat via an ABA-mediated regulatory pathway. Plant Cell Environ. 2018, 41, 79–98. [Google Scholar] [CrossRef]
- Meng, L.Y.; Liu, Z.H.; Zhang, L.L.; Hu, G.; Song, X.Y. Cytological characterization of a thermo-sensitive cytoplasmic male-sterile wheat line having K-type cytoplasm of aegilops kotschyi. Breed Sci. 2016, 66, 752–761. [Google Scholar] [CrossRef]
- Ye, J.; Duan, Y.; Hu, G.; Geng, X.; Zhang, G.; Yan, P.; Liu, Z.; Zhang, L.; Song, X. Identification of candidate genes and biosynthesis pathways related to fertility conversion by wheat KTM3315A transcriptome profiling. Front. Plant Sci. 2017, 8, 449. [Google Scholar] [CrossRef]
- Widlak, W.; Vydra, N. The role of heat shock factors in mammalian spermatogenesis. Adv. Anat. Embryol. Cell Biol. 2017, 222, 45–65. [Google Scholar] [CrossRef]
- Shinka, T.; Sato, Y.; Chen, G.; Naroda, T.; Kinoshita, K.; Unemi, Y.; Tsuji, K.; Toida, K.; Iwamoto, T.; Nakahori, Y. Molecular characterization of heat shock-like factor encoded on the human Y chromosome, and implications for male infertility. Biol. Reprod. 2004, 71, 297–306. [Google Scholar] [CrossRef]
- Zhang, X.; Li, J.; Liu, A.; Zou, J.; Zhou, X.; Xiang, J.; Rerksiri, W.; Peng, Y.; Xiong, X.; Chen, X. Expression profile in rice panicle: Insights into heat response mechanism at reproductive stage. PLoS ONE 2012, 7, e49652. [Google Scholar] [CrossRef]
- Frank, G.; Pressman, E.; Ophir, R.; Althan, L.; Shaked, R.; Freedman, M.; Shen, S.; Firon, N. Transcriptional profiling of maturing tomato (Solanum lycopersicum L.) microspores reveals the involvement of heat shock proteins, ROS scavengers, hormones, and sugars in the heat stress response. J. Exp. Bot. 2009, 60, 3891–3908. [Google Scholar] [CrossRef] [PubMed]
- Bennetzen, J.L.; Devos, K. Plant synteny, colinearity and genome evolution. eLS 2001. [Google Scholar] [CrossRef]
- Eckardt, N.A. Everything in its place: Conservation of gene order among distantly related plant species. Plant Cell 2001, 13, 723–725. [Google Scholar] [CrossRef] [PubMed]
- Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; Poland, J.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef]
- Liu, L.K.; Liu, G.Q.; Hou, N.; Liu, C.G.; Chen, J.N.; Zhang, W.H.; Fan, Y.L.; Wu, B.J. Construction and identification of HSP70 antisense RNA expression vector for genetic engineering male sterility in plant. J. Anhui Agric. Sci. 2008, 36, 6698–6700. [Google Scholar]
- YI, L.X.; Li, X.; Chen, J.N.; Xia, S.T. Creation of male sterile line in tobacco with HSP70 anti-sense fragment. Agric. Sci. Technol. 2016, 17, 2262–2266. [Google Scholar] [CrossRef]
- Czarneckaverner, E.; Yuan, C.; Scharf, K.; Englich, G.; Gurley, W.B. Plants contain a novel multi-member class of heat shock factors without transcriptional activator potential. Plant Mol. Biol. 2000, 43, 459–471. [Google Scholar] [CrossRef]
- Ikeda, M.; Mitsuda, N.; Ohmetakagi, M. Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol. 2011, 157, 1243–1254. [Google Scholar] [CrossRef]
- Liu, H.C.; Charng, Y.Y. Common and distinct functions of Arabidopsis class A1 and A2 heat shock factors in diverse abiotic stress responses and development. Plant Physiol. 2013, 163, 276–290. [Google Scholar] [CrossRef]
- Fragkostefanakis, S.; Mesihovic, A.; Simm, S.; Paupiere, M.J.; Hu, Y.; Paul, P.; Mishra, S.K.; Tschiersch, B.; Theres, K.; Bovy, A.; et al. HsfA2 controls the activity of developmentally and stress-regulated heat stress protection mechanisms in tomato male reproductive tissues. Plant Physiol. 2016, 170, 2461–2477. [Google Scholar] [CrossRef]
