Genomic Characterization and Expressional Profiles of Autophagy-Related Genes (ATGs) in Oilseed Crop Castor Bean (Ricinus communis L.)
Abstract
1. Introduction
2. Results
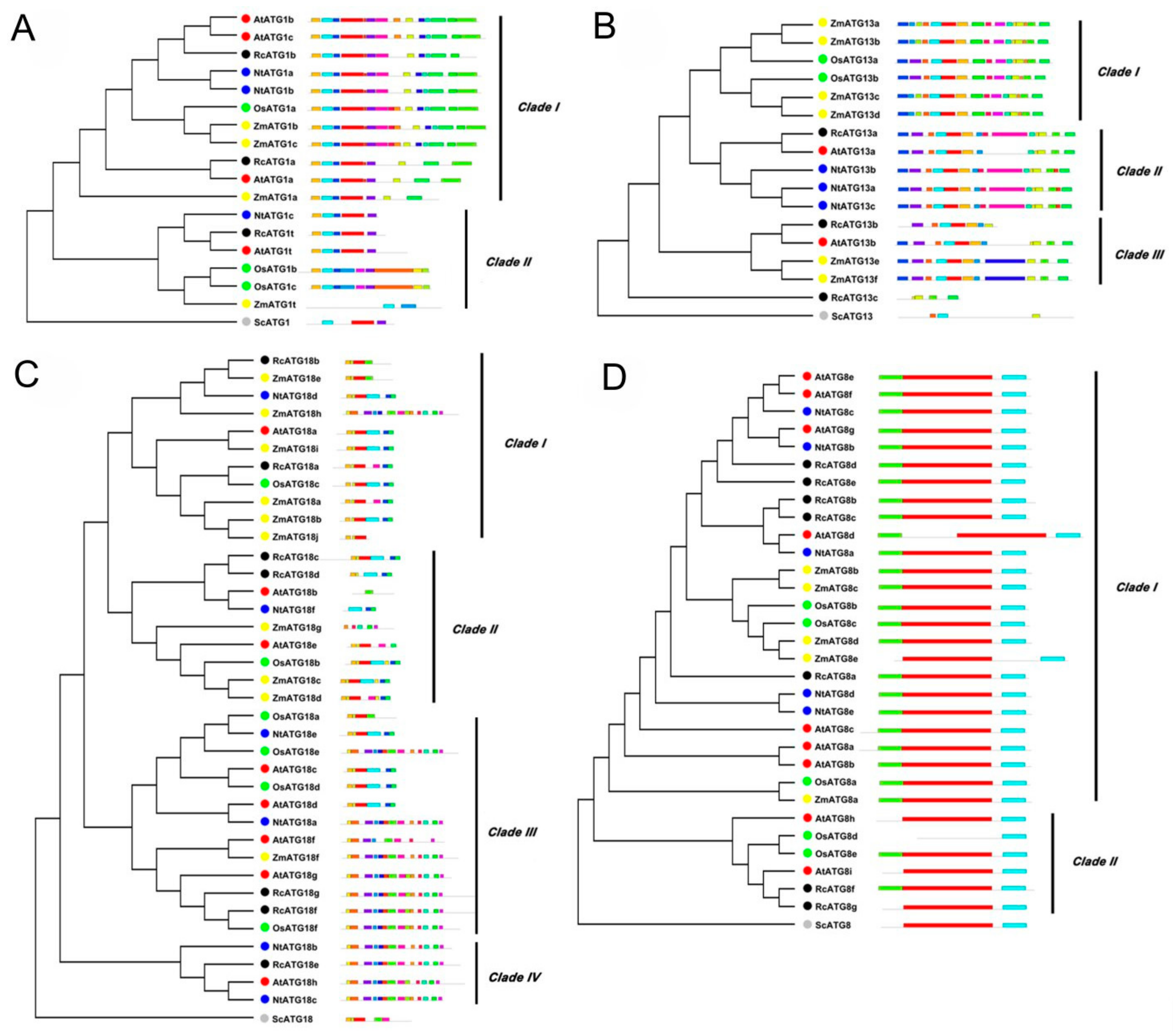
2.1. Identification of Genes Encoding ATG Proteins in Castor Bean
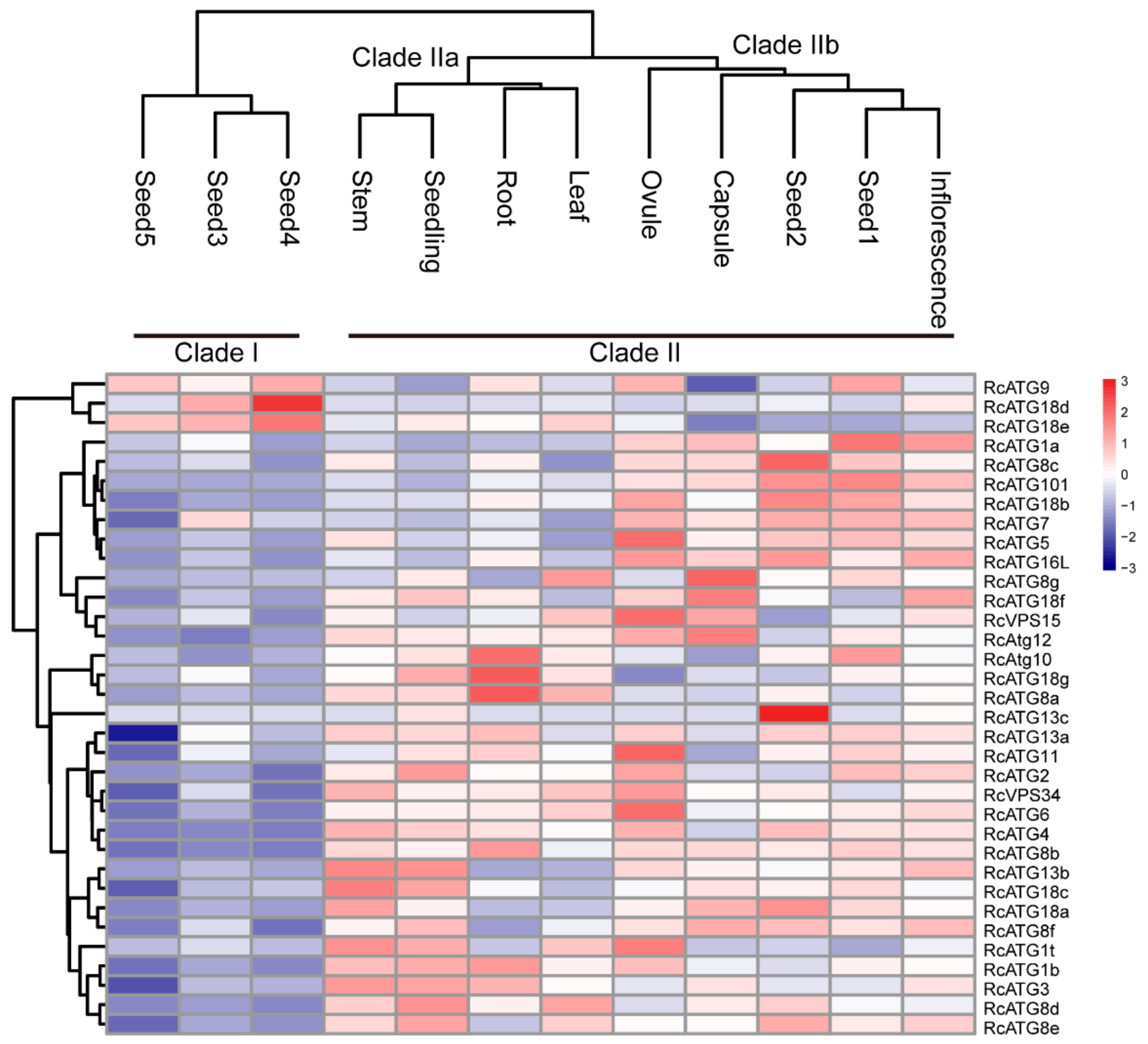
2.2. Expression Profiles of RcATGs in Various Tissues
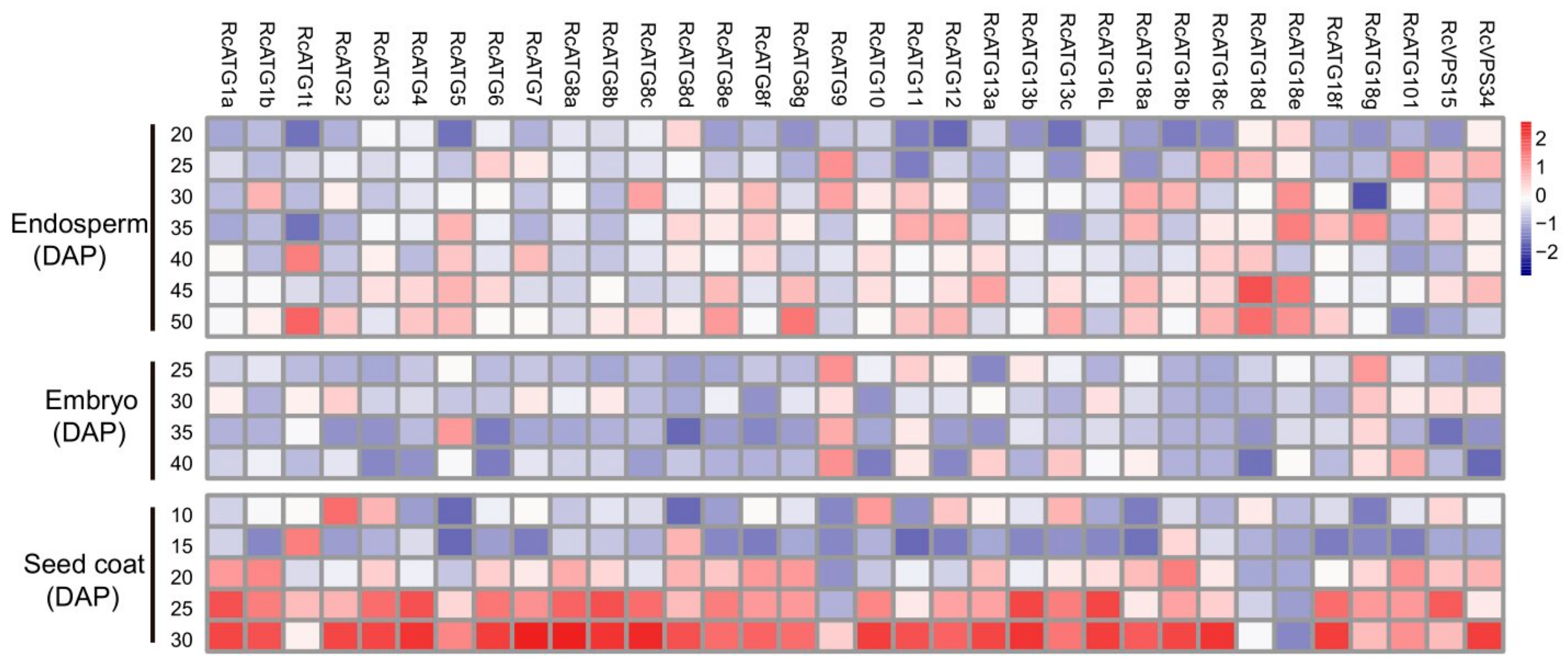
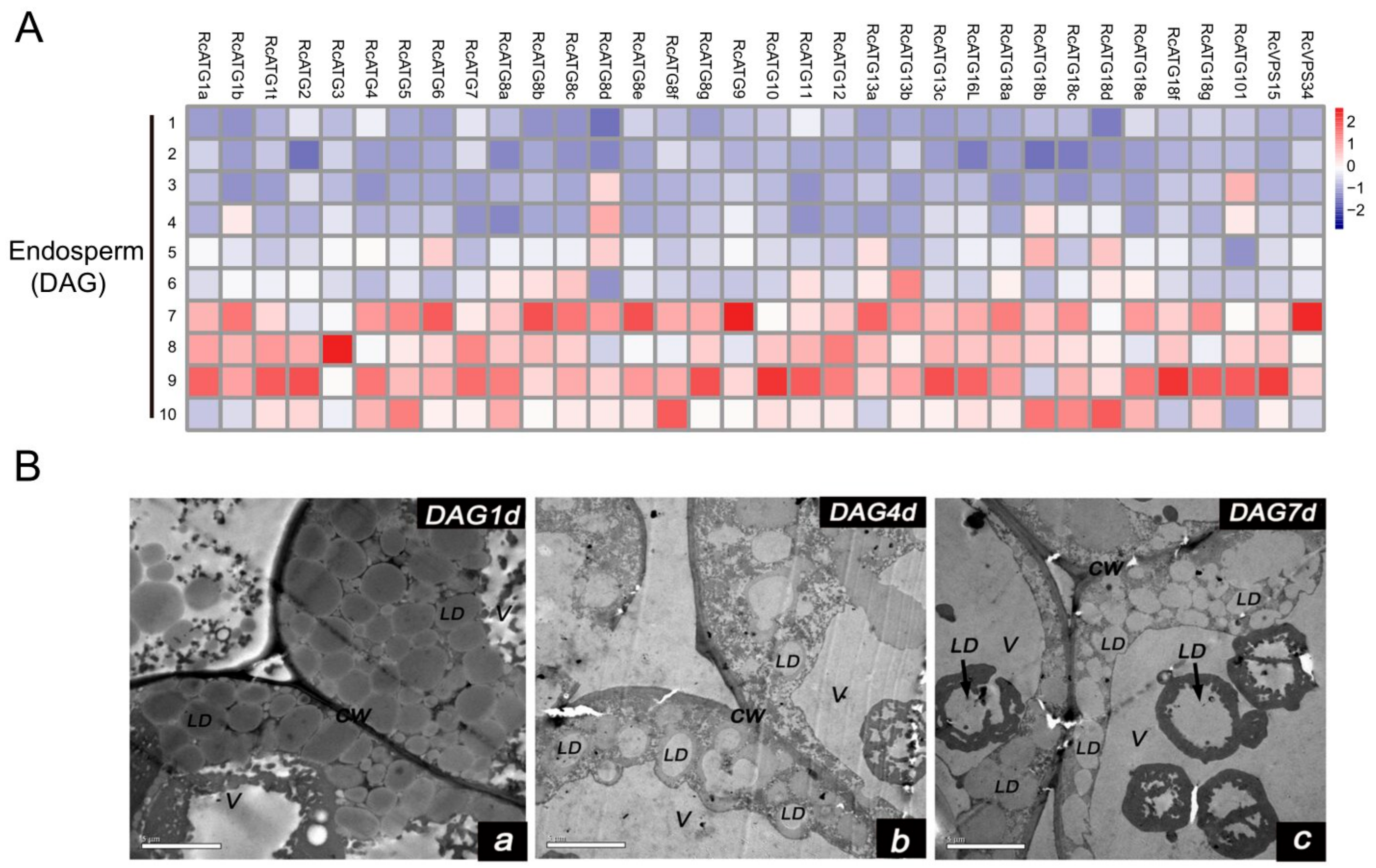
2.3. Expression Profiles of ATGs in Developing Seeds
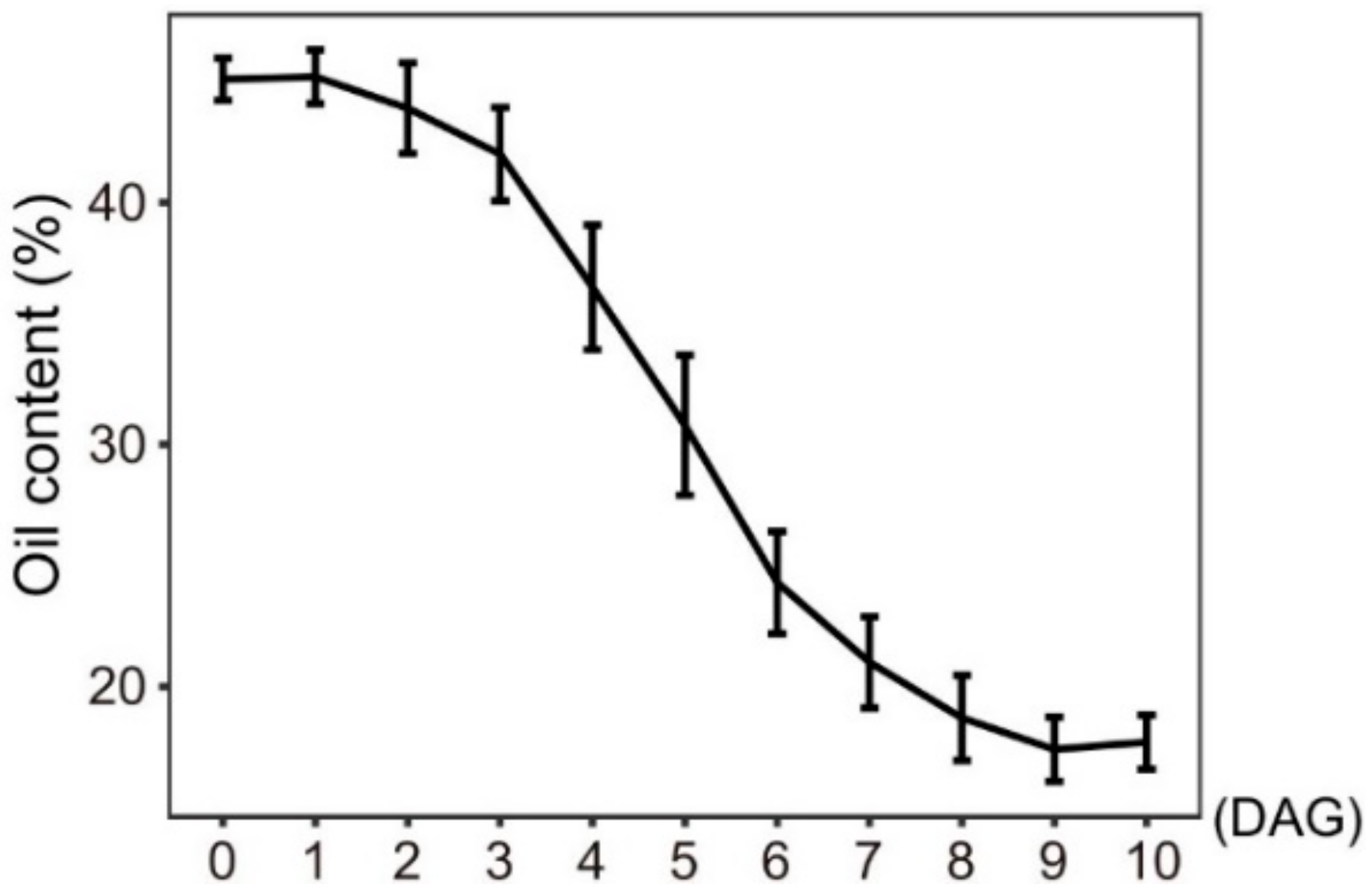
2.4. Expression and Involvement of Autophagy in Mediating Storage Lipid Decomposition during Seed Germination
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Identification of ATGs in Castor Bean
4.3. Bioinformatic Analysis and Phylogenetic Construction
4.4. Analysis of RNA-seq Data
4.5. RNA Extraction and qPCR
4.6. Determination of Lipid Content
4.7. Transmission Electron Microscopy
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Reggiori, F.; Klionsky, D.J. Autophagy in the eukaryotic cell. Eukaryot. Cell 2002, 1, 11–21. [Google Scholar] [CrossRef]
- Yang, Z.; Klionsky, D.J. An overview of the molecular mechanism of autophagy. Curr. Top. Microbiol. Immunol. 2009, 335, 1–32. [Google Scholar]
- Thompson, A.R.; Vierstra, R.D. Autophagic recycling: Lessons from yeast help define the process in plants. Curr. Opin. Plant Biol. 2005, 8, 165–173. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Meijer, A.J.; Codogno, P. Autophagy and p70S6 kinase. Autophagy 2005, 1, 59–60, discussion 60-1. [Google Scholar] [CrossRef]
- Wen, X.; Klionsky, D.J. An overview of macroautophagy in yeast. J. Mol. Biol. 2016, 428, 1681–1699. [Google Scholar] [CrossRef]
- Li, W.W.; Li, J.; Bao, J.K. Microautophagy: Lesser-known self-eating. Cell. Mol. Life Sci. CMLS 2012, 69, 1125–1136. [Google Scholar] [CrossRef]
- Wang, P.; Mugume, Y.; Bassham, D.C. New advances in autophagy in plants: Regulation, selectivity and function. Semin. Cell Dev. Biol. 2018, 80, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Slavikova, S.; Shy, G.; Yao, Y.; Glozman, R.; Levanony, H.; Pietrokovski, S.; Elazar, Z.; Galili, G. The autophagy-associated Atg8 gene family operates both under favourable growth conditions and under starvation stresses in Arabidopsis plants. J. Exp. Bot. 2005, 56, 2839–2849. [Google Scholar] [CrossRef]
