Abstract
Non-coding regulatory RNAs are generated as a core output of the eukaryotic genomes, being essential players in cell biology. At the organism level, they are key functional actors in those tissues and organs with limited proliferation capabilities such as the heart. The role of regulatory networks mediated by non-coding RNAs in the pathophysiology of cardiovascular conditions is starting to be unveiled. However, a deeper knowledge of the functional interactions among the diverse non-coding RNA families and their phenotypic consequences is required. This review presents the current knowledge about the functional crosstalk between circRNAs and other biomolecules in the framework of the cardiovascular diseases.
1. Introduction
In eukaryotic cells, circular RNAs (circRNAs) are covalently closed RNA molecules generated by non-canonical back-splicing of coding and non-coding transcriptional units [1,2]. Besides to the known viral genomes constituted by covalently closed RNA, the first evidence of the existence of non-genomic functional circular RNA molecules in eukaryotic cells was described in the 1970s by the discovery and characterization of plant viroids and virusoids, infectious circular RNAs that are replicated by the cellular transcriptional machinery and can be transferred between plant cells [3,4,5]. Eukaryotic genomes were later identified as an internal source of circRNAs, being initially considered as bystander products of incorrect splicing events [6,7]. Nowadays, the availability of high-throughput RNA sequencing methods (RNAseq) together with specialized bioinformatics algorithms devoted to circRNA analysis has enabled the identification of thousands of circRNAs along with their cell and tissue-specific expression patterns [8,9].
CircRNAs biogenesis proceeds following a unique mechanism among non-coding RNAs (ncRNAs) since they are generated as by-products during the post-transcriptional processing of other RNA species (coding or non-coding). Jeck and coworkers proposed the term back-splicing to describe the pre-mRNA process of circularization (Figure 1) and covalent joining of the upstream 5’-splice acceptor to the downstream 3’-splice donor by phosphodiester bonds to generate a covalently closed RNA [10]. Two main models are suggested to explain circRNA formation: the lariat-driven circularization and the intron-pairing-driven circularization. In the lariat-driven model, circRNAs are originated from lariat intermediates that can contain both exons and introns and are formed during exon-skipping. In the intron-pairing circularization, there is a hybridization of introns flanking the back-spliced exons. The main players in this model are cis-acting elements (short non-coding regulatory sequences, as Alu repeats) located in the flanking introns that promote intron hybridization and facilitate exon circularization [10,11]. Additionally, RNA-binding proteins (RBPs) were also shown to be involved in circRNA biogenesis by acting as trans-acting factors that recognize and bind to specific motifs located in the introns flanking the back-spliced exons. By dimerization or protein–protein interaction, the RBPs are able to bridge two exon-bordering introns together, leading to RNA circularization. For instance, muscleblind protein (MBL) and quaking protein (QKI) are two trans-acting factors that regulate the formation of several circRNAs [12,13]. Regardless of the biogenesis model, circRNA isoforms can be originated from the same locus through alternative circularization [14]. Once the circular structure is formed, circRNAs become highly resistant to nuclease action, such as RNAse R [15,16], which means that their half-life is usually higher than other linear RNA molecules [17].
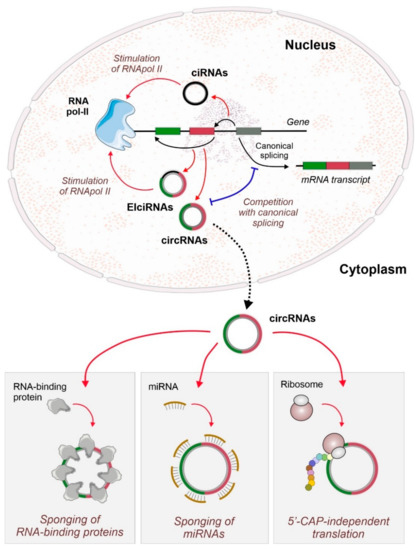
Figure 1.
Biogenesis and function of circular RNAs (circRNAs). At the nucleus, back-splicing events competing with canonical splicing could generate several families of different circRNAs. Intron-containing circRNAs (circular-intronic RNAs, ciRNAs, and exonic-intronic circular RNAs, EIcircRNAs) can stimulate the transcription of their hosting gene by direct interaction with RNA polymerase II. At the cytoplasm, circRNAs functions include their action as sponges for RNA-binding proteins or microRNAs (miRNAs, and also as scaffolds of high-range complexes. Some circRNAs can also be translated in a 5’-CAP-independent manner to generate small or micro-peptides that can have regulatory functions.
CircRNAs can be classified in three major groups: exonic circRNA (EciRNA or ecircRNA) generated by the circularization of one or more exons [10], intronic circRNA (ciRNA) originated solely from lariat-excised introns [18], and exon- and intron-containing circRNA (EIciRNA) [19]. CiRNAs and EIciRNAs are mainly found in the nucleus and are characterized as interaction partners of RNA-polymerase II, being present in the spliceosome, and even taking part in alternative splicing [19,20]. Increasing experimental pieces of evidence showed circRNAs as emerging regulatory players participating in the modulation of gene expression by sponging microRNAs (miRNAs) [2,16,21], by interacting with RBPs [12,13], and by regulating splicing and gene expression (Figure 1) [12,22].
CircRNAs, together with other ncRNA species such as miRNAs and lncRNAs, are pivotal players in the regulation of cardiovascular development and pathophysiology [23,24,25]. CircRNA-centered regulatory networks constitute a new regulatory layer of gene expression that would deserve a more detailed characterization, especially in those tissues with limited proliferation and very specialized in function such as the heart and the cardiovascular system [26].
