The Mitochondrial Outer Membrane Protein Tom70-Mediator in Protein Traffic, Membrane Contact Sites and Innate Immunity
Abstract
1. Introduction
2. Molecular Structure of Tom70
2.1. The Structure of Monomeric Tom70
2.2. Putative Preprotein Binding Sites
2.3. The Oligomeric State of Tom70
3. Evolution of Tom70
3.1. Tom70 and Its Homologs
3.2. The TPR Domain of Tom70 and Its Functional Analogs in Other Membranes
4. Functions of Tom70 in the Biogenesis of Mitochondrial Proteins
4.1. Tom70 and Chaperone Proteins
4.2. Tom70 and Targeting Signals
4.3. Tom70 and Co-Translational Protein Import
4.4. Mitochondrial Protein Import in Cooperation with the Endoplasmic Reticulum
5. Functions of Tom70 as a Mitochondrial Tether
5.1. Tom70 and the Endoplasmic Reticulum Protein Lam6/Ltc1
5.2. Additional Functions of Tom70
6. TOM70 in Health and Disease
6.1. A Role of TOM70 in Macrophages Infected by Leishmania Donovali
6.2. MAVS and RIG-I: TOM70 as a Mediator of an Innate Immune Response to Viral Infections
6.3. TOM70 in the Regulation of Mitochondrial Functions and ROS Formation in Heart Cells
6.4. Functions of TOM70 in Mitochondrial Quality Control
6.5. Mitochondriopathies Caused by Mutations in TOMM70
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AAC | ADP/ATP carrier |
| ADP | Adenosine diphosphate |
| ATP | Adenosine triphosphate |
| BN-PAGE | Blue native-polyacrylamide gel electrophoresis |
| ER | Endoplasmic reticulum |
| Hsp | Heat shock protein |
| IFN-I | Interferon type I |
| IMS | Intermembrane space |
| IP3R3 | Inositol 1,3,4-triphosphate receptor type 3 |
| Lam6 | Lipid transfer protein anchored at a membrane contact site |
| Ltc1 | Lipid transfer at contact site 1 |
| MAVS | Mitochondrial antiviral signaling protein |
| MCF | Mitochondrial carrier family |
| MCL-1 | Myeloid cell Leukemia 1 |
| Mfb1 | Mitochondria-associated F-box protein 1 |
| MIM | Mitochondrial inner membrane |
| MIM complex | Mitochondrial import complex |
| MOM | Mitochondrial outer membrane |
| RIG-I | Retinoic acid-inducible gene 1 |
| ROS | Reactive oxygen species |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| SEN | tRNA splicing endonuclease |
| SLC25 | Solute carrier family 25 |
| TIM | Translocase of the inner mitochondrial membrane |
| TOM | Translocase of the outer mitochondrial membrane |
| TOM70 | Mammalian homolog of yeast Tom70 |
References
- Ernster, L.; Schatz, G. Mitochondria: A Historical Review. J. Cell Biol. 1981, 91, 227–255. [Google Scholar] [CrossRef] [PubMed]
- Pfanner, N.; Warscheid, B.; Wiedemann, N. Mitochondrial proteins: From biogenesis to functional networks. Nat. Rev. Mol. Cell Biol. 2019, 20, 267–284. [Google Scholar] [CrossRef]
- Neupert, W.; Herrmann, J.M. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 2007, 76, 723–749. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, N.; Pfanner, N. Mitochondrial Machineries for Protein Import and Assembly. Annu. Rev. Biochem. 2017, 86, 685–714. [Google Scholar] [CrossRef]
- Pfanner, N.; Douglas, M.G.; Endo, T.; Hoogenraad, N.J.; Jensen, R.E.; Meijer, M.; Neupert, W.; Schatz, G.; Schmitz, U.K.; Shore, G.C. Uniform nomenclature for the protein transport machinery of the mitochondrial membranes. Trends Biochem. Sci. 1996, 21, 51–52. [Google Scholar] [CrossRef]
- Vögtle, F.N.; Burkhart, J.M.; Gonczarowska-Jorge, H.; Kücükköse, C.; Taskin, A.A.; Kopczynski, D.; Ahrends, R.; Mossmann, D.; Sickmann, A.; Zahedi, R.P.; et al. Landscape of submitochondrial protein distribution. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef]
- Zahedi, R.P.; Sickmann, A.; Boehm, A.M.; Winkler, C.; Zufall, N.; Schönfisch, B.; Guiard, B.; Pfanner, N.; Meisinger, C. Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Mol. Biol. Cell 2006, 17, 1436–1450. [Google Scholar] [CrossRef] [PubMed]
- Morgenstern, M.; Stiller, S.B.; Lübbert, P.; Peikert, C.D.; Dannenmaier, S.; Drepper, F.; Weill, U.; Höß, P.; Feuerstein, R.; Gebert, M.; et al. Definition of a High-Confidence Mitochondrial Proteome at Quantitative Scale. Cell Rep. 2017, 19, 2836–2852. [Google Scholar] [CrossRef]
- Riezman, H.; Hase, T.; van Loon, A.P.; Grivell, L.A.; Suda, K.; Schatz, G. Import of proteins into mitochondria: A 70 kilodalton outer membrane protein with a large carboxy-terminal deletion is still transported to the outer membrane. EMBO J. 1983, 2, 2161–2168. [Google Scholar] [CrossRef]
- Hase, T.; Riezman, H.; Suda, K.; Schatz, G. Import of proteins into mitochondria: Nucleotide sequence of the gene for a 70-kd protein of the yeast mitochondrial outer membrane. EMBO J. 1983, 2, 2169–2172. [Google Scholar] [CrossRef]
- Hase, T.; Müller, U.; Riezman, H.; Schatz, G. A 70-kd protein of the yeast mitochondrial outer membrane is targeted and anchored via its extreme amino terminus. EMBO J. 1984, 3, 3157–3164. [Google Scholar] [CrossRef] [PubMed]
- Söllner, T.; Pfaller, R.; Griffiths, G.; Pfanner, N.; Neupert, W. A mitochondrial import receptor for the ADP/ATP carrier. Cell 1990, 62, 107–115. [Google Scholar] [CrossRef][Green Version]
- Steger, H.E.; Sner, T.; Kiebler, M.; Dietmeier, K.A.; Pfaller, R.; Triilzsch, K.S.; Tropschug, M.; Neupert, W.; Planner, N. Import of ADP/ATP Carrier into Mitochondria: Two Receptors Act in Parallel. J. Cell Biol. 1990, 11, 2353–2363. [Google Scholar] [CrossRef] [PubMed]
- Hines, V.; Brandt, A.; Griffiths, G.; Horstmann, H.; Brütsch, H.; Schatz, G. Protein import into yeast mitochondria is accelerated by the outer membrane protein MAS70. EMBO J. 1990, 9, 3191–3200. [Google Scholar] [CrossRef]
- Dekker, P.J.T.; Müller, H.; Rassow, J.; Pfanner, N. Characterization of the preprotein translocase of the outer mitochondrial membrane by blue native electrophoresis. Biol. Chem. 1996, 377, 535–538. [Google Scholar] [CrossRef] [PubMed]
- Dekker, P.J.T.; Ryan, M.T.; Brix, J.; Müller, H.; Hönlinger, A.; Pfanner, N. Preprotein Translocase of the Outer Mitochondrial Membrane: Molecular Dissection and Assembly of the General Import Pore Complex. Mol. Cell. Biol. 1998, 18, 6515–6524. [Google Scholar] [CrossRef] [PubMed]
- Shiota, T.; Imai, K.; Qiu, J.; Hewitt, V.L.; Tan, K.; Shen, H.; Sakiyama, N.; Fukasawa, Y.; Hayat, S.; Kamiya, M.; et al. Molecular architecture of the active mitochondrial protein gate. Science 2015, 349, 3–8. [Google Scholar] [CrossRef]
- Haucke, V.; Horst, M.; Schatz, G.; Lithgow, T. The Mas20p and Mas70p subunits of the protein import receptor of yeast mitochondria interact via the tetratricopeptide repeat motif in Mas20p: Evidence for a single hetero-oligomeric receptor. EMBO J. 1996, 15, 1231–1237. [Google Scholar] [CrossRef]
- Ramage, L.; Junne, T.; Hahne, K.; Lithgow, T.; Schatz, G. Functional cooperation of mitochondrial protein import receptors in yeast. EMBO J. 1993, 12, 4115–4123. [Google Scholar] [CrossRef]
- Fan, A.C.Y.; Kozlov, G.; Hoegl, A.; Marcellus, R.C.; Wong, M.J.H.; Gehring, K.; Young, J.C. Interaction between the human mitochondrial import receptors Tom20 and Tom70 in vitro suggests a chaperone displacement mechanism. J. Biol. Chem. 2011, 286, 32208–32219. [Google Scholar] [CrossRef]
- Bausewein, T.; Mills, D.J.; Langer, J.D.; Nitschke, B.; Nussberger, S.; Kühlbrandt, W. Cryo-EM Structure of the TOM Core Complex from Neurospora crassa. Cell 2017, 170, 693–700. [Google Scholar] [CrossRef] [PubMed]
- Araiso, Y.; Tsutsumi, A.; Qiu, J.; Imai, K.; Shiota, T.; Song, J.; Lindau, C.; Wenz, L.S.; Sakaue, H.; Yunoki, K.; et al. Structure of the mitochondrial import gate reveals distinct preprotein paths. Nature 2019, 575, 395–401. [Google Scholar] [CrossRef]
