The Role of MicroRNAs in Mammalian Fertility: From Gametogenesis to Embryo Implantation
Abstract
1. Introduction
2. MicroRNAs During Gametogenesis
2.1. The Role of Cellular miRNAs in Follicular Development and Oocyte Maturation
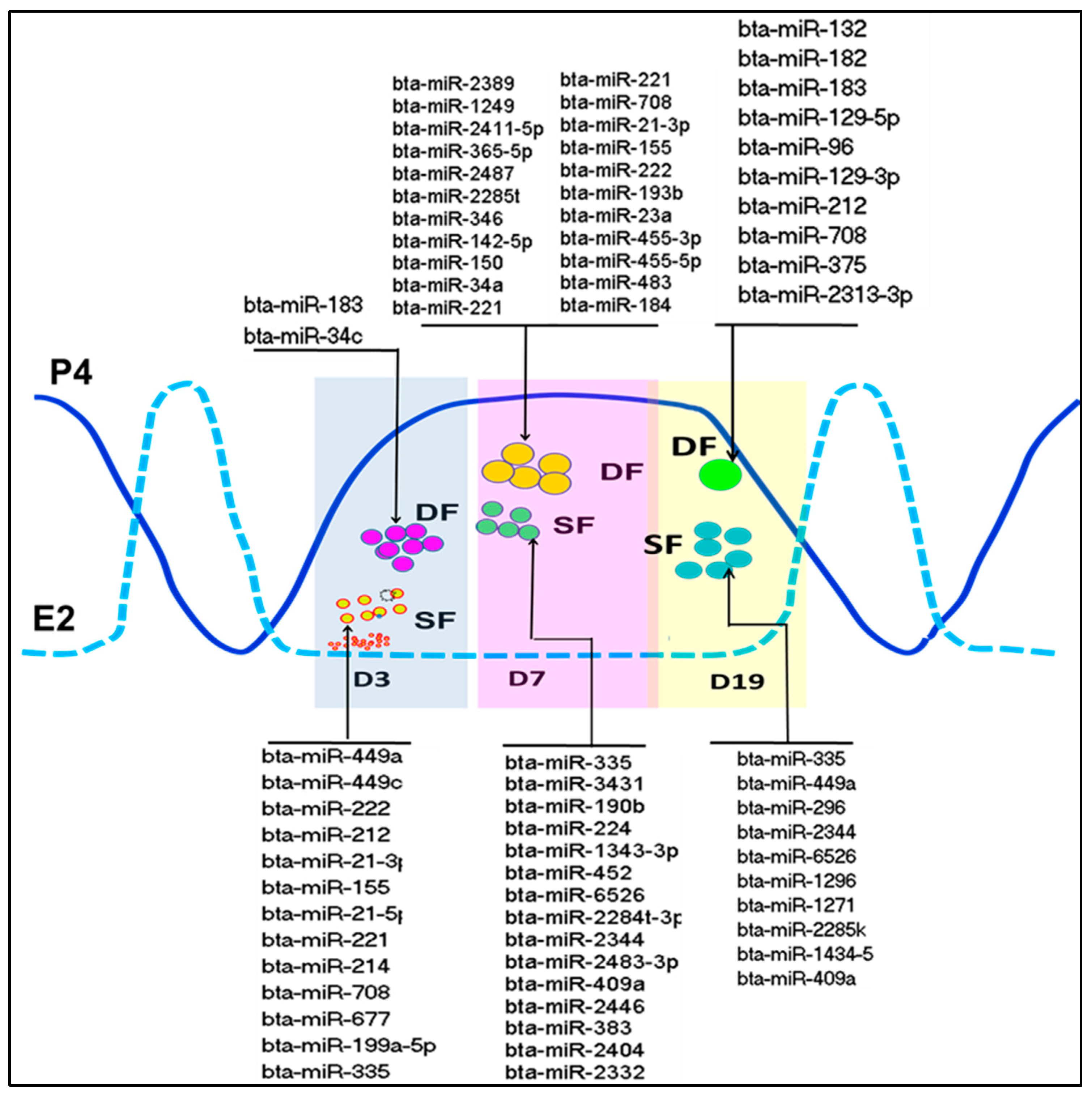
2.2. Functional Analysis of miRNAs in Oocyte Companion Somatic Cells
2.3. The Role of Extracellular Vesicles-Mediated miRNAs in Female Gametogenesis
2.4. The Role of miRNAs in Male Gametogenesis
3. The Role of miRNAs during Preimplantation Mammalian Embryo Development
3.1. The Role of Embryonic miRNAs in Embryo Implantation
3.2. The Role of Maternal miRNAs in Embryo Implantation
3.3. The Role of Extracellular Vesicle Mediated miRNAs in Embryo—Maternal Communication
4. Conclusions
Funding
Conflicts of Interest
References
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140-4. [Google Scholar] [CrossRef] [PubMed]
- Humphreys, D.T.; Westman, B.J.; Martin, D.I.K.; Preiss, T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc. Natl. Acad. Sci. USA 2005, 102, 16961–16966. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Yi, R.; Cullen, B.R. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl. Acad. Sci. USA 2003, 100, 9779–9784. [Google Scholar] [CrossRef]
- Margolis, L.; Sadovsky, Y. The biology of extracellular vesicles: The known unknowns. PLoS Biol. 2019, 17, e3000363. [Google Scholar] [CrossRef]
- Van Wolfswinkel, J.C.; Ketting, R.F. The role of small non-coding RNAs in genome stability and chromatin organization. J. Cell Sci. 2010, 123, 1825–1839. [Google Scholar] [CrossRef]
- Brennecke, J.; Hipfner, D.R.; Stark, A.; Russell, R.B.; Cohen, S.M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 2003, 113, 25–36. [Google Scholar]
- Darnell, D.K.; Kaur, S.; Stanislaw, S.; Konieczka, J.H.; Yatskievych, T.A.; Antin, P.B. MicroRNA expression during chick embryo development. Dev. Dyn. 2006, 235, 3156–3165. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef]
- Hatfield, S.D.; Shcherbata, H.R.; Fischer, K.A.; Nakahara, K.; Carthew, R.W.; Ruohola-Baker, H. Stem cell division is regulated by the microRNA pathway. Nature 2005, 435, 974–978. [Google Scholar] [CrossRef]
- Kotaja, N. MicroRNAs and spermatogenesis. Fertil. Steril. 2014, 101, 1552–1562. [Google Scholar] [CrossRef]
- Wen, J.; Friedman, J.R. miR-122 regulates hepatic lipid metabolism and tumor suppression. J. Clin. Investig. 2012, 122, 2773–2776. [Google Scholar] [CrossRef] [PubMed]
- Dahl, K.D.; Czekala, N.M.; Hsueh, A.J. Estrogen-producing ovarian granulosa cells: Use of the granulosa cell aromatase bioassay (GAB) to monitor FSH levels in body fluids. Adv. Exp. Med. Biol. 1987, 219, 275–298. [Google Scholar] [PubMed]
- Carabatsos, M.J.; Sellitto, C.; Goodenough, D.A.; Albertini, D.F. Oocyte-granulosa cell heterologous gap junctions are required for the coordination of nuclear and cytoplasmic meiotic competence. Dev. Biol. 2000, 226, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Wigglesworth, K.; Lee, K.-B.; O’Brien, M.J.; Peng, J.; Matzuk, M.M.; Eppig, J.J. Bidirectional communication between oocytes and ovarian follicular somatic cells is required for meiotic arrest of mammalian oocytes. Proc. Natl. Acad. Sci. USA 2013, 110, E3723–E3729. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, K.; Pendola, F.L.; Eppig, J.J. Oocyte control of metabolic cooperativity between oocytes and companion granulosa cells: Energy metabolism. Dev. Biol. 2005, 279, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Fayad, T.; Lévesque, V.; Sirois, J.; Silversides, D.W.; Lussier, J.G. Gene expression profiling of differentially expressed genes in granulosa cells of bovine dominant follicles using suppression subtractive hybridization. Biol. Reprod. 2004, 70, 523–533. [Google Scholar] [CrossRef]
- Evans, A.C.O.; Ireland, J.L.H.; Winn, M.E.; Lonergan, P.; Smith, G.W.; Coussens, P.M.; Ireland, J.J. Identification of genes involved in apoptosis and dominant follicle development during follicular waves in cattle. Biol. Reprod. 2004, 70, 1475–1484. [Google Scholar]
- Sisco, B.; Hagemann, L.J.; Shelling, A.N.; Pfeffer, P.L. Isolation of genes differentially expressed in dominant and subordinate bovine follicles. Endocrinology 2003, 144, 3904–3913. [Google Scholar]
- Liu, Z.; Youngquist, R.S.; Garverick, H.A.; Antoniou, E. Molecular mechanisms regulating bovine ovarian follicular selection. Mol. Reprod. Dev. 2009, 76, 351–366. [Google Scholar] [CrossRef]
- Lei, L.; Jin, S.; Gonzalez, G.; Behringer, R.R.; Woodruff, T.K. The regulatory role of Dicer in folliculogenesis in mice. Mol. Cell. Endocrinol. 2010, 315, 63–73. [Google Scholar] [CrossRef]
- Kaneda, M.; Tang, F.; O’Carroll, D.; Lao, K.; Surani, M.A. Essential role for Argonaute2 protein in mouse oogenesis. Epigenetics Chromatin 2009, 2, 9. [Google Scholar] [CrossRef]
- Muggenhumer, D.; Vesely, C.; Nimpf, S.; Tian, N.; Yongfeng, J.; Jantsch, M.F. Drosha protein levels are translationally regulated during Xenopus oocyte maturation. Mol. Biol. Cell 2014, 25, 2094–2104. [Google Scholar] [CrossRef] [PubMed]
- Tesfaye, D.; Worku, D.; Rings, F.; Phatsara, C.; Tholen, E.; Schellander, K.; Hoelker, M. Identification and expression profiling of microRNAs during bovine oocyte maturation using heterologous approach. Mol. Reprod. Dev. 2009, 76, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Jiang, H.; Gao, Y.; Zhao, Y.; Dai, L.; Xiong, Q.; Xu, Y.; Zhao, Z.; Zhang, J. Microarray analysis of differentially expressed microRNAs in non-regressed and regressed bovine corpus luteum tissue; microRNA-378 may suppress luteal cell apoptosis by targeting the interferon gamma receptor 1 gene. J. Appl. Genet. 2011, 52, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.M.; Ghanem, N.; Hoelker, M.; Rings, F.; Phatsara, C.; Tholen, E.; Schellander, K.; Tesfaye, D. Identification and characterization of miRNAs expressed in the bovine ovary. BMC Genom. 2009, 10, 443. [Google Scholar] [CrossRef]