2. CircRNAs as Active Players in the Regulation of Cardiovascular Diseases
2.1. MiRNA Sponging by circRNAs
Regulatory crosstalk between non-coding RNA species was exemplified by the characterization of competing endogenous RNA (ceRNA) networks where an anchor RNA molecule was able to capture miRNAs by sequence complementarity, removing them from the medium and decreasing their regulatory effects over target mRNA transcripts. This phenomenon was firstly described in the differentiation of muscle, where the MD1 lncRNA acts as a molecular sponge of miR-133, activating muscle-specific gene expression [27], and also in tumor cells, where the mRNA of the tumor suppressor PTEN can behave like a sponge for several miRNAs, establishing a trans-regulatory network that affects gene expression in cancer [28]. CircRNAs could also contain miRNA regulatory or response elements (MREs), sequences that complementary bind to miRNA molecules, capturing them and inhibiting their functions in the cell (Figure 2) [2,21,29,30].
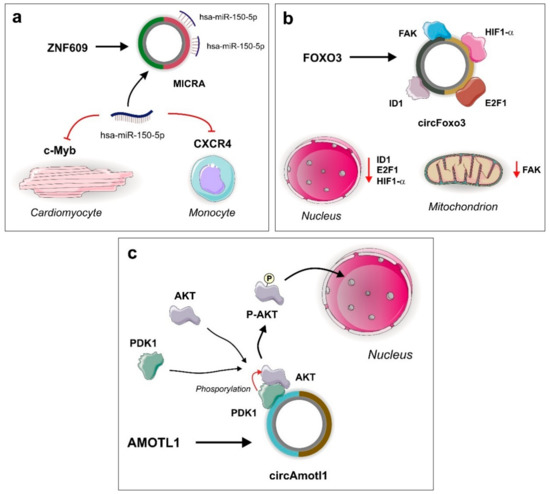
Figure 2.
Selected circRNAs and their functional roles in cardiovascular diseases acting as cytoplasmic sponges and molecular scaffolds. (a) Myocardial infarction-associated circular RNA (MICRA) circRNA generated by back-splicing of the ZNF609 gene acts as a molecular sponge of miR-150-5p. The sponging effect in cardiomyocytes is related with an increase of cellular activity by the up-regulation of its target, c-Myb [39,40]. (b) Cellular senesce related with cardiomyopathies in mouse heart is regulated by the protein-sponging function of circFoxo3 circRNA, which is able to capture the nuclear transcription factors ID1 and E2F1, and the antisenescence proteins HIF1-α and FAK, decreasing their levels and inducing cellular stress [41,42]. (c) CircAmotl1 suppress apoptosis events in cardiomyopathy-induced mouse models by facilitating the interaction between PDK1 kinase and its substrate AKT. Phosphorylated AKT originated from this scaffold-mediated interaction, is translocated to the nucleus and participates in the regulation of cell survival [43].
Several bioinformatics tools, such as CircInteractome, CircNet, Circ2Traits, and StarBase v2.0 have been of great benefit to predict miRNA binding sites in circRNAs [31,32,33,34]. Furthermore, the number of validated circRNA/miRNA interactions is increasing, being reported in practically every organ and tissue, not only in humans but also in other animal species [2,29,35,36,37]. Currently, miRNA sponging is the most well-characterized circRNA function. However, despite a large number of discovered cardiovascular expressed circRNAs [38], only a few have been reported to act as miRNA sponges (Table 1).

Table 1.
circRNAs acting as miRNAs sponges and modulating their function in cardiovascular diseases.
2.1.1. Cdr1as (ciRS-7)
Cdr1as (ciRS-7) is a very well-known circRNA that acts as a miRNA sponge in numerous and different cell types such as cardiomyocytes, neural cells, and hepatic cells [2,29,44]. Cdr1as is up-regulated in myocardial infarction (MI) mice and it was described to act as a sponge for miR-7a in cardiomyocytes [29]. It was previously shown that miR-7a protects the heart from MI in hypoxic cardiomyocytes by negatively regulating the pro-apoptotic poly (ADP-ribose) polymerase (PARP) and the zinc finger Sp1 transcription factor, thus suppressing ischemia/reperfusion(I/R)-induced apoptosis [45]. Upon interaction with cdr1as, miR-7a becomes less abundant in the cardiomyocytes and PARP levels are higher, inducing apoptosis [29,45]. Thus, higher cellular levels of this circRNA are responsible for the aggravation of MI effects [29].
2.1.2. MFACR
The mitochondrial fission and apoptosis-related circRNA (MFACR) is generated from the exon 5 of the SET and MYND domain-containing 4 (Smyd4) gene [46]. This circRNA is able to mediate mitochondrial fission and consequent activation of apoptotic events by acting as an miRNA sponge for miR-652-3p. In mammalian cells, mitochondrial fission and induced apoptosis are regulated by overexpression of mitochondrial protein 18 (MTP18) [47,48]. Similarly, Wang et al. described that MTP18 decrease led to an inhibition of mitochondrial fission and cardiomyocytes apoptosis [46]. In cultured cardiomyocytes, MTP18 expression is regulated by miR-652-3p and high levels of this miRNA in cells, contributing to the inhibition of mitochondrial fission and apoptosis. In vitro studies and bioinformatics analysis suggest that MFACR interacts with miR-652-3p in cardiomyocytes. In fact, MFACR contains 15 MREs for miR-652-3p and the overexpression of MFACR increased the protein level of MTP18 by sponging miR-652-3p. Moreover, inhibition of miR-652-3p would suppress the effect of MFACR knockdown on MTP18 [46].
2.1.3. circNCX1
CircNCX1 is generated from the second exon of the sodium/calcium exchanger 1 (ncx1) gene and also mediates cardiomyocyte apoptotic events. Higher levels of reactive oxygen species (ROS) in the myocardium are related to increased levels of circNCX1 and may intensify the effects of I/R damage. In fact, circNCX1 can act as an miRNA sponge, harboring 8 binding sites for miR-133a-3p. This miRNA was previously described as cardioprotective in hypertrophic cardiomyopathy and heart failure by targeting the cell death-inducing p53-target protein 1 (CDIP1), an apoptotic transducer. Thus, circNCX1 sponge activity may suppress miR-133a-3p, leading to an increased number of apoptotic cardiomyocytes that intensify MI effects such as myocardial damage [49,50].