- Xiong, Y.; Contento, A.L.; Bassham, D.C. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 2005, 42, 535–546. [Google Scholar] [CrossRef]
- Sheng, X.Y.; Wei, Q.; Jiang, L.P.; Li, X.; Gao, Y.; Wang, L. Different Degree in Proteasome Malfunction Has Various Effects on Root Growth Possibly through Preventing Cell Division and Promoting Autophagic Vacuolization. PLoS ONE 2012, 7, e45673. [Google Scholar] [CrossRef]
- Sankaranarayanan, S.; Samuel, M.A. A proposed role for selective autophagy in regulating auxin-dependent lateral root development under phosphate starvation in Arabidopsis. Plant Signal. Behav. 2015, 10, e989749. [Google Scholar] [CrossRef][Green Version]
- Ohsumi, Y. Historical landmarks of autophagy research. Cell Res. 2014, 24, 9–23. [Google Scholar] [CrossRef]
- Qin, G.; Ma, Z.; Zhang, L.; Xing, S.; Hou, X.; Deng, J.; Liu, J.; Chen, Z.; Qu, L.J.; Gu, H. Arabidopsis AtBECLIN 1/AtAtg6/AtVps30 is essential for pollen germination and plant development. Cell Res. 2007, 17, 249–263. [Google Scholar] [CrossRef]
- Xu, N.; Gao, X.Q.; Zhao, X.Y.; Zhu, D.Z.; Zhou, L.Z.; Zhang, X.S. Arabidopsis AtVPS15 is essential for pollen development and germination through modulating phosphatidylinositol 3-phosphate formation. Plant Mol. Biol. 2011, 77, 251–260. [Google Scholar] [CrossRef]
- Rose, T.L.; Bonneau, L.; Der, C.; Marty-Mazars, D.; Marty, F. Starvation-induced expression of autophagy-related genes in Arabidopsis. Biol. Cell 2006, 98, 53–67. [Google Scholar] [CrossRef]
- Guiboileau, A.; Yoshimoto, K.; Soulay, F.; Bataille, M.P.; Avice, J.C.; Masclaux-Daubresse, C. Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol. 2012, 194, 732–740. [Google Scholar] [CrossRef]
- Wada, S.; Hayashida, Y.; Izumi, M.; Kurusu, T.; Hanamata, S.; Kanno, K.; Kojima, S.; Yamaya, T.; Kuchitsu, K.; Makino, A.; et al. Autophagy Supports Biomass Production and Nitrogen Use Efficiency at the Vegetative Stage in Rice. Plant Physiol. 2015, 168, 60–73. [Google Scholar] [CrossRef]
