Abstract
Eosinophilic esophagitis (EoE) is a relatively new condition described as an allergic-mediated disease of the esophagus. Clinically, it is characterized by dysphagia, food impaction, and reflux-like symptoms. Multiple genome-wide association studies (GWAS) have been conducted to identify genetic loci associated with EoE. The integration of numerous studies investigating the genetic polymorphisms in EoE and the Mendelian diseases associated with EoE are discussed to provide insights into the genetic risk of EoE, notably focusing on CCL26 and CAPN14. We focus on the genetic loci investigated thus far, and their classification according to whether the function near the loci is known. The pathophysiology of EoE is described by separately presenting the known function of each cell and molecule, with the major contributors being eosinophils, Th2 cells, thymic stromal lymphopoietin (TSLP), transforming growth factor (TGF)-β1, and interleukin (IL)-13. This review aims to provide detailed descriptions of the genetics and the comprehensive pathophysiology of EoE.
1. Introduction
Eosinophilic esophagitis (EoE) is a relatively new condition first described in 1978; its pathology and phenotype were defined by Atwood et al. in 1993 and Straumann et al. in 1994 [1,2,3,4]. EoE is an allergy-mediated disease of the esophagus that is characterized by significant esophageal eosinophilia and esophageal dysfunction, such as dysphagia and food impaction [1,5,6,7]. Its diagnostic criteria are difficult to define because its symptoms are unspecific and mimic those observed in gastroesophageal reflux disease (GERD). Signs of damage to the esophageal barrier, such as tissue erosions and white exudates, are found in patients with EoE [8]. The disease occurs both in pediatric and adult populations, and is especially common in atopic males [9]. The overall prevalence of EoE seems to be increasing progressively [10]. This phenomenon is a consequence of better recognition patterns that are more likely to contribute to the increase in the incidence and prevalence of this condition [9,11]. Several environmental factors have been reported to contribute to the development of EoE. Twin studies have shown that the prevalence of EoE is more common in cold and dry climates, along with other allergic and autoimmune diseases [9,12,13]. Early life experiences such as premature delivery and antibiotic and acid-suppressant use have been identified to be associated with EoE [14,15,16].
Multiple genome-wide association studies (GWAS) have been conducted to identify genetic loci associated with EoE. Overexpression of several critical genes, including thymic stromal lymphopoietin (TSLP) and calpain 14 (CAPN-14), was found to disrupt the esophageal barrier and enhance immune-mediated inflammation [8,17,18]. We focus on the genetic loci investigated so far, and their classifications, depending on whether the function of the genes near the loci is known. Furthermore, the roles of inflammatory cells and various molecules, particularly TSLP, transforming growth factor (TGF)-β1, and interleukin (IL)-13, in EoE pathophysiology are summarized. This review aims to provide detailed and comprehensive information on both the genetics and pathophysiology of EoE.
2. Definition and Diagnosis of EoE
EoE is defined as a chronic, immune/antigen-mediated esophageal disease characterized clinically by symptoms related to esophageal dysfunction and histologically by eosinophil-predominant inflammation [19,20].
For the last decade, diagnosis of EoE required an esophageal biopsy with 15 or more eosinophils present in one high-power field even with an eight week or longer treatment with maximal dose proton pump inhibitor (PPI). The recently updated consensus criteria in 2017 removed the PPI trial for the diagnosis of EoE. Although ~50% of EoE patients show PPI-responsive esophageal eosinophilia (PPI-REE), it is agreed that as PPI is a treatment for EoE, there is no need to diagnose the disease primarily based on response to treatment [19]. As such, the updated diagnostics criteria for EoE requires symptoms of esophageal dysfunction, ≥15 eosinophils per high-power field (~60 eos/mm2) on an esophageal biopsy, and an assessment of non-EoE disorders that cause or potentially contribute to esophageal eosinophilia (Box 1) [6,19,21].
Box 1. Secondary causes of eosinophilic esophagitis (EoE).
Eosinophilic gastrointestinal diseases
Gastroesophageal reflux disease (GERD)
Celiac disease
Crohn’s disease
Infection
Hypereosinophilic syndrome
Achalasia
Drug hypersensivity
Vasculitis
Pemphigus
Connective tissue disease
Graft vs. host disease
3. Genetics
The EoE transcriptome is a distributed section throughout the human genome, which shows a conserved expression in the esophagus of patients with EoE [22,23]. The EoE transcriptome has 574 genes expressed differently in tissues of children [2,24]. Studying genetic variants in EoE transcriptome provides a deep understanding of the mechanisms of EoE, and there are still many genes remaining with an unknown role in pathophysiology. The most highly expressed gene in the EoE transcriptome is CCL26, the expression of which is induced by IL-13 [22,25,26]. The strongest transcriptional changes occur at 1q21, which encodes the epidermal differentiation complex [22]. Genes included in 1q21, such as filaggrin, show considerably minimal activity in EoE, which leads to the loss of epidermal cell differentiation and impaired barrier function [22,27,28].
3.1. Risk Genes
Candidate gene studies regarding EoE, GWAS, and analysis of its association with other monogenic disorders are the main methods used to study the genetics of EoE [9]. These studies aim to identify genetic loci that may contribute to EoE. Some of the genes have known functions, while others are still unknown [14]. Some genetic risk loci are found in most EoE patients, while some are rare even among these patients. In this review, we present identified, common, and rare genetic risk loci and their known functions.
3.1.1. Common Risk Genes and Function
Table 1 summarizes the common risk genes identified thus far that have known functions with further information on genetic risk loci, the p-value in the case of the GWAS approach, and the name of another approach if used instead of GWAS (this is highlighted inside the bracket). Some of the genes not described in this section are instead described with regard to EoE-associated diseases in Section 3.2.

Table 1.
Common risk genes with known functions.
When allergens are exposed to the esophageal epithelium, epithelial cells and basophils produce TSLP [2,29,30]. TSLP plays an important role in promoting Th2 cell differentiation by inducing the Th2-polarizing capacity of dendritic cells [22,31]. A TSLP single nucleotide polymorphism (SNP) augments the Th2 response [2]. TSLP levels are significantly higher in patients with atopic disease, including EoE [22,32]. In addition, SNPs in a gene encoding a component of the TSLP receptor, CRLF2, are associated with increased EoE risk [33]. Activated Th2 cells produce cytokines such as IL-4, IL-5, and IL-13. IL-4 promotes the differentiation of T cells into Th2 and B cells, eventually leading to IgE secretion [2,34]. IL-5 and IL-13 induce the secretion of eotaxin-3 from epithelial cells [26,29]. The eotaxin-3 gene, which induces eosinophil recruitment to the esophagus, has the strongest transcriptome expression levels, i.e., approximately 53 times higher than in controls [2,35]. IL-13 also reduces the expression levels of genes in the epidermal differentiation complex such as filaggrin and involucrin, thus weakening the barrier function of the squamous epithelium [29,36]. At the same time, locally activated eosinophils and mast cells produce TGF-β1, which triggers fibrotic changes in the esophageal wall, which in turn is mediated by fibroblasts and periostin, thus leading to smooth muscle dysfunction [24,29]. A SNP in the promoter of TGF-β1 is responsible for esophageal dysfunction [2].
CAPN encodes a proteolytic enzyme specifically in the esophagus [18,22]. IL-13 induces the activity of this enzyme, and CAPN14 invokes a pathway that alters basic epithelial cell functions, including barrier integrity [18,22]. STAT6 is known to be an IL-13-activated transcription factor; it induces CAPN expression [37]. Thus, SNPs in CAPN14 and STAT6 are common genetic risk factors in EoE. CAPN14 has also been identified as a regulator of desmoglein 1(DSG1) [38]. DSG1 regulates esophageal epithelial barrier function and immune responses [28]. DSG1 is decreased in EoE and is associated with an impaired barrier phenotype [9,28]. FLG is associated with esophageal barrier integrity maintenance [33,36,39].
LRRC32, which encodes a TGF-β binding protein, and C11orf30, which encodes EMSY, are involved in transcriptional regulation [22]. EMSY and LRRC32 are both expressed in esophageal epithelial cells; however, the roles of these proteins in EoE are yet to be reported [38].
19q13 is another genetic risk locus, and the genes ANKRD27, PDCD5, and RGS9BP are near this location. The protein ANKRD27 inhibits the activity of the SNARE complex, which could have important implications for apical transport in esophageal epithelial cells and in wound healing [14,40,41]. The PDCD5 protein is known to be involved in apoptotic pathways, transcriptional regulation, DNA damage response, and cell cycle control [42]. However, RGS9BP encodes a product not expressed in the esophagus and immune cells [14,43,44]. Further studies are needed to identify the role of these genes, and their associated proteins, in EoE.
Several other genetic risk loci were identified by GWAS, but their functions remain obscure. The locus, tag of genetic variant, and p value of each gene are specified in Table 2 [14,45,46,47,48]. These genes may have yet to be discovered functions that contribute to the genetic mechanism of EoE.