2.1.4. HRCR
The heart-related circRNA (HRCR), generated from the murine Pwwp2a gene, was the first circRNA shown to play a cardioprotective role in hypertrophy and heart failure [37]. HRCR acts as an miRNA sponge, harboring 6 binding sites for miR-223 and down-regulating its activity. MiR-223 directly suppresses the mRNA transcript of the apoptosis repressor with CARD domain protein (ARC); however, decreased levels of ARC can induce hypertrophy due to cardiomyocyte apoptosis. By regulating miR-223, HRCR is responsible for a higher rate of ARC translation causing a decrease of apoptotic cardiomyocytes and subsequent cardiomyocyte hypertrophy. Ultimately, HRCR activity lessens the odds of heart failure [37].
2.1.5. circS1c8a1
Another circRNA that plays a major role in cardiac hypertrophy is circSlc8a1. This circRNA was previously shown to be abundant in the human heart [38]. Lately, it has been confirmed that circSlc8a1 contains miRNA binding sites for miR-133a and, as mentioned before, this miRNA participates in cardioprotective processes. Inhibition of circSlc8a1 was proposed as a therapeutic approach in order to decrease the risk of dilated cardiomyopathy (DCM) and heart failure progression, resultant from pathological heart hypertrophy [51].
2.1.6. circRNA_000203
CircRNA_000203 is a nine exon circRNA transcribed from the Myosin 9a (Myo9a) gene. It contains the exons 7 to 15 and respective flanking sequences of the introns 6 and 15. Despite its large size, this circRNA was shown to only harbor two MREs for miR-26b-5p, a suppressor of Col1a2 and CTGF expression [52]. Col1a2 is a peptide found in type I collagen and CTGF is an extracellular growth factor protein responsible for the regeneration of damaged fibrotic tissue. However, higher levels of these proteins may cause fibrosis. It is noteworthy that increased levels of Col3a1 and α-SMA proteins may also be found in cardiac fibroblasts [52]. Moreover, circRNA_000203 overexpression and its ability to sponge miR-26b-5p in fibroblasts were correlated to an increased risk of diabetic cardiomyopathy due to consequent Col1a2 and CTGF up-regulation [52].
2.1.7. circRNA_010567
Another circRNA responsible for fibrotic tissue formation in diabetic cardiomyopathy conditions is circRNA_010567, which regulates the miR-141/TGF-β1 axis as an miR-141 sponge [53]. Down-regulation of this circRNA is responsible for a decrease in previously mentioned fibrotic protein levels such as Col1a2, Col3a1, and α-SMA, as well as TGF-β1, due to the suppression of circRNA_010567 sponge activity [53].
2.1.8. circZNF609
The ZNF609 locus is found to generate two different circRNAs that have distinct roles in cardiovascular diseases, the myocardial infarction-associated circular RNA (MICRA) and circZNF609. MICRA is an 874 nt long circRNA originated from exon 1 of the ZNF609 gene and proposed as a myocardial infarction biomarker [54]. Interestingly, MICRA is predicted to sponge miR-150-5p. It was shown in colon tissue that a similar circular transcript (circZNF609), derived from the same gene locus, regulates this miRNA [55]. Adding up the fact that miR-150 is up-regulated in left ventricular (LV) remodeling after acute myocardial infarction (AMI) [56], it is predicted that MICRA is able to regulate this miRNA; however, further investigation should be performed [54]. Moreover, circZNF609 was first identified to control skeletal myoblast proliferation in a knock-down-based study for the screening of circRNAs in Duchenne muscular dystrophy myoblasts, and it is originated from exon 2 of the ZNF609 gene locus [57]. In this study, circZNF609 was overexpressed in endothelial cells from diabetes mellitus, hypertension, and coronary artery disease (CAD) patients, and was associated with increased angiogenesis [58]. On the other, circZNF609 was proposed as a potential miRNA sponge by interacting with miR-615, a negative regulator of the myocyte enhancer factor 2A (MEF2A), a transcription factor largely associated with CAD and MI [59]. In fact, in hypoxic and high glucose content conditions, circZNF609 was found to be up-regulated, leading to tube formation, migration of endothelial cells, and triggering programmed cell death. Conversely, the silencing of circZNF609 reduced the effects on endothelial damage [58,60].
2.1.9. circ_000595 and circ_0010729
CircRNAs have been implicated in hypoxia-induced angiogenic events by sponging miRNAs. In an early study, hsa_circ_000595 RNA was associated with the increase of apoptosis in human aortic smooth muscle cells. Data showed that hsa_circ_000595 could target miR-19a and overexpression of the circRNA resulted in miR-19a down-regulation [61].
More recently, the circRNA hsa_circ_0010729 was described as a vascular cell apoptosis suppressor by inhibiting the activity of miR-186 [62]. In human umbilical vein endothelial cells, miR-186 appears to act as a pro-apoptotic factor down-regulating the activity of the hypoxia-inducible factor 1 alpha (HIF-1α), an anti-stress protein involved in angiogenesis under hypoxic conditions. CircRNA hsa_circ_0010729 could reverse the effects of hypoxia by regulating the vascular endothelial cell proliferation [62].
2.1.10. circDLGAP4 and circHECTD1
Ischemic stroke events and other cerebral ischemic diseases are responsible for significant and serious impairment in the brain, with increased morbidity and mortality. Rising research on circRNAs has shown that these molecules are enriched in multiple organs and are highly expressed in the brain, suggesting that circRNAs may play an important role in regulating neurophysiological and pathophysiological processes [63].
Recent research provides evidence that the circular RNA circDLGAP4, originated from the back-splicing of exons 8 to 10 of the DLGAP4 gene, may be implicated in the physiology of ischemic stroke [64]. CircDLGAP4 was found to be down-regulated in acute ischemic stroke patients and in a transient middle cerebral artery occlusion (tMCAO) mouse model. It was also demonstrated that circDLGAP4 could sponge miR-143, which can negatively modulate the expression of HECT domain E3 ubiquitin-protein ligase 1 (HECTD1). Noteworthy, it was reported that overexpression of circDLGAP4 could impact the reduction of neurological deficits and improve the damage of the blood–brain barrier, depicting the role of circDLGAP4 ischemic stroke [64].