- Huang, J.; Ju, Z.; Li, Q.; Hou, Q.; Wang, C.; Li, J.; Li, R.; Wang, L.; Sun, T.; Hang, S.; et al. Solexa sequencing of novel and differentially expressed microRNAs in testicular and ovarian tissues in Holstein cattle. Int. J. Biol. Sci. 2011, 7, 1016–1026. [Google Scholar] [CrossRef]
- Tripurani, S.K.; Xiao, C.; Salem, M.; Yao, J. Cloning and analysis of fetal ovary microRNAs in cattle. Anim. Reprod. Sci. 2010, 120, 16–22. [Google Scholar] [CrossRef]
- Sohel, M.M.H.; Hoelker, M.; Noferesti, S.S.; Salilew-Wondim, D.; Tholen, E.; Looft, C.; Rings, F.; Uddin, M.J.; Spencer, T.E.; Schellander, K.; et al. Exosomal and Non-Exosomal Transport of Extra-Cellular microRNAs in Follicular Fluid: Implications for Bovine Oocyte Developmental Competence. PLoS ONE 2013, 8, e78505. [Google Scholar] [CrossRef]
- Abd El Naby, W.S.; Hagos, T.H.; Hossain, M.M.; Salilew-Wondim, D.; Gad, A.Y.; Rings, F.; Cinar, M.U.; Tholen, E.; Looft, C.; Schellander, K.; et al. Expression analysis of regulatory microRNAs in bovine cumulus oocyte complex and preimplantation embryos. Zygote 2013, 21, 31–51. [Google Scholar] [CrossRef]
- Tscherner, A.; Gilchrist, G.; Smith, N.; Blondin, P.; Gillis, D.; LaMarre, J. MicroRNA-34 family expression in bovine gametes and preimplantation embryos. Reprod. Biol. Endocrinol. 2014, 12, 85. [Google Scholar] [CrossRef]
- Salilew-Wondim, D.; Ahmad, I.; Gebremedhn, S.; Sahadevan, S.; Hossain, M.D.; Rings, F.; Hoelker, M.; Tholen, E.; Neuhoff, C.; Looft, C.; et al. The expression pattern of microRNAs in granulosa cells of subordinate and dominant follicles during the early luteal phase of the bovine estrous cycle. PLoS ONE 2014, 9, e106795. [Google Scholar] [CrossRef]
- Gebremedhn, S.; Salilew-Wondim, D.; Ahmad, I.; Sahadevan, S.; Hossain, M.M.; Hoelker, M.; Rings, F.; Neuhoff, C.; Tholen, E.; Looft, C.; et al. MicroRNA Expression Profile in Bovine Granulosa Cells of Preovulatory Dominant and Subordinate Follicles during the Late Follicular Phase of the Estrous Cycle. PLoS ONE 2015, 10, e0125912. [Google Scholar] [CrossRef]
- Sontakke, S.D.; Mohammed, B.T.; McNeilly, A.S.; Donadeu, F.X. Characterization of microRNAs differentially expressed during bovine follicle development. Reproduction 2014, 148, 271–283. [Google Scholar] [CrossRef]
- Zayed, Y.; Qi, X.; Peng, C. Identification of Novel MicroRNAs and Characterization of MicroRNA Expression Profiles in Zebrafish Ovarian Follicular Cells. Front. Endocrinol. 2019, 10, 518. [Google Scholar] [CrossRef]
- Xu, S.; Linher-Melville, K.; Yang, B.B.; Wu, D.; Li, J. Micro-RNA378 (miR-378) regulates ovarian estradiol production by targeting aromatase. Endocrinology 2011, 152, 3941–3951. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, X.; Zhang, X.; Lu, Y.; Li, L.; Cui, S. MiRNA-143 mediates the proliferative signaling pathway of FSH and regulates estradiol production. J. Endocrinol. 2017, 234, 1–14. [Google Scholar] [CrossRef]
- Wu, S.; Sun, H.; Zhang, Q.; Jiang, Y.; Fang, T.; Cui, I.; Yan, G.; Hu, Y. MicroRNA-132 promotes estradiol synthesis in ovarian granulosa cells via translational repression of Nurr1. Reprod. Biol. Endocrinol. 2015, 13, 94. [Google Scholar] [CrossRef]
- Wang, L.; Li, C.; Li, R.; Deng, Y.; Tan, Y.; Tong, C.; Qi, H. MicroRNA-764-3p regulates 17beta-estradiol synthesis of mouse ovarian granulosa cells by targeting steroidogenic factor-1. In Vitro Cell. Dev. Biol. Anim. 2016, 52, 365–373. [Google Scholar] [CrossRef]
- Wang, M.; Liu, M.; Sun, J.; Jia, L.; Ma, S.; Gao, J.; Xu, Y.; Zhang, H.; Tsang, S.Y.; Li, X. MicroRNA-27a-3p affects estradiol and androgen imbalance by targeting Creb1 in the granulosa cells in mouse polycytic ovary syndrome model. Reprod. Biol. 2017. [Google Scholar] [CrossRef]
- Sun, X.-F.; Li, Y.-P.; Pan, B.; Wang, Y.-F.; Li, J.; Shen, W. Molecular regulation of miR-378 on the development of mouse follicle and the maturation of oocyte in vivo. Cell Cycle 2018, 17, 2230–2242. [Google Scholar] [CrossRef]
- Xiong, X.R.; Lan, D.; Li, J.; Zi, X.; Li, M. Identification of candidate miRNAs and expression profile of yak oocytes before and after in vitro maturation by high-throughput sequencing. Reprod. Domest. Anim. 2016, 51, 886–894. [Google Scholar] [CrossRef]
- Gilchrist, G.C.; Tscherner, A.; Nalpathamkalam, T.; Merico, D.; LaMarre, J. MicroRNA Expression during Bovine Oocyte Maturation and Fertilization. Int. J. Mol. Sci. 2016, 17, 396. [Google Scholar] [CrossRef]
- Song, C.; Yao, J.; Cao, C.; Liang, X.; Huang, J.; Han, Z.; Zhang, Y.; Qin, G.; Tao, C.; Li, C.; et al. PPARgamma is regulated by miR-27b-3p negatively and plays an important role in porcine oocyte maturation. Biochem. Biophys. Res. Commun. 2016, 479, 224–230. [Google Scholar] [CrossRef]
- Sinha, P.B.; Tesfaye, D.; Rings, F.; Hossien, M.; Hoelker, M.; Held, E.; Neuhoff, C.; Tholen, E.; Schellander, K.; Salilew-Wondim, D. MicroRNA-130b is involved in bovine granulosa and cumulus cells function, oocyte maturation and blastocyst formation. J. Ovarian Res. 2017, 10, 37. [Google Scholar] [CrossRef]
- Ge, W.; Deng, Q.; Guo, T.; Hong, X.; Kugler, J.M.; Yang, X.; Cohen, S.M. Regulation of pattern formation and gene amplification during Drosophila oogenesis by the miR-318 microRNA. Genetics 2015, 200, 255–265. [Google Scholar] [CrossRef]
- Gay, S.; Bugeon, J.; Bouchareb, A.; Henry, L.; Delahaye, C.; Legeai, F.; Montfort, J.; Le Cam, A.; Siegel, A.; Bobe, J.; et al. MiR-202 controls female fecundity by regulating medaka oogenesis. PLoS Genet. 2018, 14, e1007593. [Google Scholar] [CrossRef]