Table 2.
Common risk genes with unknown functions.
3.1.2. Rare Risk Genes and their Function
Studies on risk genes rarely found in EoE patients are emerging. Rochman et al. identified 39 rare variants by performing whole-exome sequencing (WES) in 33 patients; these variants have the potential to alter the biological function of EoE-associated genes [14,27]. Sherrill et al. also performed WES in 63 patients, focusing particularly on families, identifying 5 rare, damaging variants in dehydrogenase E1 and transketolase domain-containing 1 (DHTKD1) [14,49]. Conducting careful studies to identify novel rare genetic variants of EoE will provide insights into the complex pathophysiology of EoE and associated diseases.
3.2. Associated Diseases of EoE
EoE is often studied with associated Mendelian diseases. Table 3 summarizes the Mendelian diseases associated with EoE [14,22,38]. Studying these co-occurring diseases might help to identify certain genes and the corresponding pathogenic mechanisms of EoE.

Table 3.
Mendelian diseases associated with EoE.
Connective tissue disorders (CTDs) (e.g., Loeys-Dietz syndrome (LDS) and Ehlers-Danlos syndrome (hypermobility type)) are the most well-recognized diseases associated with EoE [9,22,50]. A diagnosis of EoE increases the risk of developing a CTD by eight-fold [34,38]. Increased production and/or signaling of TGF-β and dysregulated expression of collagen in the esophagus commonly occur in both CTD and EoE [22,38,51,52,53]. Specifically, LDS is caused by gain-of-function mutations in the TGF-β receptors, TGFBR1, and TGFBR2, whereas Ehlers-Danlos syndrome is caused by genetic mutations in collagen-encoding genes [9,38,54,55,56].
Severe atopy syndrome associated with metabolic wasting (SAM syndrome) also co-occurs with EoE [22]. Downregulation of DSG1 in the esophageal epithelia is reported in both SAM syndrome and EoE [2,8,38]. DSG1 is a major constituent of desmosomes; thus, downregulation of DSG1 leads to the impaired barrier phenotype [9,28].
EoE is enriched in patients with Netherton’s syndrome, which is caused by autosomal dominant loss-of-function mutations in the protease inhibitor SPINK5 [22,38,57,58]. Without SPINK5, which is a regulator of the epidermal proteases kallikrein-related peptidase KLK5 and KLK7, the skin is disrupted substantially [38,59]. The association between EoE and Netherton’s syndrome shows that barrier impairment has a central role in both diseases [38].
PTEN hamartoma tumor syndrome (PHTS) is associated with EoE [9,60]. PHTS carries a >200-fold increased risk for eosinophil-associated gastrointestinal disorders, including EoE [9,60]. PHTS is caused by mutations in the tumor suppressor PTEN, which is a critical regulator of the phosphatidylinositol-4,5-biphosphate 3-kinase (PI3K) pathway [38,61]. Moreover, eosinophils are capable of expressing PTEN [38].
Autosomal-dominant hyper-IgE syndrome, caused by deleterious mutations in STAT3, is associated with EoE [4]. Deleted function of STAT3 leads to dysregulated response to IL-6, which causes deficit in T-helper 17 cells, central T cell memory, and memory B cells [13,38]. STAT3 is activated in eosinophils following IL-5 signaling, but the role of eosinophils in hyper-IgE is yet to be discovered [13,38].
Autosomal-recessive hyper-IgE syndrome, caused by loss-of-function mutations in DOCK8, is also associated with EoE [9,62]. DOCK8, which is expressed on human eosinophils, functions in T-cell homeostasis, and in a durable secondary antibody response [38,63]. It maintains the morphological shape and nuclear integrity of T and NK cells during chemotaxis, through CDC42 and p21-activated kinase (PAK) [38,64].
In addition, ERBB2-interacting protein (ERBIN) deficiency is related to EoE [22,65]. ERBIN downregulates TGF-β signaling [22,65]. EoE is also known to be associated with esophageal granular cell tumors, but it is uncertain whether this is a disease association or consequence of EoE [22,66].
4. Pathophysiology
EoE is caused by an allergic inflammation reaction in patients that have genetic and environmental risks of EoE, and it relies on both the innate and adaptive immune pathways. Thus, the underlying pathophysiology of EoE is complex and diverse pathways are involved, with many immune cells or cytokines contributing to this disease. Figure 1 shows a compact overview of the EoE pathophysiology. Recently, approximately 50% of EoE patients were found to fall into the category of PPI-REE; therefore, the pathophysiological characteristics to distinguish PPI-REE from EoE are important future research fields [19,67].
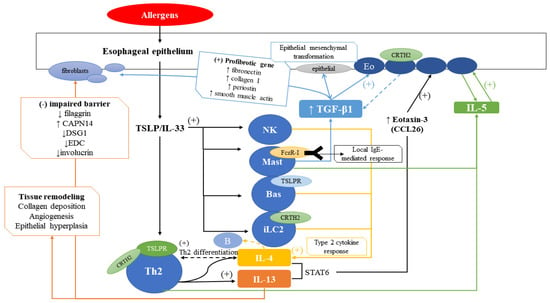
Figure 1.
Overview of EoE pathophysiology. Allergens stimulate the esophageal epithelium, inducing TSLP/IL-33, leading to stimulation of Th2 cells, NK cells, mast cells, basophils, and iLC2. Main receptors on each cell are indicated. NK cells, mast cells, basophils, iLC2, and Th2 cells induce IL-4 which induce Th2 differentiation. IL-4 and IL-13 induced by Th2 cells induce eotaxin-3 (CCL26), which stimulates eosinophils to secrete IL-5. IL-5, secreted by Th2 cells and mast cells, also stimulate eosinophils. Mast cells also induce TGF-β1 which stimulate eosinophils and fibroblasts, as outlined in the blue box. IL-13 induces impaired barrier function and tissue remodeling, as outlined in the orange box.
4.1. Role of Inflammatory Cells
4.1.1. Eosinophils
Eosinophils are recruited from the blood with local chemotaxis and they seem to be integral to EoE disease pathogenesis [2,68,69,70,71]. Eosinophils release eosinophilic peroxidase (EPO), eosinophil cationic protein (ECP), and major binding protein (MBP), which directly causes tissue damage and esophageal dysmotility [2,9,72]. ECP damages cellular membrane barriers, and MBP increases smooth muscle reactivity by causing the dysfunction of vagal muscarinic M2 receptors, while also provoking mast cell and basophil degranulation [2,9]. Eosinophils also serve as antigen-presenting cells (APCs) with MHC-II presentation as well as co-stimulatory molecules (CD40, CD28, CD86, and CD27) [1,9,73]. Eosinophils secrete a variety of cytokines (IL-2, IL-4, IL-6, IL-10, IL-12) which together can activate T cells [9,73]. Eosinophils also produce IL-1, IL-3, IL-4, IL-5, IL-13, TGF-β, eotaxin-3, RANTES, macrophage inflammatory protein 1 (MIP-1), tumor necrosis factor (TNF)-α, granulocyte-macrophage colony-stimulating factor (GM-CSF), platelet-activating factor (PAF), and leukotriene C4 (LTC4) [1,9].
The importance of eosinophils in EoE has been studied in both mouse and human models. Mice genetically engineered to lack eosinophils and mice in which eosinophils are selectively targeted via antibody treatment showed a decrease in the symptoms in many, but not all features [9,69,70,74]. This implies that an EoE-like disease is not completely dependent on eosinophils. In vivo, experimental adoptive transfer of antigen-pulsed eosinophils produced antigen-specific T cell responses, which proves T cell activation by eosinophils [9].
4.1.2. T Cells
In EoE, the numbers of CD3+, CD4+, and CD8+ T cells, as well as the CD8+/CD4+ T cell ratio, increase in the esophageal mucosa [1,9,75,76]. As observed in case of many other allergic reactions, Th2 cells are mainly involved in the inflammatory response [9]. Higher abundances of pathogenic effector Th2 cells (peTh2 cells) were detected in patients with EoE; which were chemoattractant receptor-homologous molecule-positive (CRTH2+), hematopoietic prostaglandin D synthase-positive (HPSD+), and CD161 high CD4+ T cells [22,77,78]. CRTH2 was present on peTH2 cells, eosinophils, and basophils and in response to prostaglandin D2 changed the chemotaxis of these cells [22]. Th2 cells produce type 2 cytokines such as IL-4, IL-5, and IL-13, which play a key role in the pathogenesis of EoE. In murine models, recombination activation gene 1 (RAG1) knockout mice were completely protected from experimentally derived EoE, whereas CD4 knockout mice were partially protected and CD8 and B cell knockout mice were not protected [1,9,79].
Regulatory T cells were increased in esophageal biopsies, but not in the percentage of total T cells; thus, their significance in EoE is unclear [1].