The HECTD1 locus is related to ischemic stroke development and regulation through the formation of the circular RNA circHECTD1 that contains exons 23 and 24 of the HECTD1 gene [65]. CircHECTD1 was found to be associated with down-regulation of miR-142 expression in tMCAO, in human glioblastoma A172 cells treated with oxygen–glucose deprivation–reperfusion, and in acute ischemic stroke patients. This modulation led to astrocyte activation through the expression of autophagic proteins, leading to cerebral infarction. Pieces of evidence showed that TCDD-inducible poly (ADP-ribose) polymerase (TIPARP), an enzyme linked to ischemic stroke, could be a target for miR-142. In this setting, the down-regulation of circHECTD1 was correlated with lower levels of TIPARP, resulting in a lower risk for brain infarction [65].
2.2. Interaction of circRNAs with RNA-Binding Proteins
Recent studies started to show that circRNAs may also interact and modulate the cellular roles of many RNA-binding proteins (RBPs). These interactions are very complex, as different RBP/circRNA interactions could result in diverse regulatory events. However, two main circRNA/RBP interactions can be distinguished: RBP sponging and RBP scaffolding [2,41,66].
The RBP sponging function is very similar to the previously described miRNA sponging function. Essentially, some circRNAs contain RNA-protein binding sites that present high-affinity to certain RBPs that prevent their availability and consequent roles in the cell. CircRNA/protein interactions started to be described in 2014. Drosophila melanogaster muscleblind protein (MBL) was the first RBP found to strongly regulate circRNA synthesis from the Mbl locus and was also the first protein noticed to undergo RBP/circRNA sponging. Cellular MBL protein abundance is responsible for its own pre-mRNA back-splicing and circularization. This process prevents the formation of a linear transcript and consequent MBL protein formation and accumulation, in a negative feedback mechanism. More interesting is the fact that mature circMbl molecules contain many MBL binding sites with the ability to intake these proteins and balance their cellular amount [12].
Though many circRNAs act as RBP sponges, many other circRNAs own the ability to modulate protein function by acting as scaffolds that stabilize and enable protein–protein interactions. For instance, it was previously reported that circFoxo3, generated from the forkhead box-O 3 (Foxo3) gene, acts as a sponge for certain transcription factors and antisenescence proteins but also functions as a scaffold for p21 protein. circFoxo3 contains p21 binding sites and the interaction they establish facilitates the communication between p21 and Cdk2 protein, triggering the chain of events that prevent cell cycle progression [42].
Currently, only a limited number of circRNA/RBPs interactions have been described in the context of cardiovascular pathophysiology; however, a growing number of evidence has been reported through the last few years (Table 2).

Table 2.
circRNAs involved in interactions with RNA-binding proteins and their roles in cardiovascular diseases.
2.2.1. RBP Sponging by circRNAs
Transcription of the Foxo3 gene is responsible for the generation of either linear or circRNA transcripts. The linear form is translated into Foxo3 protein, an important transcription factor involved in cell cycles, apoptosis, and autophagy processes. The levels of the circRNA circFoxo3 originated from the Foxo3 gene are higher in the hearts of older mice and humans and are responsible for controlling cell senescence. An experiment using a model of doxorubicin-induced cardiomyopathy mice demonstrated that overexpression of circFoxo3 promotes heart cell senescence. Although circFoxo3 was not shown to interact with miRNAs, it contains canonical protein binding sites that can sponge the anti-stress transcription factors ID1 (DNA-binding protein inhibitor ID-1) and E2F1 (E2F transcription factor 1) and the antisenescence proteins HIF1α (hypoxia-inducible factor 1-alpha) and FAK (focal adhesion kinase 1). The interaction between these molecules results in the retention of these proteins in the cytoplasm. The decrease of ID1, E2F1, and HIF1α in the nucleus, and FAK in the mitochondria, is responsible for cellular stress and posterior senescence (Figure 2) [41].
The CDKN2B-AS gene, located in the chromosome 9p21.3, contains the information for the antisense (long) noncoding RNA in the INK4 locus, abbreviated as ANRIL. The same locus may produce a circular RNA form, circANRIL, which is associated with atherosclerotic vascular disease (ASVD). It was shown in HEK-293 cells that this circRNA interacts with the pescadillo homolog (PES1) protein. PES1 is a component of the PeBow complex and is responsible for the maturation and formation of the 60S ribosome subunit, being crucial in cell cycle progression. In atherosclerotic plaques, PES1 sponging by circANRIL has consequences in cell proliferation suppression and a consequent increase of cell apoptosis in proliferating cells. Thus, circANRIL is responsible for atheroprotection [67,68].
2.2.2. circRNA Scaffolds
CircAmotl1 was shown to suppress apoptosis events in a doxorubicin-induced cardiomyopathy mouse model. In the cytoplasm of cardiomyocytes, circAmotl1 acts as a scaffold facilitating the interaction between the serine/threonine kinase AKT and the 3-phosphoinositide-dependent protein kinase 1 (PDK1). PDK1 phosphorylates AKT trigger the translocation of phosphorylated AKT (pAKT) into the nucleus. In the nucleus, pAKT participates in the regulation of cell survival processes. An experiment using antisense oligonucleotides for the AKT and PDK1 binding sites showed that the amount of dephosphorylated AKT in the cytoplasm was increased and therefore it could not be translocated into the nucleus. Furthermore, the use of ribonuclease A was enough to suppress the interaction between AKT and PDK1. These experiments suggested that circAmotl1 is a crucial “platform” for AKT activation and translocation into the nucleus and protects the heart from cardiomyopathy (Figure 2). Interestingly, human circAmotl1 only differs from rodent circAmotl1 due to the sequences present in their exon junctions [43].
