Cross-Talk between P2X and NMDA Receptors
Abstract
1. Introduction
2. Results
2.1. Coactivation of P2X and NMDA Receptors
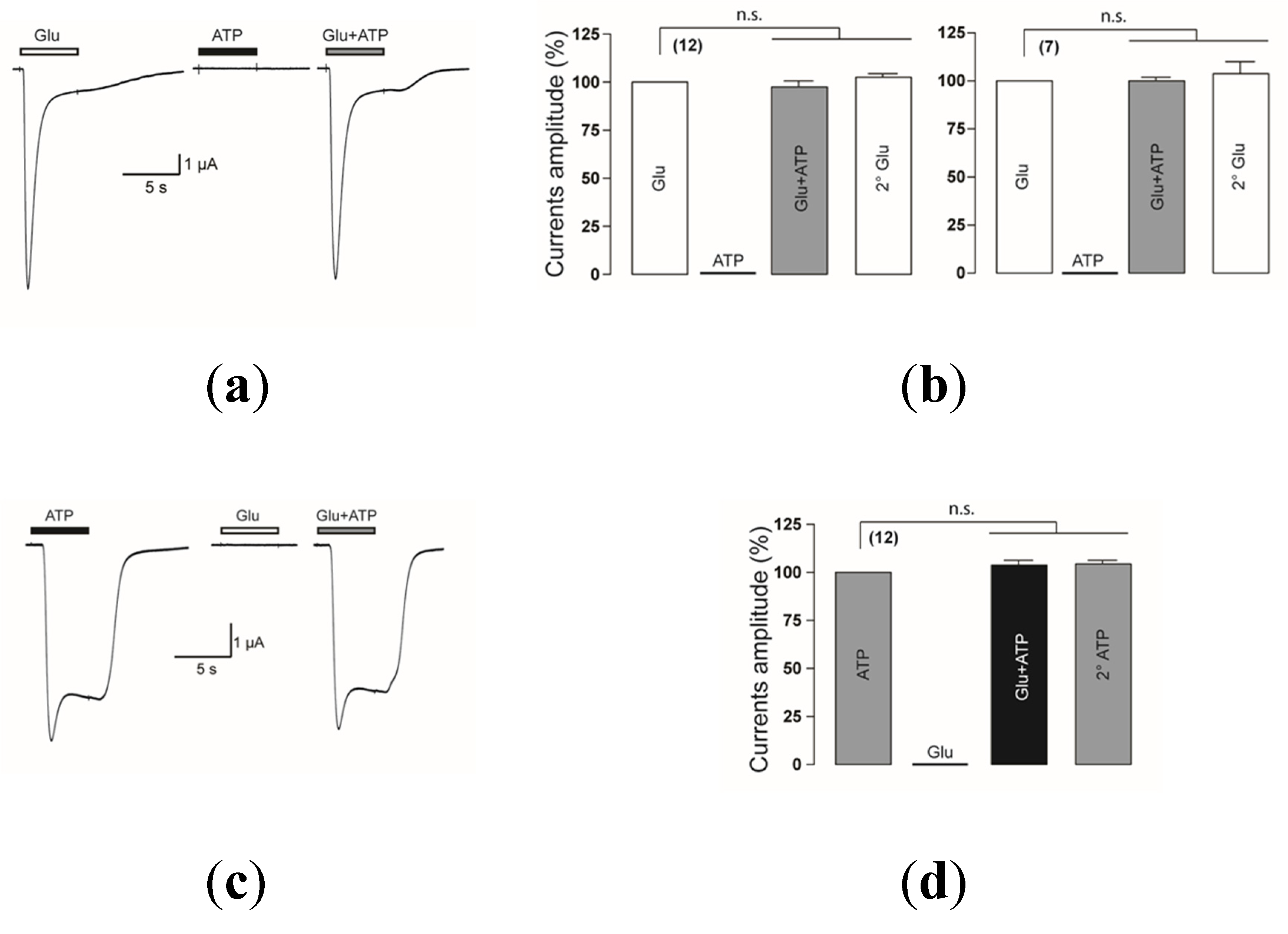
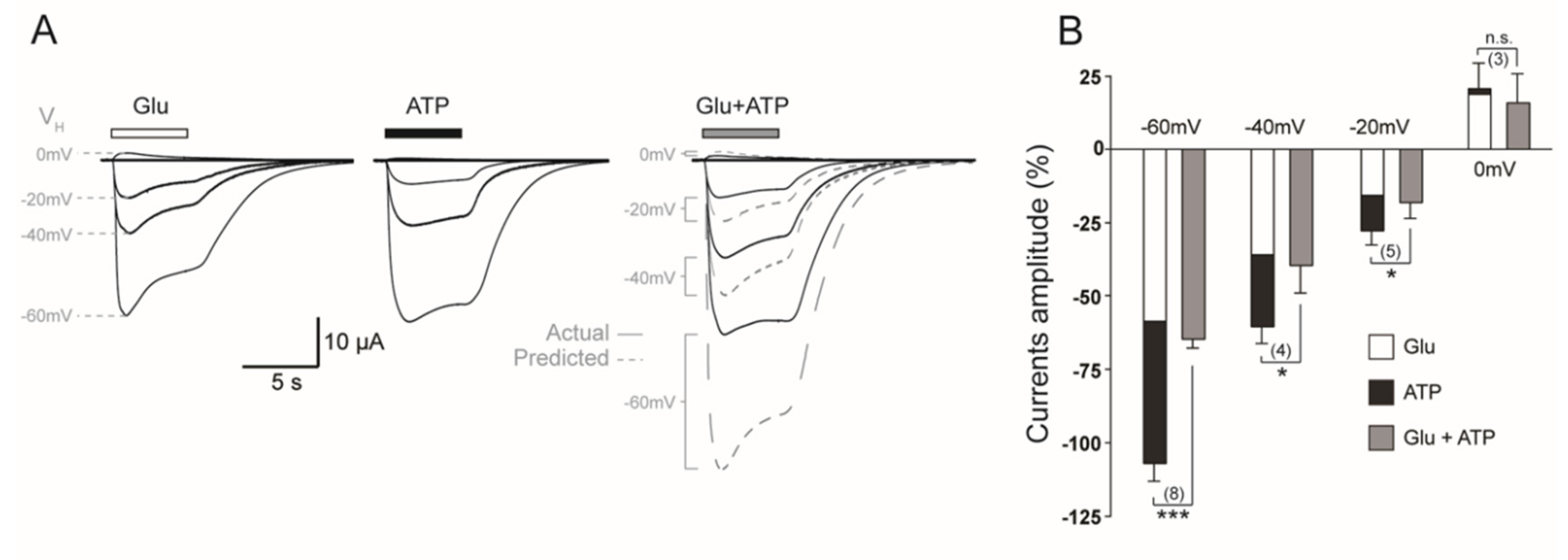
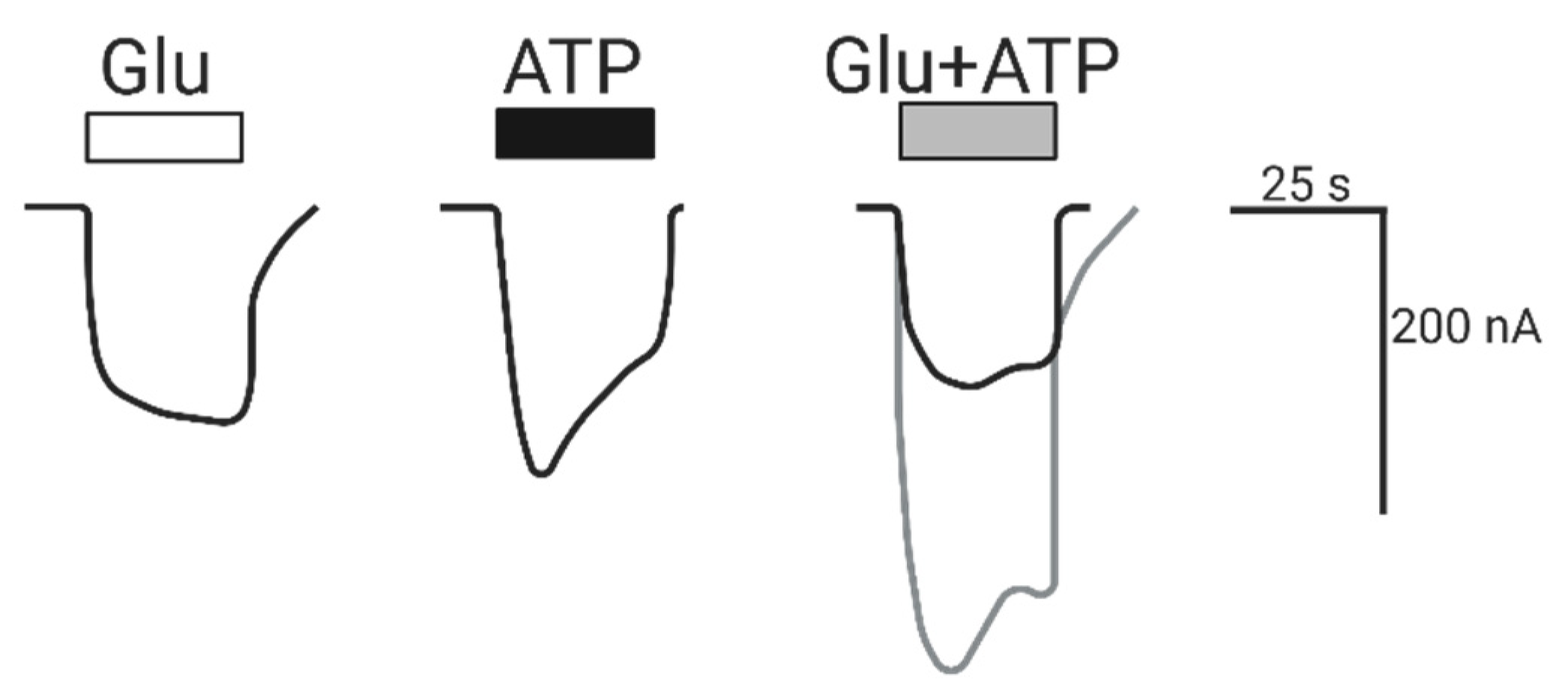
2.1.1. Coactivation of P2X4 and NMDA Receptors Produces Non-Additive (Inhibited) Responses
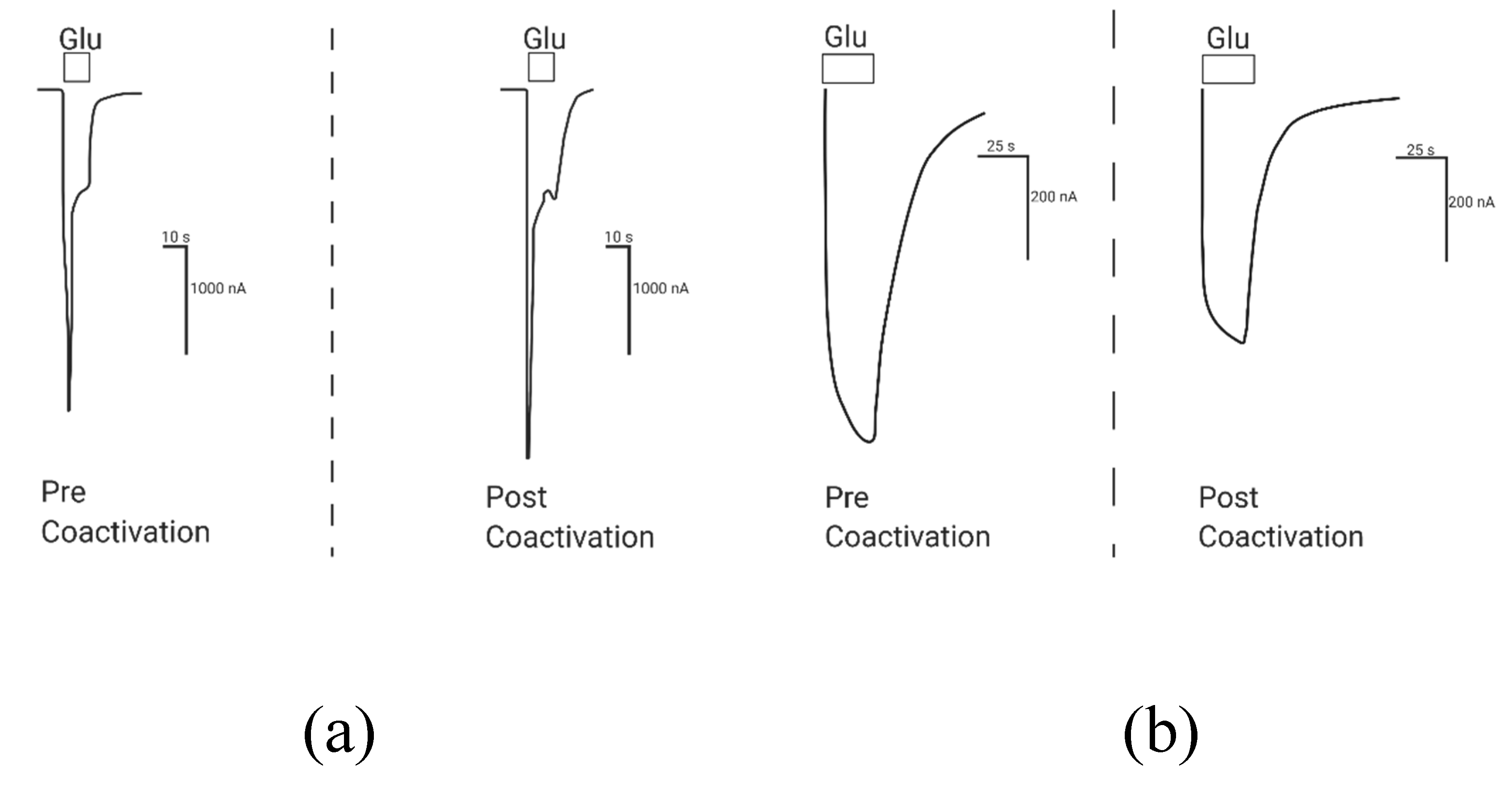
2.1.2. Coactivation of P2X2 and NMDA Receptors Produces Inhibited (Cross-Talk) Responses
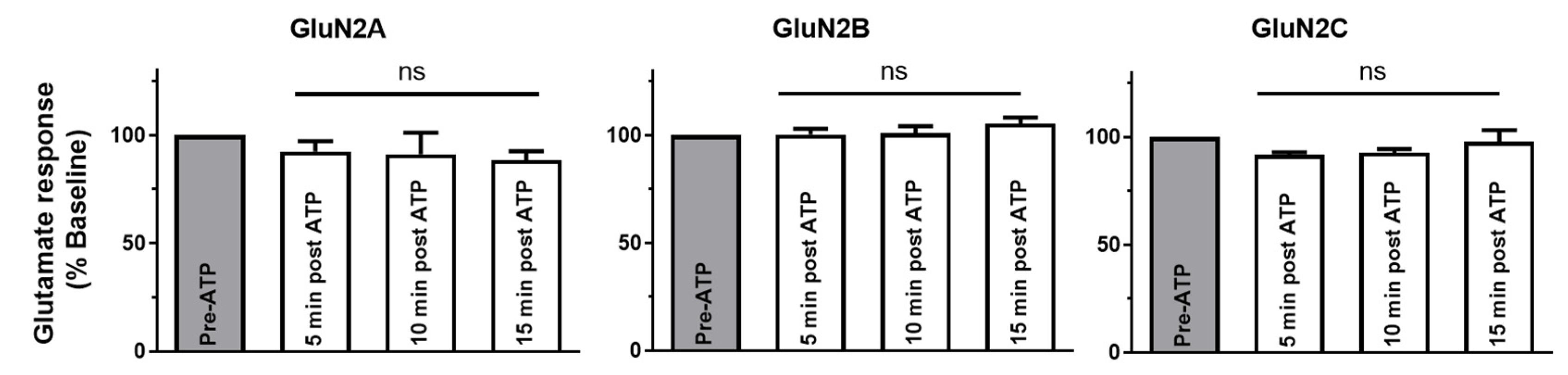
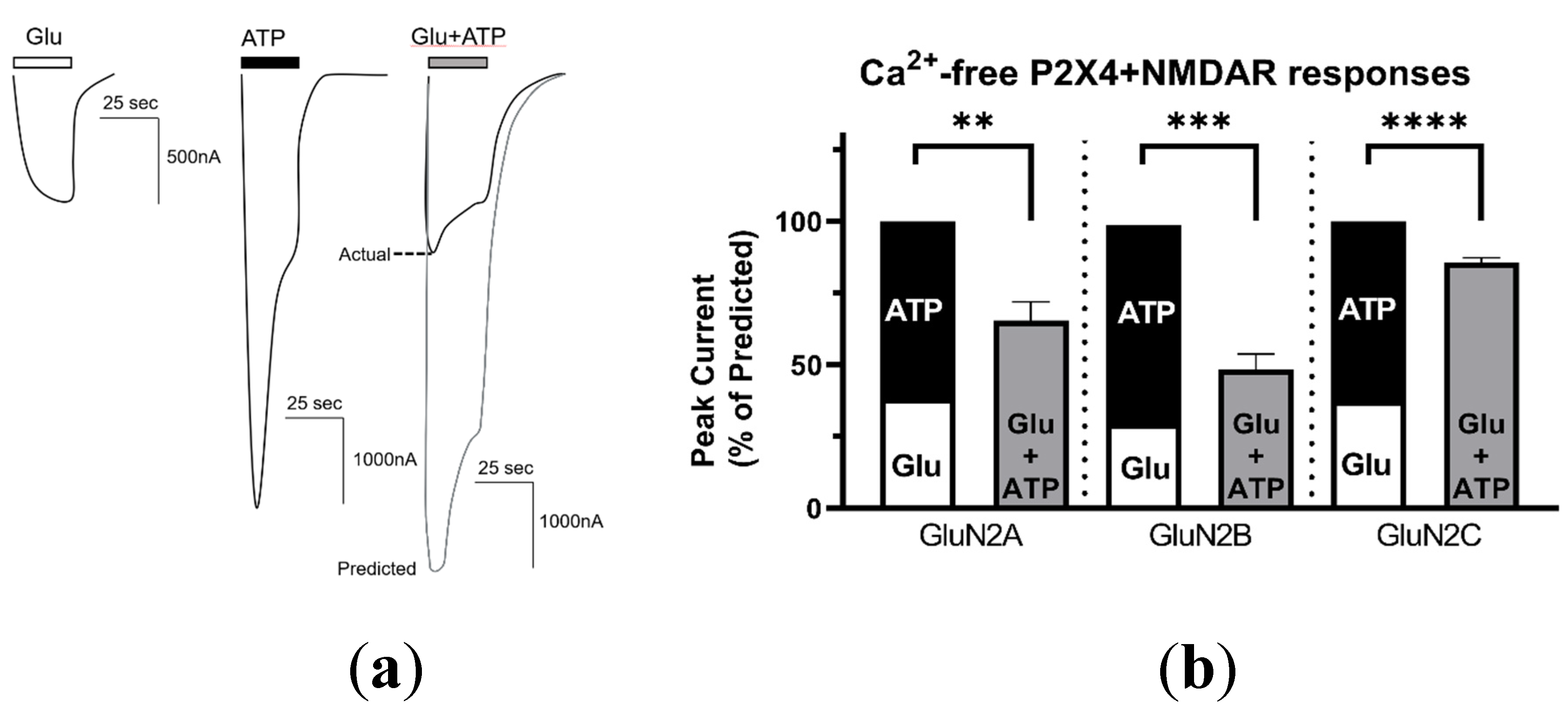
2.2. P2X4–NMDAR Interactions Are Independent of Ca2+ Influx
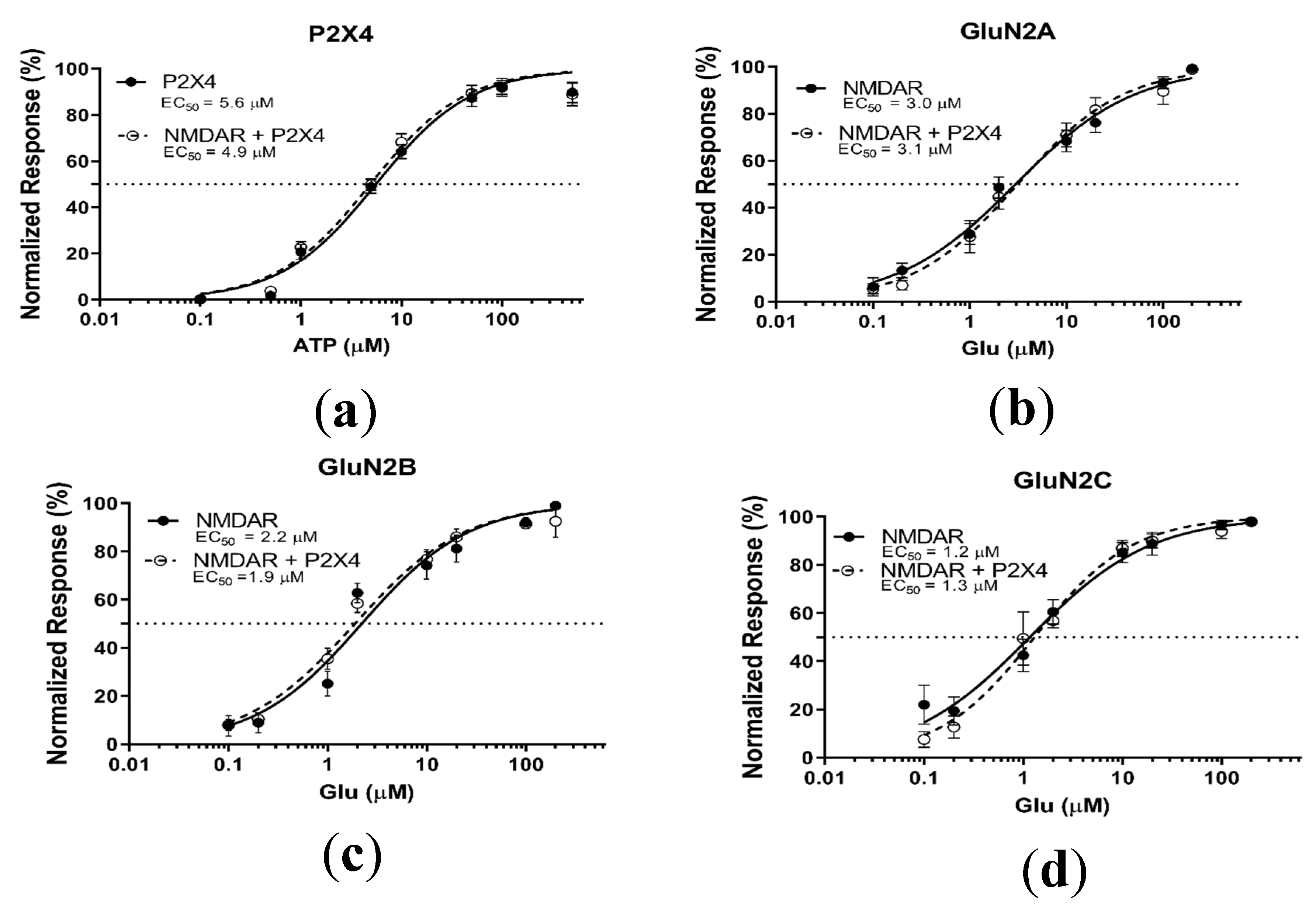