4.1.3. Mast Cells
The esophageal mast cell content increased significantly, and mast cell degranulation was detected in nearly all patients with EoE [1,33,80,81]. Genes specific to mast cells, such as those that encode carboxypeptidase 3A (CPA3), FcεR-I, and tryptase (TPSAB1), were highly expressed in the EoE transcriptome [1,9]. IgE was bound by the FcεR-I receptor in the membrane of mast cells, and IgE may have contributed to a local IgE-mediated immediate hypersensitivity response in the esophagus [9,80]. Mast cells secrete diverse products such as cytokines, proteases, and bioactive compounds, and many of these products lead to esophageal remodeling and dysmotility [9,33].
In murine models, mast cell numbers and eosinophil numbers increased as EoE symptoms increased [1,81]. Mice with depleted IgE and mast cells still showed EoE-like symptoms, but they showed reduced muscle cell hyperplasia and hypertrophy [9,81,82]. This implied that although mast cells were not required in experimentally derived EoE, they were required to increase the thickness of the muscularis mucosa [33]. Further studies in murine models have found that both IL-5 and IL-9 transgenic mice had increased numbers of mast cells, suggesting that both IL-5 and IL-9 promote mast cell activation and maturation [1,83]. A clinical trial using IL-5 antibodies showed that mast cell numbers were correlated with EoE symptom severity, while eosinophil numbers were not [9,84].
4.1.4. Basophils
Basophils are known to play a key role in allergic responses. Increased esophageal basophilia was observed in EoE, and these basophils showed increased ST2 expression [1,85,86]. Basophils have been reported to secrete various type 2 cytokines and act as antigen presenting cells (APCs) to induce Th2 cells [33,87]. In particular, basophils expressed the receptor of TSLP (TSLPR) [2,88]. Basophil-deficiency led to the prevention of EoE; furthermore, TSLP and basophils are required to maintain EoE after disease establishment [89].
By observing EoE biopsies, Siracusa et al. showed that TSLP affected basophil hematopoiesis [86]. In a murine model of EoE, Noti et al. suggested that basophils were critical to eosinophil recruitment via TSLP [1,90]. In addition, in a TSLP-dependent, experimentally derived EoE model, an increased number of basophils were recruited in an ST2-dependent manner, implying that IL-33 may induce basophil recruitment [33,91].
4.1.5. Dendritic Cells
Dendritic cells normally reside in the esophageal epithelium and an increased number of these cells were found in patients with EoE [33,75,76]. There was evidence that dendritic cells may present antigens in EoE [9,79,92,93]. Langerhans cells, a type of dendritic cells found in the esophagus, expressed FcεRI, which was correlated with the Th2 response level [9,94]. These Th2 responses were induced by allergens and/or environmental adjuvants, likely via communication between resident stromal and dendritic cells [9,95,96].
4.1.6. Innate Lymphoid Cells
Innate lymphoid cells (ILCs) are resident immune cells in tissues that may serve as sources of type 2 cytokines [9,33,97,98,99]. Group 2 ILCs (ILC2s) expressed CRTH2 and secreted large quantities of type 2 cytokines in response to IL-25, IL-33, and TSLP [9].
In murine models, ILCs were important in infection and inflammation responses, as well as in tissue repair of EoE [9,99]. Doherty et al. reported that ILC2s were present in EoE biopsies, with an increased level in active EoE, and this correlated with the number of eosinophils found in biopsies [9,99].
4.1.7. Invariant Natural Killer T Cells (iNKT Cells)
Invariant natural killer T cells (iNKT cells) recognize lipid and glycolipid antigens that are presented by CD1d molecules, and they have the capacity to produce type 2 cytokines [9,33,100,101,102]. Mucosal iNKT tolerance to environmental antigens can mediate allergic sensitization and tissue inflammation in the absence of tolerance [9,103].
In murine models, CD1d-deficient mice were protected from experimental EoE, and activation of iNKT was sufficient to induce EoE. Furthermore, iNKT neutralized mice were also protected from experimental EoE [9,104,105]. Recently, a possible role of iNKT cells in protecting from EoE-specific pathologies was shown in RAG1-deficient mice [9]. Fewer iNKT cells were found in the peripheral blood of patients, while increased numbers of iNKT were found in the esophagus [33,105,106]. iNKTs from patients expanded more readily and produced more IL-13 in response to stimulation [9,106].
4.1.8. B Cells
Mouse models of EoE showed that B cell-deficient mice still developed EoE, suggesting that EoE does not rely on B cells [1,79].
4.2. Role of Various Molecules
4.2.1. TSLP
TSLP and its receptor TSLPR are implicated in various EoE pathways. TSLP expression was increased in esophageal tissues in patients with EoE [89]. TSLP mainly induced a type 2 immune response [89]. TSLPR-deficient mice were protected from experimentally derived EoE [89,90].
4.2.2. TGF-β1
TGF-β1, produced by mast cells, eosinophils, and esophageal epithelial cells, is a key cytokine for epithelial fibrosis and epithelial cell transformation [9]. Elevated expression of TGF-β1 was found in the esophageal biopsy samples of patients with active EoE when compared to that in samples from control patients or patients with GERD [9,70,107,108,109].
TGF-β1 can have profibrotic effects on esophageal fibroblasts [89] and TGF-β1 can directly induce the expression of profibrotic genes such as fibronectin, collagen I, periostin, and smooth muscle actin in EoE fibroblasts [89]. Studies on murine models support this function; the TGF-β1 pathway mediator SMAD2/3 was important in esophageal fibrosis, and SMAD3-deficient mice were partially protected from EoE-associated fibrosis [68,110]. TGF-β1 also affects associated cellular functions. TGF-β1 can alter the contraction of collagen gels in esophageal smooth muscle cells [52,53,68]. This function has recently been found to rely on the expression of phospholamban (PLN), a protein that regulates calcium flux [52,68]. EoE esophageal smooth muscles expressed PLN, while the absence of PLN was found in controls [52,68]. Inhibition of PLN expression and signaling through TGF-β receptor I both decreased esophageal smooth muscle contraction in response to TGF-β1 [52,68].
Studies of human epithelial cells showed that TGF-β1 induced epithelial mesenchymal transformation, with increased vimentin expression [68,111]. The degree of epithelial mesenchymal transformation correlated positively with both TGF-β1 expression and eosinophil numbers [53,68,107].
4.2.3. IL-4
IL-4 is secreted by Th2 cells, NK cells, and TSLP-dependent basophils [2,34]. IL-4, but not IL-13, induced Th2 cell differentiation, differentiation of naive T cells into Th2 and active B cells class switching to produce IgE [9].
4.2.4. IL-5
IL-5 is secreted by Th2 cells, mast cells, and eosinophils [9,112]. IL-5 promotes eosinophil proliferation, survival, activation, and chemotaxis [1]. IL-5 and its receptor were expressed in esophageal tissue in EoE, and it increased the IL-5 mRNA and protein expression levels, which were observed in the esophagus of EoE patients [89,93]. Previous studies of anti-IL-5 therapy in humans were not effective in improving the symptoms of EoE, although protective effects against esophageal eosinophilia were observed [9,109].
4.2.5. IL-13
IL-13, a key cytokine and the most studied cytokine in EoE pathogenesis, is secreted by Th2 cells and activates eosinophils [9,93,113,114]. Esophageal IL-13 overexpression by Th2 cells induces CCL26, eotaxin-3, and periostin expression, eosinophilic recruitment by upregulation of an eosinophil chemokine, and CAPN14 expression, which was responsible for STAT6 and IL-33 production [2,34]. IL-13 downregulates the expression of DSG-1, filaggrin, EDC, and involucrin, which are proteins important in epithelial integrity and barrier function [9,28,36]. In addition, independent of eosinophilia, IL-13 induces tissue remodeling by promoting collagen deposition, angiogenesis, and epithelial hyperplasia [9,71].
In murine models, IL-13 induced EoE and tissue remodeling whereas IL-13-deficient mice showed improvements in esophageal symptoms [9,71,115]. In human trials, anti-IL-13 reduces tissue eosinophilia [1,116,117].
4.2.6. IL-15
An increased amount of IL-15 is observed in patients with EoE and in murine models [9]. IL-15 contributes to CD4+ T and iNKT cell growth, including the synthesis of IL-5 and IL-13 in EoE [9,25,36,70,118].
4.2.7. Eotaxin-3 (CCL26)
Eotaxin-3, mainly produced by esophageal epithelial cells through the IL-13 signaling pathway, and is implicated in eosinophil trafficking to the esophagus in patients with EoE [9,26]. Eotaxin-3 is the most abundant EoE chemokine regardless of the age, sex, and atopic status of the patient [1,26,119]. Microarray analysis showed that eotaxin-3 has the largest fold change in mRNA expression level between patients with EoE and controls [89].
In murine models, mice lacking the eotaxin receptor CCR3 were protected from developing experimental EoE [9,26].
4.2.8. IgE and IgG4
IgE is a cytokine that contributes to many atopic pathways; however, growing evidence suggests that IgE has no direct role in EoE [89]. B cell-deficient mice still developed EoE [1,79]. IgE was not elevated in all patients with EoE and omalizumab, an anti-IgE monoclonal antibody, was ineffective in the treatment of EoE [89,120].
