Genetic Dissection of Grain Yield and Agronomic Traits in Maize under Optimum and Low-Nitrogen Stressed Environments
Abstract
1. Introduction
2. Results
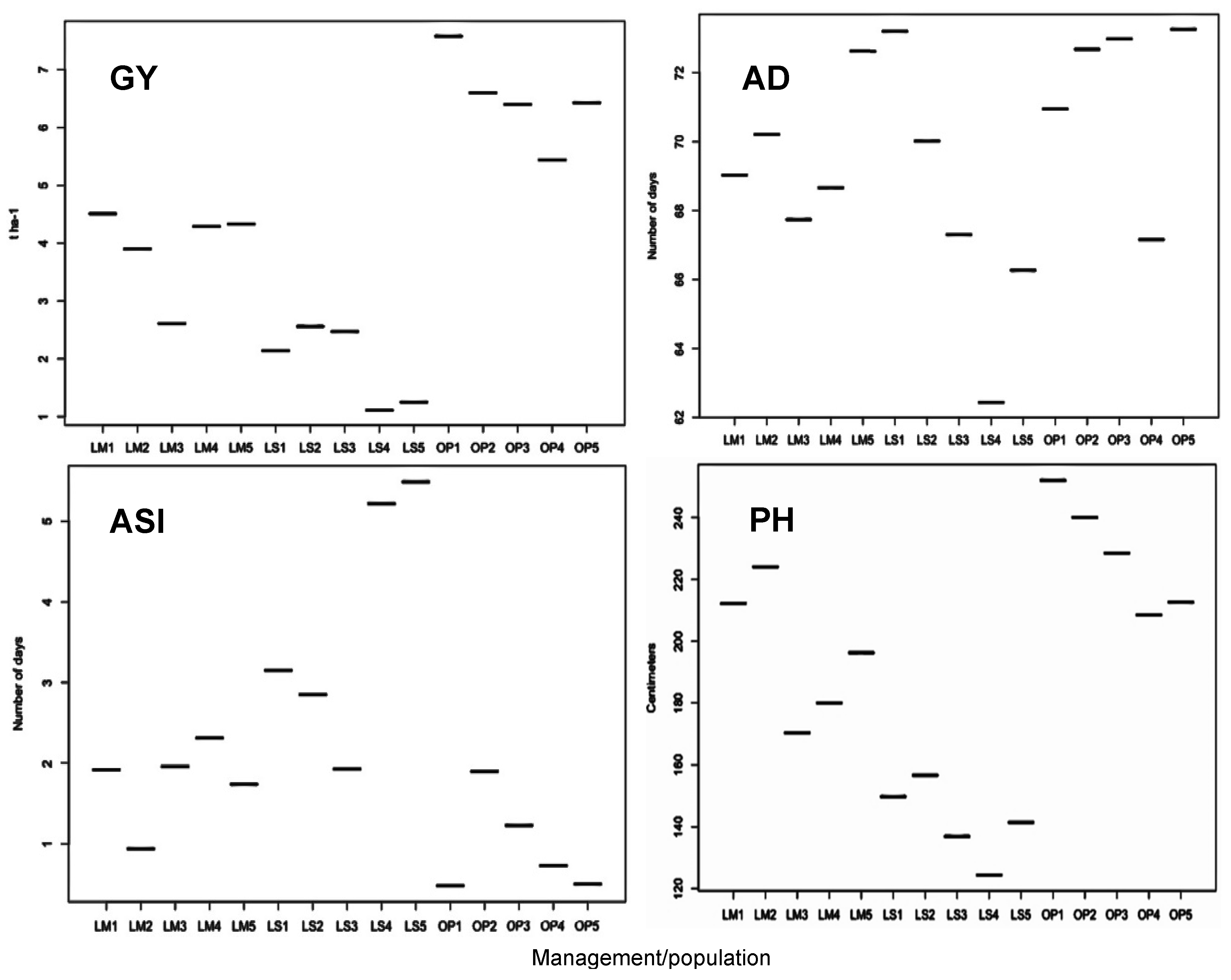
2.1. Trial Mean, Genetic Variance, and Heritability of Traits
2.2. QTL Mapping in Five DH Populations
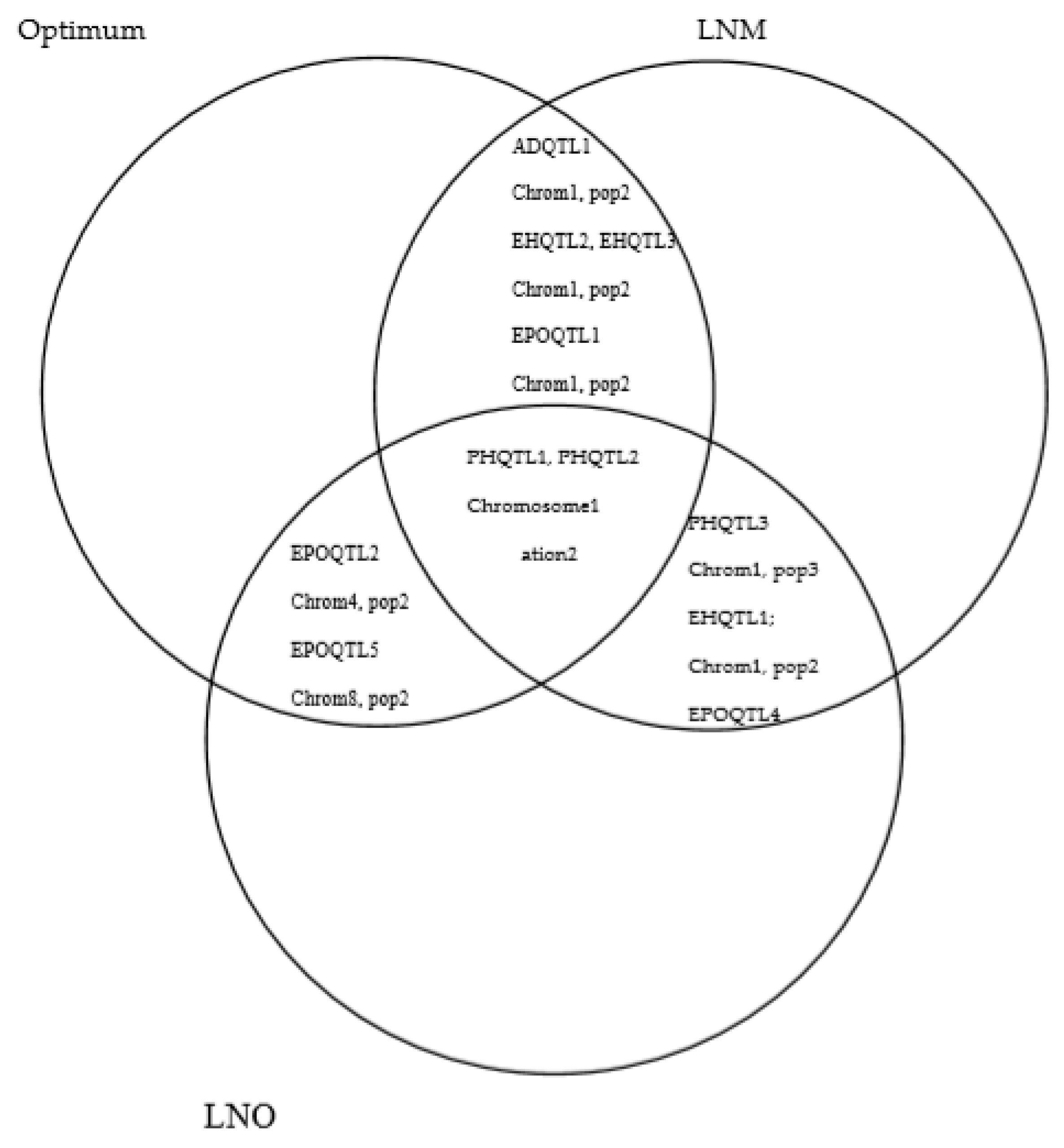
2.3. QTL Overlapping among Management Conditions for Each Trait
2.4. QTL for Multiple Traits in One/Different Population
3. Discussion
3.1. Yield Reduction, Variances, and Heritability
3.2. QTL for GY and Secondary Traits under Optimum and Low-N Conditions
4. Materials and Methods
4.1. Plant Materials
4.2. Field Experiments and Data Analysis
4.3. Genotyping, Genetic Maps, and QTL Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lafitte, H.R.; Edmeades, G.O. Improvement for tolerance to low soil nitrogen in tropical maize I. Selection criteria. Field Crop. Res. 1994, 39, 1–14. [Google Scholar] [CrossRef]
- Bänziger, M.; Betran, F.J.; Lafitte, H.R. Efficiency of high-nitrogen selection environments for improving maize for low-nitrogen target environments. Crop Sci. 1997, 37, 1103–1109. [Google Scholar] [CrossRef]
- Worku, M.; Bänziger, M.; Friesen, D.; Schulte, G.; Horst, W.J.; Vivek, B.S. Relative importance of general combining ability and specific combining ability among tropical maize (Zea mays L.) inbreds under contrasting nitrogen environments. Maydica 2008, 53, 279–288. [Google Scholar]
- Agrama, H.A. Application of molecular markers in breeding for nitrogen use efficiency. J. Crop Improv. 2006, 15, 97–125. [Google Scholar] [CrossRef]
- Ribaut, J.M.; Hoisington, D.A.; Deutsch, J.A.; Jiang, C.; Gonzalez-De-Leon, D. Identification of quantitative trait loci under drought conditions in tropical maize. Flowering parameters and the anthesis-silking interval. Theor. Appl. Genet. 1996, 92, 905–914. [Google Scholar] [CrossRef] [PubMed]