- Chen, Y.B.; Wang, Y.; Zhang, H.L. Identification and expression analysis of heat shock factor (Hsf) gene family in tomato (Solanum lycopersicum). J. Agric. Biotechnol. 2015, 23, 492–501. [Google Scholar] [CrossRef]
- Song, X.; Liu, G.; Duan, W.; Liu, T.; Huang, Z.; Ren, J.; Li, Y.; Hou, X. Genome-wide identification, classification and expression analysis of the heat shock transcription factor family in Chinese cabbage. Mol. Genet. Genom. 2014, 289, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.; Liu, B.; Zhang, Y.; Li, G.; Guo, X. Genome-wide identification and abiotic stress-responsive pattern of heat shock transcription factor family in Triticum aestivum L. BMC Genom. 2019, 20, 257. [Google Scholar] [CrossRef] [PubMed]
- Mauro, D.; Terry, S. An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics 2002, 18, 617–625. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45. [Google Scholar] [CrossRef]
- Freeling, M. Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 2009, 60, 433–453. [Google Scholar] [CrossRef]
- Edger, P.P.; Pires, J.C. Gene and genome duplications: The impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 2009, 17, 699–717. [Google Scholar] [CrossRef]
- Birchler, J.A.; Bhadra, U.; Bhadra, M.P.; Auger, D.L. Dosage-dependent gene regulation in multicellular eukaryotes: Implications for dosage compensation, aneuploid syndromes, and quantitative traits. Dev. Biol. 2001, 234, 275–288. [Google Scholar] [CrossRef]
- Papp, B.; Pal, C.; Hurst, L.D. Dosage sensitivity and the evolution of gene families in yeast. Nature 2003, 424, 194–197. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Michelmore, R.W.; Meyers, B.C. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 1998, 8, 1113–1130. [Google Scholar] [CrossRef] [PubMed]
- Maere, S.; De Bodt, S.; Raes, J.; Casneuf, T.; Van Montagu, M.; Kuiper, M.; Van de Peer, Y. Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 5454–5459. [Google Scholar] [CrossRef] [PubMed]
- Rose, A.B. Intron-mediated regulation of gene expression. Curr. Top. Microbiol. Immunol. 2008, 326, 277–290. [Google Scholar] [CrossRef]
- Barrett, L.W.; Fletcher, S.; Wilton, S.D. Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell. Mol. Life Sci. 2012, 69, 3613–3634. [Google Scholar] [CrossRef]
- Vu, L.D.; Gevaert, K.; De Smet, I. Feeling the heat: Searching for plant thermosensors. Trends Plant Sci. 2019, 24, 210–219. [Google Scholar] [CrossRef]
- Reňák, D.; Gibalová, A.; Solcová, K.; Honys, D. A new link between stress response and nucleolar function during pollen development in Arabidopsis mediated by AtREN1 protein. Plant Cell Environ. 2014, 37, 670–683. [Google Scholar] [CrossRef]
- Tuncozdemir, M.; Tang, C.; Ishka, M.R.; Brown, E.; Groves, N.R.; Myers, C.T.; Rato, C.; Poulsen, L.R.; McDowell, S.C.; Miller, G.; et al. A cyclic nucleotide-gated channel (CNGC16) in pollen is critical for stress tolerance in pollen reproductive development. Plant Physiol. 2013, 161, 1010–1020. [Google Scholar] [CrossRef]
- Bukau, B.; Weissman, J.S.; Horwich, A.L. Molecular chaperones and protein quality control. Cell 2006, 125, 443–451. [Google Scholar] [CrossRef]
- Neumann, D.; Nover, L.; Parthier, B.; Rieger, R.; Scharf, K.D.; Wollgiehn, R.; zur Nieden, U. Heat shock and other stress response systems of plants. Results Probl Cell Differ. 1989, 16, 1–155. [Google Scholar] [PubMed]