Increased levels of IgG4 were observed in EoE esophageal tissues [89,120,121]. However, the specific contribution of IgG4 in EoE is yet to be discovered [89].
4.2.9. Prostaglandins
Prostaglandins affect the eosinophil pathway in the esophagus [9,122]. Prostaglandin inhibitors showed protective effects against EoE-like inflammation [9,123]. The chemoattractant receptor (CRTH2) expressed on Th2 cells is the receptor for prostaglandin D2 (PGD2) and mediated the chemotaxis of Th2 cells, eosinophils, and basophils [9,123].
4.2.10. Additional Cytokines
Several studies have identified additional cytokines in the EoE pathway. Collison et al. found that TNF-related, apoptosis-inducing ligand (TRAIL) controlled MID1 and TSLP expression, inflammation, fibrosis, smooth muscle hypertrophy, and expression of cytokines in experimentally derived EoE. These included cytokines such as TSLP, CCL11, CCL20, CCL24, IL-5, IL-13, IL-25, and TGF-β [9,124]. De Souza et al. identified that macrophage migration inhibitory factor (MIF) induced eosinophil infiltration and remodeling in EoE in a murine model [9,125]. Dutt et al. identified that allergen-induced IL-18 promoted IL-5-and iNKT-dependent EoE pathology [9,126].
5. Summary
Eosinophilic esophagitis (EoE) is a relatively new allergy-mediated condition. In this comprehensive review, we focused on providing a detailed description of both common and rare genetic risk loci of EoE. The function of various common and rare genetic loci remains unknown (Table 2, Section 3.1.2.), and further genetic studies should focus on revealing these roles. The pathophysiology of EoE is complex, with a network of various cells and molecules contributing to it, especially eosinophils, Th2 cells, TSLP, TGF-β1, and IL-13. Molecules that have not yet been identified may contribute to the mechanism of EoE. Moreover, although patients with PPI-REE account for about 50% of patients with EoE, the distinguishing genetics and pathophysiology have not yet been identified and should be investigated further.
Author Contributions
All authors have read and agreed to the published version of the manuscript. All authors made substantial contributions to all of the following: (1) conception and design of the study, data acquisition, and analysis and interpretation of data; (2) drafting or critical revision of the article for intellectual content; and (3) final approval of version to be submitted.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Clayton, F.; Peterson, K. Eosinophilic Esophagitis: Pathophysiology and Definition. Gastrointest. Endosc. Clin. N. Am. 2018, 28, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Vinit, C.; Dieme, A.; Courbage, S.; Dehaine, C.; Dufeu, C.M.; Jacquemot, S.; Lajus, M.; Montigny, L.; Payen, E.; Yang, D.D.; et al. Eosinophilic esophagitis: Pathophysiology, diagnosis, and management. Arch. Pediatr. 2019, 26, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Landres, R.T.; Kuster, G.G.; Strum, W.B. Eosinophilic esophagitis in a patient with vigorous achalasia. Gastroenterology 1978, 74, 1298–1301. [Google Scholar] [CrossRef]
- Straumann, A.; Spichtin, H.P.; Bernoulli, R.; Loosli, J.; Vogtlin, J. Idiopathic eosinophilic esophagitis: A frequently overlooked disease with typical clinical aspects and discrete endoscopic findings. Schweiz. Med. Wochenschr. 1994, 124, 1419–1429. [Google Scholar] [PubMed]
- Lehman, H.K.; Lam, W. Eosinophilic Esophagitis. Pediatr. Clin. N. Am. 2019, 66, 955–965. [Google Scholar] [CrossRef] [PubMed]
- Dellon, E.S.; Gonsalves, N.; Hirano, I.; Furuta, G.T.; Liacouras, C.A.; Katzka, D.A.; American College of Gastroenterology. ACG clinical guideline: Evidenced based approach to the diagnosis and management of esophageal eosinophilia and eosinophilic esophagitis (EoE). Am. J. Gastroenterol. 2013, 108, 679–692, quiz 693. [Google Scholar] [CrossRef]
- Moawad, F.J.; Cheng, E.; Schoepfer, A.; Al-Haddad, S.; Bellizzi, A.M.; Dawson, H.; El-Zimaity, H.; Guindi, M.; Penagini, R.; Safrooneva, E.; et al. Eosinophilic esophagitis: Current perspectives from diagnosis to management. Ann. N. Y. Acad. Sci. 2016, 1380, 204–217. [Google Scholar] [CrossRef]
- Pesek, R.D.; Gupta, S.K. Emerging drugs for eosinophilic esophagitis. Expert Opin. Emerg. Drugs 2018, 23, 173–183. [Google Scholar] [CrossRef]
- Davis, B.P. Pathophysiology of Eosinophilic Esophagitis. Clin. Rev. Allergy Immunol. 2018, 55, 19–42. [Google Scholar] [CrossRef]
- Navarro, P.; Arias, A.; Arias-Gonzalez, L.; Laserna-Mendieta, E.J.; Ruiz-Ponce, M.; Lucendo, A.J. Systematic review with meta-analysis: The growing incidence and prevalence of eosinophilic oesophagitis in children and adults in population-based studies. Aliment. Pharmacol. Ther. 2019, 49, 1116–1125. [Google Scholar] [CrossRef]
- DeBrosse, C.W.; Collins, M.H.; Buckmeier Butz, B.K.; Allen, C.L.; King, E.C.; Assa’ad, A.H.; Abonia, J.P.; Putnam, P.E.; Rothenberg, M.E.; Franciosi, J.P. Identification, epidemiology, and chronicity of pediatric esophageal eosinophilia, 1982–1999. J. Allergy Clin. Immunol. 2010, 126, 112–119. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hurrell, J.M.; Genta, R.M.; Dellon, E.S. Prevalence of esophageal eosinophilia varies by climate zone in the United States. Am. J. Gastroenterol. 2012, 107, 698–706. [Google Scholar] [CrossRef] [PubMed]
- Spergel, J.M.; Book, W.M.; Mays, E.; Song, L.; Shah, S.S.; Talley, N.J.; Bonis, P.A. Variation in prevalence, diagnostic criteria, and initial management options for eosinophilic gastrointestinal diseases in the United States. J. Pediatr. Gastroenterol. Nutr. 2011, 52, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Kottyan, L.C.; Parameswaran, S.; Weirauch, M.T.; Rothenberg, M.E.; Martin, L.J. The genetic etiology of eosinophilic esophagitis. J. Allergy Clin. Immunol. 2020, 145, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Furuta, G.T.; Katzka, D.A. Eosinophilic Esophagitis. N. Engl. J. Med. 2015, 373, 1640–1648. [Google Scholar] [CrossRef]
- Jensen, E.T.; Kuhl, J.T.; Martin, L.J.; Rothenberg, M.E.; Dellon, E.S. Prenatal, intrapartum, and postnatal factors are associated with pediatric eosinophilic esophagitis. J. Allergy Clin. Immunol. 2018, 141, 214–222. [Google Scholar] [CrossRef]
- Chandramouleeswaran, P.M.; Shen, D.; Lee, A.J.; Benitez, A.; Dods, K.; Gambanga, F.; Wilkins, B.J.; Merves, J.; Noah, Y.; Toltzis, S.; et al. Preferential Secretion of Thymic Stromal Lymphopoietin (TSLP) by Terminally Differentiated Esophageal Epithelial Cells: Relevance to Eosinophilic Esophagitis (EoE). PLoS ONE 2016, 11, e0150968. [Google Scholar] [CrossRef]
- Litosh, V.A.; Rochman, M.; Rymer, J.K.; Porollo, A.; Kottyan, L.C.; Rothenberg, M.E. Calpain-14 and its association with eosinophilic esophagitis. J. Allergy Clin. Immunol. 2017, 139, 1762–1771.e1767. [Google Scholar] [CrossRef]
- Dellon, E.S.; Liacouras, C.A.; Molina-Infante, J.; Furuta, G.T.; Spergel, J.M.; Zevit, N.; Spechler, S.J.; Attwood, S.E.; Straumann, A.; Aceves, S.S.; et al. Updated International Consensus Diagnostic Criteria for Eosinophilic Esophagitis: Proceedings of the AGREE Conference. Gastroenterology 2018, 155, 1022–1033.e10. [Google Scholar] [CrossRef]
- Lucendo, A.J.; Molina-Infante, J.; Arias, A.; von Arnim, U.; Bredenoord, A.J.; Bussmann, C.; Amil Dias, J.; Bove, M.; Gonzalez-Cervera, J.; Larsson, H.; et al. Guidelines on eosinophilic esophagitis: Evidence-based statements and recommendations for diagnosis and management in children and adults. United Eur. Gastroenterol. J. 2017, 5, 335–358. [Google Scholar] [CrossRef]
- Singla, M.B.; Moawad, F.J. An Overview of the Diagnosis and Management of Eosinophilic Esophagitis. Clin. Transl. Gastroenterol. 2016, 7, e155. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, K.M.; Aceves, S.S.; Dellon, E.S.; Gupta, S.K.; Spergel, J.M.; Furuta, G.T.; Rothenberg, M.E. Pathophysiology of Eosinophilic Esophagitis. Gastroenterology 2018, 154, 333–345. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.K.; Fang, R.; Wagner, B.D.; Choe, H.N.; Kelly, C.J.; Schroeder, S.; Moore, W.; Stevens, M.J.; Yeckes, A.; Amsden, K.; et al. Esophageal microbiome in eosinophilic esophagitis. PLoS ONE 2015, 10, e0128346. [Google Scholar] [CrossRef] [PubMed]