- Tucker, K.; Park, E. Cryo-EM structure of the mitochondrial protein-import channel TOM complex at near-atomic resolution. Nat. Struct. Mol. Biol. 2019, 26, 1158–1166. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Sha, B. Crystal structure of yeast mitochondrial outer membrane translocon member Tom70p. Nat. Struct. Mol. Biol. 2006, 13, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Young, J.C.; Hoogenraad, N.J.; Hartl, F.U. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 2003, 112, 41–50. [Google Scholar] [CrossRef]
- Schmidt, O.; Harbauer, A.B.; Rao, S.; Eyrich, B.; Zahedi, R.P.; Stojanovski, D.; Schönfisch, B.; Guiard, B.; Sickmann, A.; Pfanner, N.; et al. Regulation of mitochondrial protein import by cytosolic kinases. Cell 2011, 144, 227–239. [Google Scholar] [CrossRef]
- Liu, X.Y.; Wei, B.; Shi, H.X.; Shan, Y.F.; Wang, C. Tom70 mediates activation of interferon regulatory factor 3 on mitochondria. Cell Res. 2010, 20, 994–1011. [Google Scholar] [CrossRef]
- Jiang, H.-W.; Zhang, H.-N.; Meng, Q.-F.; Xie, J.; Li, Y.; Chen, H.; Zheng, Y.-X.; Wang, X.-N.; Qi, H.; Zhang, J.; et al. SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cell. Mol. Immunol. 2020, 17, 998–1000. [Google Scholar] [CrossRef]
- Kreimendahl, S.; Schwichtenberg, J.; Günnewig, K.; Brandherm, L.; Rassow, J. The selectivity filter of the mitochondrial protein import machinery. 2020; submitted for publication. [Google Scholar]
- Becker, T.; Pfannschmidt, S.; Guiard, B.; Stojanovski, D.; Milenkovic, D.; Kutik, S.; Pfanner, N.; Meisinger, C.; Wiedemann, N. Biogenesis of the mitochondrial TOM complex: Mim1 promotes insertion and assembly of signal-anchored receptors. J. Biol. Chem. 2008, 283, 120–127. [Google Scholar] [CrossRef]
- Schlossmann, J.; Neupert, W. Assembly of the preprotein receptor MOM72/MAS70 into the protein import complex of the outer membrane of mitochondria. J. Biol. Chem. 1995, 270, 27116–27121. [Google Scholar] [CrossRef]
- Scheufler, C.; Brinker, A.; Bourenkov, G.; Pegoraro, S.; Moroder, L.; Bartunik, H.; Hartl, F.U.; Moarefi, I. Structure of TPR domain-peptide complexes: Critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 2000, 101, 199–210. [Google Scholar] [CrossRef]
- Bushell, S.R.; Bottomley, S.P.; Rossjohn, J.; Beddoe, T. Tracking the unfolding pathway of a multirepeat protein via tryptophan scanning: Evidence of localized instability in the mitochondrial import receptor Tom70. J. Biol. Chem. 2006, 281, 24345–24350. [Google Scholar] [CrossRef] [PubMed]
- Bömer, U.; Pfanner, N.; Dietmeier, K. Identification of a third yeast mitochondrial Tom protein with tetratrico peptide repeats. FEBS Lett. 1996, 382, 153–158. [Google Scholar] [CrossRef]
- Schlossmann, J.; Lill, R.; Neupert, W.; Court, D.A. Tom71, a novel homologue of the mitochondrial preprotein receptor Tom70. J. Biol. Chem. 1996, 271, 17890–17895. [Google Scholar] [CrossRef]
- Koh, J.Y.; Hájek, P.; Bedwell, D.M. Overproduction of PDR3 Suppresses Mitochondrial Import Defects Associated with a TOM70 Null Mutation by Increasing the Expression of TOM72 inSaccharomyces cerevisiae. Mol. Cell. Biol. 2001, 21, 7576–7586. [Google Scholar] [CrossRef]
- Li, J.; Qian, X.; Hu, J.; Sha, B. Molecular Chaperone Hsp70/Hsp90 Prepares the Mitochondrial Outer Membrane Translocon Receptor Tom71 for Preprotein Loading. J. Biol. Chem. 2009, 284, 23852–23859. [Google Scholar] [CrossRef]
- Li, J.; Cui, W.; Sha, B. The structural plasticity of Tom71 for mitochondrial precursor translocations. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 985–989. [Google Scholar] [CrossRef]
- Mills, R.D.; Trewhella, J.; Qiu, T.W.; Welte, T.; Ryan, T.M.; Hanley, T.; Knott, R.B.; Lithgow, T.; Mulhern, T.D. Domain Organization of the Monomeric Form of the Tom70 Mitochondrial Import Receptor. J. Mol. Biol. 2009, 388, 1043–1058. [Google Scholar] [CrossRef]
- Melin, J.; Kilisch, M.; Neumann, P.; Lytovchenko, O.; Gomkale, R.; Schendzielorz, A.; Schmidt, B.; Liepold, T.; Ficner, R.; Jahn, O.; et al. A presequence-binding groove in Tom70 supports import of Mdl1 into mitochondria. Biochim. Biophys. Acta-Mol. Cell Res. 2015, 1853, 1850–1859. [Google Scholar] [CrossRef]
- Brix, J.; Ziegler, G.A.; Dietmeier, K.; Schneider-Mergener, J.; Schulz, G.E.; Pfanner, N. The mitochondrial import receptor Tom70: Identification of a 25 kDa core domain with a specific binding site for preproteins. J. Mol. Biol. 2000, 303, 479–488. [Google Scholar] [CrossRef]
- Beddoe, T.; Bushell, S.R.; Perugini, M.A.; Lithgow, T.; Mulhern, T.D.; Bottomley, S.P.; Rossjohn, J. A biophysical analysis of the tetratricopeptide repeat-rich mitochondrial import receptor, Tom70, reveals an elongated monomer that is inherently flexible, unstable, and unfolds via a multistate pathway. J. Biol. Chem. 2004, 279, 46448–46454. [Google Scholar] [CrossRef] [PubMed]
- Truscott, K.N.; Wiedemann, N.; Rehling, P.; Müller, H.; Meisinger, C.; Pfanner, N.; Guiard, B. Mitochondrial Import of the ADP/ATP Carrier: The Essential TIM Complex of the Intermembrane Space Is Required for Precursor Release from the TOM Complex. Mol. Cell. Biol. 2002, 22, 7780–7789. [Google Scholar] [CrossRef] [PubMed]
- Söllner, T.; Rassow, J.; Wiedmann, M.; Schlossmann, J.; Keil, P.; Neupert, W.; Pfanner, N. Mapping of the protein import machinery in the mitochondrial outer membrane by crosslinking of translocation intermediates. Nature 1992, 355, 84–87. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wiedemann, N.; Pfanner, N.; Ryan, M.T. The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J. 2001, 20, 951–960. [Google Scholar] [CrossRef]
- Millar, D.G.; Shore, G.C. The signal anchor sequence of mitochondrial Mas70p contains an oligomerization domain. J. Biol. Chem. 1993, 268, 18403–18406. [Google Scholar] [PubMed]
- Millar, D.G.; Shore, G.C. Mitochondrial Mas70p signal anchor sequence. Mutations in the transmembrane domain that disrupt dimerization but not targeting or membrane insertion. J. Biol. Chem. 1994, 269, 12229–12232. [Google Scholar]
- Fan, A.C.Y.; Gava, L.M.; Ramos, C.H.I.; Young, J.C. Human mitochondrial import receptor Tom70 functions as a monomer. Biochem. J. 2010, 429, 553–563. [Google Scholar] [CrossRef]
- Hewitt, V.; Alcock, F.; Lithgow, T. Minor modifications and major adaptations: The evolution of molecular machines driving mitochondrial protein import. Biochim. Biophys. Acta-Biomembr. 2011, 1808, 947–954. [Google Scholar] [CrossRef]
- Zeytuni, N.; Zarivach, R. Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure 2012, 20, 397–405. [Google Scholar] [CrossRef]
- Chan, N.C.; Likić, V.A.; Waller, R.F.; Mulhern, T.D.; Lithgow, T. The C-terminal TPR Domain of Tom70 Defines a Family of Mitochondrial Protein Import Receptors Found only in Animals and Fungi. J. Mol. Biol. 2006, 358, 1010–1022. [Google Scholar] [CrossRef]
- Sokol, A.M.; Sztolsztener, M.E.; Wasilewski, M.; Heinz, E.; Chacinska, A. Mitochondrial protein translocases for survival and wellbeing. FEBS Lett. 2014, 588, 2484–2495. [Google Scholar] [CrossRef] [PubMed]
- Cavalier-Smith, T.; Chao, E.E.; Snell, E.A.; Berney, C.; Fiore-Donno, A.M.; Lewis, R. Multigene eukaryote phylogeny reveals the likely protozoan ancestors of opisthokonts (animals, fungi, choanozoans) and Amoebozoa. Mol. Phylogenet. Evol. 2014, 81, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Maeda, M.; Mihara, K. Characterization of rat TOM70 as a receptor of the preprotein translocase of the mitochondrial outer membrane. J. Cell Sci. 2002, 115, 1895–1905. [Google Scholar] [PubMed]