- Hirel, B.; Bertin, P.; Quilleré, I.; Bourdoncle, W.; Attagnant, C.; Dellay, C.; Gouy, A.; Cadiou, S.; Retailliau, C.; Falque, M.; et al. Towards a better understanding of the genetic and physiological basis for nitrogen use efficiency in maize. Plant Physiol. 2001, 125, 1258–1270. [Google Scholar] [CrossRef]
- Malosetti, M.; Ribaut, J.M.; Vargas, M.; Crossa, J.; Eeuwijk, F.A. A multi-trait multi-environment QTL mixed model with an application to drought and nitrogen stress trials in maize (Zea mays L.). Euphytica 2007, 161, 241–257. [Google Scholar] [CrossRef]
- Almeida, G.D.; Makumbi, D.; Magorokosho, C.; Nair, S.; Borém, A.; Ribaut, J.M.; Bänziger, M.; Prasanna, B.M.; Crossa, J.; Babu, R. QTL mapping in three tropical maize populations reveals a set of constitutive and adaptive genomic regions for drought tolerance. Theor. Appl. Genet. 2013, 126, 583–600. [Google Scholar] [CrossRef]
- Almeida, G.D.; Nair, S.; Borem, A.; Cairns, J.; Trachsel, S.; Ribaut, J.M.; Bänziger, M.; Prasanna, B.M.; Crossa, J.; Babu, R. Molecular mapping across three populations reveals a QTL hotspot region on chromosome 3 for secondary traits associated with drought tolerance in tropical maize. Mol. Breed. 2014, 34, 701–715. [Google Scholar] [CrossRef]
- Semagn, K.; Beyene, Y.; Warburton, M.L.; Tarekegne, A.; Mugo, S.; Meisel, B.; Sehabiague, P.; Prasanna, B.M. Meta-analyses of QTL for grain yield and anthesis silking interval in 18 maize populations evaluated under water-stressed and well-watered environments. BMC Genom. 2013, 14, 313. [Google Scholar] [CrossRef]
- Fan, Y.; Shabala, S.; Ma, Y.; Xu, R.; Zhou, M. Using QTL mapping to investigate the relationships between abiotic stress tolerance (drought and salinity) and agronomic and physiological traits. BMC Genom. 2015, 16, 43. [Google Scholar] [CrossRef] [PubMed]
- Ribaut, J.; Frachebound, Y.; Monneveux, P.; Bänziger, M.; Vargas, M.; Jiang, C. Quantitative trait loci for yield and correlated traits under high and low nitrogen conditions in tropical maize. Mol. Breed. 2007, 20, 15–29. [Google Scholar] [CrossRef]