- Sherrill, J.D.; Rothenberg, M.E. Genetic dissection of eosinophilic esophagitis provides insight into disease pathogenesis and treatment strategies. J. Allergy Clin. Immunol. 2011, 128, 23–32, quiz 33-24. [Google Scholar] [CrossRef]
- Blanchard, C.; Mingler, M.K.; Vicario, M.; Abonia, J.P.; Wu, Y.Y.; Lu, T.X.; Collins, M.H.; Putnam, P.E.; Wells, S.I.; Rothenberg, M.E. IL-13 involvement in eosinophilic esophagitis: Transcriptome analysis and reversibility with glucocorticoids. J. Allergy Clin. Immunol. 2007, 120, 1292–1300. [Google Scholar] [CrossRef]
- Blanchard, C.; Wang, N.; Stringer, K.F.; Mishra, A.; Fulkerson, P.C.; Abonia, J.P.; Jameson, S.C.; Kirby, C.; Konikoff, M.R.; Collins, M.H.; et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J. Clin. Investig. 2006, 116, 536–547. [Google Scholar] [CrossRef]
- Rochman, M.; Travers, J.; Miracle, C.E.; Bedard, M.C.; Wen, T.; Azouz, N.P.; Caldwell, J.M.; Kc, K.; Sherrill, J.D.; Davis, B.P.; et al. Profound loss of esophageal tissue differentiation in patients with eosinophilic esophagitis. J. Allergy Clin. Immunol. 2017, 140, 738–749.e733. [Google Scholar] [CrossRef]
- Sherrill, J.D.; Kc, K.; Wu, D.; Djukic, Z.; Caldwell, J.M.; Stucke, E.M.; Kemme, K.A.; Costello, M.S.; Mingler, M.K.; Blanchard, C.; et al. Desmoglein-1 regulates esophageal epithelial barrier function and immune responses in eosinophilic esophagitis. Mucosal Immunol. 2014, 7, 718–729. [Google Scholar] [CrossRef]
- Abe, Y.; Sasaki, Y.; Yagi, M.; Yaoita, T.; Nishise, S.; Ueno, Y. Diagnosis and treatment of eosinophilic esophagitis in clinical practice. Clin. J. Gastroenterol. 2017, 10, 87–102. [Google Scholar] [CrossRef]
- Comeau, M.R.; Ziegler, S.F. The influence of TSLP on the allergic response. Mucosal Immunol. 2010, 3, 138–147. [Google Scholar] [CrossRef]
- Kitajima, M.; Lee, H.C.; Nakayama, T.; Ziegler, S.F. TSLP enhances the function of helper type 2 cells. Eur. J. Immunol. 2011, 41, 1862–1871. [Google Scholar] [CrossRef] [PubMed]
- Hui, C.C.; Rusta-Sallehy, S.; Asher, I.; Heroux, D.; Denburg, J.A. The effects of thymic stromal lymphopoietin and IL-3 on human eosinophil-basophil lineage commitment: Relevance to atopic sensitization. Immun. Inflamm. Dis. 2014, 2, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, J.M.; Paul, M.; Rothenberg, M.E. Novel immunologic mechanisms in eosinophilic esophagitis. Curr. Opin. Immunol. 2017, 48, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Rothenberg, M.E. Molecular, genetic, and cellular bases for treating eosinophilic esophagitis. Gastroenterology 2015, 148, 1143–1157. [Google Scholar] [CrossRef]
- Blanchard, C.; Wang, N.; Rothenberg, M.E. Eosinophilic esophagitis: Pathogenesis, genetics, and therapy. J. Allergy Clin. Immunol. 2006, 118, 1054–1059. [Google Scholar] [CrossRef]
- Blanchard, C.; Stucke, E.M.; Burwinkel, K.; Caldwell, J.M.; Collins, M.H.; Ahrens, A.; Buckmeier, B.K.; Jameson, S.C.; Greenberg, A.; Kaul, A.; et al. Coordinate interaction between IL-13 and epithelial differentiation cluster genes in eosinophilic esophagitis. J. Immunol. 2010, 184, 4033–4041. [Google Scholar] [CrossRef]
- Miller, D.E.; Forney, C.; Rochman, M.; Cranert, S.; Habel, J.; Rymer, J.; Lynch, A.; Schroeder, C.; Lee, J.; Sauder, A.; et al. Genetic, Inflammatory, and Epithelial Cell Differentiation Factors Control Expression of Human Calpain-14. G3 (Bethesda) 2019, 9, 729–736. [Google Scholar] [CrossRef]
- Kottyan, L.C.; Rothenberg, M.E. Genetics of eosinophilic esophagitis. Mucosal Immunol. 2017, 10, 580–588. [Google Scholar] [CrossRef]
- Simon, D.; Radonjic-Hosli, S.; Straumann, A.; Yousefi, S.; Simon, H.U. Active eosinophilic esophagitis is characterized by epithelial barrier defects and eosinophil extracellular trap formation. Allergy 2015, 70, 443–452. [Google Scholar] [CrossRef]
- Tamura, K.; Ohbayashi, N.; Ishibashi, K.; Fukuda, M. Structure-function analysis of VPS9-ankyrin-repeat protein (Varp) in the trafficking of tyrosinase-related protein 1 in melanocytes. J. Biol. Chem. 2011, 286, 7507–7521. [Google Scholar] [CrossRef]
- Fukuda, M. Multiple Roles of VARP in Endosomal Trafficking: Rabs, Retromer Components and R-SNARE VAMP7 Meet on VARP. Traffic 2016, 17, 709–719. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Ma, D.; Chen, Y. Cellular functions of programmed cell death 5. Biochim. Biophys. Acta 2016, 1863, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Wensel, T.G. R9AP, a membrane anchor for the photoreceptor GTPase accelerating protein, RGS9-1. Proc. Natl. Acad. Sci. USA 2002, 99, 9755–9760. [Google Scholar] [CrossRef] [PubMed]
- Sundermeier, T.R.; Vinberg, F.; Mustafi, D.; Bai, X.; Kefalov, V.J.; Palczewski, K. R9AP overexpression alters phototransduction kinetics in iCre75 mice. Investig. Ophthalmol. Vis. Sci. 2014, 55, 1339–1347. [Google Scholar] [CrossRef] [PubMed]
- Kottyan, L.C.; Maddox, A.; Braxton, J.R.; Stucke, E.M.; Mukkada, V.; Putnam, P.E.; Abonia, J.P.; Chehade, M.; Wood, R.A.; Pesek, R.D.; et al. Genetic variants at the 16p13 locus confer risk for eosinophilic esophagitis. Genes Immun. 2019, 20, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Rothenberg, M.E.; Spergel, J.M.; Sherrill, J.D.; Annaiah, K.; Martin, L.J.; Cianferoni, A.; Gober, L.; Kim, C.; Glessner, J.; Frackelton, E.; et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nat. Genet. 2010, 42, 289–291. [Google Scholar] [CrossRef]
- Sleiman, P.M.; Wang, M.L.; Cianferoni, A.; Aceves, S.; Gonsalves, N.; Nadeau, K.; Bredenoord, A.J.; Furuta, G.T.; Spergel, J.M.; Hakonarson, H. GWAS identifies four novel eosinophilic esophagitis loci. Nat. Commun. 2014, 5, 5593. [Google Scholar] [CrossRef]
- Kottyan, L.C.; Davis, B.P.; Sherrill, J.D.; Liu, K.; Rochman, M.; Kaufman, K.; Weirauch, M.T.; Vaughn, S.; Lazaro, S.; Rupert, A.M.; et al. Genome-wide association analysis of eosinophilic esophagitis provides insight into the tissue specificity of this allergic disease. Nat. Genet. 2014, 46, 895–900. [Google Scholar] [CrossRef]
- Sherrill, J.D.; Kc, K.; Wang, X.; Wen, T.; Chamberlin, A.; Stucke, E.M.; Collins, M.H.; Abonia, J.P.; Peng, Y.; Wu, Q.; et al. Whole-exome sequencing uncovers oxidoreductases DHTKD1 and OGDHL as linkers between mitochondrial dysfunction and eosinophilic esophagitis. JCI Insight 2018, 3, e99922. [Google Scholar] [CrossRef]
- Abonia, J.P.; Wen, T.; Stucke, E.M.; Grotjan, T.; Griffith, M.S.; Kemme, K.A.; Collins, M.H.; Putnam, P.E.; Franciosi, J.P.; von Tiehl, K.F.; et al. High prevalence of eosinophilic esophagitis in patients with inherited connective tissue disorders. J. Allergy Clin. Immunol. 2013, 132, 378–386. [Google Scholar] [CrossRef]
- Frischmeyer-Guerrerio, P.A.; Guerrerio, A.L.; Oswald, G.; Chichester, K.; Myers, L.; Halushka, M.K.; Oliva-Hemker, M.; Wood, R.A.; Dietz, H.C. TGFbeta receptor mutations impose a strong predisposition for human allergic disease. Sci. Transl. Med. 2013, 5, 195ra194. [Google Scholar] [CrossRef] [PubMed]
- Beppu, L.Y.; Anilkumar, A.A.; Newbury, R.O.; Dohil, R.; Broide, D.H.; Aceves, S.S. TGF-beta1-induced phospholamban expression alters esophageal smooth muscle cell contraction in patients with eosinophilic esophagitis. J. Allergy Clin. Immunol. 2014, 134, 1100–1107.e1104. [Google Scholar] [CrossRef] [PubMed]
- Aceves, S.S.; Chen, D.; Newbury, R.O.; Dohil, R.; Bastian, J.F.; Broide, D.H. Mast cells infiltrate the esophageal smooth muscle in patients with eosinophilic esophagitis, express TGF-beta1, and increase esophageal smooth muscle contraction. J. Allergy Clin. Immunol. 2010, 126, 1198–1204.e1194. [Google Scholar] [CrossRef] [PubMed]
- Sobey, G. Ehlers-Danlos syndrome: How to diagnose and when to perform genetic tests. Arch. Dis. Child. 2015, 100, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Syx, D.; Van Damme, T.; Symoens, S.; Maiburg, M.C.; van de Laar, I.; Morton, J.; Suri, M.; Del Campo, M.; Hausser, I.; Hermanns-Le, T.; et al. Genetic heterogeneity and clinical variability in musculocontractural Ehlers-Danlos syndrome caused by impaired dermatan sulfate biosynthesis. Hum. Mutat. 2015, 36, 535–547. [Google Scholar] [CrossRef]
- Dietz, H. Marfan Syndrome. In GeneReviews; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Stephens, K., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 2001. Available online: https://www.ncbi.nlm.nih.gov/books/NBK1335/ (accessed on 30 September 2020).