2.2.3. Other circRNA-Mediated Interactions
The titin (TTN) gene is the largest coding gene found in mammals. The RNA that results from its transcription may contain up to 363 exons [69]. Some of the resultant TTN isoforms are crucial in muscle contraction, regulation of calcium content in the muscle fiber, and cell cycle upkeep [69]. The TTN gene may also produce circular RNA forms [70]. However, while most RNA circles are single-exon, circTTN isoforms are very long and may contain up to 153 exons [38]. Like their linear counterparts, different circTTN secondary structures are yet to be described. circTTN 105–111 is a downstream effector of quaking isoform 5 (QKI5). QKI5 and other QKI isoforms suppress I/R-induced apoptosis and attenuate cellular senescence in doxorubicin-induced cardiomyocytes. As such, lower levels of circTTN 105-111 are responsible for a significant decrease in active QKI5 levels and cells are more prone to senescence. In short, although the circTTN 105–111 effect on QKI5 is still not very well understood, it plays a cardioprotective role alongside this RBP [71].
3. CircRNAs in Biofluids as Biomarkers of Cardiovascular Diseases
NcRNAs and other nucleic acids are actively secreted by cells, being present in all body biofluids and offering their potential use as disease biomarkers. The biochemical nature of ncRNAs offer better stability and flexible sample storage conditions, and increased sensitivity when compared with classical biomarkers [72]. Previous studies were successful to show the importance of circulating circRNAs as disease biomarkers and their potential uses as biological parameters that could be correlated with the onset, progression, or therapeutic response of cardiovascular diseases [54,73,74]. Detection and quantification of circRNAs in biofluids have been routinely performed either by hybridization microarrays for screening purposes, or by RT-qPCR with the use of divergent primers for individual quantification. Next-generation sequencing techniques are also starting to be applied but with a more limited extent when compared with microarrays or qPCR since they are dependent on the quality of the RNA samples and more expensive [75]. Due to their continuous covalently closed structure and high resistance to exoribonucleases, circRNAs are highly stable molecules in comparison with other RNAs that can be detected in biofluids such as miRNAs. In fact, some extracellular circRNA have been proposed and as possible disease biomarkers by using small cohorts of patients [8,54,75,76]. For instance, circulating hsa_circ_0001785 has been proposed as a detection biomarker for breast cancer [77] and hsa_circ_0124644 could be employed as a diagnosis biomarker for coronary artery disease (CAD) [76]. However, the clinical implementation of circulating RNAs, including circRNAs, as biomarkers in the medical practice of cardiovascular diseases is still in its infancy mainly due to technical limitations and lack of validation in extended patient cohorts.
3.1. MICRA
MICRA, previously mentioned in the Section 2.1.8 as being transcribed from the first exon of the ZNF609 gene, was found in peripheral blood samples of acute myocardial infarction (AMI) patients. Two recent reports emphasized the possibility of using MICRA from blood as a biomarker to infer on left ventricular (LV) dysfunction and remodeling four months after AMI. MICRA’s blood levels were shown to be low in a cohort of MI patients when compared to healthy volunteers. Salgado-Somoza et al. also reported that lower levels of MICRA in the blood of patients at reperfusion time will present a reduced heart ejection fraction and that higher levels of blood MICRA are indicative of a mid-range or preserved ejection fraction [54,78].
3.2. circRNA_081881
Another circRNA that may be used to diagnose AMI is circRNA_081881. This molecule is down-regulated in the plasma of AMI patients alongside the peroxisome proliferator-activated receptor gamma (PPARγ) transcription factor. Deng et al. [73] performed siRNA-mediated knockdown assays for circRNA_081881 and found a decrease of blood PPARγ, suggesting that circRNA_081881 is directly related to PPARγ expression. In fact, the same group found seven MREs for miR-548, PPARγ being one of its targets [73].
4. Conclusions and Further Perspectives
The dynamic output of the human genome in the form of RNA transcripts contains a wide range of molecular species with different functions. The non-coding transcriptome constitutes by far the more diverse part of the genomic output, harboring regulatory ncRNAs that can control the information flow from the coding genome. The intricated functional relationships established between the different species belonging to the non-coding transcriptome are starting to be characterized in cell biology, but also in human disease. Among the ncRNAs, lncRNAs and circRNAs are very diversified in their functions due to their abilities to interact with different biomolecules including DNA, RNA, and proteins. In cardiovascular diseases, circRNAs have been characterized as important players in the control of the cellular levels of miRNAs and RNA-binding proteins, with different functional and pathological consequences ranging to direct involvement in pathology to protection from a cardiovascular condition.
Despite of the importance of circRNAs and the established regulatory networks, the availability of data about their roles in cardiovascular diseases is still limited due to the lack of knowledge about the rules governing their biogenesis from splicing events, the limitations of laboratory models, and also the lack of consensus within the scientific community in circRNA annotation and databases. A stronger effort is required to circumvent these limitations, since the central position of circRNAs in the regulatory events leading to some cardiovascular conditions may lead to potential new therapeutic and diagnostic strategies for better healthcare.
Funding
This work is supported by COST (European Cooperation in Science and Technology) Action EU-CardioRNA CA17129 and Portuguese Foundation for Science and Technology (FCT) under the framework of the research grant PTDC-MED-GEN-29389-2017.