- Edmonson, A.M.; Mayfield, D.K.; Vervoort, V.; DuPont, B.R.; Argyropoulos, G. Characterization of a human import component of the mitochondrial outer membrane, TOMM70A. Cell Commun. Adhes. 2002, 9, 15–27. [Google Scholar] [CrossRef]
- Alvarez-Dolado, M.; González-Moreno, M.; Valencia, A.; Zenke, M.; Bernal, J.; Muñoz, A. Identification of a mammalian homologue of the fungal Tom70 mitochondrial precursor protein import receptor as a thyroid hormone- regulated gene in specific brain regions. J. Neurochem. 1999, 73, 2240–2249. [Google Scholar] [CrossRef] [PubMed]
- Hata, S.; Koyama, S.; Kawahara, H.; Doi, N.; Maeda, T.; Toyama-Sorimachi, N.; Abe, K.; Suzuki, K.; Sorimachi, H. Stomach-specific calpain, nCL-2, localizes in mucus cells and proteolyzes the β-subunit of coatomer complex, β-COP. J. Biol. Chem. 2006, 281, 11214–11224. [Google Scholar] [CrossRef]
- Filadi, R.; Leal, N.S.; Schreiner, B.; Rossi, A.; Dentoni, G.; Pinho, C.M.; Wiehager, B.; Cieri, D.; Calì, T.; Pizzo, P.; et al. TOM70 Sustains Cell Bioenergetics by Promoting IP3R3-Mediated ER to Mitochondria Ca2+ Transfer. Curr. Biol. 2018, 28, 369–382. [Google Scholar] [CrossRef]
- Wolfe, K. Robustness-It’s not where you think it is. Nat. Genet. 2000, 25, 3–4. [Google Scholar] [CrossRef]
- Burki, F.; Roger, A.J.; Brown, M.W.; Simpson, A.G.B. The New Tree of Eukaryotes. Trends Ecol. Evol. 2020, 35, 43–55. [Google Scholar] [CrossRef]
- Wojtkowska, M.; Szczech, N.; Stobienia, O.; Jarmuszkiewicz, W.; Budzinska, M.; Kmita, H. An inception report on the TOM complex of the amoeba Acanthamoeba castellanii, a simple model protozoan in mitochondria studies. J. Bioenerg. Biomembr. 2005, 37, 261–268. [Google Scholar] [CrossRef]
- Wojtkowska, M.; Buczek, D.; Stobienia, O.; Karachitos, A.; Antoniewicz, M.; Slocinska, M.; Makałowski, W.; Kmita, H. The TOM Complex of Amoebozoans: The Cases of the Amoeba Acanthamoeba castellanii and the Slime Mold Dictyostelium discoideum. Protist 2015, 166, 349–362. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mani, J.; Desy, S.; Niemann, M.; Chanfon, A.; Oeljeklaus, S.; Pusnik, M.; Schmidt, O.; Gerbeth, C.; Meisinger, C.; Warscheid, B.; et al. Mitochondrial protein import receptors in Kinetoplastids reveal convergent evolution over large phylogenetic distances. Nat. Commun. 2015, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Makki, A.; Rada, P.; Žárský, V.; Kereïche, S.; Kováčik, L.; Novotný, M.; Jores, T.; Rapaport, D.; Tachezy, J. Triplet-pore structure of a highly divergent TOM complex of hydrogenosomes in Trichomonas vaginalis. PLoS Biol. 2019, 17, 1–32. [Google Scholar] [CrossRef]
- Tsaousis, A.D.; Gaston, D.; Stechmann, A.; Walker, P.B.; Lithgow, T.; Roger, A.J. A Functional Tom70 in the human parasite Blastocystis sp.: Implications for the evolution of the mitochondrial import apparatus. Mol. Biol. Evol. 2011, 28, 781–791. [Google Scholar] [CrossRef]
- Dolezal, P.; Likic, V.; Tachezy, J.; Lithgow, T. Evolution of the molecular machines for protein import into mitochondria. Science 2006, 313, 314–318. [Google Scholar] [CrossRef] [PubMed]
- Schlegel, T.; Mirus, O.; Von Haeseler, A.; Schleiff, E. The tetratricopeptide repeats of receptors involved in protein translocation across membranes. Mol. Biol. Evol. 2007, 24, 2763–2774. [Google Scholar] [CrossRef]
- Schwenkert, S.; Dittmer, S.; Soll, J. Structural components involved in plastid protein import. Essays Biochem. 2018, 62, 65–75. [Google Scholar] [CrossRef]
- Duncan, O.; Murcha, M.W.; Whelan, J. Unique components of the plant mitochondrial protein import apparatus. Biochim. Biophys. Acta-Mol. Cell Res. 2013, 1833, 304–313. [Google Scholar] [CrossRef]
- Hirano, T.; Kinoshita, N.; Morikawa, K.; Yanagida, M. Snap helix with knob and hole: Essential repeats in S. pombe nuclear protein nuc2+. Cell 1990, 60, 319–328. [Google Scholar] [CrossRef]
- Sikorski, R.S.; Boguski, M.S.; Goebl, M.; Hieter, P. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell 1990, 60, 307–317. [Google Scholar] [CrossRef]
- Perez-Riba, A.; Itzhaki, L.S. The tetratricopeptide-repeat motif is a versatile platform that enables diverse modes of molecular recognition. Curr. Opin. Struct. Biol. 2019, 54, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Marshall-Carlson, L.; Carlson, M. The N-terminal TPR region is the functional domain of SSN6, a nuclear phosphoprotein of Saccharomyces cerevisiae. Mol. Cell. Biol. 1990, 10, 4744–4756. [Google Scholar] [CrossRef] [PubMed]
- Das, A.K.; Cohen, P.T.W.; Barford, D. The structure of the tetratricopeptide repeats of protein phosphatase 5: Implications for TPR-mediated protein-protein interactions. EMBO J. 1998, 17, 1192–1199. [Google Scholar] [CrossRef] [PubMed]
- Blatch, G.L.; Lässle, M. The tetratricopeptide repeat: A structural motif mediating protein-protein interactions. BioEssays 1999, 21, 932–939. [Google Scholar] [CrossRef]
- D’Andrea, L.D.; Regan, L. TPR proteins: The versatile helix. Trends Biochem. Sci. 2003, 28, 655–662. [Google Scholar] [CrossRef]
- Tripathi, A.; Mandon, E.C.; Gilmore, R.; Rapoport, T.A. Two alternative binding mechanisms connect the protein translocation Sec71-Sec72 complex with heat shock proteins. J. Biol. Chem. 2017, 292, 8007–8018. [Google Scholar] [CrossRef]
- Chew, O.; Lister, R.; Qbadou, S.; Heazlewood, J.L.; Soll, J.; Schleiff, E.; Millar, A.H.; Whelan, J. A plant outer mitochondrial membrane protein with high amino acid sequence identity to a chloroplast protein import receptor. FEBS Lett. 2004, 557, 109–114. [Google Scholar] [CrossRef]
- Lister, R.; Carrie, C.; Duncan, O.; Ho, L.H.M.; Howell, K.A.; Murcha, M.W.; Whelan, J. Functional definition of outer membrane proteins involved in preprotein import into mitochondria. Plant Cell 2007, 19, 3739–3759. [Google Scholar] [CrossRef]
- Nickel, C.; Horneff, R.; Heermann, R.; Neumann, B.; Jung, K.; Soll, J.; Schwenkert, S. Phosphorylation of the outer membrane mitochondrial protein OM64 influences protein import into mitochondria. Mitochondrion 2019, 44, 93–102. [Google Scholar] [CrossRef]
- Sohrt, K.; Soll, J. Toc64, a new component of the protein translocon of chloroplasts. J. Cell Biol. 2000, 148, 1213–1221. [Google Scholar] [CrossRef]
- Qbadou, S.; Becker, T.; Mirus, O.; Tews, I.; Soll, J.; Schleiff, E. The molecular chaperone Hsp90 delivers precursor proteins to the chloroplast import receptor Toc64. EMBO J. 2006, 25, 1836–1847. [Google Scholar] [CrossRef] [PubMed]
- Mirus, O.; Bionda, T.; von Haeseler, A.; Schleiff, E. Evolutionarily evolved discriminators in the 3-TPR domain of the Toc64 family involved in protein translocation at the outer membrane of chloroplasts and mitochondria. J. Mol. Model. 2009, 15, 971–982. [Google Scholar] [CrossRef] [PubMed]
- Panigrahi, R.; Adina-Zada, A.; Whelan, J.; Vrielink, A. Ligand recognition by the TPR domain of the import factor Toc64 from Arabidopsis thaliana. PLoS ONE 2013, 8, e83461. [Google Scholar] [CrossRef] [PubMed]
- Aronsson, H.; Boij, P.; Patel, R.; Wardle, A.; Töpel, M.; Jarvis, P. Toc64/OEP64 is not essential for the efficient import of proteins into chloroplasts in Arabidopsis thaliana. Plant J. 2007, 52, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Gatto, G.J.; Geisbrecht, B.V.; Gould, S.J.; Berg, J.M. A proposed model for the PEX5-peroxisomal targeting signal-1 recognition complex. Proteins Struct. Funct. Genet. 2000, 38, 241–246. [Google Scholar] [CrossRef]