- Paluel-Marmont, C.; Bellon, N.; Barbet, P.; Leclerc-Mercier, S.; Hadj-Rabia, S.; Dupont, C.; Bodemer, C. Eosinophilic esophagitis and colonic mucosal eosinophilia in Netherton syndrome. J. Allergy Clin. Immunol. 2017, 139, 2003–2005.e2001. [Google Scholar] [CrossRef]
- Brown, S.J.; McLean, W.H. Eczema genetics: Current state of knowledge and future goals. J. Investig. Dermatol. 2009, 129, 543–552. [Google Scholar] [CrossRef]
- Furio, L.; Pampalakis, G.; Michael, I.P.; Nagy, A.; Sotiropoulou, G.; Hovnanian, A. KLK5 Inactivation Reverses Cutaneous Hallmarks of Netherton Syndrome. PLoS Genet. 2015, 11, e1005389. [Google Scholar] [CrossRef]
- Stewart, M.J.; Shaffer, E.; Urbanski, S.J.; Beck, P.L.; Storr, M.A. The association between celiac disease and eosinophilic esophagitis in children and adults. BMC Gastroenterol. 2013, 13, 96. [Google Scholar] [CrossRef]
- Milella, M.; Falcone, I.; Conciatori, F.; Cesta Incani, U.; Del Curatolo, A.; Inzerilli, N.; Nuzzo, C.M.; Vaccaro, V.; Vari, S.; Cognetti, F.; et al. PTEN: Multiple Functions in Human Malignant Tumors. Front. Oncol. 2015, 5, 24. [Google Scholar] [CrossRef]
- Lyons, J.J.; Sun, G.; Stone, K.D.; Nelson, C.; Wisch, L.; O’Brien, M.; Jones, N.; Lindsley, A.; Komarow, H.D.; Bai, Y.; et al. Mendelian inheritance of elevated serum tryptase associated with atopy and connective tissue abnormalities. J. Allergy Clin. Immunol. 2014, 133, 1471–1474. [Google Scholar] [CrossRef] [PubMed]
- Randall, K.L.; Lambe, T.; Johnson, A.L.; Treanor, B.; Kucharska, E.; Domaschenz, H.; Whittle, B.; Tze, L.E.; Enders, A.; Crockford, T.L.; et al. Dock8 mutations cripple B cell immunological synapses, germinal centers and long-lived antibody production. Nat. Immunol. 2009, 10, 1283–1291. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Dove, C.G.; Hor, J.L.; Murdock, H.M.; Strauss-Albee, D.M.; Garcia, J.A.; Mandl, J.N.; Grodick, R.A.; Jing, H.; Chandler-Brown, D.B.; et al. DOCK8 regulates lymphocyte shape integrity for skin antiviral immunity. J. Exp. Med. 2014, 211, 2549–2566. [Google Scholar] [CrossRef] [PubMed]
- Lyons, J.J.; Liu, Y.; Ma, C.A.; Yu, X.; O’Connell, M.P.; Lawrence, M.G.; Zhang, Y.; Karpe, K.; Zhao, M.; Siegel, A.M.; et al. ERBIN deficiency links STAT3 and TGF-beta pathway defects with atopy in humans. J. Exp. Med. 2017, 214, 669–680. [Google Scholar] [CrossRef]
- Riffle, M.E.; Polydorides, A.D.; Niakan, J.; Chehade, M. Eosinophilic Esophagitis and Esophageal Granular Cell Tumor: An Unexpected Association. Am. J. Surg. Pathol. 2017, 41, 616–621. [Google Scholar] [CrossRef]
- Shoda, T.; Matsuda, A.; Nomura, I.; Okada, N.; Orihara, K.; Mikami, H.; Ishimura, N.; Ishihara, S.; Matsumoto, K.; Kinoshita, Y. Eosinophilic esophagitis versus proton pump inhibitor-responsive esophageal eosinophilia: Transcriptome analysis. J. Allergy Clin. Immunol. 2017, 139, 2010–2013.e2014. [Google Scholar] [CrossRef][Green Version]
- Aceves, S.S. Eosinophilic esophagitis. Immunol. Allergy Clin. N. Am. 2015, 35, 145–159. [Google Scholar] [CrossRef]
- Mavi, P.; Rajavelu, P.; Rayapudi, M.; Paul, R.J.; Mishra, A. Esophageal functional impairments in experimental eosinophilic esophagitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 302, G1347–G1355. [Google Scholar] [CrossRef]
- Mishra, A.; Wang, M.; Pemmaraju, V.R.; Collins, M.H.; Fulkerson, P.C.; Abonia, J.P.; Blanchard, C.; Putnam, P.E.; Rothenberg, M.E. Esophageal remodeling develops as a consequence of tissue specific IL-5-induced eosinophilia. Gastroenterology 2008, 134, 204–214. [Google Scholar] [CrossRef]
- Zuo, L.; Fulkerson, P.C.; Finkelman, F.D.; Mingler, M.; Fischetti, C.A.; Blanchard, C.; Rothenberg, M.E. IL-13 induces esophageal remodeling and gene expression by an eosinophil-independent, IL-13R alpha 2-inhibited pathway. J. Immunol. 2010, 185, 660–669. [Google Scholar] [CrossRef]
- Rieder, F.; Nonevski, I.; Ma, J.; Ouyang, Z.; West, G.; Protheroe, C.; DePetris, G.; Schirbel, A.; Lapinski, J.; Goldblum, J.; et al. T-helper 2 cytokines, transforming growth factor beta1, and eosinophil products induce fibrogenesis and alter muscle motility in patients with eosinophilic esophagitis. Gastroenterology 2014, 146, 1266–1277.e1261–e1269. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.P.; Rothenberg, M.E. Antigen presentation by eosinophils in eosinophilic esophagitis? J. Pediatr. Gastroenterol. Nutr. 2013, 56, 242. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, E.; Cho, J.Y.; Rosenthal, P.; Chao, J.; Miller, M.; Pham, A.; Aceves, S.S.; Varki, A.; Broide, D.H. Siglec-F inhibition reduces esophageal eosinophilia and angiogenesis in a mouse model of eosinophilic esophagitis. J. Pediatr. Gastroenterol. Nutr. 2011, 53, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Lucendo, A.J.; Navarro, M.; Comas, C.; Pascual, J.M.; Burgos, E.; Santamaria, L.; Larrauri, J. Immunophenotypic characterization and quantification of the epithelial inflammatory infiltrate in eosinophilic esophagitis through stereology: An analysis of the cellular mechanisms of the disease and the immunologic capacity of the esophagus. Am. J. Surg. Pathol. 2007, 31, 598–606. [Google Scholar] [CrossRef] [PubMed]
- Teitelbaum, J.E.; Fox, V.L.; Twarog, F.J.; Nurko, S.; Antonioli, D.; Gleich, G.; Badizadegan, K.; Furuta, G.T. Eosinophilic esophagitis in children: Immunopathological analysis and response to fluticasone propionate. Gastroenterology 2002, 122, 1216–1225. [Google Scholar] [CrossRef] [PubMed]
- Wen, T.; Rothenberg, M.E.; Wang, Y.H. Hematopoietic prostaglandin D synthase: Linking pathogenic effector CD4+ TH2 cells to proeosinophilic inflammation in patients with gastrointestinal allergic disorders. J. Allergy Clin. Immunol. 2016, 137, 919–921. [Google Scholar] [CrossRef]
- Mitson-Salazar, A.; Yin, Y.; Wansley, D.L.; Young, M.; Bolan, H.; Arceo, S.; Ho, N.; Koh, C.; Milner, J.D.; Stone, K.D.; et al. Hematopoietic prostaglandin D synthase defines a proeosinophilic pathogenic effector human T(H)2 cell subpopulation with enhanced function. J. Allergy Clin. Immunol. 2016, 137, 907–918.e909. [Google Scholar] [CrossRef]
- Mishra, A.; Schlotman, J.; Wang, M.; Rothenberg, M.E. Critical role for adaptive T cell immunity in experimental eosinophilic esophagitis in mice. J. Leukoc. Biol. 2007, 81, 916–924. [Google Scholar] [CrossRef]