- Zhou, X.M.; Zhao, P.; Wang, W.; Zou, J.; Cheng, T.H.; Peng, X.B.; Sun, M.X. A comprehensive, genome-wide analysis of autophagy-related genes identified in tobacco suggests a central role of autophagy in plant response to various environmental cues. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2015, 22, 245–257. [Google Scholar] [CrossRef]
- Li, F.; Chung, T.; Pennington, J.G.; Federico, M.L.; Kaeppler, H.F.; Kaeppler, S.M.; Otegui, M.S.; Vierstra, R.D. Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell 2015, 27, 1389–1408. [Google Scholar] [CrossRef] [PubMed]
- Lapierre, L.R.; Silvestrini, M.J.; Nunez, L.; Ames, K.; Wong, S.; Le, T.T.; Hansen, M.; Melendez, A. Autophagy genes are required for normal lipid levels in C. elegans. Autophagy 2013, 9, 278–286. [Google Scholar] [CrossRef]
- Martinez-Lopez, N.; Singh, R. Autophagy and Lipid Droplets in the Liver. Annu. Rev. Nutr. 2015, 35, 215–237. [Google Scholar] [CrossRef]
- Kurusu, T.; Koyano, T.; Hanamata, S.; Kubo, T.; Noguchi, Y.; Yagi, C.; Nagata, N.; Yamamoto, T.; Ohnishi, T.; Okazaki, Y.; et al. OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy 2014, 10, 878–888. [Google Scholar] [CrossRef]
- Thompson, A.R.; Doelling, J.H.; Suttangkakul, A.; Vierstra, R.D. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 2005, 138, 2097–2110. [Google Scholar] [CrossRef]
- Bassham, D.C.; Laporte, M.; Marty, F.; Moriyasu, Y.; Ohsumi, Y.; Olsen, L.J.; Yoshimoto, K. Autophagy in development and stress responses of plants. Autophagy 2006, 2, 2–11. [Google Scholar] [CrossRef]
- Seay, M.D.; Patel, S.; Dinesh-Kumar, S.P. Autophagy and plant innate immunity. Cell. Microbiol. 2006, 8, 899–906. [Google Scholar] [CrossRef] [PubMed]
- Bassham, D.C. Plant autophagy-more than a starvation response. Curr. Opin. Plant Biol. 2007, 10, 587–593. [Google Scholar] [CrossRef]
- Xia, K.; Liu, T.; Ouyang, J.; Wang, R.; Fan, T.; Zhang, M. Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.). DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2011, 18, 363–377. [Google Scholar] [CrossRef]
- Zhai, Y.F.; Guo, M.; Wang, H.; Lu, J.P.; Liu, J.H.; Zhang, C.; Gong, Z.H.; Lu, M.H. Autophagy, a Conserved Mechanism for Protein Degradation, Responds to Heat, and Other Abiotic Stresses in Capsicum annuum L. Front. Plant Sci. 2016, 7, 131. [Google Scholar] [CrossRef]