- Song, J.; Li, W.; Zhao, H.; Gao, L.; Fan, Y.; Zhou, S. The microRNAs let-7 and miR-278 regulate insect metamorphosis and oogenesis by targeting the juvenile hormone early-response gene Kruppel-homolog 1. Development 2018, 145. [Google Scholar] [CrossRef]
- Pan, B.; Toms, D.; Shen, W.; Li, J. MicroRNA-378 regulates oocyte maturation via the suppression of aromatase in porcine cumulus cells. Am. J. Physiol. Endocrinol. Metab. 2015, 308, E525–E534. [Google Scholar] [CrossRef]
- Grossman, H.; Har-Paz, E.; Gindi, N.; Levi, M.; Miller, I.; Nevo, N.; Galiani, D.; Dekel, N.; Shalgi, R. Regulation of GVBD in mouse oocytes by miR-125a-3p and Fyn kinase through modulation of actin filaments. Sci. Rep. 2017, 7, 2238. [Google Scholar] [CrossRef]
- Li, X.; Wang, H.; Sheng, Y.; Wang, Z. MicroRNA-224 delays oocyte maturation through targeting Ptx3 in cumulus cells. Mech. Dev. 2017, 143, 20–25. [Google Scholar] [CrossRef]
- Zhang, J.; Guan, Y.; Shen, C.; Zhang, L.; Wang, X. MicroRNA-375 regulates oocyte in vitro maturation by targeting ADAMTS1 and PGR in bovine cumulus cells. Biomed. Pharmacother. 2019, 118, 109350. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Zheng, Y.; Tang, X.; Gao, H.; Liu, N.; Gao, Y.; Hao, L.; Liu, S.; Jiang, Z. miR-21-3p inhibits autophagy of bovine granulosa cells by targeting VEGFA via PI3K/AKT signaling. Reproduction 2019. [Google Scholar] [CrossRef]
- Pan, B.; Toms, D.; Li, J. MicroRNA-574 suppresses oocyte maturation via targeting hyaluronan synthase 2 in porcine cumulus cells. Am. J. Physiol. Cell Physiol. 2018, 314, C268–C277. [Google Scholar] [CrossRef]
- Li, Z.; Li, W. [miR-383 inhibits proliferation of granulosa cells by down-regulation of cell cycle-related proteins in mice]. Xibao Yu Fenzi Mianyixue Zazhi 2019, 35, 518–525. [Google Scholar]
- Gebremedhn, S.; Salilew-Wondim, D.; Hoelker, M.; Rings, F.; Neuhoff, C.; Tholen, E.; Schellander, K.; Tesfaye, D. MicroRNA-183-96-182 Cluster Regulates Bovine Granulosa Cell Proliferation and Cell Cycle Transition by Coordinately Targeting FOXO1. Biol. Reprod. 2016, 94, 127. [Google Scholar] [CrossRef]
- Andreas, E.; Hoelker, M.; Neuhoff, C.; Tholen, E.; Schellander, K.; Tesfaye, D.; Salilew-Wondim, D. MicroRNA 17-92 cluster regulates proliferation and differentiation of bovine granulosa cells by targeting PTEN and BMPR2 genes. Cell Tissue Res. 2016, 366, 219–230. [Google Scholar] [CrossRef]
- Pande, H.O.; Tesfaye, D.; Hoelker, M.; Gebremedhn, S.; Held, E.; Neuhoff, C.; Tholen, E.; Schellander, K.; Wondim, D.S. MicroRNA-424/503 cluster members regulate bovine granulosa cell proliferation and cell cycle progression by targeting SMAD7 gene through activin signalling pathway. J. Ovarian Res. 2018, 11, 34. [Google Scholar] [CrossRef]
- Chen, H.; Liu, C.; Jiang, H.; Gao, Y.; Xu, M.; Wang, J.; Liu, S.; Fu, Y.; Sun, X.; Xu, J.; et al. Regulatory Role of miRNA-375 in Expression of BMP15/GDF9 Receptors and its Effect on Proliferation and Apoptosis of Bovine Cumulus Cells. Cell. Physiol. Biochem. 2017, 41, 439–450. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, C.-Z.; Xu, M.-Q.; Zhang, L.-Q.; Liu, J.-B.; Gao, Y.; Jiang, H.; Yuan, B.; Zhang, J.-B. MiR-31 and miR-143 affect steroid hormone synthesis and inhibit cell apoptosis in bovine granulosa cells through FSHR. Theriogenology 2019, 123, 45–53. [Google Scholar] [CrossRef]
- Yao, Y.; Niu, J.; Sizhu, S.; Li, B.; Chen, Y.; Li, R.; Yangzong, Q.; Li, Q.; Xu, Y. microRNA-125b Regulates Apoptosis by Targeting Bone Morphogenetic Protein Receptor 1B in Yak Granulosa Cells. DNA Cell Biol. 2018, 37, 878–887. [Google Scholar] [CrossRef]
- Han, X.; Xue, R.; Yuan, H.-J.; Wang, T.-Y.; Lin, J.; Zhang, J.; Liang, B.; Tan, J.-H. MicroRNA-21 plays a pivotal role in the oocyte-secreted factor-induced suppression of cumulus cell apoptosis. Biol. Reprod. 2017, 96, 1167–1180. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, B.T.; Sontakke, S.D.; Ioannidis, J.; Duncan, W.C.; Donadeu, F.X. The Adequate Corpus Luteum: MiR-96 Promotes Luteal Cell Survival and Progesterone Production. J. Clin. Endocrinol. Metab. 2017, 102, 2188–2198. [Google Scholar] [CrossRef]
- Yao, L.; Li, M.; Hu, J.; Wang, W.; Gao, M. MiRNA-335-5p negatively regulates granulosa cell proliferation via SGK3 in PCOS. Reproduction 2018, 156, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Sui, C.; Xun, Y.; Lai, Q.; Jin, L. MiRNA-99a can regulate proliferation and apoptosis of human granulosa cells via targeting IGF-1R in polycystic ovary syndrome. J. Assist. Reprod. Genet. 2019, 36, 211–221. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, Q.; Hu, Y.; Xu, L.; Jiang, Y.; Zhang, C.; Ding, L.; Jiang, R.; Sun, J.; Sun, H.; et al. miR-181a increases FoxO1 acetylation and promotes granulosa cell apoptosis via SIRT1 downregulation. Cell Death Dis. 2017, 8, e3088. [Google Scholar] [CrossRef]
- Nie, M.; Yu, S.; Peng, S.; Fang, Y.; Wang, H.; Yang, X. miR-23a and miR-27a promote human granulosa cell apoptosis by targeting SMAD5. Biol. Reprod. 2015, 93, 98. [Google Scholar] [CrossRef] [PubMed]
- Sirotkin, A.V.; Kisova, G.; Brenaut, P.; Ovcharenko, D.; Grossmann, R.; Mlyncek, M. Involvement of microRNA Mir15a in control of human ovarian granulosa cell proliferation, apoptosis, steroidogenesis, and response to FSH. Microrna 2014, 3, 29–36. [Google Scholar] [CrossRef]
- Yuan, X.; Deng, X.; Zhou, X.; Zhang, A.; Xing, Y.; Zhang, Z.; Zhang, H.; Li, J. MiR-126-3p promotes the cell proliferation and inhibits the cell apoptosis by targeting TSC1 in the porcine granulosa cells. In Vitro Cell. Dev. Biol. Anim. 2018, 54, 715–724. [Google Scholar] [CrossRef]