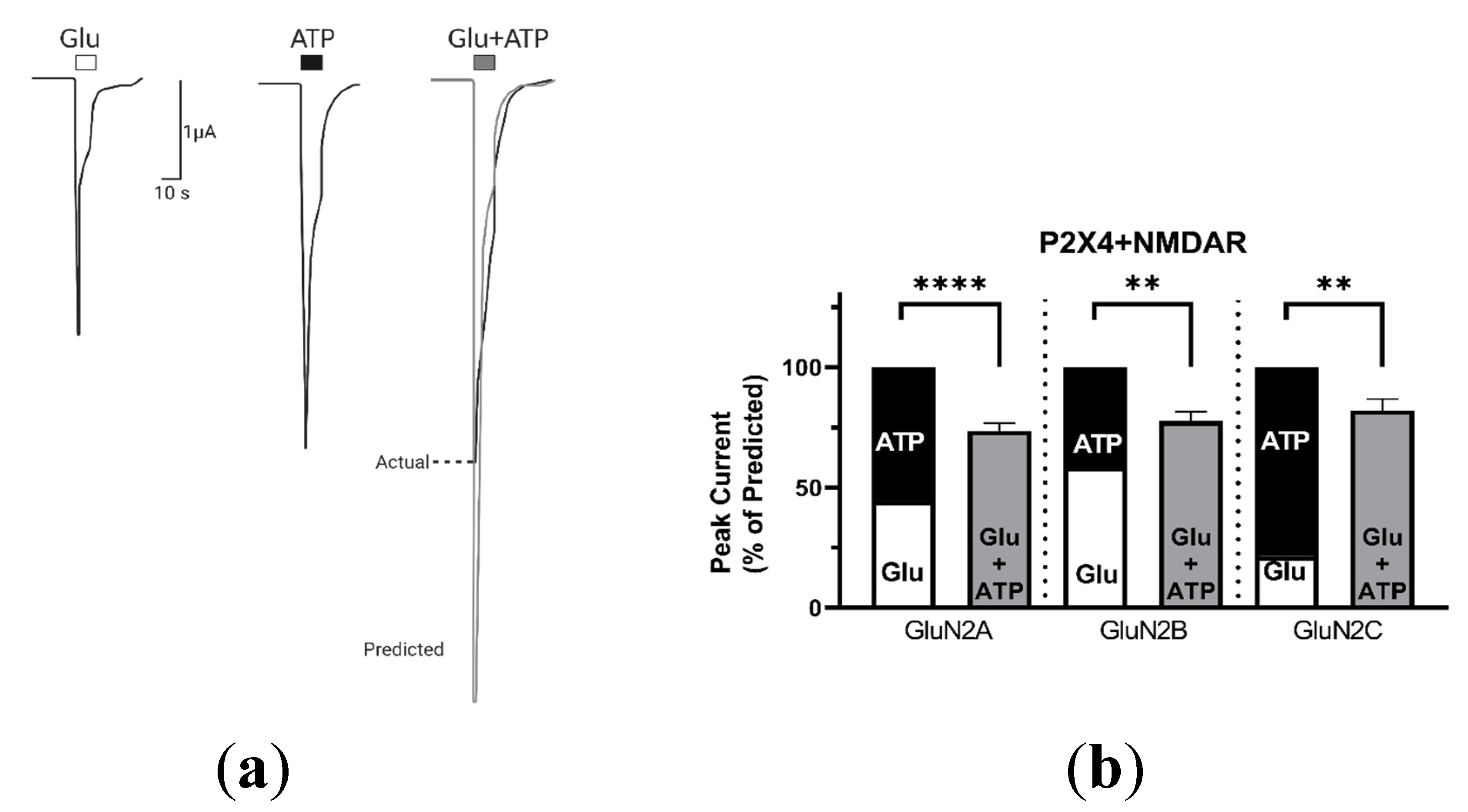
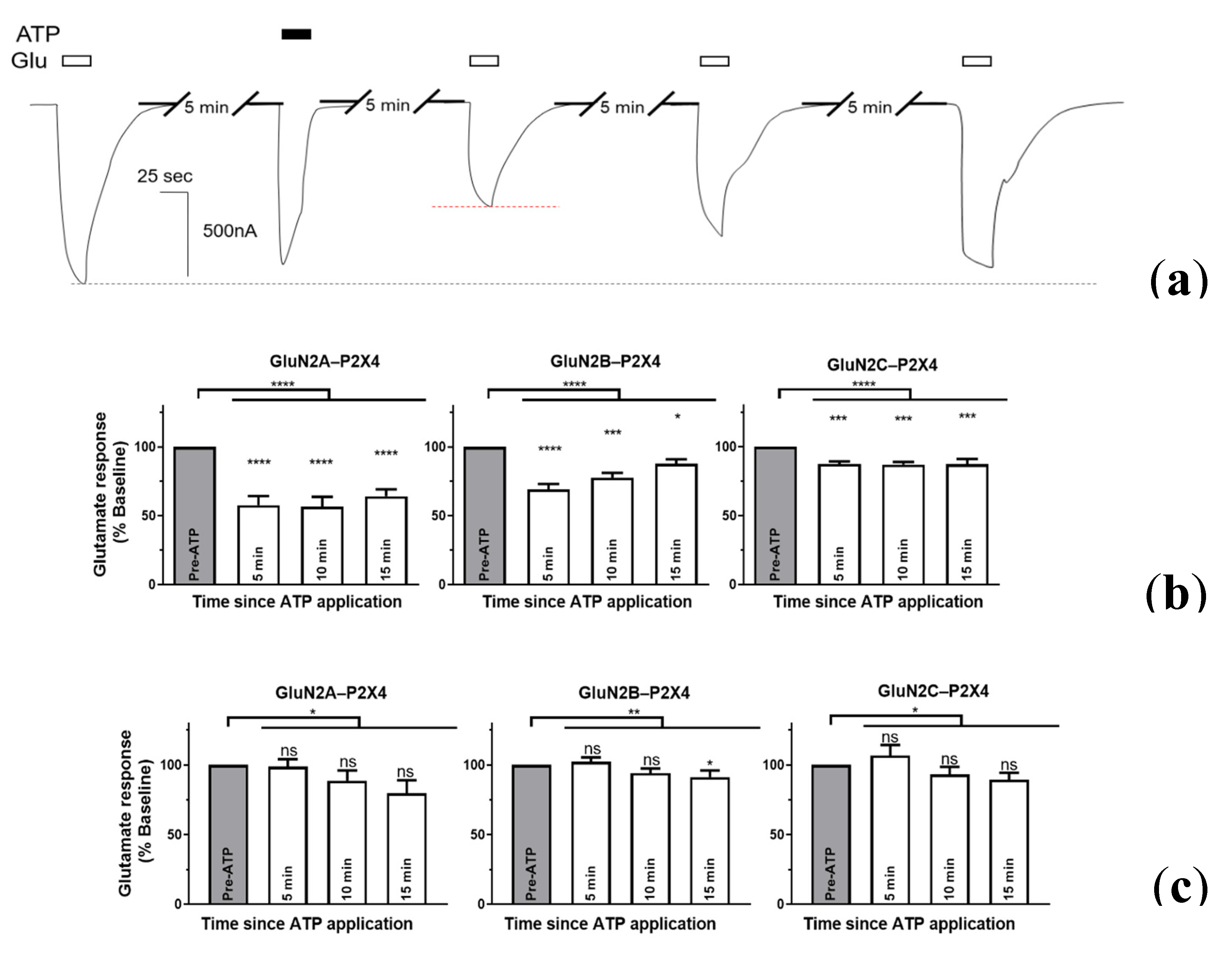
2.3. P2X4-Induced a Long-Lasting Inhibition of NMDAR Is GluN2-Subunit Dependent
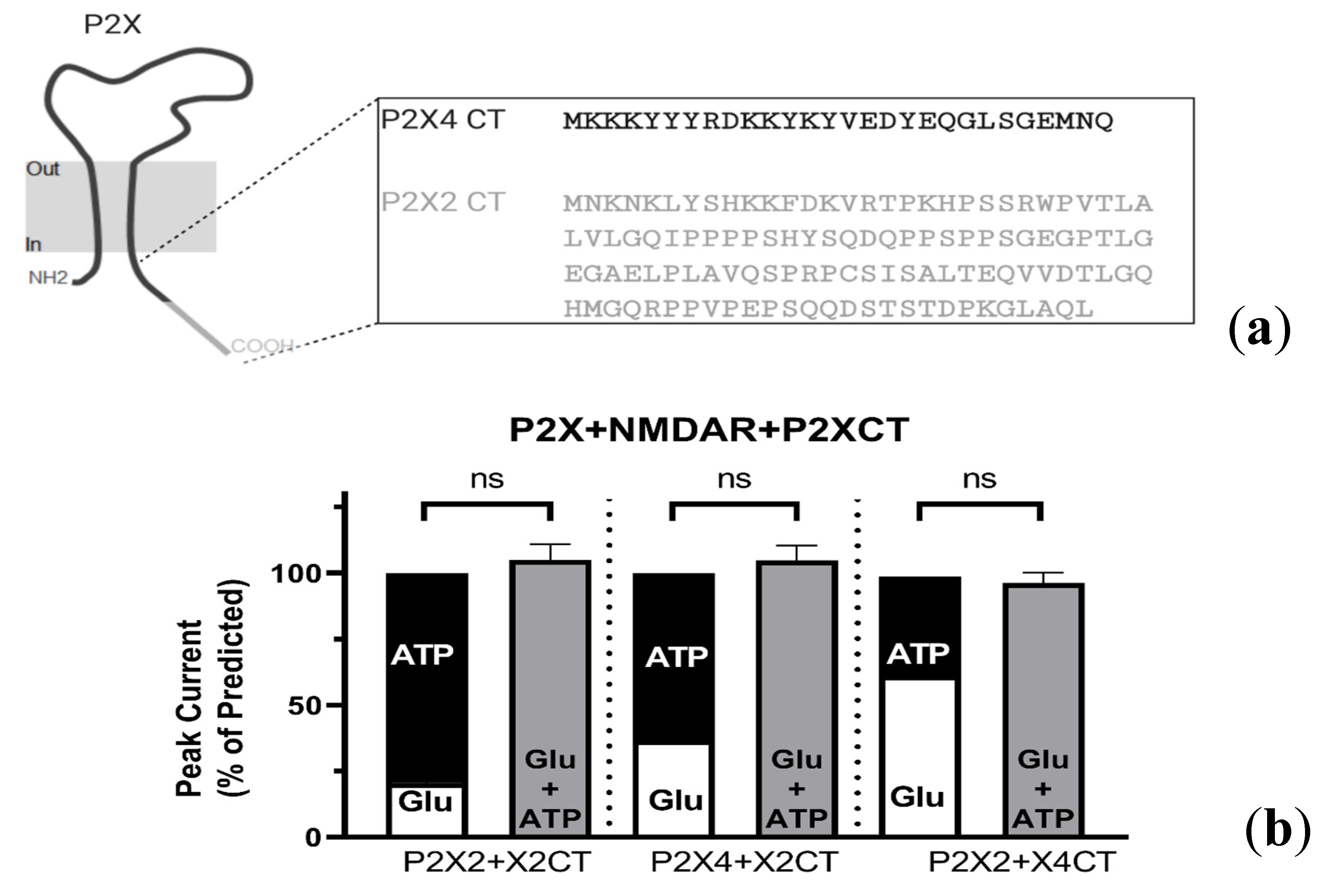
2.4. Intracellular P2X Domains Mediate NMDAR Interactions
2.4.1. P2X-NMDAR Inhibited Interactions Depend on C-Terminal P2X Domains
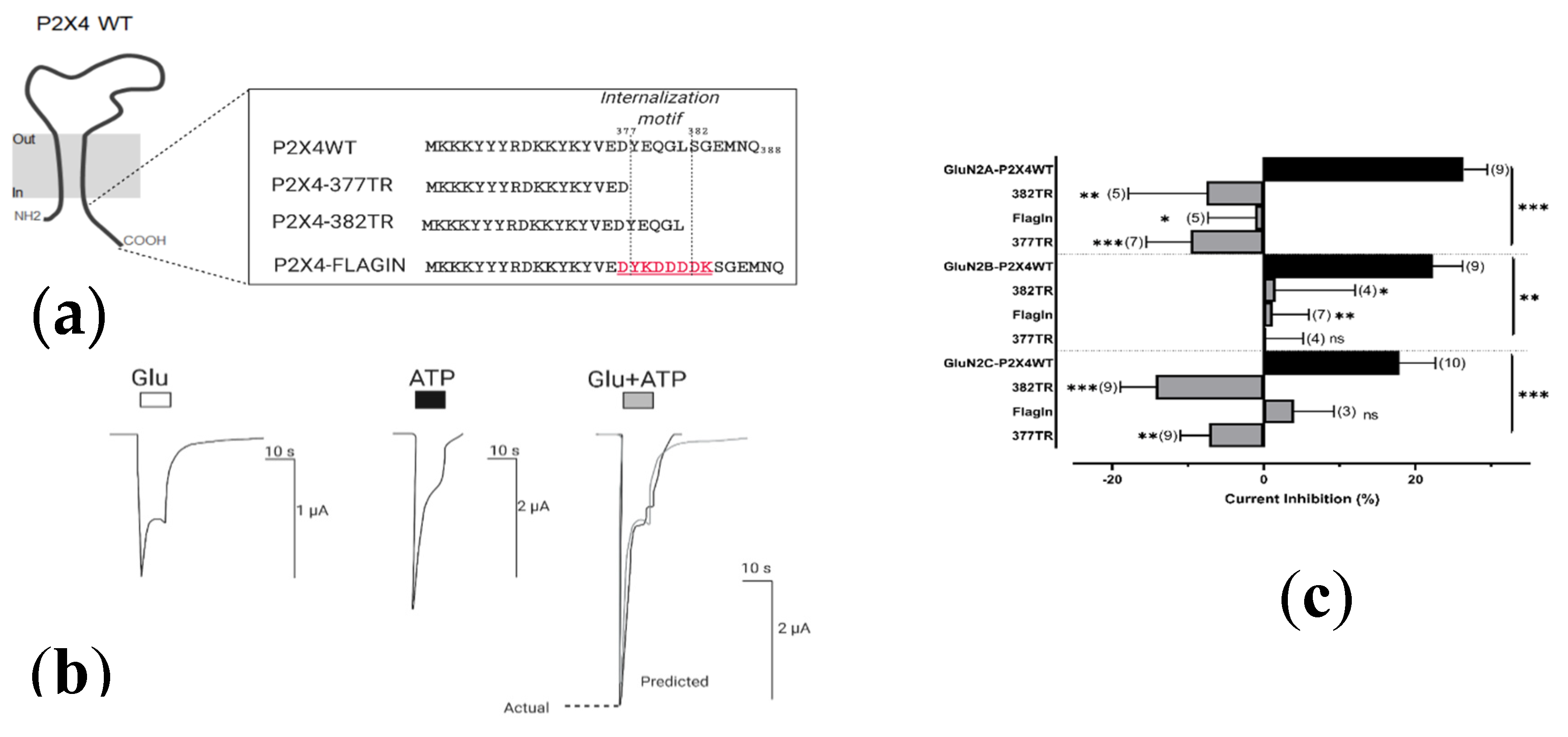
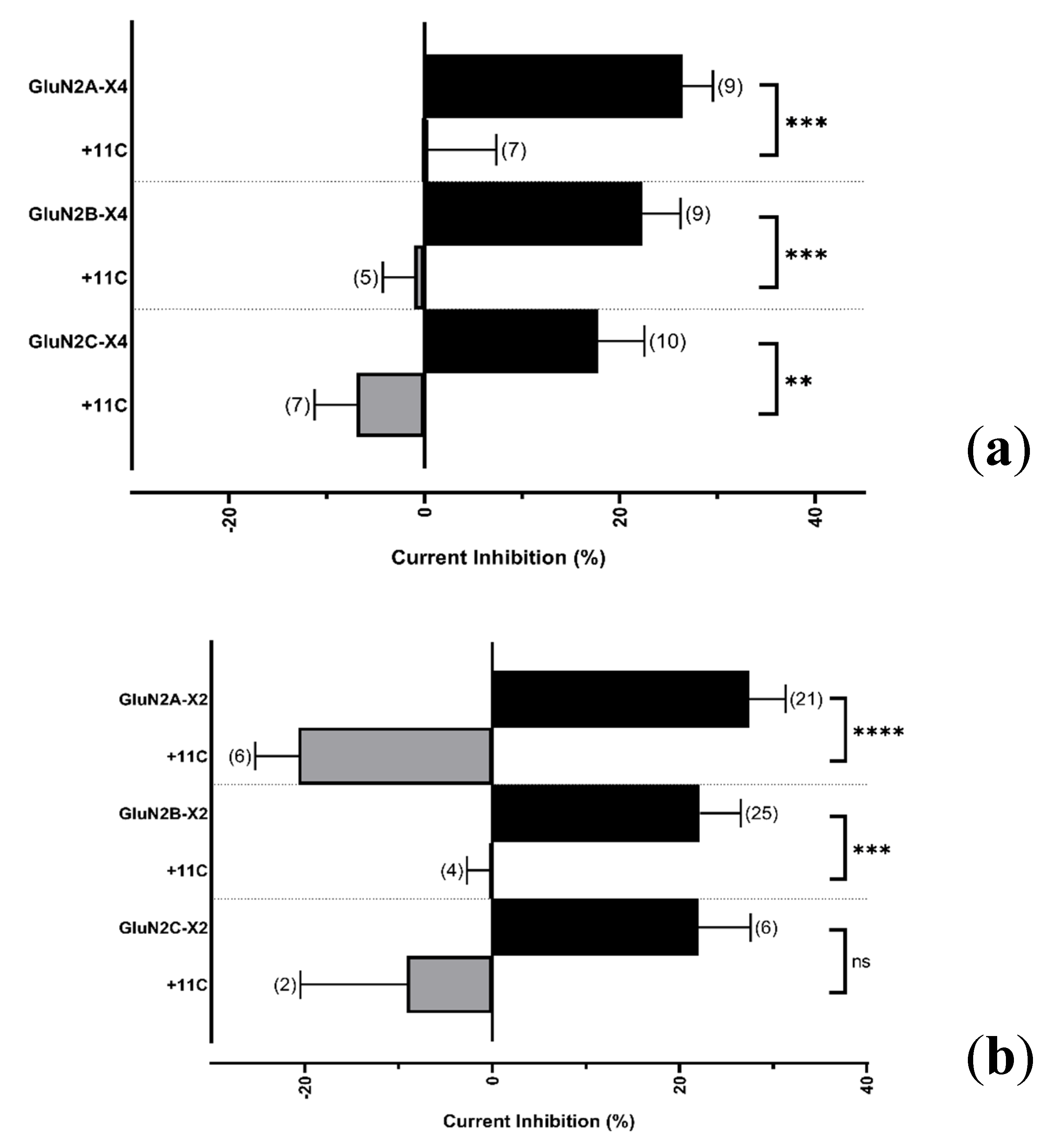
2.4.2. Resolving the P2X4 CT Domain Responsible for NMDAR Inhibition
2.4.3. Resolving the P2X4 CT Domain Responsible for NMDAR Inhibition
3. Discussion
3.1. P2X Modulation of NMDA Receptors
3.2. P2X Intracellular Domains Mediate NMDAR Cross-Talk