- El Magraoui, F.; Brinkmeier, R.; Mastalski, T.; Hupperich, A.; Strehl, C.; Schwerter, D.; Girzalsky, W.; Meyer, H.E.; Warscheid, B.; Erdmann, R.; et al. The deubiquitination of the PTS1-import receptor Pex5p is required for peroxisomal matrix protein import. Biochim. Biophys. Acta-Mol. Cell Res. 2019, 1866, 199–213. [Google Scholar] [CrossRef]
- Erdmann, R.; Schliebs, W. Peroxisomal matrix protein import: The transient pore model. Nat. Rev. Mol. Cell Biol. 2005, 6, 738–742. [Google Scholar] [CrossRef]
- Hines, V.; Schatz, G. Precursor Binding to Yeast Mitochondria. J. Biol. Chem. 1993, 268, 449–454. [Google Scholar]
- Dietmeier, K.; Zara, V.; Palmisanoll, A.; Palmieri, F.; Voos, W.; Schlossmann, J.; Moczko, M.; Kispal, G.; Pfanner, N. Targeting and Translocation of the Phosphate Carrier/p32 to the Inner Membrane of Yeast Mitochondria. J. Biol. Chem. 1993, 268, 25958–25964. [Google Scholar]
- Palmisano, A.; Zara, V.; Hönlinger, A.; Vozza, A.; Dekker, P.J.T.; Pfanner, N.; Palmieri, F. Targeting and assembly of the oxoglutarate carrier: General principles for biogenesis of carrier proteins of the mitochondrial inner membrane. Biochem. J. 1998, 333, 151–158. [Google Scholar] [CrossRef]
- Palmieri, F.; Monné, M. Discoveries, metabolic roles and diseases of mitochondrial carriers: A review. Biochim. Biophys. Acta-Mol. Cell Res. 2016, 1863, 2362–2378. [Google Scholar] [CrossRef] [PubMed]
- Habib, S.J.; Waizenegger, T.; Lech, M.; Neupert, W.; Rapaport, D. Assembly of the TOB complex of mitochondria. J. Biol. Chem. 2005, 280, 6434–6440. [Google Scholar] [CrossRef] [PubMed]
- Rampelt, H.; Sucec, I.; Bersch, B.; Horten, P.; Perschil, I.; Martinou, J.C.; Van Der Laan, M.; Wiedemann, N.; Schanda, P.; Pfanner, N. The mitochondrial carrier pathway transports non-canonical substrates with an odd number of transmembrane segments. BMC Biol. 2020, 18, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Vitali, D.G.; Drwesh, L.; Cichocki, B.A.; Kolb, A.; Rapaport, D. The Biogenesis of Mitochondrial Outer Membrane Proteins Show Variable Dependence on Import Factors. iScience 2020, 23, 100779. [Google Scholar] [CrossRef] [PubMed]
- Papić, D.; Krumpe, K.; Dukanovic, J.; Dimmer, K.S.; Rapaport, D. Multispan mitochondrial outer membrane protein Ugo1 follows a unique Mim1-dependent import pathway. J. Cell Biol. 2011, 194, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Becker, T.; Wenz, L.S.; Krüger, V.; Lehmann, W.; Müller, J.M.; Goroncy, L.; Zufall, N.; Lithgow, T.; Guiard, B.; Chacinska, A.; et al. The mitochondrial import protein Mim1 promotes biogenesis of multispanning outer membrane proteins. J. Cell Biol. 2011, 194, 387–395. [Google Scholar] [CrossRef]
- Doan, K.N.; Grevel, A.; Mårtensson, C.U.; Ellenrieder, L.; Thornton, N.; Wenz, L.S.; Opaliński, Ł.; Guiard, B.; Pfanner, N.; Becker, T. The Mitochondrial Import Complex MIM Functions as Main Translocase for α-Helical Outer Membrane Proteins. Cell Rep. 2020, 31, 107567. [Google Scholar] [CrossRef]
- Pebay-Peyroula, E.; Dahout-Gonzalez, C.; Kahn, R.; Trézéguet, V.; Lauquin, G.J.M.; Brandolin, G. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 2003, 426, 39–44. [Google Scholar] [CrossRef]
- Ruprecht, J.J.; King, M.S.; Zögg, T.; Aleksandrova, A.A.; Pardon, E.; Crichton, P.G.; Steyaert, J.; Kunji, E.R.S. The Molecular Mechanism of Transport by the Mitochondrial ADP/ATP Carrier. Cell 2019, 176, 435–447.e15. [Google Scholar] [CrossRef]
- Komiya, T.; Rospert, S.; Schatz, G.; Mihara, K. Binding of mitochondrial precursor proteins to the cytoplasmic domains of the import receptors Tom70 and Tom20 is determined by cytoplasmic chaperones. EMBO J. 1997, 16, 4267–4275. [Google Scholar] [CrossRef]
- Fan, A.C.Y.; Bhangoo, M.K.; Young, J.C. Hsp90 functions in the targeting and outer membrane translocation steps of Tom70-mediated mitochondrial import. J. Biol. Chem. 2006, 281, 33313–33324. [Google Scholar] [CrossRef] [PubMed]
- Zanphorlin, L.M.; Lima, T.B.; Wong, M.J.; Balbuena, T.S.; Minetti, C.A.S.A.; Remeta, D.P.; Young, J.C.; Barbosa, L.R.S.; Gozzo, F.C.; Ramos, C.H.I. Heat shock protein 90 kDa (Hsp90) has a second functional interaction site with the mitochondrial import receptor Tom70. J. Biol. Chem. 2016, 291, 18620–18631. [Google Scholar] [CrossRef] [PubMed]
- Gava, L.M.; Gonçalves, D.C.; Borges, J.C.; Ramos, C.H.I. Stoichiometry and thermodynamics of the interaction between the C-terminus of human 90 kDa heat shock protein Hsp90 and the mitochondrial translocase of outer membrane Tom70. Arch. Biochem. Biophys. 2011, 513, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Bykov, Y.S.; Rapaport, D.; Herrmann, J.M.; Schuldiner, M. Cytosolic Events in the Biogenesis of Mitochondrial Proteins. Trends Biochem. Sci. 2020, 45, 650–667. [Google Scholar] [CrossRef] [PubMed]
- Becker, T.; Song, J.; Pfanner, N. Versatility of Preprotein Transfer from the Cytosol to Mitochondria. Trends Cell Biol. 2019, 29, 534–548. [Google Scholar] [CrossRef]
- Opaliński, Ł.; Song, J.; Priesnitz, C.; Wenz, L.S.; Oeljeklaus, S.; Warscheid, B.; Pfanner, N.; Becker, T. Recruitment of Cytosolic J-Proteins by TOM Receptors Promotes Mitochondrial Protein Biogenesis. Cell Rep. 2018, 25, 2036–2043. [Google Scholar] [CrossRef]
- Papic, D.; Elbaz-Alon, Y.; Koerdt, S.N.; Leopold, K.; Worm, D.; Jung, M.; Schuldiner, M.; Rapaport, D. The Role of Djp1 in Import of the Mitochondrial Protein Mim1 Demonstrates Specificity between a Cochaperone and Its Substrate Protein. Mol. Cell. Biol. 2013, 33, 4083–4094. [Google Scholar] [CrossRef]
- Jores, T.; Lawatscheck, J.; Beke, V.; Franz-Wachtel, M.; Yunoki, K.; Fitzgerald, J.C.; Macek, B.; Endo, T.; Kalbacher, H.; Buchner, J.; et al. Cytosolic Hsp70 and Hsp40 chaperones enable the biogenesis of mitochondrial β-barrel proteins. J. Cell Biol. 2018, 217, 3091–3108. [Google Scholar] [CrossRef]
- Hoseini, H.; Pandey, S.; Jores, T.; Schmitt, A.; Franz-Wachtel, M.; Macek, B.; Buchner, J.; Dimmer, K.S.; Rapaport, D. The cytosolic cochaperone Sti1 is relevant for mitochondrial biogenesis and morphology. FEBS J. 2016, 283, 3338–3352. [Google Scholar] [CrossRef]
- Bhangoo, M.K.; Tzankov, S.; Fan, A.C.Y.; Dejgaard, K.; Thomas, D.Y.; Young, J.C. Multiple 40-kDa Heat-Shock Protein Chaperones Function in Tom70-dependent Mitochondrial Import. Mol. Biol. Cell 2007, 18, 3414–3428. [Google Scholar] [CrossRef]
- Ferramosca, A.; Zara, V. Biogenesis of mitochondrial carrier proteins: Molecular mechanisms of import into mitochondria. Biochim. Biophys. Acta 2013, 1833, 494–502. [Google Scholar] [CrossRef] [PubMed]
- Zara, V.; Ferramosca, A.; Capoblanco, L.; Baltz, K.M.; Randel, O.; Rassow, J.; Palmieri, F.; Papatheodorou, P. Biogenesis of yeast dicarboxylate carrier: The carrier signature facilities translocation across the mitochondrial outer membrane. J. Cell Sci. 2007, 120, 4099–4106. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zara, V.; Ferramosca, A.; Papatheodorou, P.; Palmieri, F.; Rassow, J. Import of rat mitochondrial citrate carrier (CIC) at increasing salt concentrations promotes presequence binding to import receptor Tom20 and inhibits membrane translocation. J. Cell Sci. 2005, 118, 3985–3995. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zara, V.; Palmieri, F.; Mahlke, K.; Pfanner, N. The cleavable presequence is not essential for import and assembly of the phosphate carrier of mammalian mitochondria but enhances the specificity and efficiency of import. J. Biol. Chem. 1992, 267, 12077–12081. [Google Scholar] [PubMed]