- Liu, J.; Yao, W.; Yao, Y.; Du, X.; Zhou, J.; Ma, B.; Liu, H.; Li, Q.; Pan, Z. MiR-92a inhibits porcine ovarian granulosa cell apoptosis by targeting Smad7 gene. FEBS Lett. 2014, 588, 4497–4503. [Google Scholar] [CrossRef]
- Liu, J.; Li, X.; Yao, Y.; Li, Q.; Pan, Z.; Li, Q. miR-1275 controls granulosa cell apoptosis and estradiol synthesis by impairing LRH-1/CYP19A1 axis. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 246–257. [Google Scholar] [CrossRef]
- Machtinger, R.; Laurent, L.C.; Baccarelli, A.A. Extracellular vesicles: Roles in gamete maturation, fertilization and embryo implantation. Hum. Reprod. Update 2016, 22, 182–193. [Google Scholar] [CrossRef] [PubMed]
- Da, S.J.C.; Veeramachaneni, D.N.; Winger, Q.A.; Carnevale, E.M.; Bouma, G.J. Cell-secreted vesicles in equine ovarian follicular fluid contain miRNAs and proteins: A possible new form of cell communication within the ovarian follicle. Biol. Reprod. 2012, 86, 71. [Google Scholar]
- Hailay, T.; Hoelker, M.; Poirier, M.; Gebremedhn, S.; Rings, F.; Saeed-Zidane, M.; Salilew-Wondim, D.; Dauben, C.; Tholen, E.; Neuhoff, C.; et al. Extracellular vesicle-coupled miRNA profiles in follicular fluid of cows with divergent post-calving metabolic status. Sci. Rep. 2019, 9, 12851. [Google Scholar] [CrossRef] [PubMed]
- Santonocito, M.; Vento, M.; Guglielmino, M.R.; Battaglia, R.; Wahlgren, J.; Ragusa, M.; Barbagallo, D.; Borzi, P.; Rizzari, S.; Maugeri, M.; et al. Molecular characterization of exosomes and their microRNA cargo in human follicular fluid: Bioinformatic analysis reveals that exosomal microRNAs control pathways involved in follicular maturation. Fertil. Steril. 2014, 102, 1751–1761.e1. [Google Scholar] [CrossRef]
- Matsuno, Y.; Kanke, T.; Maruyama, N.; Fujii, W.; Naito, K.; Sugiura, K. Characterization of mRNA profiles of the exosome-like vesicles in porcine follicular fluid. PLoS ONE 2019, 14, e0217760. [Google Scholar] [CrossRef]
- Navakanitworakul, R.; Hung, W.T.; Gunewardena, S.; Davis, J.S.; Chotigeat, W.; Christenson, L.K. Characterization and Small RNA Content of Extracellular Vesicles in Follicular Fluid of Developing Bovine Antral Follicles. Sci. Rep. 2016, 6, 25486. [Google Scholar] [CrossRef]
- Da Silveira, J.C.; Carnevale, E.M.; Winger, Q.A.; Bouma, G.J. Regulation of ACVR1 and ID2 by cell-secreted exosomes during follicle maturation in the mare. Reprod. Biol. Endocrinol. 2014, 12, 44. [Google Scholar] [CrossRef]
- Martinez, R.M.; Liang, L.; Racowsky, C.; Dioni, L.; Mansur, A.; Adir, M.; Bollati, V.; Baccarelli, A.A.; Hauser, R.; Machtinger, R. Extracellular microRNAs profile in human follicular fluid and IVF outcomes. Sci. Rep. 2018, 8, 17036. [Google Scholar] [CrossRef]
- Rodrigues, T.A.; Tuna, K.M.; Alli, A.A.; Tribulo, P.; Hansen, P.J.; Koh, J.; Paula-Lopes, F.F. Follicular fluid exosomes act on the bovine oocyte to improve oocyte competence to support development and survival to heat shock. Reprod. Fertil. Dev. 2019, 31, 888–897. [Google Scholar] [CrossRef]
- Morales Dalanezi, F.; Mogollon Garcia, H.D.; de Andrade Ferrazza, R.; Fagali Franchi, F.; Kubo Fontes, P.; de Souza Castilho, A.C.; Gouveia Nogueira, M.F.; Dos Santos Schmidt, E.M.; Sartori, R.; Pinheiro Ferreira, J.C. Extracellular vesicles of follicular fluid from heat-stressed cows modify the gene expression of in vitro-matured oocytes. Anim. Reprod. Sci. 2019, 205, 94–104. [Google Scholar] [CrossRef]
- Laiho, A.; Kotaja, N.; Gyenesei, A.; Sironen, A. Transcriptome profiling of the murine testis during the first wave of spermatogenesis. PLoS ONE 2013, 8, e61558. [Google Scholar] [CrossRef] [PubMed]
- Eddy, E.M. Regulation of gene expression during spermatogenesis. Semin. Cell Dev. Biol. 1998, 9, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Kasimanickam, V.R.; Kasimanickam, R.K. Differential expression of microRNAs in sexually immature and mature canine testes. Theriogenology 2015, 83, 394–398.e1. [Google Scholar] [CrossRef] [PubMed]
- Sree, S.; Radhakrishnan, K.; Indu, S.; Kumar, P.G. Dramatic changes in 67 miRNAs during initiation of first wave of spermatogenesis in Mus musculus testis: Global regulatory insights generated by miRNA-mRNA network analysis. Biol. Reprod. 2014, 91, 69. [Google Scholar] [CrossRef] [PubMed]
- Buchold, G.M.; Coarfa, C.; Kim, J.; Milosavljevic, A.; Gunaratne, P.H.; Matzuk, M.M. Analysis of microRNA expression in the prepubertal testis. PLoS ONE 2010, 5, e15317. [Google Scholar] [CrossRef]
- Wainwright, E.N.; Jorgensen, J.S.; Kim, Y.; Truong, V.; Bagheri-Fam, S.; Davidson, T.; Svingen, T.; Fernandez-Valverde, S.L.; McClelland, K.S.; Taft, R.J.; et al. SOX9 regulates microRNA miR-202-5p/3p expression during mouse testis differentiation. Biol. Reprod. 2013, 89, 34. [Google Scholar] [CrossRef]
- Yuan, S.; Schuster, A.; Tang, C.; Yu, T.; Ortogero, N.; Bao, J.; Zheng, H.; Yan, W. Sperm-borne miRNAs and endo-siRNAs are important for fertilization and preimplantation embryonic development. Development 2016, 143, 635–647. [Google Scholar] [CrossRef]
- Ma, J.; Fan, Y.; Zhang, J.; Feng, S.; Hu, Z.; Qiu, W.; Long, K.; Jin, L.; Tang, Q.; Wang, X.; et al. Testosterone-Dependent miR-26a-5p and let-7g-5p Act as Signaling Mediators to Regulate Sperm Apoptosis via Targeting PTEN and PMAIP1. Int. J. Mol. Sci. 2018, 19, 1233. [Google Scholar] [CrossRef]
- Kasimanickam, V.; Kastelic, J. MicroRNA in sperm from Duroc, Landrace and Yorkshire boars. Sci. Rep. 2016, 6, 32954. [Google Scholar] [CrossRef]
- Gao, H.; Wen, H.; Cao, C.; Dong, D.; Yang, C.; Xie, S.; Zhang, J.; Huang, X.; Huang, X.; Yuan, S.; et al. Overexpression of MicroRNA-10a in Germ Cells Causes Male Infertility by Targeting Rad51 in Mouse and Human. Front. Physiol. 2019, 10, 765. [Google Scholar] [CrossRef]