3.3. Resolving the Function of P2X in the Brain
4. Materials and Methods
4.1. Molecular Biology
4.2. Xenopus Laevis Oocyte Injection and Electrophysiology
4.3. Data Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| P2X | Purinergic receptors |
| NMDARs | N-methyl-d-aspartate receptors |
| GABA | γ-aminobutyric acid |
| AMPA | α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid |
| CT | Carboxy-terminal tail |
| LGICS | Ligand-gated ion channels |
| ATP | Adenosine triphosphate |
| CNS | Central nervous system |
| P2X4 KO | P2X4 knockout |
| LTP | Long-term potentiation |
| LTD | Long-term depression |
| TEVC | Two-electrode voltage clamp |
| Glu | Glutamate |
| CfRS | Calcium-free Ringers’ solution |
| P2X4-377TR | P2X4 receptor truncated at residue 377 |
| P2X4-382TR | P2X4 receptor truncated at residue 382 |
| P2X4-FlagIN | P2X4 receptor with a FLAG epitope in place of internalization domain (YEQGL) |
| 11C | Peptide consisting of the distal C-terminal 11 amino-acids of P2X4 |
| 5-HT3A | 5-Hydroxytryptamine Receptor 3A |
Appendix A
Appendix B
References
- Barria, A.; Malinow, R. Subunit-Specific NMDA Receptor Trafficking to Synapses. Neuron 2002, 35, 345–353. [Google Scholar] [CrossRef]