- Collins, N.C.; Tardieu, F.; Tuberosa, R. Quantitative trait loci and crop performance under abiotic stress: Where do we stand? Plant Physiol. 2008, 147, 469–486. [Google Scholar] [CrossRef] [PubMed]
- Agrama, H.A.; Zakaria, A.; Said, F.; Tuinstra, M. Identification of quantitative trait loci for nitrogen use efficiency in maize. Mol. Breed. 1999, 5, 187–195. [Google Scholar] [CrossRef]
- Gallais, A.; Hirel, B. An approach to the genetics of nitrogen use efficiency in maize. J. Exp. Bot. 2004, 55, 295–306. [Google Scholar] [CrossRef]
- Bänziger, M.; Edmeades, G.O.; Beck, D.; Bellon, M. Breeding for Drought and Nitrogen Stress Tolerance in Maize: From Theory to Practice; CIMMYT: Mexico City, Mexico, 2000. [Google Scholar]
- Ribeiro, P.F.; Badu-Apraku, B.; Gracen, V.E.; Danquah, E.Y.; Garcia-Oliviera, A.L.; Asante, M.D.; Afriyie-Debrah, C.; Gedil, M. Identification of quantitative trait loci for grain yield and other traits in tropical maize under high and low soil nitrogen conditions. Crop Sci. 2018, 58, 321–330. [Google Scholar] [CrossRef]
- Worku, M.; Bänziger, M.; Erley, G.S.A.; Friesen, D.; Diallo, A.O.; Horst, W.J. Nitrogen uptake and utilization in contrasting nitrogen efficient tropical maize hybrids. Identification of quantitative trait loci for grain yield and other traits in tropical maize under high and low soil-nitrogen environments. Crop Sci. 2007, 47, 519–528. [Google Scholar] [CrossRef]
- Presterl, T.; Seitz, G.; Landbeck, M.; Thiemt, E.M.; Schmidt, W.; Geiger, H.H. Improving nitrogen-use efficiency in European maize: Estimation of quantitative genetic parameters. Crop Sci. 2003, 43, 1259–1265. [Google Scholar] [CrossRef]
- Gallais, A.; Coque, M. Genetic variation and selection for nitrogen use efficiency in maize: A synthesis. Maydica 2005, 50, 531–547. [Google Scholar]
- Geiger, H.H.; Gordillo, G.A. Doubled haploids in hybrid maize breeding. Maydica 2009, 54, 485–499. [Google Scholar]
- Patterson, H.D.; Williams, E.R. A new class of resolvable incomplete block designs. Biometrika 1976, 63, 83–92. [Google Scholar] [CrossRef]