- Dabaja, A.A.; Mielnik, A.; Robinson, B.D.; Wosnitzer, M.S.; Schlegel, P.N.; Paduch, D.A. Possible germ cell-Sertoli cell interactions are critical for establishing appropriate expression levels for the Sertoli cell-specific MicroRNA, miR-202-5p, in human testis. Basic Clin. Androl. 2015, 25, 2. [Google Scholar] [CrossRef] [PubMed]
- Menezes, E.S.B.; Badial, P.R.; El Debaky, H.; Husna, A.U.; Ugur, M.R.; Kaya, A.; Topper, E.; Bulla, C.; Grant, K.E.; Bolden-Tiller, O.; et al. Sperm miR-15a and miR-29b are associated with bull fertility. Andrologia 2019, e13412. [Google Scholar] [CrossRef] [PubMed]
- Griswold, M.D. The central role of Sertoli cells in spermatogenesis. Semin. Cell Dev. Biol. 1998, 9, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Song, H.; Yu, L.; Guan, K.; Hu, P.; Li, Y.; Xia, X.; Li, J.; Jiang, S.; Li, F. miR-762 promotes porcine immature Sertoli cell growth via the ring finger protein 4 (RNF4) gene. Sci. Rep. 2016, 6, 32783. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Guan, K.; Feng, Y.; Ma, C.; Song, H.; Li, Y.; Xia, X.; Li, J.; Li, F. miR-638 Inhibits immature Sertoli cell growth by indirectly inactivating PI3K/AKT pathway via SPAG1 gene. Cell Cycle 2017, 16, 2290–2300. [Google Scholar] [CrossRef] [PubMed]
- Duranthon, V.; Watson, A.J.; Lonergan, P. Preimplantation embryo programming: Transcription, epigenetics, and culture environment. Reproduction 2008, 135, 141–150. [Google Scholar] [CrossRef]
- Wang, S.; Cowan, C.A.; Chipperfield, H.; Powers, R.D. Gene expression in the preimplantation embryo: In-vitro developmental changes. Reprod. Biomed. Online 2005, 10, 607–616. [Google Scholar] [CrossRef]
- Kues, W.A.; Sudheer, S.; Herrmann, D.; Carnwath, J.W.; Havlicek, V.; Besenfelder, U.; Lehrach, H.; Adjaye, J.; Niemann, H. Genome-wide expression profiling reveals distinct clusters of transcriptional regulation during bovine preimplantation development in vivo. Proc. Natl. Acad. Sci. USA 2008, 105, 19768–19773. [Google Scholar] [CrossRef]
- Ozawa, M.; Sakatani, M.; Yao, J.; Shanker, S.; Yu, F.; Yamashita, R.; Wakabayashi, S.; Nakai, K.; Dobbs, K.B.; Sudano, M.J.; et al. Global gene expression of the inner cell mass and trophectoderm of the bovine blastocyst. BMC Dev. Biol. 2012, 12, 33. [Google Scholar] [CrossRef]
- Hamatani, T.; Daikoku, T.; Wang, H.; Matsumoto, H.; Carter, M.G.; Ko, M.S.H.; Dey, S.K. Global gene expression analysis identifies molecular pathways distinguishing blastocyst dormancy and activation. Proc. Natl. Acad. Sci. USA 2004, 101, 10326–10331. [Google Scholar] [CrossRef]
- El-Sayed, A.; Hoelker, M.; Rings, F.; Salilew, D.; Jennen, D.; Tholen, E.; Sirard, M.A.; Schellander, K.; Tesfaye, D. Large-scale transcriptional analysis of bovine embryo biopsies in relation to pregnancy success after transfer to recipients. Physiol. Genom. 2006, 28, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Parks, J.C.; McCallie, B.R.; Janesch, A.M.; Schoolcraft, W.B.; Katz-Jaffe, M.G. Blastocyst gene expression correlates with implantation potential. Fertil. Steril. 2011, 95, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, E.; Kim, S.Y.; Carmell, M.A.; Murchison, E.P.; Alcorn, H.; Li, M.Z.; Mills, A.A.; Elledge, S.J.; Anderson, K.V.; Hannon, G.J. Dicer is essential for mouse development. Nat. Genet. 2003, 35, 215–217. [Google Scholar] [CrossRef]
- Alisch, R.S.; Jin, P.; Epstein, M.; Caspary, T.; Warren, S.T. Argonaute2 is essential for mammalian gastrulation and proper mesoderm formation. PLoS Genet. 2007, 3, e227. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Kaneda, M.; O’Carroll, D.; Hajkova, P.; Barton, S.C.; Sun, Y.A.; Lee, C.; Tarakhovsky, A.; Lao, K.; Surani, M.A. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007, 21, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Cheong, A.W.Y.; Pang, R.T.K.; Liu, W.-M.; Kottawatta, K.S.A.; Lee, K.-F.; Yeung, W.S.B. MicroRNA Let-7a and dicer are important in the activation and implantation of delayed implanting mouse embryos. Hum. Reprod. 2014, 29, 750–762. [Google Scholar] [CrossRef] [PubMed]
- Mondou, E.; Dufort, I.; Gohin, M.; Fournier, E.; Sirard, M.-A. Analysis of microRNAs and their precursors in bovine early embryonic development. Mol. Hum. Reprod. 2012, 18, 425–434. [Google Scholar] [CrossRef]
- Goossens, K.; Mestdagh, P.; Lefever, S.; van Poucke, M.; van Zeveren, A.; van Soom, A.; Vandesompele, J.; Peelman, L. Regulatory microRNA network identification in bovine blastocyst development. Stem Cells Dev. 2013, 22, 1907–1920. [Google Scholar] [CrossRef]
- Viswanathan, S.R.; Mermel, C.H.; Lu, J.; Lu, C.W.; Golub, T.R.; Daley, G.Q. microRNA expression during trophectoderm specification. PLoS ONE 2009, 4, e6143. [Google Scholar]
- Houbaviy, H.B.; Murray, M.F.; Sharp, P.A. Embryonic stem cell-specific MicroRNAs. Dev. Cell 2003, 5, 351–358. [Google Scholar] [CrossRef]
- Kropp, J.; Khatib, H. Characterization of microRNA in bovine in vitro culture media associated with embryo quality and development. J. Dairy Sci. 2015, 98, 6552–6563. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Papagiannakopoulos, T.; Pan, G.; Thomson, J.A.; Kosik, K.S. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 2009, 137, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Qian, K.; Hu, L.; Chen, H.; Li, H.; Liu, N.; Li, Y.; Ai, J.; Zhu, G.; Tang, Z.; Zhang, H. Hsa-miR-222 is involved in differentiation of endometrial stromal cells in vitro. Endocrinology 2009, 150, 4734–4743. [Google Scholar] [CrossRef] [PubMed]