- Kopec, C.D.; Li, B.; Wei, W.; Boehm, J.; Malinow, R. Glutamate Receptor Exocytosis and Spine Enlargement during Chemically Induced Long-Term Potentiation. J. Neurosci. 2006, 26, 2000–2009. [Google Scholar] [CrossRef]
- Khakh, B.S.; North, R.A. Neuromodulation by Extracellular ATP and P2X Receptors in the CNS. Neuron 2012, 76, 51–69. [Google Scholar] [CrossRef]
- Mori, M.; Heuss, C.; Gähwiler, B.H.; Gerber, U. Fast synaptic transmission mediated by P2X receptors in CA3 pyramidal cells of rat hippocampal slice cultures. J. Physiol. 2001, 535, 115–123. [Google Scholar] [CrossRef]
- Jo, Y.-H.; Schlichter, R. Synaptic corelease of ATP and GABA in cultured spinal neurons. Nat. Neurosci. 1999, 2, 241–245. [Google Scholar] [CrossRef]
- Sim, J.A.; Chaumont, S.; Jo, J.; Ulmann, L.; Young, M.T.; Cho, K.; Buell, G.; North, R.A.; Rassendren, F. Altered Hippocampal Synaptic Potentiation in P2X4 Knock-Out Mice. J. Neurosci. 2006, 26, 9006–9009. [Google Scholar] [CrossRef]
- Baxter, A.W.; Choi, S.J.; Sim, J.A.; North, R.A. Role of P2X4 receptors in synaptic strengthening in mouse CA1 hippocampal neurons. Eur. J. Neurosci. 2011, 34, 213–220. [Google Scholar] [CrossRef]
- Lalo, U.; Palygin, O.; Verkhratsky, A.; Grant, S.G.; Pankratov, Y. ATP from synaptic terminals and astrocytes regulates NMDA receptors and synaptic plasticity through PSD-95 multi-protein complex. Sci. Rep. 2016, 6, 33609. [Google Scholar] [CrossRef]