- Zara, V.; Ferramosca, A.; Palmisano, I.; Palmieri, F.; Rassow, J. Biogenesis of rat mitochondrial citrate carrier (CIC): The N-terminal presequence facilitates the solubility of the preprotein but does not act as a targeting signal. J. Mol. Biol. 2003. [Google Scholar] [CrossRef]
- Cheng, T.L.; Liao, C.C.; Tsai, W.H.; Lin, C.C.; Yeh, C.W.; Teng, C.F.; Chang, W.T. Identification and characterization of the mitochondrial targeting sequence and mechanism in human citrate synthase. J. Cell. Biochem. 2009, 107, 1002–1015. [Google Scholar] [CrossRef] [PubMed]
- Brix, J.; Ru, S.; Bukau, B.; Schneider-Mergener, J.; Pfanner, N. Distribution of Binding Sequences for the Mitochondrial Import Receptors Tom20, Tom22, and Tom70 in a Presequence-carrying Preprotein and a Non-cleavable Preprotein. J. Biol. Chem. 1999, 274, 16522–16530. [Google Scholar] [CrossRef]
- Künkele, K.P.; Heins, S.; Dembowski, M.; Nargang, F.E.; Benz, R.; Thieffry, M.; Walz, J.; Lill, R.; Nussberger, S.; Neupert, W. The preprotein translocation channel of the outer membrane of mitochondria. Cell 1998, 93, 1009–1019. [Google Scholar] [CrossRef]
- Hill, K.; Model, K.; Ryan, M.T.; Dietmeier, K.; Martint, F.; Wagner, R.; Pfanner, N. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature 1998, 395, 516–521. [Google Scholar] [CrossRef]
- Künkele, K.P.; Juin, P.; Pompa, C.; Nargang, F.E.; Henry, J.P.; Neupert, W.; Lill, R.; Thieffry, M. The isolated complex of the translocase of the outer membrane of mitochondria. Characterization of the cation-selective and voltage-gated preprotein-conducting pore. J. Biol. Chem. 1998, 273, 31032–31039. [Google Scholar] [CrossRef]
- Rehling, P.; Model, K.; Brandner, K.; Kovermann, P.; Sickmann, A.; Meyer, H.E.; Kühlbrandt, W.; Wagner, R.; Truscott, K.N.; Pfanner, N. Protein insertion into the mitochondrial inner membrane by a twin-pore translocase. Science 2003, 299, 1747–1751. [Google Scholar] [CrossRef] [PubMed]
- Weinhäupl, K.; Lindau, C.; Hessel, A.; Wang, Y.; Schütze, C.; Jores, T.; Melchionda, L.; Schönfisch, B.; Kalbacher, H.; Bersch, B.; et al. Structural Basis of Membrane Protein Chaperoning through the Mitochondrial Intermembrane Space. Cell 2018, 175, 1365–1379. [Google Scholar] [CrossRef] [PubMed]
- Gebert, N.; Chacinska, A.; Wagner, K.; Guiard, B.; Koehler, C.M.; Rehling, P.; Pfanner, N.; Wiedemann, N. Assembly of the three small Tim proteins precedes docking to the mitochondrial carrier translocase. EMBO Rep. 2008, 9, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Sirrenberg, C.; Bauer, M.F.; Guiard, B.; Neupert, W.; Brunner, M. Import of carrier proteins into the mitochondrial inner membrane mediated by Tim22. Nature 1996, 384, 582–585. [Google Scholar] [CrossRef]
- Koehler, C.M.; Jarosch, E.; Tokatlidis, K.; Schmid, K.; Schweyen, R.J.; Schatz, G. Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science 1998, 279, 369–373. [Google Scholar] [CrossRef]
- Kübrich, M.; Rassow, J.; Voos, W.; Pfanner, N.; Honlinger, A. The import route of ADP/ATP carrier into mitochondria separates from the general import pathway of cleavable preproteins at the trans side of the outer membrane. J. Biol. Chem. 1998, 273, 16374–16381. [Google Scholar] [CrossRef] [PubMed]
- Endres, M.; Neupert, W.; Brunner, M. Transport of the ADP/ATP carrier of mitochondria from the TOM complex to the TIM22.54 complex. EMBO J. 1999, 18, 3214–3221. [Google Scholar] [CrossRef]
- Ryan, M.T.; Müller, H.; Pfanner, N. Functional staging of ADP/ATP carrier translocation across the outer mitochondrial membrane. J. Biol. Chem. 1999, 274, 20619–20627. [Google Scholar] [CrossRef]
- Asai, T.; Takahashi, T.; Esaki, M.; Nishikawa, S.I.; Ohtsuka, K.; Nakai, M.; Endo, T. Reinvestigation of the Requirement of Cytosolic ATP for Mitochondrial Protein Import. J. Biol. Chem. 2004, 279, 19464–19470. [Google Scholar] [CrossRef]
- Otera, H.; Taira, Y.; Horie, C.; Suzuki, Y.; Suzuki, H.; Setoguchi, K.; Kato, H.; Oka, T.; Mihara, K. A novel insertion pathway of mitochondrial outer membrane proteins with multiple transmembrane segments. J. Cell Biol. 2007, 179, 1355–1363. [Google Scholar] [CrossRef]
- Coonrod, E.M.; Karren, M.A.; Shaw, J.M. Ugo1p is a multipass transmembrane protein with a single carrier domain required for mitochondrial fusion. Traffic 2007, 8, 500–511. [Google Scholar] [CrossRef] [PubMed]
- Jores, T.; Klinger, A.; Groß, L.E.; Kawano, S.; Flinner, N.; Duchardt-Ferner, E.; Wöhnert, J.; Kalbacher, H.; Endo, T.; Schleiff, E.; et al. Characterization of the targeting signal in mitochondrial β-barrel proteins. Nat. Commun. 2016, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Fukui, K.; Takahashi, H.; Kitamura, S.; Shiota, T.; Terao, K.; Uchida, M.; Esaki, M.; Nishikawa, S.I.; Yoshihisa, T.; et al. Roles of Tom70 in import of presequence-containing mitochondrial proteins. J. Biol. Chem. 2009, 284, 31635–31646. [Google Scholar] [CrossRef] [PubMed]
- Horvath, S.E.; Böttinger, L.; Vögtle, F.N.; Wiedemann, N.; Meisinger, C.; Becker, T.; Daum, G. Processing and topology of the yeast mitochondrial phosphatidylserine decarboxylase. J. Biol. Chem. 2012, 287, 36744–36755. [Google Scholar] [CrossRef]
- Sinzel, M.; Tan, T.; Wendling, P.; Kalbacher, H.; Özbalci, C.; Chelius, X.; Westermann, B.; Brügger, B.; Rapaport, D.; Dimmer, K.S. Mcp3 is a novel mitochondrial outer membrane protein that follows a unique IMP-dependent biogenesis pathway. EMBO Rep. 2017, 18, 1869. [Google Scholar] [CrossRef]
- Backes, S.; Hess, S.; Boos, F.; Woellhaf, M.W.; Gödel, S.; Jung, M.; Mühlhaus, T.; Herrmann, J.M. Tom70 enhances mitochondrial preprotein import efficiency by binding to internal targeting sequences. J. Cell Biol. 2018, 217, 1369–1382. [Google Scholar] [CrossRef]
- Mårtensson, C.U.; Priesnitz, C.; Song, J.; Ellenrieder, L.; Doan, K.N.; Boos, F.; Floerchinger, A.; Zufall, N.; Oeljeklaus, S.; Warscheid, B.; et al. Mitochondrial protein translocation-associated degradation. Nature 2019, 569, 679–683. [Google Scholar] [CrossRef]
- Pfaller, R.; Pfanner, N.; Neupert, W. Mitochondrial protein import. Bypass of proteinaceous surface receptors can occur with low specificity and efficiency. J. Biol. Chem. 1989, 264, 34–39. [Google Scholar]
- Lithgow, T.; Junne, T.; Wachter, C.; Schatz, G. Yeast mitochondria lacking the two import receptors Mas20p and Mas70p can efficiently and specifically import precursor proteins. J. Biol. Chem. 1994, 269, 15325–15330. [Google Scholar]
- Kellems, R.E.; Butow, R.A. Cytoplasmic-type 80 S Ribosomes Associated with Yeast Mitochondria. J. Biol. Chem. 1972, 247, 8043–8050. [Google Scholar]
- Ades, I.Z.; Butow, R.A. The products of mitochondria-bound cytoplasmic polysomes in yeast. J. Biol. Chem. 1980, 255, 9918–9924. [Google Scholar] [PubMed]
- Hallermayer, G.; Zimmermann, R.; Neupert, W. Kinetic Studies on the Transport of Cytoplasmically Synthesized Proteins into the Mitochondria in Intact Cells of Neurospora crassa. Eur. J. Biochem. 1977, 81, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Maccecchini, M.L.; Rudin, Y.; Blobel, G.; Schatz, G. Import of proteins into mitochondria: Precursor forms of the extramitochondrially made F1-ATPase subunits in yeast. Proc. Natl. Acad. Sci. USA 1979, 76, 343–347. [Google Scholar] [CrossRef] [PubMed]