- Szekeres-Bartho, J.; Sucurovic, S.; Mulac-Jericevic, B. The Role of Extracellular Vesicles and PIBF in Embryo-Maternal Immune-Interactions. Front. Immunol. 2018, 9, 2890. [Google Scholar] [CrossRef] [PubMed]
- Salilew-Wondim, D.; Schellander, K.; Hoelker, M.; Tesfaye, D. Oviductal, endometrial and embryonic gene expression patterns as molecular clues for pregnancy establishment. Anim. Reprod. Sci. 2012, 134, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Spruce, T.; Pernaute, B.; Di-Gregorio, A.; Cobb, B.S.; Merkenschlager, M.; Manzanares, M.; Rodriguez, T.A. An early developmental role for miRNAs in the maintenance of extraembryonic stem cells in the mouse embryo. Dev. Cell 2010, 19, 207–219. [Google Scholar] [CrossRef]
- Morales-Prieto, D.M.; Chaiwangyen, W.; Ospina-Prieto, S.; Schneider, U.; Herrmann, J.; Gruhn, B.; Markert, U.R. MicroRNA expression profiles of trophoblastic cells. Placenta 2012, 33, 725–734. [Google Scholar] [CrossRef]
- Foshay, K.M.; Gallicano, G.I. miR-17 family miRNAs are expressed during early mammalian development and regulate stem cell differentiation. Dev. Biol. 2009, 326, 431–443. [Google Scholar] [CrossRef]
- Kim, J.; Lee, J.; Jun, J.H. Identification of differentially expressed microRNAs in outgrowth embryos compared with blastocysts and non-outgrowth embryos in mice. Reprod. Fertil. Dev. 2019, 31, 645–657. [Google Scholar] [CrossRef]
- Berg, D.K.; Pfeffer, P.L. MicroRNA expression in bovine preimplantation embryos. Reprod. Fertil. Dev. 2018, 30, 546–554. [Google Scholar] [CrossRef]
- Liu, W.-M.; Pang, R.T.K.; Cheong, A.W.Y.; Ng, E.H.Y.; Lao, K.; Lee, K.-F.; Yeung, W.S.B. Involvement of microRNA lethal-7a in the regulation of embryo implantation in mice. PLoS ONE 2012, 7, e37039. [Google Scholar] [CrossRef] [PubMed]
- Bazer, F.W.; Wu, G.; Spencer, T.E.; Johnson, G.A.; Burghardt, R.C.; Bayless, K. Novel pathways for implantation and establishment and maintenance of pregnancy in mammals. Mol. Hum. Reprod. 2010, 16, 135–152. [Google Scholar] [CrossRef]
- Spencer, T.E.; Johnson, G.A.; Bazer, F.W.; Burghardt, R.C.; Palmarini, M. Pregnancy recognition and conceptus implantation in domestic ruminants: Roles of progesterone, interferons and endogenous retroviruses. Reprod. Fertil. Dev. 2007, 19, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Salilew-Wondim, D.; Holker, M.; Rings, F.; Ghanem, N.; Ulas-Cinar, M.; Peippo, J.; Tholen, E.; Looft, C.; Schellander, K.; Tesfaye, D. Bovine pretransfer endometrium and embryo transcriptome fingerprints as predictors of pregnancy success after embryo transfer. Physiol. Genom. 2010, 42, 201–218. [Google Scholar] [CrossRef] [PubMed]
- Bazer, F.W. Pregnancy recognition signaling mechanisms in ruminants and pigs. J. Anim. Sci. Biotechnol. 2013, 4, 23. [Google Scholar] [CrossRef]
- Spencer, T.E.; Hansen, T.R. Implantation and Establishment of Pregnancy in Ruminants. Adv. Anat. Embryol. Cell Biol. 2015, 216, 105–135. [Google Scholar] [CrossRef]
- Yelian, F.D.; Edgeworth, N.A.; Dong, L.J.; Chung, A.E.; Armant, D.R. Recombinant entactin promotes mouse primary trophoblast cell adhesion and migration through the Arg-Gly-Asp (RGD) recognition sequence. J. Cell Biol. 1993, 121, 923–929. [Google Scholar] [CrossRef][Green Version]
- Armant, D.R.; Kaplan, H.A.; Mover, H.; Lennarz, W.J. The effect of hexapeptides on attachment and outgrowth of mouse blastocysts cultured in vitro: Evidence for the involvement of the cell recognition tripeptide Arg-Gly-Asp. Proc. Natl. Acad. Sci. USA 1986, 83, 6751–6755. [Google Scholar] [CrossRef]
- Armant, D.R.; Kaplan, H.A.; Lennarz, W.J. Fibronectin and laminin promote in vitro attachment and outgrowth of mouse blastocysts. Dev. Biol. 1986, 116, 519–523. [Google Scholar] [CrossRef]
- Vilella, F.; Moreno-Moya, J.M.; Balaguer, N.; Grasso, A.; Herrero, M.; Martinez, S.; Marcilla, A.; Simon, C. Hsa-miR-30d, secreted by the human endometrium, is taken up by the pre-implantation embryo and might modify its transcriptome. Development 2015, 142, 3210–3221. [Google Scholar] [CrossRef]
- Revel, A.; Achache, H.; Stevens, J.; Smith, Y.; Reich, R. MicroRNAs are associated with human embryo implantation defects. Hum. Reprod. 2011, 26, 2830–2840. [Google Scholar] [CrossRef] [PubMed]
- Sha, A.-G.; Liu, J.-L.; Jiang, X.-M.; Ren, J.-Z.; Ma, C.-H.; Lei, W.; Su, R.-W.; Yang, Z.-M. Genome-wide identification of micro-ribonucleic acids associated with human endometrial receptivity in natural and stimulated cycles by deep sequencing. Fertil. Steril. 2011, 96, 150–155.e5. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.-Q.; Zhao, M.; Huang, M.-Y.; Liu, J.-L. Comparative Transcriptomic Analysis of Embryo Implantation in Mice and Rats. Cell. Physiol. Biochem. 2018, 50, 668–678. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.-J.; Ren, G.; Liu, J.-L.; Zhao, Z.-A.; Yu, Y.-S.; Su, R.-W.; Ma, X.-H.; Ni, H.; Lei, W.; Yang, Z.-M. MicroRNA expression and regulation in mouse uterus during embryo implantation. J. Biol. Chem. 2008, 283, 23473–23484. [Google Scholar] [CrossRef]
- Geng, Y.; He, J.; Ding, Y.; Chen, X.; Zhou, Y.; Liu, S.; Liu, X.; Wang, Y. The differential expression of microRNAs between implantation sites and interimplantation sites in early pregnancy in mice and their potential functions. Reprod. Sci. 2014, 21, 1296–1306. [Google Scholar] [CrossRef]
- Ponsuksili, S.; Tesfaye, D.; Schellander, K.; Hoelker, M.; Hadlich, F.; Schwerin, M.; Wimmers, K. Differential expression of miRNAs and their target mRNAs in endometria prior to maternal recognition of pregnancy associates with endometrial receptivity for in vivo- and in vitro-produced bovine embryos. Biol. Reprod. 2014, 91, 135. [Google Scholar] [CrossRef]