- Semagn, K. Leaf tissue sampling and DNA extraction protocols. Methods in Molecular Biology 2014, 1115, 53–67. [Google Scholar] [PubMed]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ye, G.; Wang, J. A modified algorithm for the improvement of composite interval mapping. Genetics 2007, 374, 361–374. [Google Scholar] [CrossRef]
- Kosambi, D. The estimation of map distance from recombination values. Ann. Eugen. 1944, 12, 172–175. [Google Scholar] [CrossRef]
| Chr. | CML494/CML550 | CML504/CML550 | CML511/CML550 | CML505/LaPostaSeqC7-F64-2-6-2-2-B-B | CML536/LaPostaSeqC7-F64-2-6-2-2-B-B | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNPs | Distance (cM) | SNPs | Distance (cM) | SNPs | Distance (cM) | SNPs | Distance (cM) | SNPs | Distance (cM) | |
| 1 | 371 | 644.9 | 404 | 511.5 | 285 | 642.5 | 314 | 1068.4 | 311 | 512.5 |
| 2 | 242 | 432.4 | 310 | 460.6 | 237 | 428.5 | 216 | 841.4 | 256 | 270.9 |
| 3 | 233 | 417.7 | 286 | 538.8 | 197 | 434.7 | 259 | 961.6 | 237 | 489 |
| 4 | 238 | 359.3 | 325 | 336.7 | 244 | 470.7 | 254 | 796.0 | 277 | 381.5 |
| 5 | 211 | 502.7 | 283 | 539.4 | 193 | 568.4 | 118 | 581.7 | 211 | 311.1 |
| 6 | 138 | 249.6 | 195 | 320.3 | 180 | 225.2 | 172 | 808.5 | 177 | 312.4 |
| 7 | 172 | 420.8 | 228 | 375.4 | 153 | 318.4 | 151 | 671.1 | 152 | 432.7 |
| 8 | 195 | 379.9 | 250 | 441.6 | 177 | 322.5 | 170 | 506.0 | 176 | 247.9 |
| 9 | 162 | 103.9 | 221 | 294.7 | 139 | 199.5 | 162 | 543.7 | 140 | 236 |
| 10 | 142 | 177.1 | 197 | 185.6 | 157 | 261.5 | 169 | 414.7 | 149 | 232.4 |
| Total | 2104 | 3688.3 | 2699 | 4004.6 | 1962 | 3871.9 | 1985 | 7193.1 | 2086 | 3426.4 |
| Chr. | LNM | LNO | OPT | Total | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AD | ASI | EH | EPO | GY | PH | SEN | Tot | AD | ASI | EH | EPO | GY | PH | SEN | Tot | AD | ASI | EH | EPO | GY | PH | SEN | Total | ||
| 1 | 5 | 1 | 4 | 1 | 4 | 1 | 16 | 7 | 1 | 3 | 4 | 2 | 3 | 20 | 3 | 5 | 2 | 2 | 3 | 15 | 51 | ||||
| 2 | 2 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | 8 | ||||||||||||||