- Shangguan, L.; Fang, X.; Chen, L.; Cui, L.; Fang, J. Genome-wide analysis of autophagy-related genes (ARGs) in grapevine and plant tolerance to copper stress. Planta 2018, 247, 1449–1463. [Google Scholar] [CrossRef]
- De Souza Costa, E.T.; Guilherme, L.R.; De Melo, E.E.; Ribeiro, B.T.; Dos Santos, B.I.E.; Da Costa Severiano, E.; Faquin, V.; Hale, B.A. Assessing the tolerance of castor bean to Cd and Pb for phytoremediation purposes. Biol. Trace Elem. Res. 2012, 145, 93–100. [Google Scholar] [CrossRef]
- Patel, V.R.; Dumancas, G.G.; Kasi Viswanath, L.C.; Maples, R.; Subong, B.J. Castor Oil: Properties, Uses, and Optimization of Processing Parameters in Commercial Production. Lipid Insights 2016, 9, 1–12. [Google Scholar] [CrossRef]
- Ogunniyi, D.S. Castor oil: A vital industrial raw material. Bioresour. Technol. 2006, 97, 1086–1091. [Google Scholar] [CrossRef]
- Greenwood, J.S.; Bewley, J.D. Seed Development In Ricinus-Communis Cv Hale (Castor Bean) .3. Pattern Of Storage Protein And Phytin Accumulation In the Endosperm. Can. J. Bot. 1985, 63, 2121–2128. [Google Scholar] [CrossRef]
- Houston, N.L.; Hajduch, M.; Thelen, J.J. Quantitative Proteomics of Seed Filling in Castor: Comparison with Soybean and Rapeseed Reveals Differences between Photosynthetic and Nonphotosynthetic Seed Metabolism. Plant Physiol. 2009, 151, 857–868. [Google Scholar] [CrossRef]
- Nogueira, F.C.S.; Palmisano, G.; Schwammle, V.; Campos, F.A.P.; Larsen, M.R.; Domont, G.B.; Roepstorff, P. Performance of Isobaric and Isotopic Labeling in Quantitative Plant Proteomics. J. Proteome Res. 2012, 11, 3046–3052. [Google Scholar] [CrossRef]
- Xu, W.; Dai, M.Y.; Li, F.; Liu, A.Z. Genomic imprinting, methylation and parent-of-origin effects in reciprocal hybrid endosperm of castor bean. Nucleic Acids Res. 2014, 42, 6987–6998. [Google Scholar] [CrossRef]
- Chan, A.P.; Crabtree, J.; Zhao, Q.; Lorenzi, H.; Orvis, J.; Puiu, D.; Melake-Berhan, A.; Jones, K.M.; Redman, J.; Chen, G.; et al. Draft genome sequence of the oilseed species Ricinus communis. Nat. Biotechnol. 2010, 28, 951–956. [Google Scholar] [CrossRef]