- Abonia, J.P.; Blanchard, C.; Butz, B.B.; Rainey, H.F.; Collins, M.H.; Stringer, K.; Putnam, P.E.; Rothenberg, M.E. Involvement of mast cells in eosinophilic esophagitis. J. Allergy Clin. Immunol. 2010, 126, 140–149. [Google Scholar] [CrossRef]
- Niranjan, R.; Mavi, P.; Rayapudi, M.; Dynda, S.; Mishra, A. Pathogenic role of mast cells in experimental eosinophilic esophagitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 304, G1087–G1094. [Google Scholar] [CrossRef]
- Maggadottir, S.M.; Hill, D.A.; Ruymann, K.; Brown-Whitehorn, T.F.; Cianferoni, A.; Shuker, M.; Wang, M.L.; Chikwava, K.; Verma, R.; Liacouras, C.A.; et al. Resolution of acute IgE-mediated allergy with development of eosinophilic esophagitis triggered by the same food. J. Allergy Clin. Immunol. 2014, 133, 1487–1489.e1481. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.H.; Hogan, S.P.; Fulkerson, P.C.; Abonia, J.P.; Rothenberg, M.E. Expanding the paradigm of eosinophilic esophagitis: Mast cells and IL-9. J. Allergy Clin. Immunol. 2013, 131, 1583–1585. [Google Scholar] [CrossRef] [PubMed]
- Otani, I.M.; Anilkumar, A.A.; Newbury, R.O.; Bhagat, M.; Beppu, L.Y.; Dohil, R.; Broide, D.H.; Aceves, S.S. Anti-IL-5 therapy reduces mast cell and IL-9 cell numbers in pediatric patients with eosinophilic esophagitis. J. Allergy Clin. Immunol. 2013, 131, 1576–1582. [Google Scholar] [CrossRef] [PubMed]
- Allen-Brady, K.; Firszt, R.; Fang, J.C.; Wong, J.; Smith, K.R.; Peterson, K.A. Population-based familial aggregation of eosinophilic esophagitis suggests a genetic contribution. J. Allergy Clin. Immunol. 2017, 140, 1138–1143. [Google Scholar] [CrossRef]
- Siracusa, M.C.; Saenz, S.A.; Hill, D.A.; Kim, B.S.; Headley, M.B.; Doering, T.A.; Wherry, E.J.; Jessup, H.K.; Siegel, L.A.; Kambayashi, T.; et al. TSLP promotes interleukin-3-independent basophil haematopoiesis and type 2 inflammation. Nature 2011, 477, 229–233. [Google Scholar] [CrossRef]
- Voehringer, D. Protective and pathological roles of mast cells and basophils. Nat. Rev. Immunol. 2013, 13, 362–375. [Google Scholar] [CrossRef]
- Davis, B.P.; Rothenberg, M.E. Mechanisms of Disease of Eosinophilic Esophagitis. Annu. Rev. Pathol. 2016, 11, 365–393. [Google Scholar] [CrossRef]
- Inage, E.; Furuta, G.T.; Menard-Katcher, C.; Masterson, J.C. Eosinophilic esophagitis: Pathophysiology and its clinical implications. Am. J. Physiol. Gastrointest. Liver Physiol. 2018, 315, G879–G886. [Google Scholar] [CrossRef]
- Noti, M.; Wojno, E.D.; Kim, B.S.; Siracusa, M.C.; Giacomin, P.R.; Nair, M.G.; Benitez, A.J.; Ruymann, K.R.; Muir, A.B.; Hill, D.A.; et al. Thymic stromal lymphopoietin-elicited basophil responses promote eosinophilic esophagitis. Nat. Med. 2013, 19, 1005–1013. [Google Scholar] [CrossRef]
- Venturelli, N.; Lexmond, W.S.; Ohsaki, A.; Nurko, S.; Karasuyama, H.; Fiebiger, E.; Oyoshi, M.K. Allergic skin sensitization promotes eosinophilic esophagitis through the IL-33-basophil axis in mice. J. Allergy Clin. Immunol. 2016, 138, 1367–1380.e1365. [Google Scholar] [CrossRef]
- Tantibhaedhyangkul, U.; Tatevian, N.; Gilger, M.A.; Major, A.M.; Davis, C.M. Increased esophageal regulatory T cells and eosinophil characteristics in children with eosinophilic esophagitis and gastroesophageal reflux disease. Ann. Clin. Lab. Sci. 2009, 39, 99–107. [Google Scholar] [PubMed]
- Straumann, A.; Bauer, M.; Fischer, B.; Blaser, K.; Simon, H.U. Idiopathic eosinophilic esophagitis is associated with a T(H)2-type allergic inflammatory response. J. Allergy Clin. Immunol. 2001, 108, 954–961. [Google Scholar] [CrossRef] [PubMed]
- De Fraissinette, A.; Schmitt, D.; Thivolet, J. Langerhans cells of human mucosa. J. Dermatol. 1989, 16, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Eiwegger, T.; Akdis, C.A. IL-33 links tissue cells, dendritic cells and Th2 cell development in a mouse model of asthma. Eur. J. Immunol. 2011, 41, 1535–1538. [Google Scholar] [CrossRef] [PubMed]
- Hammad, H.; Chieppa, M.; Perros, F.; Willart, M.A.; Germain, R.N.; Lambrecht, B.N. House dust mite allergen induces asthma via Toll-like receptor 4 triggering of airway structural cells. Nat. Med. 2009, 15, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Christianson, C.A.; Goplen, N.P.; Zafar, I.; Irvin, C.; Good, J.T., Jr.; Rollins, D.R.; Gorentla, B.; Liu, W.; Gorska, M.M.; Chu, H.; et al. Persistence of asthma requires multiple feedback circuits involving type 2 innate lymphoid cells and IL-33. J. Allergy Clin. Immunol. 2015, 136, 59–68.e14. [Google Scholar] [CrossRef]
- Mjosberg, J.M.; Trifari, S.; Crellin, N.K.; Peters, C.P.; van Drunen, C.M.; Piet, B.; Fokkens, W.J.; Cupedo, T.; Spits, H. Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat. Immunol. 2011, 12, 1055–1062. [Google Scholar] [CrossRef]
- Doherty, T.A.; Baum, R.; Newbury, R.O.; Yang, T.; Dohil, R.; Aquino, M.; Doshi, A.; Walford, H.H.; Kurten, R.C.; Broide, D.H.; et al. Group 2 innate lymphocytes (ILC2) are enriched in active eosinophilic esophagitis. J. Allergy Clin. Immunol. 2015, 136, 792–794.e793. [Google Scholar] [CrossRef]
- Berin, M.C.; Shreffler, W.G. T(H)2 adjuvants: Implications for food allergy. J. Allergy Clin. Immunol. 2008, 121, 1311–1320, quiz 1321-1312. [Google Scholar] [CrossRef]
- Montalvillo, E.; Garrote, J.A.; Bernardo, D.; Arranz, E. Innate lymphoid cells and natural killer T cells in the gastrointestinal tract immune system. Rev. Esp. Enferm. Dig. 2014, 106, 334–345. [Google Scholar]
- Bendelac, A.; Savage, P.B.; Teyton, L. The biology of NKT cells. Annu. Rev. Immunol. 2007, 25, 297–336. [Google Scholar] [CrossRef] [PubMed]
- Olszak, T.; An, D.; Zeissig, S.; Vera, M.P.; Richter, J.; Franke, A.; Glickman, J.N.; Siebert, R.; Baron, R.M.; Kasper, D.L.; et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 2012, 336, 489–493. [Google Scholar] [CrossRef] [PubMed]
- Rajavelu, P.; Rayapudi, M.; Moffitt, M.; Mishra, A.; Mishra, A. Significance of para-esophageal lymph nodes in food or aeroallergen-induced iNKT cell-mediated experimental eosinophilic esophagitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 302, G645–G654. [Google Scholar] [CrossRef] [PubMed]