- Verner, K. Co-translational protein import into mitochondria: An alternative view. Trends Biochem. Sci. 1993, 18, 366–371. [Google Scholar] [CrossRef]
- Quenault, T.; Lithgow, T.; Traven, A. PUF proteins: Repression, activation and mRNA localization. Trends Cell Biol. 2011, 21, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Lesnik, C.; Golani-Armon, A.; Arava, Y. Localized translation near the mitochondrial outer membrane: An update. RNA Biol. 2015, 12, 801–809. [Google Scholar] [CrossRef]
- Vardi-Oknin, D.; Arava, Y. Characterization of Factors Involved in Localized Translation Near Mitochondria by Ribosome-Proximity Labeling. Front. Cell Dev. Biol. 2019, 7, 1–9. [Google Scholar] [CrossRef]
- Gadir, N.; Haim-Vilmovsky, L.; Kraut-Cohen, J.; Gerst, J.E. Localization of mRNAs coding for mitochondrial proteins in the yeast Saccharomyces cerevisiae. RNA 2011, 17, 1551–1565. [Google Scholar] [CrossRef]
- Eliyahu, E.; Lesnik, C.; Arava, Y. The protein chaperone Ssa1 affects mRNA localization to the mitochondria. FEBS Lett. 2012, 586, 64–69. [Google Scholar] [CrossRef]
- Chen, Y.; Umanah, G.K.E.; Dephoure, N.; Andrabi, S.A.; Gygi, S.P.; Dawson, T.M.; Dawson, V.L.; Rutter, J. Msp1/ ATAD 1 maintains mitochondrial function by facilitating the degradation of mislocalized tail-anchored proteins. EMBO J. 2014, 33, 1548–1564. [Google Scholar] [CrossRef]
- Williams, C.C.; Jan, C.H.; Weissman, J.S. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science 2014, 346, 748–751. [Google Scholar] [CrossRef] [PubMed]
- Okreglak, V.; Walter, P. The conserved AAA-ATPase Msp1 confers organelle specificity to tail-anchored proteins. Proc. Natl. Acad. Sci. USA 2014, 111, 8019–8024. [Google Scholar] [CrossRef] [PubMed]
- Gamerdinger, M.; Hanebuth, M.A.; Frickey, T.; Deuerling, E. The principle of antagonism ensures protein targeting specificity at the endoplasmic reticulum. Science 2015, 348, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Costa, E.A.; Subramanian, K.; Nunnari, J.; Weissman, J.S. Defining the physiological role of SRP in protein-targeting efficiency and specificity. Science 2018, 359, 689–692. [Google Scholar] [CrossRef]
- Hansen, K.G.; Aviram, N.; Laborenz, J.; Bibi, C.; Meyer, M.; Spang, A.; Schuldiner, M.; Herrmann, J.M. An ER surface retrieval pathway safeguards the import of mitochondrial membrane proteins in yeast. Science 2018, 361, 1118–1122. [Google Scholar] [CrossRef]
- Kurz, M.; Martin, H.; Rassow, J.; Pfanner, N.; Ryan, M.T. Biogenesis of Tim proteins of the mitochondrial carrier import pathway: Differential targeting mechanisms and crossing over with the main import pathway. Mol. Biol. Cell 1999, 10, 2461–2474. [Google Scholar] [CrossRef][Green Version]
- Wan, Y.; Hopper, A.K. From powerhouse to processing plant: Conserved roles of mitochondrial outer membrane proteins in tRNA splicing. Genes Dev. 2018, 32, 1309–1314. [Google Scholar] [CrossRef]
- Kondo-Okamoto, N.; Shaw, J.M.; Okamoto, K. Tetratricopeptide repeat proteins Tom70 and Tom71 mediate yeast mitochondrial morphogenesis. EMBO Rep. 2008, 9, 63–69. [Google Scholar] [CrossRef]
- Murley, A.; Sarsam, R.D.; Toulmay, A.; Yamada, J.; Prinz, W.A.; Nunnari, J. Ltc1 is an ER-localized sterol transporter and a component of ER-mitochondria and ER-vacuole contacts. J. Cell Biol. 2015, 209, 539–548. [Google Scholar] [CrossRef]
- Elbaz-Alon, Y.; Eisenberg-Bord, M.; Shinder, V.; Stiller, S.B.; Shimoni, E.; Wiedemann, N.; Geiger, T.; Schuldiner, M. Lam6 Regulates the Extent of Contacts between Organelles. Cell Rep. 2015, 12, 7–14. [Google Scholar] [CrossRef]
- Kato, H.; Lu, Q.; Rapaport, D.; Kozjak-Pavlovic, V. Tom70 Is Essential for PINK1 Import into Mitochondria. PLoS ONE 2013, 8, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Takahashi, Y.; He, H.; Hattori, T.; Chen, C.; Liang, X.; Chen, H.; Young, M.M.; Wang, H.G. TOM40 Targets Atg2 to Mitochondria-Associated ER Membranes for Phagophore Expansion. Cell Rep. 2019, 28, 1744–1757.e5. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Manik, M.K.; Im, Y.J. Structural basis of sterol recognition and nonvesicular transport by lipid transfer proteins anchored at membrane contact sites. Proc. Natl. Acad. Sci. USA 2018, 115, E856–E865. [Google Scholar] [CrossRef] [PubMed]
- Horenkamp, F.A.; Valverde, D.P.; Nunnari, J.; Reinisch, K.M. Molecular basis for sterol transport by StART-like lipid transfer domains. EMBO J. 2018, 37, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg-Bord, M.; Tsui, H.S.; Antunes, D.; Fernández-del-Río, L.; Bradley, M.C.; Dunn, C.D.; Nguyen, T.P.T.; Rapaport, D.; Clarke, C.F.; Schuldiner, M. The Endoplasmic Reticulum-Mitochondria Encounter Structure Complex Coordinates Coenzyme Q Biosynthesis. Contact 2019, 2, 251525641882540. [Google Scholar] [CrossRef] [PubMed]
- Gatta, A.T.; Wong, L.H.; Sere, Y.Y.; Calderón-Noreña, D.M.; Cockcroft, S.; Menon, A.K.; Levine, T.P. A new family of StART domain proteins at membrane contact sites has a role in ER-PM sterol transport. Elife 2015, 4, 1–46. [Google Scholar] [CrossRef]
- Wu, J.; Bao, A.; Chatterjee, K.; Wan, Y.; Hopper, A.K. Genome-wide screen uncovers novel pathways for tRNA processing and nuclear–cytoplasmic dynamics. Genes Dev. 2015, 29, 2633–2644. [Google Scholar] [CrossRef]
- Dürr, M.; Escobar-Henriques, M.; Merz, S.; Geimer, S.; Langer, T.; Westermann, B. Nonredundant roles of mitochondria-associated F-box proteins Mfb1 and Mdm30 in maintenance of mitochondrial morphology in yeast. Mol. Biol. Cell 2006, 17, 3745–3755. [Google Scholar] [CrossRef][Green Version]
- Grad, L.I.; Descheneau, A.T.; Neupert, W.; Lill, R.; Nargang, F.E. Inactivation of the Neurospora crassa mitochondrial outer membrane protein TOM70 by repeat-induced point mutation (RIP) causes defects in mitochondrial protein import and morphology. Curr. Genet. 1999, 36, 137–146. [Google Scholar] [CrossRef]
- Jackson, T.D.; Palmer, C.S.; Stojanovski, D. Mitochondrial diseases caused by dysfunctional mitochondrial protein import. Biochem. Soc. Trans. 2018, 46, 1225–1238. [Google Scholar] [CrossRef]
- Dutta, D.; Briere, L.C.; Kanca, O.; Marcogliese, P.C.; Walker, M.A.; High, F.A.; Vanderver, A.; Krier, J.; Carmichael, N.; Callahan, C.; et al. De novo mutations in TOMM70, a receptor of the mitochondrial import translocase, cause neurological impairment. Hum. Mol. Genet. 2020, 29, 1568–1579. [Google Scholar] [CrossRef] [PubMed]
- Bornstein, R.; Gonzalez, B.; Johnson, S.C. Mitochondrial pathways in human health and aging. Mitochondrion 2020, 54, 72–84. [Google Scholar] [CrossRef] [PubMed]
- Picard, M.; Wallace, D.C.; Burelle, Y. The rise of mitochondria in medicine. Mitochondrion 2016, 30, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Giri, J.; Srivastav, S.; Basu, M.; Palit, S.; Gupta, P.; Ukil, A. Leishmania donovani exploits myeloid cell Leukemia 1 (MCL-1) protein to prevent mitochondria-dependent host cell apoptosis. J. Biol. Chem. 2016, 291, 3496–3507. [Google Scholar] [CrossRef]
- Li, J.; Qi, M.; Li, C.; Shi, D.; Zhang, D.; Xie, D.; Yuan, T.; Feng, J.; Liu, Y.; Liang, D.; et al. Tom70 serves as a molecular switch to determine pathological cardiac hypertrophy. Cell Res. 2014, 24, 977–993. [Google Scholar] [CrossRef]
- Pei, H.F.; Hou, J.N.; Wei, F.P.; Xue, Q.; Zhang, F.; Peng, C.F.; Yang, Y.; Tian, Y.; Feng, J.; Du, J.; et al. Melatonin attenuates postmyocardial infarction injury via increasing Tom70 expression. J. Pineal Res. 2017, 62, 1–13. [Google Scholar] [CrossRef]