- Wan, L.L.; Xia, Q.; Qiu, X.; Selvaraj, G. Early stages of seed development in Brassica napus: A seed coat-specific cysteine proteinase associated with programmed cell death of the inner integument. Plant J. 2002, 30, 1–10. [Google Scholar] [CrossRef]
- Liu, K.; Czaja, M.J. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ. 2013, 20, 3–11. [Google Scholar] [CrossRef]
- Elander, P.H.; Minina, E.A.; Bozhkov, P.V. Autophagy in turnover of lipid stores: Trans-kingdom comparison. J. Exp. Bot. 2018, 69, 1301–1311. [Google Scholar] [CrossRef]
- Zhao, L.; Dai, J.; Wu, Q. Autophagy-like processes are involved in lipid droplet degradation in Auxenochlorella protothecoides during the heterotrophy-autotrophy transition. Front. Plant Sci. 2014, 5, 400. [Google Scholar] [CrossRef]
- Van Zutphen, T.; Todde, V.; De Boer, R.; Kreim, M.; Hofbauer, H.F.; Wolinski, H.; Veenhuis, M.; Van der Klei, I.J.; Kohlwein, S.D. Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 2014, 25, 290–301. [Google Scholar] [CrossRef]
- Batoko, H.; Dagdas, Y.; Baluska, F.; Sirko, A. Understanding and exploiting autophagy signaling in plants. Essays Biochem. 2017, 61, 675–685. [Google Scholar]
- Ustun, S.; Hafren, A.; Hofius, D. Autophagy as a mediator of life and death in plants. Curr. Opin. Plant Biol. 2017, 40, 122–130. [Google Scholar] [CrossRef]
- Il Kwon, S.; Park, O.K. Autophagy in Plants. J. Plant Biol. 2008, 51, 313–320. [Google Scholar] [CrossRef]
- Pottier, M.; Masclaux-Daubresse, C.; Yoshimoto, K.; Thomine, S. Autophagy as a possible mechanism for micronutrient remobilization from leaves to seeds. Front. Plant Sci. 2014, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Di Berardino, J.; Marmagne, A.; Berger, A.; Yoshimoto, K.; Cueff, G.; Chardon, F.; Masclaux-Daubresse, C.; Reisdorf-Cren, M. Autophagy controls resource allocation and protein storage accumulation in Arabidopsis seeds. J. Exp. Bot. 2018, 69, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Raviv, B.; Aghajanyan, L.; Granot, G.; Makover, V.; Frenkel, O.; Gutterman, Y.; Grafi, G. The dead seed coat functions as a long-term storage for active hydrolytic enzymes. PLoS ONE 2017, 12, e0181102. [Google Scholar] [CrossRef]