- Rayapudi, M.; Rajavelu, P.; Zhu, X.; Kaul, A.; Niranjan, R.; Dynda, S.; Mishra, A.; Mattner, J.; Zaidi, A.; Dutt, P.; et al. Invariant natural killer T-cell neutralization is a possible novel therapy for human eosinophilic esophagitis. Clin. Transl. Immunol. 2014, 3, e9. [Google Scholar] [CrossRef] [PubMed]
- Jyonouchi, S.; Smith, C.L.; Saretta, F.; Abraham, V.; Ruymann, K.R.; Modayur-Chandramouleeswaran, P.; Wang, M.L.; Spergel, J.M.; Cianferoni, A. Invariant natural killer T cells in children with eosinophilic esophagitis. Clin. Exp. Allergy 2014, 44, 58–68. [Google Scholar] [CrossRef]
- Aceves, S.S.; Newbury, R.O.; Dohil, R.; Bastian, J.F.; Broide, D.H. Esophageal remodeling in pediatric eosinophilic esophagitis. J. Allergy Clin. Immunol. 2007, 119, 206–212. [Google Scholar] [CrossRef]
- Straumann, A.; Conus, S.; Degen, L.; Felder, S.; Kummer, M.; Engel, H.; Bussmann, C.; Beglinger, C.; Schoepfer, A.; Simon, H.U. Budesonide is effective in adolescent and adult patients with active eosinophilic esophagitis. Gastroenterology 2010, 139, 1526–1537.e1521. [Google Scholar] [CrossRef]
- Straumann, A.; Conus, S.; Grzonka, P.; Kita, H.; Kephart, G.; Bussmann, C.; Beglinger, C.; Smith, D.A.; Patel, J.; Byrne, M.; et al. Anti-interleukin-5 antibody treatment (mepolizumab) in active eosinophilic oesophagitis: A randomised, placebo-controlled, double-blind trial. Gut 2010, 59, 21–30. [Google Scholar] [CrossRef]
- Cho, J.Y.; Doshi, A.; Rosenthal, P.; Beppu, A.; Miller, M.; Aceves, S.; Broide, D. Smad3-deficient mice have reduced esophageal fibrosis and angiogenesis in a model of egg-induced eosinophilic esophagitis. J. Pediatr. Gastroenterol. Nutr. 2014, 59, 10–16. [Google Scholar] [CrossRef]
- Kagalwalla, A.F.; Akhtar, N.; Woodruff, S.A.; Rea, B.A.; Masterson, J.C.; Mukkada, V.; Parashette, K.R.; Du, J.; Fillon, S.; Protheroe, C.A.; et al. Eosinophilic esophagitis: Epithelial mesenchymal transition contributes to esophageal remodeling and reverses with treatment. J. Allergy Clin. Immunol. 2012, 129, 1387–1396.e1387. [Google Scholar] [CrossRef]
- O’Byrne, P.M.; Inman, M.D.; Parameswaran, K. The trials and tribulations of IL-5, eosinophils, and allergic asthma. J. Allergy Clin. Immunol. 2001, 108, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Straumann, A.; Kristl, J.; Conus, S.; Vassina, E.; Spichtin, H.P.; Beglinger, C.; Simon, H.U. Cytokine expression in healthy and inflamed mucosa: Probing the role of eosinophils in the digestive tract. Inflamm. Bowel Dis. 2005, 11, 720–726. [Google Scholar] [CrossRef]
- Yamazaki, K.; Murray, J.A.; Arora, A.S.; Alexander, J.A.; Smyrk, T.C.; Butterfield, J.H.; Kita, H. Allergen-specific in vitro cytokine production in adult patients with eosinophilic esophagitis. Dig. Dis. Sci. 2006, 51, 1934–1941. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, C.; Mishra, A.; Saito-Akei, H.; Monk, P.; Anderson, I.; Rothenberg, M.E. Inhibition of human interleukin-13-induced respiratory and oesophageal inflammation by anti-human-interleukin-13 antibody (CAT-354). Clin. Exp. Allergy 2005, 35, 1096–1103. [Google Scholar] [CrossRef] [PubMed]
- Rothenberg, M.E.; Wen, T.; Greenberg, A.; Alpan, O.; Enav, B.; Hirano, I.; Nadeau, K.; Kaiser, S.; Peters, T.; Perez, A.; et al. Intravenous anti-IL-13 mAb QAX576 for the treatment of eosinophilic esophagitis. J. Allergy Clin. Immunol. 2015, 135, 500–507. [Google Scholar] [CrossRef] [PubMed]
- Niranjan, R.; Rayapudi, M.; Mishra, A.; Dutt, P.; Dynda, S.; Mishra, A. Pathogenesis of allergen-induced eosinophilic esophagitis is independent of interleukin (IL)-13. Immunol. Cell Biol. 2013, 91, 408–415. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, M.; Mavi, P.; Rayapudi, M.; Pandey, A.K.; Kaul, A.; Putnam, P.E.; Rothenberg, M.E.; Mishra, A. Interleukin-15 expression is increased in human eosinophilic esophagitis and mediates pathogenesis in mice. Gastroenterology 2010, 139, 182–193.e187. [Google Scholar] [CrossRef]
- Sherrill, J.D.; Kiran, K.C.; Blanchard, C.; Stucke, E.M.; Kemme, K.A.; Collins, M.H.; Abonia, J.P.; Putnam, P.E.; Mukkada, V.A.; Kaul, A.; et al. Analysis and expansion of the eosinophilic esophagitis transcriptome by RNA sequencing. Genes Immun. 2014, 15, 361–369. [Google Scholar] [CrossRef]
- Clayton, F.; Fang, J.C.; Gleich, G.J.; Lucendo, A.J.; Olalla, J.M.; Vinson, L.A.; Lowichik, A.; Chen, X.; Emerson, L.; Cox, K.; et al. Eosinophilic esophagitis in adults is associated with IgG4 and not mediated by IgE. Gastroenterology 2014, 147, 602–609. [Google Scholar] [CrossRef]
- Schuyler, A.J.; Wilson, J.M.; Tripathi, A.; Commins, S.P.; Ogbogu, P.U.; Kruzsewski, P.G.; Barnes, B.H.; McGowan, E.C.; Workman, L.J.; Lidholm, J.; et al. Specific IgG4 antibodies to cow’s milk proteins in pediatric patients with eosinophilic esophagitis. J. Allergy Clin. Immunol. 2018, 142, 139–148.e112. [Google Scholar] [CrossRef]
- Zhang, S.; Wu, X.; Yu, S. Prostaglandin D2 receptor D-type prostanoid receptor 2 mediates eosinophil trafficking into the esophagus. Dis. Esophagus 2014, 27, 601–606. [Google Scholar] [CrossRef] [PubMed]
- Pettipher, R.; Hansel, T.T.; Armer, R. Antagonism of the prostaglandin D2 receptors DP1 and CRTH2 as an approach to treat allergic diseases. Nat. Rev. Drug Discov. 2007, 6, 313–325. [Google Scholar] [CrossRef] [PubMed]
- Collison, A.M.; Sokulsky, L.A.; Sherrill, J.D.; Nightingale, S.; Hatchwell, L.; Talley, N.J.; Walker, M.M.; Rothenberg, M.E.; Mattes, J. TNF-related apoptosis-inducing ligand (TRAIL) regulates midline-1, thymic stromal lymphopoietin, inflammation, and remodeling in experimental eosinophilic esophagitis. J. Allergy Clin. Immunol. 2015, 136, 971–982. [Google Scholar] [CrossRef] [PubMed]
- De Souza, H.S.; Tortori, C.A.; Lintomen, L.; Figueiredo, R.T.; Bernardazzi, C.; Leng, L.; Bucala, R.; Madi, K.; Buongusto, F.; Elia, C.C.; et al. Macrophage migration inhibitory factor promotes eosinophil accumulation and tissue remodeling in eosinophilic esophagitis. Mucosal Immunol. 2015, 8, 1154–1165. [Google Scholar] [CrossRef] [PubMed]
- Dutt, P.; Shukla, J.S.; Ventateshaiah, S.U.; Mariswamy, S.J.; Mattner, J.; Shukla, A.; Mishra, A. Allergen-induced interleukin-18 promotes experimental eosinophilic oesophagitis in mice. Immunol. Cell Biol. 2015, 93, 849–857. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).