Acknowledgments
The authors would like to acknowledge Francisco Enguita Jr. for excellent technical assistance and support for designing of the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest. The funders have no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Diener, T.O. Potato spindle tuber “virus”. IV. A replicating, low molecular weight RNA. Virology 1971, 45, 411–428. [Google Scholar] [CrossRef]
- Symons, R.H. The intriguing viroids and virusoids: What is their information content and how did they evolve? Mol. Plant Microbe Interact. 1991, 4, 111–121. [Google Scholar] [CrossRef]
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef]
- Cocquerelle, C.; Mascrez, B.; Hetuin, D.; Bailleul, B. Mis-splicing yields circular RNA molecules. FASEB J. 1993, 7, 155–160. [Google Scholar] [CrossRef]
- Hsu, M.T.; Coca-Prados, M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 1979, 280, 339–340. [Google Scholar] [CrossRef]
- Maass, P.G.; Glazar, P.; Memczak, S.; Dittmar, G.; Hollfinger, I.; Schreyer, L.; Sauer, A.V.; Toka, O.; Aiuti, A.; Luft, F.C.; et al. A map of human circular RNAs in clinically relevant tissues. J. Mol. Med. 2017, 95, 1179–1189. [Google Scholar] [CrossRef]
- Guo, J.U.; Agarwal, V.; Guo, H.; Bartel, D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014, 15, 409. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Liang, D.; Wilusz, J.E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014, 28, 2233–2247. [Google Scholar] [CrossRef] [PubMed]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.O.; Dong, R.; Zhang, Y.; Zhang, J.L.; Luo, Z.; Zhang, J.; Chen, L.L.; Yang, L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016, 26, 1277–1287. [Google Scholar] [CrossRef] [PubMed]
- Salzman, J.; Chen, R.E.; Olsen, M.N.; Wang, P.L.; Brown, P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013, 9, e1003777. [Google Scholar] [CrossRef]
- Bachmayr-Heyda, A.; Reiner, A.T.; Auer, K.; Sukhbaatar, N.; Aust, S.; Bachleitner-Hofmann, T.; Mesteri, I.; Grunt, T.W.; Zeillinger, R.; Pils, D. Correlation of circular RNA abundance with proliferation-exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci. Rep. 2015, 5, 8057. [Google Scholar] [CrossRef]
- Enuka, Y.; Lauriola, M.; Feldman, M.E.; Sas-Chen, A.; Ulitsky, I.; Yarden, Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016, 44, 1370–1383. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.O.; Chen, T.; Xiang, J.F.; Yin, Q.F.; Xing, Y.H.; Zhu, S.; Yang, L.; Chen, L.L. Circular intronic long noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef]
- Talhouarne, G.J.; Gall, J.G. Lariat intronic RNAs in the cytoplasm of Xenopus tropicalis oocytes. RNA 2014, 20, 1476–1487. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, K.; Panda, A.C.; Munk, R.; Grammatikakis, I.; Dudekula, D.B.; De, S.; Kim, J.; Noh, J.H.; Kim, K.M.; Martindale, J.L.; et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017, 14, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, L.; Wang, Y.; Ding, H.; Xue, S.; Qi, H.; Li, P. MicroRNAs or Long Noncoding RNAs in Diagnosis and Prognosis of Coronary Artery Disease. Aging Dis. 2019, 10, 353–366. [Google Scholar] [CrossRef] [PubMed]
- Thum, T.; Condorelli, G. Long noncoding RNAs and microRNAs in cardiovascular pathophysiology. Circ. Res. 2015, 116, 751–762. [Google Scholar] [CrossRef]
- Rotini, A.; Martinez-Sarra, E.; Pozzo, E.; Sampaolesi, M. Interactions between microRNAs and long non-coding RNAs in cardiac development and repair. Pharmacol. Res. 2018, 127, 58–66. [Google Scholar] [CrossRef]
- Aufiero, S.; Reckman, Y.J.; Pinto, Y.M.; Creemers, E.E. Circular RNAs open a new chapter in cardiovascular biology. Nat. Rev. Cardiol. 2019, 16, 503–514. [Google Scholar] [CrossRef]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar] [CrossRef]
- Tay, Y.; Kats, L.; Salmena, L.; Weiss, D.; Tan, S.M.; Ala, U.; Karreth, F.; Poliseno, L.; Provero, P.; Di Cunto, F.; et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 2011, 147, 344–357. [Google Scholar] [CrossRef]
- Geng, H.H.; Li, R.; Su, Y.M.; Xiao, J.; Pan, M.; Cai, X.X.; Ji, X.P. The Circular RNA Cdr1as Promotes Myocardial Infarction by Mediating the Regulation of miR-7a on Its Target Genes Expression. PLoS ONE 2016, 11, e0151753. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Dudekula, D.B.; Panda, A.C.; Grammatikakis, I.; De, S.; Abdelmohsen, K.; Gorospe, M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016, 13, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.C.; Li, J.R.; Sun, C.H.; Andrews, E.; Chao, R.F.; Lin, F.M.; Weng, S.L.; Hsu, S.D.; Huang, C.C.; Cheng, C.; et al. CircNet: A database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2016, 44, D209–D215. [Google Scholar] [CrossRef] [PubMed]
- Ghosal, S.; Das, S.; Sen, R.; Basak, P.; Chakrabarti, J. Circ2Traits: A comprehensive database for circular RNA potentially associated with disease and traits. Front. Genet. 2013, 4, 283. [Google Scholar] [CrossRef] [PubMed]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef] [PubMed]
- Kulcheski, F.R.; Christoff, A.P.; Margis, R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J. Biotechnol. 2016, 238, 42–51. [Google Scholar] [CrossRef]