- Hong, L.; Liu, R.; Qiao, X.; Wang, X.; Wang, S.; Li, J.; Wu, Z.; Zhang, H. Differential microRNA Expression in Porcine Endometrium Involved in Remodeling and Angiogenesis That Contributes to Embryonic Implantation. Front. Genet. 2019, 10, 661. [Google Scholar] [CrossRef]
- Tian, S.; Su, X.; Qi, L.; Jin, X.-H.; Hu, Y.; Wang, C.-L.; Ma, X.; Xia, H.-F. MiR-143 and rat embryo implantation. Biochim. Biophys. Acta 2015, 1850, 708–721. [Google Scholar] [CrossRef]
- Balaguer, N.; Moreno, I.; Herrero, M.; Gonzalez-Monfort, M.; Vilella, F.; Simon, C. MicroRNA-30d deficiency during preconception affects endometrial receptivity by decreasing implantation rates and impairing fetal growth. Am. J. Obstet. Gynecol. 2019, 221, 46.e1–46.e16. [Google Scholar] [CrossRef]
- Shen, L.-J.; He, J.-L.; Yang, D.-H.; Ding, Y.-B.; Chen, X.-M.; Geng, Y.-Q.; Liu, S.-J.; Liu, X.-Q.; Wang, Y.-X. Mmu-microRNA-200a overexpression leads to implantation defect by targeting phosphatase and tensin homolog in mouse uterus. Reprod. Sci. 2013, 20, 1518–1528. [Google Scholar] [CrossRef]
- Li, Z.; Gou, J.; Jia, J.; Zhao, X. MicroRNA-429 functions as a regulator of epithelial-mesenchymal transition by targeting Pcdh8 during murine embryo implantation. Hum. Reprod. 2015, 30, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.-J.; Lees, M.; Matthews, L.C.; Kimber, S.J.; Forbes, K.; Aplin, J.D. MiR-145 suppresses embryo-epithelial juxtacrine communication at implantation by modulating maternal IGF1R. J. Cell Sci. 2015, 128, 804–814. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; He, J.; Chen, X.; Ding, Y.; Wang, Y.; Long, C.; Shen, L.; Liu, X. Mmu-miR-193 is involved in embryo implantation in mouse uterus by regulating GRB7 gene expression. Reprod. Sci. 2014, 21, 733–742. [Google Scholar] [CrossRef] [PubMed]
- Chu, B.; Zhong, L.; Dou, S.; Wang, J.; Li, J.; Wang, M.; Shi, Q.; Mei, Y.; Wu, M. miRNA-181 regulates embryo implantation in mice through targeting leukemia inhibitory factor. J. Mol. Cell Biol. 2015, 7, 12–22. [Google Scholar] [CrossRef]
- Liu, X.; Gao, R.; Chen, X.; Zhang, H.; Zheng, A.; Yang, D.; Ding, Y.; Wang, Y.; He, J. Possible roles of mmu-miR-141 in the endometrium of mice in early pregnancy following embryo implantation. PLoS ONE 2013, 8, e67382. [Google Scholar] [CrossRef]
- Li, Z.; Jia, J.; Gou, J.; Zhao, X.; Yi, T. MicroRNA-451 plays a role in murine embryo implantation through targeting Ankrd46, as implicated by a microarray-based analysis. Fertil. Steril. 2015, 103, 834–844.e4. [Google Scholar] [CrossRef]
- Liang, S.; Nie, Z.-W.; Guo, J.; Niu, Y.-J.; Shin, K.-T.; Ock, S.A.; Cui, X.-S. Overexpression of MicroRNA-29b Decreases Expression of DNA Methyltransferases and Improves Quality of the Blastocysts Derived from Somatic Cell Nuclear Transfer in Cattle. Microsc. Microanal. 2018, 24, 29–37. [Google Scholar] [CrossRef]
- Sun, M.; Chen, H.; Liu, J.; Tong, C.; Meng, T. MicroRNA-34a inhibits human trophoblast cell invasion by targeting MYC. BMC Cell Biol. 2015, 16, 21. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Yang, L.; Wang, H. miR-520 promotes DNA-damage-induced trophoblast cell apoptosis by targeting PARP1 in recurrent spontaneous abortion (RSA). Gynecol. Endocrinol. 2017, 33, 274–278. [Google Scholar] [CrossRef]
- Li, Z.; Jia, J.; Gou, J.; Tong, A.; Liu, X.; Zhao, X.; Yi, T. Mmu-miR-126a-3p plays a role in murine embryo implantation by regulating Itga11. Reprod. Biomed. Online 2015, 31, 384–393. [Google Scholar] [CrossRef]
- Ma, H.-L.; Gong, F.; Tang, Y.; Li, X.; Li, X.; Yang, X.; Lu, G. Inhibition of Endometrial Tiam1/Rac1 Signals Induced by miR-22 Up-Regulation Leads to the Failure of Embryo Implantation During the Implantation Window in Pregnant Mice. Biol. Reprod. 2015, 92, 152. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.-F.; Cao, J.-L.; Jin, X.-H.; Ma, X. MiR199a is implicated in embryo implantation by regulating Grb10 in rat. Reproduction 2014, 147, 91–99. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nair, S.; Salomon, C. Extracellular vesicles and their immunomodulatory functions in pregnancy. Semin. Immunopathol. 2018, 40, 425–437. [Google Scholar] [CrossRef] [PubMed]
- Kurian, N.K.; Modi, D. Extracellular vesicle mediated embryo-endometrial cross talk during implantation and in pregnancy. J. Assist. Reprod. Genet. 2019, 36, 189–198. [Google Scholar] [CrossRef]
- Alminana, C.; Bauersachs, S. Extracellular Vesicles in the Oviduct: Progress, Challenges and Implications for the Reproductive Success. Bioengineering 2019, 6, 32. [Google Scholar] [CrossRef]
- Alminana, C.; Tsikis, G.; Labas, V.; Uzbekov, R.; da Silveira, J.C.; Bauersachs, S.; Mermillod, P. Deciphering the oviductal extracellular vesicles content across the estrous cycle: Implications for the gametes-oviduct interactions and the environment of the potential embryo. BMC Genom. 2018, 19, 622. [Google Scholar] [CrossRef]
- Al-Dossary, A.A.; Strehler, E.E.; Martin-Deleon, P.A. Expression and secretion of plasma membrane Ca2+-ATPase 4a (PMCA4a) during murine estrus: Association with oviductal exosomes and uptake in sperm. PLoS ONE 2013, 8, e80181. [Google Scholar] [CrossRef]
- Bathala, P.; Fereshteh, Z.; Li, K.; Al-Dossary, A.A.; Galileo, D.S.; Martin-Deleon, P.A. Oviductal extracellular vesicles (oviductosomes, OVS) are conserved in humans: Murine OVS play a pivotal role in sperm capacitation and fertility. Mol. Hum. Reprod. 2018, 24, 143–157. [Google Scholar] [CrossRef]
- Lopera-Vasquez, R.; Hamdi, M.; Fernandez-Fuertes, B.; Maillo, V.; Beltran-Brena, P.; Calle, A.; Redruello, A.; Lopez-Martin, S.; Gutierrez-Adan, A.; Yanez-Mo, M.; et al. Extracellular Vesicles from BOEC in in Vitro Embryo Development and Quality. PLoS ONE 2016, 11, e0148083. [Google Scholar] [CrossRef]