| 3 | 1 | 1 | 3 | 2 | 7 | 3 | 3 | 2 | 2 | 10 | 3 | 2 | 1 | 1 | 1 | 1 | 9 | 26 | |||||||
| 4 | 1 | 2 | 3 | 1 | 1 | 1 | 3 | 3 | 2 | 5 | 11 | ||||||||||||||
| 5 | 2 | 2 | 1 | 1 | 2 | 3 | 1 | 1 | 1 | 6 | 10 | ||||||||||||||
| 6 | 1 | 1 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 4 | 8 | |||||||||||||
| 7 | 1 | 2 | 1 | 1 | 5 | 1 | 1 | 1 | 1 | 2 | 8 | ||||||||||||||
| 8 | 2 | 1 | 2 | 5 | 1 | 1 | 2 | 2 | 1 | 2 | 9 | 4 | 1 | 1 | 6 | 20 | |||||||||
| 9 | 1 | 1 | 1 | 1 | 1 | 3 | 1 | 1 | 2 | 6 | |||||||||||||||
| 10 | 2 | 1 | 1 | 1 | 5 | 2 | 2 | 7 | |||||||||||||||||
| Total | 13 | 3 | 15 | 4 | 2 | 10 | 2 | 49 | 12 | 5 | 5 | 8 | 8 | 8 | 5 | 51 | 18 | 5 | 10 | 9 | 3 | 7 | 3 | 55 | 155 |
| Population | Management | Chr. | Pos (cM) | Left Marker | Right Marker | LOD | PVE (%) | TPVE (%) | Add | Fav Allele |
|---|---|---|---|---|---|---|---|---|---|---|
| CML550 × CML494 | OPT | 2 | 318 | S2_15120146 | S2_15909091 | 4.48 | 17.23 | 16.68 | 0.12 | CML550 |
| CML550 × CML504 | OPT | 1 | 46 | S1_283186611 | S1_280222332 | 4.43 | 6.31 | 39.17 | −0.12 | CML504 |
| OPT | 1 | 183 | S1_219232023 | S1_217114738 | 10.24 | 15.04 | −0.18 | CML504 | ||
| LNM | 7 | 278 | S7_106325823 | S7_105221050 | 3.39 | 6.84 | 11.50 | −0.04 | CML504 | |
| LNM | 10 | 143 | S10_11189892 | S10_10138689 | 3.06 | 6.05 | 0.04 | CML550 | ||
| LNO | 2 | 173 | S2_200745112 | S2_202138908 | 3.10 | 6.66 | 11.55 | −0.09 | CML504 | |
| CML550 × CML511 | LNO | 3 | 228 | S3_127100888 | S3_128233351 | 4.69 | 17.55 | 23.34 | −0.10 | CML511 |
| LNO | 4 | 177 | S4_165623425 | S4_162255436 | 3.71 | 13.56 | −0.09 | CML511 | ||
| CML505/LaPostaSeqC7-F64-2-6-2-2-B-B | LNO | 1 | 531 | S1_177877619 | S1_183811363 | 3.91 | 10.14 | 22.30 | 0.06 | LP |
| LNO | 3 | 169 | S3_204998702 | S3_203869859 | 4.38 | 10.34 | −0.05 | CML505 | ||
| CML536/LaPostaSeqC7-F64-2-6-2-2-B-B | LNO | 1 | 439 | S1_41220359 | S1_39739703 | 3.64 | 9.90 | 38.54 | 0.08 | LP |
| LNO | 3 | 345 | S3_38439419 | S3_31449087 | 3.68 | 10.10 | 0.08 | LP | ||
| LNO | 8 | 157 | S8_166372615 | S8_168274395 | 3.77 | 10.54 | 0.08 | LP |