- Schmid, M.; Simpson, D.; Kalousek, F.; Gietl, C. A cysteine endopeptidase with a C-terminal KDEL motif isolated from castor bean endosperm is a marker enzyme for the ricinosome, a putative lytic compartment. Planta 1998, 206, 466–475. [Google Scholar] [CrossRef]
- Schmid, M.; Simpson, D.J.; Sarioglu, H.; Lottspeich, F.; Gietl, C. The ricinosomes of senescing plant tissue bud from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 2001, 98, 5353–5358. [Google Scholar] [CrossRef] [PubMed]
- Kurusu, T.; Kuchitsu, K. Autophagy, programmed cell death and reactive oxygen species in sexual reproduction in plants. J. Plant Res. 2017, 130, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.B.; Lu, Q.; Cheng, S.Y.; Wang, X.C.; Zhang, H. Autophagy activity contributes to programmed cell death in Caenorhabditis elegans. Autophagy 2013, 9, 1975–1982. [Google Scholar] [CrossRef] [PubMed]
- Minina, E.A.; Bozhkov, P.V.; Hofius, D. Autophagy as initiator or executioner of cell death. Trends Plant Sci. 2014, 19, 692–697. [Google Scholar] [CrossRef]
- Toyoshi, F.; Parton, R.G. Not just fat: The structure and function of the lipid droplet. Cold Spring Harb. Perspect. Biol. 2011, 3, a004838. [Google Scholar]
- Gao, M.J.; Yi, X.; Yang, W.B.; Lam, S.M.; Tong, X.H.; Liu, J.Y.; Wang, X.; Li, Q.; Shui, G.H.; He, Z.H. GDSL lipases modulate immunity through lipid homeostasis in rice. PLoS Pathog. 2017, 13, e1006724. [Google Scholar] [CrossRef]
- Singh, R.; Kaushik, S.; Wang, Y.J.; Xiang, Y.Q.; Novak, I.; Komatsu, M.; Tanaka, K.; Cuervo, A.M.; Czaja, M.J. Autophagy regulates lipid metabolism. Nature 2009, 458, 1131–1135. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Xu, R.H.; Wang, R.L.; Liu, A.Z. Expression profiles of genes involved in fatty acid and triacylglycerol synthesis in developing seeds of Jatropha (Jatropha curcas L.). Biomass Bioenerg. 2011, 35, 1683–1692. [Google Scholar] [CrossRef]
- Jiang, Q.; Zhao, L.; Dai, J.B.; Wu, Q.Y. Analysis of Autophagy Genes in Microalgae: Chlorella as a Potential Model to Study Mechanism of Autophagy. PLoS ONE 2012, 7, e41826. [Google Scholar] [CrossRef] [PubMed]

| Gene | Arabidopsis ID | Gene | Castor Bean ID | No. of Amino Acid Residues | Mw(kDa) | PI | E-value to Arabidopsis |