- Ng, Y.H.; Rome, S.; Jalabert, A.; Forterre, A.; Singh, H.; Hincks, C.L.; Salamonsen, L.A. Endometrial exosomes/microvesicles in the uterine microenvironment: A new paradigm for embryo-endometrial cross talk at implantation. PLoS ONE 2013, 8, e58502. [Google Scholar] [CrossRef]
- Stenqvist, A.-C.; Nagaeva, O.; Baranov, V.; Mincheva-Nilsson, L. Exosomes secreted by human placenta carry functional Fas ligand and TRAIL molecules and convey apoptosis in activated immune cells, suggesting exosome-mediated immune privilege of the fetus. J. Immunol. 2013, 191, 5515–5523. [Google Scholar] [CrossRef]
| MiRNA | Function | Author/s |
|---|---|---|
| miR-202 | Oogenesis and fecundity in fish | [46] |
| let-7 miR-278 | Ovarian arrest and blocked oocyte maturation in adult insects | [47] |
| miR-318 | Oogenesis in drosophila | [45] |
| miR-224 | Porcine oocyte maturation | [50] |
| miR-378 | Porcine oocyte maturation and mouse in vivo follicular development and oocyte in vitro maturation | [40,48] |
| miR-274 | Oocyte maturation in pig | [53] |
| miR-375 | Bovine oocyte maturation | [51] |
| miR-125a-3p | Mouse oocyte germinal vesicle breakdown | |
| miR-21-3p | Inhibits bovine granulosa cell autophagy | [52] |
| miR-383 | Cell proliferation in bovine granulosa in mice | [54] |
| miR-183-96-182 cluster | Promote bovine granulosa cells proliferation and cell cycle transition | [55] |
| mir-130b | Bovine granulosa and cumulus cell proliferation and oocyte maturation | [44] |
| mir-17-92 cluster | Bovine granulosa cell proliferation and differentiation | [56] |
| miR-424/503 | Bovine granulosa cell proliferation and cell cycle progression | [57] |
| miR-375 | Proliferation and apoptosis in bovine cumulus cells | [58] |
| miR-31 miR-143 | Regulate steroid hormone synthesis and inhibit bovine granulosa cells apoptosis by targeting follicle stimulating hormone receptor (FSHR) gene | [59] |
| miR-125b | Regulates granulosa cells apoptosis in the yak ovary | [60] |
| miR-21 | Prevented apoptosis via the PI3K/Akt signaling in bovine cumulus cells | [61] |
| miR-96 | Regulates and progesterone production and luteal development in human luteinized granulosa cells | [62] |
| miR-335-5p | Regulates human granulosa cells proliferation by targeting serum/glucocorticoid regulated kinase family member 3 (SGK3) expression | [63] |
| MiR-99a | Regulation of granulosa cell proliferation and apoptosis in woman affected by polycystic ovary syndrome PCOS | [64] |
| miR-143 | Promotes follicle-stimulating hormone (FSH) -induced estradiol production and granulosa cell proliferation in humans | [36] |
| miR-181a | Regulate oxidative stress-induced Forkhead box protein O1 (FOXO1) acetylation and granulosa cell apoptosis in mice | [65] |
| miR-23a. miR-27a | Apoptosis in human granulosa cells | [66] |
| miR-15a | Regulates cell proliferation and steroid hormone production in human ovarian granulosa cells | [67] |
| miR-126-3p | Promoting cell proliferation and inhibiting cell apoptosis of porcine granulosa cells | [68] |
| miR-92a | Regulates porcine ovarian granulosa cell apoptosis | [69] |
| miR-1275 | Implicated in porcine granulosa cell apoptosis and estradiol production | [70] |
| miR-378 | Estradiol production in porcine granulosa cells | [35] |
| miRNAs | Potential Function | Author/s |
|---|---|---|
| miR-143 | Human endometrial stromal cells cell proliferation, migration and invasion | [138] |
| miR-193 | Regulation of implantation rate in mouse uterus | [143] |
| miR-181 | Inhibit mouse embryo implantation | [144] |
| miR-141 | Mouse endometrial cell proliferation and apoptosis | [145] |
| miR-451 | Regulate embryo implantations sites | [146] |
| miR-429 | Mouse embryo implantation by suppressing epithelial-mesenchymal transition | [141] |
| miR-29b | Bovine somatic cell nuclear transfer (SCNT) embryo development | [147] |
| Let-7a | Implantation of the activated mouse blastocysts, regulation of mouse blastocyst attachment and outgrowth | [106,121] |
| miR-34a | Suppress human trophoblast cell invasion | [148] |
| MiR-520 | Regulating human trophoblast cell apoptosis | [149] |
| miR-126a-3p | Determining the number of implantation sites in murine endometrium | [150] |
| miR-22 | Regulates embryo implantation in mice by targeting Tiam1/Rac1 pathways | [151] |
| miR-199 | Promote rat endometrial stromal cells cell proliferation and apoptosis | [152] |
| miR-200a | Regulation of implantation rate in mouse uterus | [140] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salilew-Wondim, D.; Gebremedhn, S.; Hoelker, M.; Tholen, E.; Hailay, T.; Tesfaye, D. The Role of MicroRNAs in Mammalian Fertility: From Gametogenesis to Embryo Implantation. Int. J. Mol. Sci. 2020, 21, 585. https://doi.org/10.3390/ijms21020585
Salilew-Wondim D, Gebremedhn S, Hoelker M, Tholen E, Hailay T, Tesfaye D. The Role of MicroRNAs in Mammalian Fertility: From Gametogenesis to Embryo Implantation. International Journal of Molecular Sciences. 2020; 21(2):585. https://doi.org/10.3390/ijms21020585
Chicago/Turabian StyleSalilew-Wondim, Dessie, Samuel Gebremedhn, Michael Hoelker, Ernst Tholen, Tsige Hailay, and Dawit Tesfaye. 2020. "The Role of MicroRNAs in Mammalian Fertility: From Gametogenesis to Embryo Implantation" International Journal of Molecular Sciences 21, no. 2: 585. https://doi.org/10.3390/ijms21020585
APA StyleSalilew-Wondim, D., Gebremedhn, S., Hoelker, M., Tholen, E., Hailay, T., & Tesfaye, D. (2020). The Role of MicroRNAs in Mammalian Fertility: From Gametogenesis to Embryo Implantation. International Journal of Molecular Sciences, 21(2), 585. https://doi.org/10.3390/ijms21020585