| Characteristic | Population | Management | Number of QTL | TPVE (%) |
|---|---|---|---|---|
| CML550/CML494 | OPT | 8 | 71.31 | |
| Anthesis date | LNM | 2 | 28.86 | |
| LNO | 4 | 47.27 | ||
| CML550/CML504 | OPT | 8 | 46.88 | |
| LNM | 6 | 58.04 | ||
| LNO | 2 | 33.58 | ||
| CML550/CML511 | OPT | 1 | 29.02 | |
| LNM | 1 | 12.04 | ||
| LNO | 2 | 46.11 | ||
| CML505/LaPostaSeqC7-F64-2-6-2-2-B-B | OPT | 1 | 13.36 | |
| LNM | 1 | 8.00 | ||
| LNO | 2 | 25.45 | ||
| CML536/LaPostaSeqC7-F64-2-6-2-2-B-B | LNM | 1 | 37.71 | |
| LNO | 1 | 29.69 | ||
| Anthesis- silking | CML550/CML504 | OPT | 4 | 31.26 |
| interval | LNM | 3 | 24.10 | |
| LNO | 1 | 12.84 | ||
| CML505/LaPostaSeqC7- | OPT | 3 | 11.70 | |
| F64-2-6-2-2-B-B | LNO | 2 | 19.68 | |
| CML536/LaPostaSeqC7- F64-2-6-2-2-B-B | LNO | 2 | 39.41 | |
| Plant height | CML550/CML494 | LNM | 1 | 20.40 |
| LNO | 2 | 26.45 | ||
| CML550/CML504 | OPT | 7 | 59.82 | |
| LNM | 6 | 61.72 | ||
| LNO | 3 | 49.52 | ||
| CML550/CML511 | LNM | 3 | 44.52 | |
| LNO | 1 | 24.33 | ||
| CML505/LaPostaSeqC7-F64-2-6-2-2-B-B | LNO | 1 | 8.05 | |
| CML536/LaPostaSeqC7-F64-2-6-2-2-B-B | LNO | 1 | 13.39 | |
| Ear height | CML550/CML494 | LNM | 5 | 53.16 |
| LNO | 1 | 24.29 | ||
| CML550/CML504 | OPT | 7 | 56.64 | |
| LNM | 6 | 52.26 | ||
| LNO | 3 | 27.42 | ||
| CML550/CML511 | OPT | 2 | 22.87 | |
| LNM | 2 | 23.70 | ||
| CML505/LaPostaSeqC7-F64-2-6-2-2-B-B | OPT | 1 | 14.79 | |
| LNM | 1 | |||
| CML536/LaPostaSeqC7-F64-2-6-2-2-B-B | LNM | 1 | 23.10 | |
| LNO | 1 | 11.40 | ||
| Ear position | CML550/CML494 | OPT | 1 | 10.53 |
| LNM | 1 | 23.30 | ||
| LNO | 1 | 28.75 | ||
| CML550/CML504 | OPT | 5 | 38.35 | |
| LNM | 3 | 33.54 | ||
| LNO | 5 | 38.34 | ||
| CML550/CML511 | OPT | 2 | 28.05 | |
| CML505/LaPostaSeqC7-F64-2-6-2-2-B-B | OPT | 1 | 15.30 | |
| CML536/LaPostaSeqC7-F64-2-6-2-2-B-B | LNO | 2 | 31.04 | |
| Senescence | CML550/CML504 | OPT | 3 | 23.65 |
| LNM | 2 | 15.06 | ||
| LNO | 5 | 45.87 |
| Trial Name | Site | Year * | Nr of Entries | Nr of Checks | Management | Rep | Mean GY |