- Baniwal, S.K.; Bharti, K.; Chan, K.Y.; Fauth, M.; Ganguli, A.; Kotak, S.; Mishra, S.K.; Nover, L.; Port, M.; Scharf, K.D.; et al. Heat stress response in plants: A complex game with chaperones and more than twenty heat stress transcription factors. J. Biosci. 2004, 29, 471–487. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2018, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Jaina, M.; Robert, D.F.; Sean, R.E.; Alex, B.; Marco, P. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef]
- La Cour, T.; Kiemer, L.; Molgaard, A.; Gupta, R.; Skriver, K.; Brunak, S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 2004, 17, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, S.; Hasebe, M.; Tomita, M.; Yanagawa, H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. USA 2009, 106, 10171–10176. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Guo, J.; Jain, R.; Yang, P.; Fan, R.; Kwoh, C.K.; Zheng, J. Reliable and fast estimation of recombination rates by convergence diagnosis and parallel markov chain monte carlo. IEEE/ACM Trans. Comput. Biol. Bioinform. 2014, 11, 63–72. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.E.; Birol, I.; Connors, J.M.; Gascoyne, R.D.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Timothy, L.B.; Mikael, B.; Fabian, A.B.; Martin, F.; Charles, E.G.; Luca, C.; Ren, J.Y.; Wilfred, W.L.; William, S.N. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Chen, C.; Xia, R.; Chen, H.; He, Y. TBtools, a toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv 2018. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Clavijo, B.J.; Venturini, L.; Schudoma, C.; Accinelli, G.G.; Kaithakottil, G.; Wright, J.; Borrill, P.; Kettleborough, G.; Heavens, D.; Chapman, H.D.; et al. An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations. Genome Res. 2017, 27, 885–896. [Google Scholar] [CrossRef]
- Liu, Z.; Xin, M.; Qin, J.; Peng, H.; Ni, Z.; Yao, Y.; Sun, Q. Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biol. 2015, 15, 152. [Google Scholar] [CrossRef]
- Schmittgen, T.; Livak, K. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]



| Homoeologous Group (A:B:D) | All Wheat Genes | Classes | Number of Groups | Number of Genes | % of Total TaHSFs | ||
|---|---|---|---|---|---|---|---|
| A | B | C | |||||
| 1:1:1 | 35.8% | 7 | 4 | 3 | 14 | 42 | 68.8 |
| 1:1:0/1:0:1 | 13.2% | 2 | 1 | 1 | 4 | 8 | 13.1 |
| Orphans/singletons | 37.1% | 1 | 1 | 2 | 2 | 3.3 | |
| Others | - | 1 | 1 | 2 | 9 | 14.8 | |
| Total | - | 11 | 5 | 6 | 20 | 61 | 100 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, J.; Yang, X.; Hu, G.; Liu, Q.; Li, W.; Zhang, L.; Song, X. Genome-Wide Investigation of Heat Shock Transcription Factor Family in Wheat (Triticum aestivum L.) and Possible Roles in Anther Development. Int. J. Mol. Sci. 2020, 21, 608. https://doi.org/10.3390/ijms21020608
Ye J, Yang X, Hu G, Liu Q, Li W, Zhang L, Song X. Genome-Wide Investigation of Heat Shock Transcription Factor Family in Wheat (Triticum aestivum L.) and Possible Roles in Anther Development. International Journal of Molecular Sciences. 2020; 21(2):608. https://doi.org/10.3390/ijms21020608
Chicago/Turabian StyleYe, Jiali, Xuetong Yang, Gan Hu, Qi Liu, Wei Li, Lingli Zhang, and Xiyue Song. 2020. "Genome-Wide Investigation of Heat Shock Transcription Factor Family in Wheat (Triticum aestivum L.) and Possible Roles in Anther Development" International Journal of Molecular Sciences 21, no. 2: 608. https://doi.org/10.3390/ijms21020608
APA StyleYe, J., Yang, X., Hu, G., Liu, Q., Li, W., Zhang, L., & Song, X. (2020). Genome-Wide Investigation of Heat Shock Transcription Factor Family in Wheat (Triticum aestivum L.) and Possible Roles in Anther Development. International Journal of Molecular Sciences, 21(2), 608. https://doi.org/10.3390/ijms21020608