- Wang, P.L.; Bao, Y.; Yee, M.C.; Barrett, S.P.; Hogan, G.J.; Olsen, M.N.; Dinneny, J.R.; Brown, P.O.; Salzman, J. Circular RNA is expressed across the eukaryotic tree of life. PLoS ONE 2014, 9, e90859. [Google Scholar] [CrossRef]
- Wang, K.; Long, B.; Liu, F.; Wang, J.X.; Liu, C.Y.; Zhao, B.; Zhou, L.Y.; Sun, T.; Wang, M.; Yu, T.; et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016, 37, 2602–2611. [Google Scholar] [CrossRef]
- Tan, W.L.; Lim, B.T.; Anene-Nzelu, C.G.; Ackers-Johnson, M.; Dashi, A.; See, K.; Tiang, Z.; Lee, D.P.; Chua, W.W.; Luu, T.D.; et al. A landscape of circular RNA expression in the human heart. Cardiovasc. Res. 2017, 113, 298–309. [Google Scholar] [CrossRef]
- Deng, P.; Chen, L.; Liu, Z.; Ye, P.; Wang, S.; Wu, J.; Yao, Y.; Sun, Y.; Huang, X.; Ren, L.; et al. MicroRNA-150 Inhibits the Activation of Cardiac Fibroblasts by Regulating c-Myb. Cell. Physiol. Biochem. 2016, 38, 2103–2122. [Google Scholar] [CrossRef]
- Liu, Z.; Ye, P.; Wang, S.; Wu, J.; Sun, Y.; Zhang, A.; Ren, L.; Cheng, C.; Huang, X.; Wang, K.; et al. MicroRNA-150 protects the heart from injury by inhibiting monocyte accumulation in a mouse model of acute myocardial infarction. Circ. Cardiovasc. Genet. 2015, 8, 11–20. [Google Scholar] [CrossRef]
- Du, W.W.; Yang, W.N.; Chen, Y.; Wu, Z.K.; Foster, F.S.; Yang, Z.G.; Li, X.M.; Yang, B.B. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 2017, 38, 1402–1412. [Google Scholar] [CrossRef] [PubMed]
- Du, W.W.; Yang, W.; Liu, E.; Yang, Z.; Dhaliwal, P.; Yang, B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016, 44, 2846–2858. [Google Scholar] [CrossRef]
- Zeng, Y.; Du, W.W.; Wu, Y.Y.; Yang, Z.G.; Awan, F.M.; Li, X.M.; Yang, W.N.; Zhang, C.; Yang, Q.; Yee, A.; et al. A Circular RNA Binds To and Activates AKT Phosphorylation and Nuclear Localization Reducing Apoptosis and Enhancing Cardiac Repair. Theranostics 2017, 7, 3842–3855. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zhang, M.; Zheng, X.; Yi, P.; Lan, C.; Xu, M. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 2017, 143, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Li, R.; Zhang, C.; Bian, H.J.; Wang, F.; Xiao, J.; Liu, S.W.; Yi, W.; Zhang, M.X.; Wang, S.X.; et al. MicroRNA-7a/b protects against cardiac myocyte injury in ischemia/reperfusion by targeting poly(ADP-ribose) polymerase. PLoS ONE 2014, 9, e90096. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Gan, T.Y.; Li, N.; Liu, C.Y.; Zhou, L.Y.; Gao, J.N.; Chen, C.; Yan, K.W.; Ponnusamy, M.; Zhang, Y.H.; et al. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 2017, 24, 1111–1120. [Google Scholar] [CrossRef]
- Tondera, D.; Czauderna, F.; Paulick, K.; Schwarzer, R.; Kaufmann, J.; Santel, A. The mitochondrial protein MTP18 contributes to mitochondrial fission in mammalian cells. J. Cell Sci. 2005, 118, 3049–3059. [Google Scholar] [CrossRef]
- Tondera, D.; Santel, A.; Schwarzer, R.; Dames, S.; Giese, K.; Klippel, A.; Kaufmann, J. Knockdown of MTP18, a novel phosphatidylinositol 3-kinase-dependent protein, affects mitochondrial morphology and induces apoptosis. J. Biol. Chem. 2004, 279, 31544–31555. [Google Scholar] [CrossRef]
- Divakaran, V.; Mann, D.L. The emerging role of microRNAs in cardiac remodeling and heart failure. Circ. Res. 2008, 103, 1072–1083. [Google Scholar] [CrossRef]
- Li, M.; Ding, W.; Tariq, M.A.; Chang, W.; Zhang, X.; Xu, W.; Hou, L.; Wang, Y.; Wang, J. A circular transcript of ncx1 gene mediates ischemic myocardial injury by targeting miR-133a-3p. Theranostics 2018, 8, 5855–5869. [Google Scholar] [CrossRef]
- Lim, T.B.; Aliwarga, E.; Luu, T.D.A.; Li, Y.P.; Ng, S.L.; Annadoray, L.; Sian, S.; Ackers-Johnson, M.A.; Foo, R.S. Targeting the highly abundant circular RNA circSlc8a1 in cardiomyocytes attenuates pressure overload induced hypertrophy. Cardiovasc. Res. 2019, 115, 1998–2007. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.M.; Zhang, M.; Huang, L.; Hu, Z.Q.; Zhu, J.N.; Xiao, Z.; Zhang, Z.; Lin, Q.X.; Zheng, X.L.; Yang, M.; et al. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci. Rep. 2017, 7, 40342. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Yu, J.W. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-beta1. Biochem. Biophys. Res. Commun. 2017, 487, 769–775. [Google Scholar] [CrossRef] [PubMed]
- Salgado-Somoza, A.; Zhang, L.; Vausort, M.; Devaux, Y. The circular RNA MICRA for risk stratification after myocardial infarction. Int. J. Cardiol. Heart Vasc. 2017, 17, 33–36. [Google Scholar] [CrossRef]
- Peng, L.; Chen, G.; Zhu, Z.; Shen, Z.; Du, C.; Zang, R.; Su, Y.; Xie, H.; Li, H.; Xu, X.; et al. Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprung’s disease. Oncotarget 2017, 8, 808–818. [Google Scholar]
- Devaux, Y.; Vausort, M.; McCann, G.P.; Zangrando, J.; Kelly, D.; Razvi, N.; Zhang, L.; Ng, L.L.; Wagner, D.R.; Squire, I.B. MicroRNA-150: A novel marker of left ventricular remodeling after acute myocardial infarction. Circ. Cardiovasc. Genet. 2013, 6, 290–298. [Google Scholar] [CrossRef]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37. [Google Scholar] [CrossRef]
- Boeckel, J.N.; Jae, N.; Heumuller, A.W.; Chen, W.; Boon, R.A.; Stellos, K.; Zeiher, A.M.; John, D.; Uchida, S.; Dimmeler, S. Identification and Characterization of Hypoxia-Regulated Endothelial Circular RNA. Circ. Res. 2015, 117, 884–890. [Google Scholar] [CrossRef]