|---|---|---|---|---|---|---|---|
| CML494/CML550; Tester: CML312 | |||||||
| Population Size: 108 | |||||||
| 15B-EMB-8 | Embu | 2015B | 110 | 2 | LNO | 2 | 2.24 |
| 15A-KKM-9 | Kakamega | 2015A | 110 | 2 | Opt | 2 | 7.13 |
| 15A-RWA-3 | Rwanda | 2015A | 110 | 2 | Opt | 2 | 6.72 |
| 15A-KBK-1 | Kiboko | 2015A | 110 | 2 | Opt | 2 | 8.88 |
| 15A-KBK-2 | Kiboko | 2015A | 110 | 2 | LNM | 2 | 4.00 |
| 15B-KBK-5 | Kiboko | 2015B | 110 | 2 | LNO | 2 | 2.05 |
| 15A-KIT-7 | Kitale | 2015A | 110 | 2 | LNM | 2 | 5.43 |
| 15A-MTW-4 | Mtwapa | 2015A | 110 | 2 | LNM | 2 | 4.10 |
| CML504/CML550; Tester: CML312/CML443 | |||||||
| Population Size: 219 | |||||||
| 14A-ALU-9 | Alupe | 2014A | 224 | 5 | LNM | 2 | 3.60 |
| 14A-EMB-5 | Embu | 2014A | 224 | 5 | LNM | 2 | 4.22 |
| 14A-KKM-3 | Kakamega | 2014A | 224 | 5 | LNO | 2 | 2.40 |
| 14A-KKM-4 | Kakamega | 2014A | 224 | 5 | Opt | 2 | 5.35 |
| 14B-KBK-1 | Kiboko | 2014B | 224 | 5 | LNO | 3 | 2.92 |
| 14B-KBK-2 | Kiboko | 2014B | 224 | 5 | LNO | 3 | 2.39 |
| 14A-KBK-1 | Kiboko | 2014A | 224 | 5 | Opt | 2 | 6.49 |
| 14A-KBK-2 | Kiboko | 2014A | 224 | 5 | Opt | 2 | 10.60 |
| 14A-KIT-10 | Kitale | 2014A | 224 | 5 | Opt | 2 | 5.67 |
| 14A-KIT-8 | Kitale | 2014A | 224 | 5 | Opt | 2 | 4.91 |
| CML511/CML550; Tester: CML312/CML443 | |||||||
| Population Size: 111 | |||||||
| 14A-ALU-9 | Alupe | 2014A | 116 | 6 | LNM | 2 | 2.65 |
| 14A-KKM-3 | Kakamega | 2014A | 116 | 6 | LNM | 2 | 2.72 |
| 14A-KKM-4 | Kakamega | 2014A | 116 | 6 | Opt | 2 | 4.99 |
| 14B-KBK-1 | Kiboko | 2014B | 116 | 6 | LNO | 3 | 2.12 |
| 14B-KBK-2 | Kiboko | 2014B | 116 | 6 | LNO | 3 | 2.83 |
| 14A-KBK-2 | Kiboko | 2014A | 116 | 6 | Opt | 2 | 10.40 |
| 14A-KBK-5 | Kiboko | 2014A | 116 | 6 | Opt | 2 | 6.07 |
| 14A-KIT-10 | Kitale | 2014A | 116 | 6 | Opt | 2 | 5.19 |
| 14A-KIT-8 | Kitale | 2014A | 116 | 6 | Opt | 2 | 5.35 |
| 14A-MTW-6 | Mtwapa | 2014A | 116 | 6 | LNM | 2 | 2.47 |
| CML505/LaPostaSeqC7-F64-2-6-2-2-B-B; Tester: CML395/CML444 | |||||||
| Population size: 159 | |||||||