- Heo, J.M.; Harper, N.J.; Paulo, J.A.; Li, M.; Xu, Q.; Coughlin, M.; Elledge, S.J.; Wade Harper, J. Integrated proteogenetic analysis reveals the landscape of a mitochondrial-autophagosome synapse during PARK2-dependent mitophagy. Sci. Adv. 2019, 5, eaay4624. [Google Scholar] [CrossRef]
- Orrenius, S.; Gogvadze, V.; Zhivotovsky, B. Calcium and mitochondria in the regulation of cell death. Biochem. Biophys. Res. Commun. 2015, 460, 72–81. [Google Scholar] [CrossRef]
- Motz, C.; Martin, H.; Krimmer, T.; Rassow, J. Bcl-2 and porin follow different pathways of TOM-dependent insertion into the mitochondrial outer membrane. J. Mol. Biol. 2002, 323, 729–738. [Google Scholar] [CrossRef]
- Ott, M.; Norberg, E.; Walter, K.M.; Schreiner, P.; Kemper, C.; Rapaport, D.; Zhivotovsky, B.; Orrenius, S. The mitochondrial TOM complex is required for tBid/Bax-induced cytochrome c release. J. Biol. Chem. 2007, 282, 27633–27639. [Google Scholar] [CrossRef]
- Bellot, G.; Cartron, P.F.; Er, E.; Oliver, L.; Juin, P.; Armstrong, L.C.; Bornstein, P.; Mihara, K.; Manon, S.; Vallette, F.M. TOM22, a core component of the mitochondria outer membrane protein translocation pore, is a mitochondrial receptor for the proapoptotic protein Bax. Cell Death Differ. 2007, 14, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Cartron, P.F.; Bellot, G.; Oliver, L.; Grandier-Vazeille, X.; Manon, S.; Vallette, F.M. Bax inserts into the mitochondrial outer membrane by different mechanisms. FEBS Lett. 2008, 582, 3045–3051. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.O.; Dengjel, J.; Wilfling, F.; Kozjak-Pavlovic, V.; Häcker, G.; Weber, A. The pro-apoptotic BH3-only protein bim interacts with components of the translocase of the outer mitochondrial membrane (TOM). PLoS ONE 2015, 10, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Ross, K.; Rudel, T.; Kozjak-Pavlovic, V. TOM-independent complex formation of Bax and Bak in mammalian mitochondria during TNFα-induced apoptosis. Cell Death Differ. 2009, 16, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Colin, J.; Garibal, J.; Mignotte, B.; Guénal, I. The mitochondrial TOM complex modulates bax-induced apoptosis in Drosophila. Biochem. Biophys. Res. Commun. 2009, 379, 939–943. [Google Scholar] [CrossRef]
- Chou, C.-H.; Lee, R.-S.; Yang-Yen, H.-F. An Internal EELD Domain Facilitates Mitochondrial Targeting of Mcl-1 via a Tom70-dependent Pathway. Mol. Biol. Cell 2006, 17, 3952–3963. [Google Scholar] [CrossRef][Green Version]
- Sly, L.M.; Hingley-Wilson, S.M.; Reiner, N.E.; McMaster, W.R. Survival of Mycobacterium tuberculosis in Host Macrophages Involves Resistance to Apoptosis Dependent upon Induction of Antiapoptotic Bcl-2 Family Member Mcl-1. J. Immunol. 2003, 170, 430–437. [Google Scholar] [CrossRef]
- Rajalingam, K.; Sharma, M.; Lohmann, C.; Oswald, M.; Thieck, O.; Froelich, C.J.; Rudel, T. Mcl-1 Is a Key Regulator of Apoptosis Resistance in Chlamydia trachomatis-Infected Cells. PLoS ONE 2008, 3, e3102. [Google Scholar] [CrossRef]
- Koziel, J.; Kmiecik, K.; Chmiest, D.; Maresz, K.; Mizgalska, D.; Maciag-gudowska, A.; Mydel, P.; Potempa, J. The Role of Mcl-1 in S. aureus-Induced Cytoprotection of Infected Macrophages. Mediators Inflamm. 2013, 2013, 1–12. [Google Scholar] [CrossRef]
- Xiang, W.; Yang, C.Y.; Bai, L. MCL-1 inhibition in cancer treatment. Onco. Targets. Ther. 2018, 11, 7301. [Google Scholar] [CrossRef]
- Yoneyama, M.; Kikuchi, M.; Natsukawa, T.; Shinobu, N.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Akira, S.; Fujita, T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004, 5, 730–737. [Google Scholar] [CrossRef]
- Seth, R.B.; Sun, L.; Ea, C.K.; Chen, Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF3. Cell 2005, 122, 669–682. [Google Scholar] [CrossRef]
- Wei, B.; Cui, Y.; Huang, Y.; Liu, H.; Li, L.; Li, M.; Ruan, K.-C.; Zhou, Q.; Wang, C. Tom70 Mediates Sendai Virus-Induced Apoptosis on Mitochondria. J. Virol. 2015, 89, 3804–3818. [Google Scholar] [CrossRef]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.C.; Uhl, S.; Hoagland, D.; Møller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045.e9. [Google Scholar] [CrossRef]
- Lamers, M.M.; Beumer, J.; van der Vaart, J.; Knoops, K.; Puschhof, J.; Breugem, T.I.; Ravelli, R.B.G.; Paul van Schayck, J.; Mykytyn, A.Z.; Duimel, H.Q.; et al. SARS-CoV-2 productively infects human gut enterocytes. Science 2020, 369, 50–54. [Google Scholar] [CrossRef]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Pinton, P.; Giorgi, C.; Siviero, R.; Zecchin, E.; Rizzuto, R. Calcium and apoptosis: ER-mitochondria Ca2+ transfer in the control of apoptosis. Oncogene 2010, 50, 6407–6418. [Google Scholar] [CrossRef]
- Cárdenas, C.; Miller, R.A.; Smith, I.; Bui, T.; Molgó, J.; Müller, M.; Vais, H.; Cheung, K.H.; Yang, J.; Parker, I.; et al. Essential Regulation of Cell Bioenergetics by Constitutive InsP3 Receptor Ca2+ Transfer to Mitochondria. Cell 2010, 142, 270–283. [Google Scholar] [CrossRef]
- Zhang, A.; Williamson, C.D.; Wong, D.S.; Bullough, M.D.; Brown, K.J.; Hathout, Y.; Colberg-Poley, A.M. Quantitative proteomic analyses of human cytomegalovirus-induced restructuring of endoplasmic reticulum-mitochondrial contacts at late times of infection. Mol. Cell. Proteomics 2011, 10, 1–17. [Google Scholar] [CrossRef]
- Xue, Q.; Pei, H.; Liu, Q.; Zhao, M.; Sun, J.; Gao, E.; Ma, X.; Tao, L. MICU1 protects against myocardial ischemia/reperfusion injury and its control by the importer receptor Tom70. Cell Death Dis. 2017, 8, e2923. [Google Scholar] [CrossRef]
- Wang, P.; Wang, D.; Yang, Y.; Hou, J.; Wan, J.; Ran, F.; Dai, X.; Zhou, P.; Yang, Y. Tom70 protects against diabetic cardiomyopathy through its antioxidant and antiapoptotic properties. Hypertens. Res. 2020, 43, 1047–1056. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Qin, J.; Osei, W.; Hu, K. Regulation of protein kinase C-epsilon and its age-dependence. Biochem. Biophys. Res. Commun. 2017, 482, 1201–1206. [Google Scholar] [CrossRef] [PubMed]
- Giorgianni, F.; Khan, M.U.; Weber, K.T.; Gerling, I.C.; Beranova-Giorgianni, S. Phosphoproteome mapping of cardiomyocyte mitochondria in a rat model of heart failure. Mol. Cell. Biochem. 2014, 389, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Sena, L.A.; Chandel, N.S. Physiological roles of mitochondrial reactive oxygen species. Mol. Cell 2012, 48, 158–167. [Google Scholar] [CrossRef]
- Kanki, T.; Klionsky, D.J.; Okamoto, K. Mitochondria autophagy in yeast. Antioxid. Redox Signal. 2011, 14, 1989–2001. [Google Scholar] [CrossRef]
- Morciano, G.; Patergnani, S.; Bonora, M.; Pedriali, G.; Tarocco, A.; Bouhamida, E.; Marchi, S.; Ancora, G.; Anania, G.; Wieckowski, M.R.; et al. Mitophagy in Cardiovascular Diseases. J. Clin. Med. 2020, 9, 892. [Google Scholar] [CrossRef]
- Lou, G.; Palikaras, K.; Lautrup, S.; Scheibye-Knudsen, M.; Tavernarakis, N.; Fang, E.F. Mitophagy and Neuroprotection. Trends Mol. Med. 2020, 26, 8–20. [Google Scholar] [CrossRef]
- Chai, Y.L.; Xing, H.; Chong, J.R.; Francis, P.T.; Ballard, C.G.; Chen, C.P.; Lai, M.K.P. Mitochondrial Translocase of the Outer Membrane Alterations May Underlie Dysfunctional Oxidative Phosphorylation in Alzheimer’s Disease. J. Alzheimer’s Dis. 2017, 61, 793–801. [Google Scholar] [CrossRef]
- Bader, V.; Winklhofer, K.F. PINK1 and Parkin: Team players in stress-induced mitophagy. Biol. Chem. 2020, 401, 891–899. [Google Scholar] [CrossRef]