- Wang, J.; Liu, L.; Sun, Y.; Xue, Y.; Qu, J.; Pan, S.; Li, H.; Qu, H.; Wang, J.; Zhang, J. miR-615-3p promotes proliferation and migration and inhibits apoptosis through its potential target CELF2 in gastric cancer. Biomed. Pharmacother. 2018, 101, 406–413. [Google Scholar] [CrossRef]
- Liu, C.; Yao, M.D.; Li, C.P.; Shan, K.; Yang, H.; Wang, J.J.; Liu, B.; Li, X.M.; Yao, J.; Jiang, Q.; et al. Silencing Of Circular RNA-ZNF609 Ameliorates Vascular Endothelial Dysfunction. Theranostics 2017, 7, 2863–2877. [Google Scholar] [CrossRef]
- Zheng, C.; Niu, H.; Li, M.; Zhang, H.; Yang, Z.; Tian, L.; Wu, Z.; Li, D.; Chen, X. Cyclic RNA hsacirc000595 regulates apoptosis of aortic smooth muscle cells. Mol. Med. Rep. 2015, 12, 6656–6662. [Google Scholar] [CrossRef] [PubMed]
- Dang, R.Y.; Liu, F.L.; Li, Y. Circular RNA hsa_circ_0010729 regulates vascular endothelial cell proliferation and apoptosis by targeting the miR-186/HIF-1alpha axis. Biochem. Biophys. Res. Commun. 2017, 490, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Rybak-Wolf, A.; Stottmeister, C.; Glazar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol. Cell 2015, 58, 870–885. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Zhang, Y.; Han, B.; Yang, L.; Chen, X.; Huang, R.; Wu, F.; Chao, J.; Liu, P.; Hu, G.; et al. Circular RNA DLGAP4 Ameliorates Ischemic Stroke Outcomes by Targeting miR-143 to Regulate Endothelial-Mesenchymal Transition Associated with Blood-Brain Barrier Integrity. J. Neurosci. 2018, 38, 32–50. [Google Scholar]
- Han, B.; Zhang, Y.; Zhang, Y.; Bai, Y.; Chen, X.; Huang, R.; Wu, F.; Leng, S.; Chao, J.; Zhang, J.H.; et al. Novel insight into circular RNA HECTD1 in astrocyte activation via autophagy by targeting MIR142-TIPARP: Implications for cerebral ischemic stroke. Autophagy 2018, 14, 1164–1184. [Google Scholar] [CrossRef]
- Greene, J.; Baird, A.M.; Brady, L.; Lim, M.; Gray, S.G.; McDermott, R.; Finn, S.P. Circular RNAs: Biogenesis, Function and Role in Human Diseases. Front. Mol. Biosci. 2017, 4, 38. [Google Scholar] [CrossRef]
- Burd, C.E.; Jeck, W.R.; Liu, Y.; Sanoff, H.K.; Wang, Z.; Sharpless, N.E. Expression of Linear and Novel Circular Forms of an INK4/ARF-Associated Non-Coding RNA Correlates with Atherosclerosis Risk. PLoS Genet. 2010, 6, e1001233. [Google Scholar] [CrossRef] [PubMed]
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.H.; Nicolaou, A.; et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016, 7, 12429. [Google Scholar] [CrossRef] [PubMed]
- Bang, M.L.; Centner, T.; Fornoff, F.; Geach, A.J.; Gotthardt, M.; McNabb, M.; Witt, C.C.; Labeit, D.; Gregorio, C.C.; Granzier, H.; et al. The complete gene sequence of titin, expression of an unusual approximately 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ. Res. 2001, 89, 1065–1072. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Reckman, Y.J.; Aufiero, S.; van den Hoogenhof, M.M.; van der Made, I.; Beqqali, A.; Koolbergen, D.R.; Rasmussen, T.B.; van der Velden, J.; Creemers, E.E.; et al. RBM20 Regulates Circular RNA Production From the Titin Gene. Circ. Res. 2016, 119, 996–1003. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Garg, A.; Bar, C.; Chatterjee, S.; Foinquinos, A.; Milting, H.; Streckfuss-Bomeke, K.; Fiedler, J.; Thum, T. Quaking Inhibits Doxorubicin-Mediated Cardiotoxicity through Regulation of Cardiac Circular RNA Expression. Circ. Res. 2018, 122, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Stepien, E.; Costa, M.C.; Kurc, S.; Drozdz, A.; Cortez-Dias, N.; Enguita, F.J. The circulating non-coding RNA landscape for biomarker research: Lessons and prospects from cardiovascular diseases. Acta Pharm. Sin. 2018, 39, 1085–1099. [Google Scholar]
- Deng, Y.; Zhang, W.; She, J.; Zhang, L.; Chen, T.; Zhou, J.; Yuan, Z. GW27-e1167 Circular RNA Related to PPARγ Function as ceRNA of microRNA in Human Acute Myocardial Infarction. J. Am. Coll. Cardiol. 2016, 68, 51–52. [Google Scholar] [CrossRef]
- Bazan, H.A.; Hatfield, S.A.; Brug, A.; Brooks, A.J.; Lightell, D.J., Jr.; Woods, T.C. Carotid Plaque Rupture Is Accompanied by an Increase in the Ratio of Serum circR-284 to miR-221 Levels. Circ. Cardiovasc. Genet. 2017, 10, e001720. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, T.; Xiao, J. Circular RNAs: Promising Biomarkers for Human Diseases. EBioMedicine 2018, 34, 267–274. [Google Scholar] [CrossRef]
- Zhao, Z.; Li, X.; Gao, C.; Jian, D.; Hao, P.; Rao, L.; Li, M. Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci. Rep. 2017, 7, 39918. [Google Scholar] [CrossRef]
- Yin, W.B.; Yan, M.G.; Fang, X.; Guo, J.J.; Xiong, W.; Zhang, R.P. Circulating circular RNA hsa_circ_0001785 acts as a diagnostic biomarker for breast cancer detection. Clin. Chim. Acta 2018, 487, 363–368. [Google Scholar] [CrossRef]
- Vausort, M.; Salgado-Somoza, A.; Zhang, L.; Leszek, P.; Scholz, M.; Teren, A.; Burkhardt, R.; Thiery, J.; Wagner, D.R.; Devaux, Y. Myocardial Infarction-Associated Circular RNA Predicting Left Ventricular Dysfunction. J. Am. Coll. Cardiol. 2016, 68, 1247–1248. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).