|---|---|---|---|---|---|---|---|
| ATG1/13 Kinase Complex | |||||||
| AtATG1a | AT3G61960 | RcATG1a | 30076.m004465 | 676 | 75.1 | 5.90 | 4.4 × 10−66 |
| AtATG1b | AT3G53930 | RcATG1b | 29973.m000397 | 694 | 76.9 | 6.44 | 7.6 × 10−62 |
| AtATG1c | AT2G37840 | ||||||
| AtATG1t | AT1G49180 | RcATG1t | 29659.m000143 | 321 | 36.6 | 8.29 | 1.2 × 10−88 |
| AtATG13a | AT3G49590 | RcATG13a | 30098.m001725 | 621 | 69.4 | 8.81 | 3.5 × 10−103 |
| AtATG13b | AT3G18770 | RcATG13b | 30206.m000754 | 342 | 39.5 | 9.59 | 3 × 10−83 |
| RcATG13c | 30206.m000753 | 220 | 23.9 | 5.42 | 2.1 × 10−26 | ||
| AtATG11 | AT4G30790 | RcATG11 | 30114.m000524 | 1145 | 129.2 | 5.73 | 0 |
| AtATG101 | AT5G66930 | RcATG101 | 29676.m001649 | 205 | 24.1 | 5.94 | 7.9 × 10−76 |
| PI3 Kinase Complex | |||||||
| AtVPS34 | AT1G60490 | RcVPS34 | 29631.m001016 | 813 | 93 | 6.48 | 0 |
| AtATG6 | AT3G61710 | RcATG6 | 29742.m001430 | 523 | 59.9 | 5.85 | 2.1 × 10−201 |
| AtVPS15 | AT4G29380 | RcVPS15 | 29912.m005424 | 1455 | 163.5 | 7.00 | 0 |
| ATG9/2/18 Complex | |||||||
| AtATG2 | AT3G19190 | RcATG2 | 29801.m003197 | 1989 | 218.4 | 5.43 | 0 |
| AtATG9 | AT2G31260 | RcATG9 | 29584.m000241 | 864 | 99.7 | 6.32 | 8.4 × 10−301 |
| AtATG18a | AT3G62770 | RcATG18a | 30129.m000356 | 447 | 49 | 6.59 | 3.6 × 10−32 |
| AtATG18b | AT4G30510 | RcATG18b | 28612.m000123 | 349 | 38 | 8.86 | 7.1 × 10−146 |
| AtATG18c | AT2G40810 | RcATG18c | 29729.m002396 | 598 | 67 | 7.56 | 1.2 × 10−81 |
| AtATG18d | AT3G56440 | RcATG18d | 30128.m008811 | 330 | 36.6 | 9.33 | 1.6 × 10−70 |
| AtATG18e | AT5G05150 | RcATG18e | 28166.m001060 | 891 | 96.9 | 6.44 | 0 |
| AtATG18f | AT5G54730 | RcATG18f | 29703.m001515 | 1016 | 110.7 | 6.25 | 3 × 10−115 |
| AtATG18g | AT1G03380 | RcATG18g | 30128.m008700 | 991 | 107.6 | 5.44 | 1 × 10−113 |
| AtATG18h | AT1G54710 | ||||||
| ATG8/12 Conjugation System | |||||||
| AtATG3 | AT5G61500 | RcATG3 | 30147.m014252 | 314 | 35.5 | 4.79 | 3.2 × 10−150 |
| AtATG4a | AT2G44140 | RcATG4 | 28966.m000547 | 489 | 53.9 | 5.13 | 2.3 × 10−140 |
| AtATG4b | AT3G59950 | ||||||
| AtATG5 | AT5G17290 | RcATG5 | 30171.m000414 | 368 | 41.9 | 5.28 | 3.2 × 10−115 |