| WET15A-EVALITC-08-1 | Kakamega | 2015A | 174 | 6 | Opt | 2 | 7.53 |
| WET15A-EVALITC-08-2 | Kiboko | 2015A | 174 | 6 | Opt | 2 | 5.81 |
| WET15A-EVALITC-08-5 | Kiboko_LN | 2015A | 174 | 6 | LNM | 2 | 4.29 |
| WET15A-EVALITC-08-6 | Kiboko2_LN | 2015A | 174 | 6 | Opt | 2 | 5.12 |
| WET15A-EVALITC-08-8 | Kiboko3_LN | 2015B | 174 | 6 | LNO | 2 | 1.11 |
| CML536/LaPostaSeqC7-F64-2-6-2-2-B-B; Tester: CML395/CML444 | |||||||
| Population Size: 109 | |||||||
| WET15A-EVALITC-11-1 | Kakamega | 2015A | 130 | 8 | Opt | 2 | 8.58 |
| WET15A-EVALITC-11-2 | Kiboko | 2015A | 130 | 8 | Opt | 2 | 5.00 |
| WET15A-EVALITC-11-5 | Kiboko_LN | 2015A | 130 | 8 | LNM | 2 | 4.33 |
| WET15A-EVALITC-11-6 | Kiboko2_LN | 2015A | 130 | 8 | Opt | 2 | 5.71 |
| WET15A-EVALITC-11-8 | Kiboko3_LN | 2015B | 130 | 8 | LNO | 2 | 1.25 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tadesse Ertiro, B.; Olsen, M.; Das, B.; Gowda, M.; Labuschagne, M. Genetic Dissection of Grain Yield and Agronomic Traits in Maize under Optimum and Low-Nitrogen Stressed Environments. Int. J. Mol. Sci. 2020, 21, 543. https://doi.org/10.3390/ijms21020543
Tadesse Ertiro B, Olsen M, Das B, Gowda M, Labuschagne M. Genetic Dissection of Grain Yield and Agronomic Traits in Maize under Optimum and Low-Nitrogen Stressed Environments. International Journal of Molecular Sciences. 2020; 21(2):543. https://doi.org/10.3390/ijms21020543
Chicago/Turabian StyleTadesse Ertiro, Berhanu, Michael Olsen, Biswanath Das, Manje Gowda, and Maryke Labuschagne. 2020. "Genetic Dissection of Grain Yield and Agronomic Traits in Maize under Optimum and Low-Nitrogen Stressed Environments" International Journal of Molecular Sciences 21, no. 2: 543. https://doi.org/10.3390/ijms21020543
APA StyleTadesse Ertiro, B., Olsen, M., Das, B., Gowda, M., & Labuschagne, M. (2020). Genetic Dissection of Grain Yield and Agronomic Traits in Maize under Optimum and Low-Nitrogen Stressed Environments. International Journal of Molecular Sciences, 21(2), 543. https://doi.org/10.3390/ijms21020543