- Rüb, C.; Wilkening, A.; Voos, W. Mitochondrial quality control by the Pink1/Parkin system. Cell Tissue Res. 2017, 367, 111–123. [Google Scholar] [CrossRef]
- Pickles, S.; Vigié, P.; Youle, R.J. Mitophagy and Quality Control Mechanisms in Mitochondrial Maintenance. Curr. Biol. 2018, 28, R170–R185. [Google Scholar] [CrossRef] [PubMed]
- Sarraf, S.A.; Raman, M.; Guarani-Pereira, V.; Sowa, M.E.; Huttlin, E.L.; Gygi, S.P.; Harper, J.W. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 2013, 496, 372–376. [Google Scholar] [CrossRef] [PubMed]
- Bertolin, G.; Ferrando-Miguel, R.; Jacoupy, M.; Traver, S.; Grenier, K.; Greene, A.W.; Dauphin, A.; Waharte, F.; Bayot, A.; Salamero, J.; et al. The TOMM machinery is a molecular switch in PINK1 and PARK2/PARKIN- dependent mitochondrial clearance. Autophagy 2013, 9, 1801–1817. [Google Scholar] [CrossRef] [PubMed]
- Jacoupy, M.; Hamon-Keromen, E.; Ordureau, A.; Erpapazoglou, Z.; Coge, F.; Corvol, J.C.; Nosjean, O.; Mannoury la Cour, C.; Millan, M.J.; Boutin, J.A.; et al. The PINK1 kinase-driven ubiquitin ligase Parkin promotes mitochondrial protein import through the presequence pathway in living cells. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef]
- Sugiura, A.; McLelland, G.; Fon, E.A.; McBride, H.M. A new pathway for mitochondrial quality control: Mitochondrial-derived vesicles. EMBO J. 2014, 33, 2142–2156. [Google Scholar] [CrossRef]
- Soubannier, V.; McLelland, G.L.; Zunino, R.; Braschi, E.; Rippstein, P.; Fon, E.A.; McBride, H.M. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr. Biol. 2012, 22, 135–141. [Google Scholar] [CrossRef]
- Hughes, A.L.; Hughes, C.E.; Henderson, K.A.; Yazvenko, N.; Gottschling, D.E. Selective sorting and destruction of mitochondrial membrane proteins in aged yeast. Elife 2016, 5, 1–25. [Google Scholar] [CrossRef]
- Finsterer, J. Mitochondriopathies. Eur. J. Neurol. 2004, 11, 163–186. [Google Scholar] [CrossRef]
- Wei, X.; Du, M.; Xie, J.; Luo, T.; Zhou, Y.; Zhang, K.; Li, J.; Chen, D.; Xu, P.; Jia, M.; et al. Mutations in TOMM70 lead to multi-OXPHOS deficiencies and cause severe anemia, lactic acidosis, and developmental delay. J. Hum. Genet. 2020, 65, 231–240. [Google Scholar] [CrossRef]
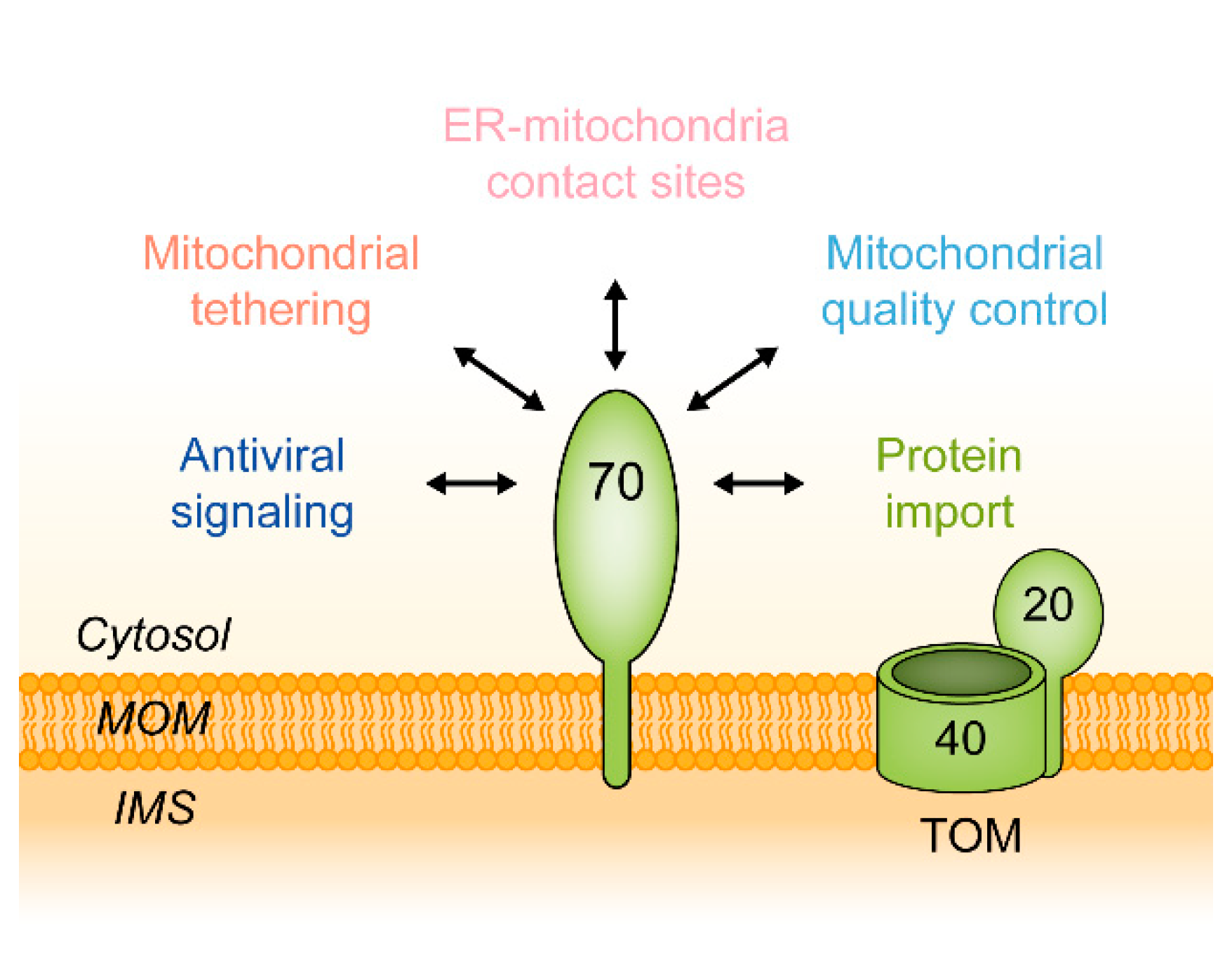


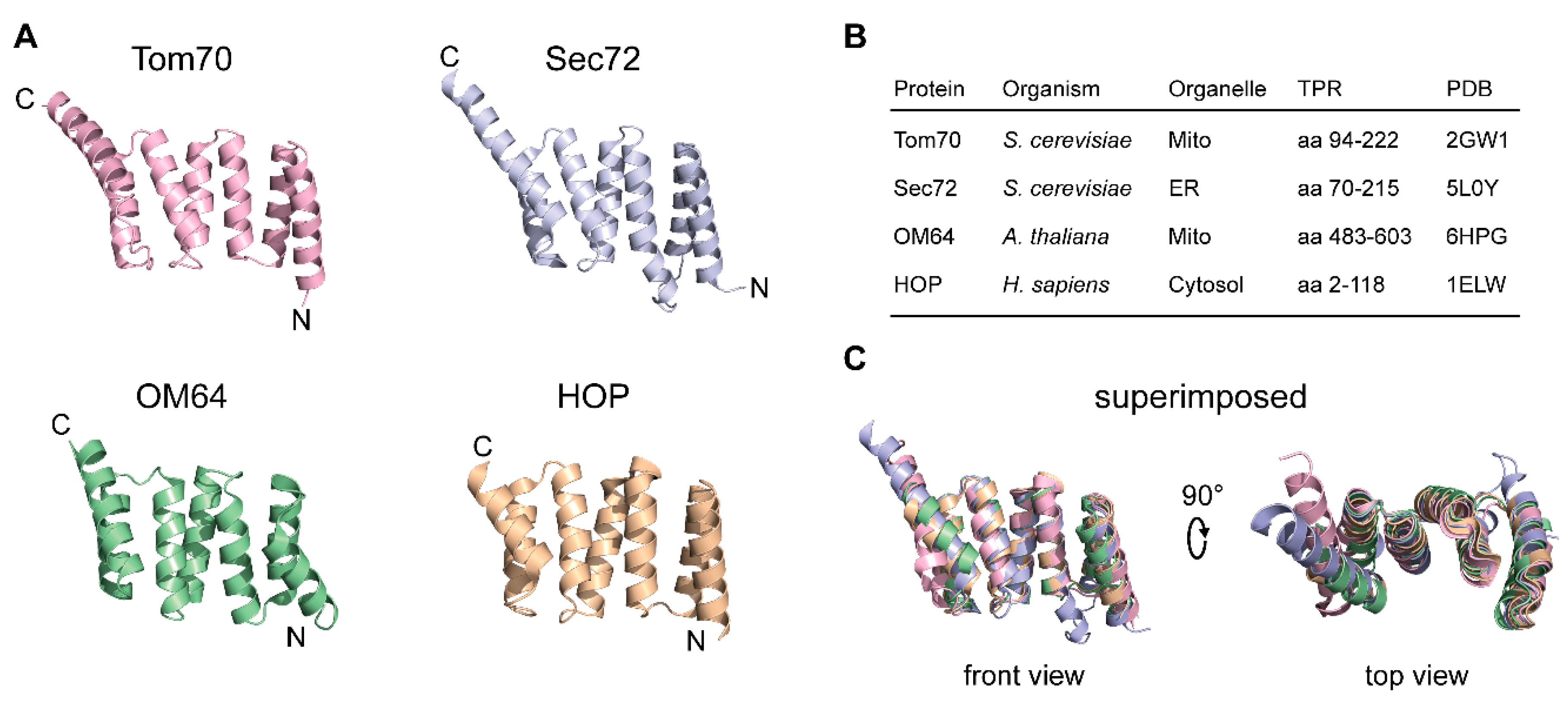



| Receptor | Cellular Context | Substrate Class | Example | Reference | Subcellular Location | Function |
|---|---|---|---|---|---|---|
| S. c. Tom70 | Protein import | Mitochondrial carrier family (MCF) | AAC | [92,112] | MOM, MIM | Translocation of metabolites across membranes |
| β-Barrel proteins | Porin | [109] | MOM | Metabolite translocation across the MOM, VDAC in humans | ||
| Single-membrane-spanning proteins | Atg32 | [95] | MOM | Essential receptor for mitophagy in yeast | ||
| Multi-membrane-spanning proteins | Ugo1 | [96,97] | MOM | Required for mitochondrial fusion | ||
| MPC | [94] | MIM | Translocation of pyruvate across the MIM | |||
| Presequence-containing proteins | Aco1 | [134] | Matrix | Isomerization of citrate to isocitrate via cis-aconitate in the TCA cycle | ||
| iMTS proteins | Atp1, Atp25 | [134,137] | MIM | Subunits of the F1FO-ATPase, contain internal mitochondrial targeting signals (iMTS) | ||
| Ubx2 | [138] | ER, MOM | Recruitment of Cdc48 for removal of arrested proteins from the TOM channel | |||
| TIM complex subunits | Tim54 | [157] | MIM | Recruitment of small Tim proteins | ||
| Cooperation with cytosolic proteins and nucleic acids | Chaperones | Hsp70 | [25] | Cytosol | Mediates association of hydrophobic preproteins with TPR receptors | |
| Co-chaperones | Djp1 | [107] | ER, Cytosol | Co-chaperone of the Hsp40 family | ||
| Other cytosolic factors | SEN subunits | [158] | Cytosol | Mediates tRNA splicing at the MOM | ||
| Mfb1 | [159] | Cytosol | Recruitment of Mfb1 by Tom70/71 is crucial for mitochondrial morphogenesis | |||
| mRNA | [149] | Cytosol | Co-translational protein import | |||
| Cooperation with other membranes | ER proteins | Lam6/ Ltc1 | [160,161] | ER | Formation of ER-mitochondria contact sites via binding to Tom70/71, involved in sterol transfer | |
| Djp1 | [156] | ER, Cytosol | ER surface retrieval pathway (ER-SURF) | |||
| H. s. TOM70 | Protein import | Solute carrier family (SLC25) | ANT1 | [111] | MIM | Mammalian homolog of the AAC |
| Multi-membrane- spanning proteins | PBR | [131] | MOM | Cholesterol import into the MIM | ||
| Presequence-containing proteins | PINK1 | [162] | MOM, Matrix | Induction of mitophagy after mitochondrial depolarization in mammals | ||
| Cooperation with cytosolic proteins and nucleic acids | Chaperones | Hsp70, Hsp90 | [25] | Cytosol | Mediate association of hydrophobic preproteins with N-terminal TPR domain of TOM70 | |
| Co-chaperones | Hsp40 family | [111] | Enhance binding of chaperones to TOM70 | |||
| Autophagy machinery | Atg2 | [163] | Cytosol | Crucial for autophagosome formation | ||
| Other cytosolic factors | mRNA | [148] | Cytosol | Co-translational protein import | ||
| Cooperation with other membranes | ER proteins | IP3R3 | [58] | ER | Ca2+ transfer via ER-mitochondria contact sites formed by IP3R3 and TOM70 | |
| Signaling | Antiviral signaling | MAVS | [27] | MOM | Involved in antiviral signaling cascade triggering an innate immune response | |
| Viral proteins | Orf9b | [28] | MOM | Alternative ORF of nucleocapsid (N) gene of SARS-CoV-2, suppresses IFN-I response via binding to TOM70 |
| Implication | Associated Disease | Involvement of TOM70 | Reference |
|---|---|---|---|
| Leishmania donovani infection | Leishmaniosis | Suppression of apoptosis by mediating import of anti-apoptotic protein MCL-1 | [175] |
| SARS-CoV-2 infection | COVID-19 | Suppression of IFN-I inducing antiviral RIG-I/MAVS cascade through inhibition of TOM70 by Orf9b | [28] |
| Cell survival | Cancer | Ca2+ transfer from ER to mitochondria by binding to IP3R3 | [58] |
| Pathological hypertrophy | Heart failure | Downregulation of TOMM70 leads to increased ROS levels and diminished Opa1 import | [176] |
| Post-MI injury | Heart failure | TOM70 is essential for melatonin-induced protection against post-MI injury | [177] |
| Mitochondrial quality control | PD, ALS, AD | TOM70 is involved in PINK1 import and formation of the TOM/PINK1/Parkin complex upon mitochondrial depolarization | [162,178] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kreimendahl, S.; Rassow, J. The Mitochondrial Outer Membrane Protein Tom70-Mediator in Protein Traffic, Membrane Contact Sites and Innate Immunity. Int. J. Mol. Sci. 2020, 21, 7262. https://doi.org/10.3390/ijms21197262
Kreimendahl S, Rassow J. The Mitochondrial Outer Membrane Protein Tom70-Mediator in Protein Traffic, Membrane Contact Sites and Innate Immunity. International Journal of Molecular Sciences. 2020; 21(19):7262. https://doi.org/10.3390/ijms21197262
Chicago/Turabian StyleKreimendahl, Sebastian, and Joachim Rassow. 2020. "The Mitochondrial Outer Membrane Protein Tom70-Mediator in Protein Traffic, Membrane Contact Sites and Innate Immunity" International Journal of Molecular Sciences 21, no. 19: 7262. https://doi.org/10.3390/ijms21197262
APA StyleKreimendahl, S., & Rassow, J. (2020). The Mitochondrial Outer Membrane Protein Tom70-Mediator in Protein Traffic, Membrane Contact Sites and Innate Immunity. International Journal of Molecular Sciences, 21(19), 7262. https://doi.org/10.3390/ijms21197262