| AtATG7 | AT5G45900 | RcATG7 | 29900.m001609 | 710 | 77.9 | 5.35 | 3.3 × 10−235 |
| AtATG8a | AT4G21980 | RcATG8a | 29842.m003584 | 120 | 13.8 | 7.88 | 3.7 × 10−53 |
| AtATG8b | AT4G04620 | RcATG8b | 30064.m000482 | 125 | 14.3 | 7.89 | 5.5 × 10−52 |
| AtATG8c | AT1G62040 | RcATG8c | 30064.m000481 | 120 | 13.9 | 7.89 | 7 × 10−52 |
| AtATG8d | AT2G05630 | RcATG8d | 29636.m000773 | 122 | 14.1 | 7.85 | 5.8 × 10−48 |
| AtATG8e | AT2G45170 | RcATG8e | 30174.m008829 | 117 | 13.5 | 8.75 | 1.2 × 10−45 |
| AtATG8f | AT4G16520 | RcATG8f | 30076.m004580 | 125 | 14.5 | 6.73 | 1.2 × 10−33 |
| AtATG8g | AT3G60640 | RcATG8g | 29889.m003380 | 115 | 13.2 | 9.30 | 2 × 10−31 |
| AtATG8h | AT3G06420 | ||||||
| AtATG8i | AT3G15580 | ||||||
| AtATG10 | AT3G07525 | RcAtg10 | 30178.m000879 | 233 | 26.9 | 5.25 | 7.3 × 10−57 |
| AtATG12a | AT1G54210 | RcAtg12 | 29706.m001289 | 154 | 17.4 | 8.58 | 1.3 × 10−20 |
| AtATG12b | AT3G13970 | ||||||
| AtATG16L | AT5G50230 | RcATG16L | 30147.m013828 | 520 | 57.6 | 6.06 | 1.3 × 10−203 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, B.; Xu, H.; Feng, Y.; Xu, W.; Cui, Q.; Liu, A. Genomic Characterization and Expressional Profiles of Autophagy-Related Genes (ATGs) in Oilseed Crop Castor Bean (Ricinus communis L.). Int. J. Mol. Sci. 2020, 21, 562. https://doi.org/10.3390/ijms21020562
Han B, Xu H, Feng Y, Xu W, Cui Q, Liu A. Genomic Characterization and Expressional Profiles of Autophagy-Related Genes (ATGs) in Oilseed Crop Castor Bean (Ricinus communis L.). International Journal of Molecular Sciences. 2020; 21(2):562. https://doi.org/10.3390/ijms21020562
Chicago/Turabian StyleHan, Bing, Hui Xu, Yingting Feng, Wei Xu, Qinghua Cui, and Aizhong Liu. 2020. "Genomic Characterization and Expressional Profiles of Autophagy-Related Genes (ATGs) in Oilseed Crop Castor Bean (Ricinus communis L.)" International Journal of Molecular Sciences 21, no. 2: 562. https://doi.org/10.3390/ijms21020562
APA StyleHan, B., Xu, H., Feng, Y., Xu, W., Cui, Q., & Liu, A. (2020). Genomic Characterization and Expressional Profiles of Autophagy-Related Genes (ATGs) in Oilseed Crop Castor Bean (Ricinus communis L.). International Journal of Molecular Sciences, 21(2), 562. https://doi.org/10.3390/ijms21020562