- Wyatt, L.; Finn, D.A.; Khoja, S.; Yardley, M.M.; Asatryan, L.; Alkana, R.L.; Davies, D.L. Contribution of P2X4 receptors to ethanol intake in male C57BL/6 mice. Neurochem. Res. 2014, 39, 1127–1139. [Google Scholar] [CrossRef]
- Wyatt, L.; Godar, S.C.; Khoja, S.; Jakowec, M.W.; Alkana, R.L.; Bortolato, M.; Davies, D.L. Sociocommunicative and Sensorimotor Impairments in Male P2X4-Deficient Mice. Neuropsychopharmacology 2013, 38, 1993–2002. [Google Scholar] [CrossRef]
- Khoja, S.; Shah, V.; Garcia, D.; Asatryan, L.; Jakowec, M.W.; Davies, D.L. Role of purinergic P2X4 receptors in regulating striatal dopamine homeostasis and dependent behaviors. J. Neurochem. 2016, 139, 134–148. [Google Scholar] [CrossRef] [PubMed]
- Stokes, L.; Bidula, S.; Bibič, L.; Allum, E. To Inhibit or Enhance? Is There a Benefit to Positive Allosteric Modulation of P2X Receptors? Front. Pharmacol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Yardley, M.M.; Wyatt, L.; Khoja, S.; Asatryan, L.; Ramaker, M.J.; Finn, D.A.; Alkana, R.L.; Huynh, N.; Louie, S.G.; Petasis, N.A.; et al. Ivermectin reduces alcohol intake and preference in mice. Neuropharmacology 2012, 63, 190–201. [Google Scholar] [CrossRef] [PubMed]
- Huynh, N.; Arabian, N.; Naito, A.; Louie, S.; Jakowec, M.W.; Asatryan, L.; Davies, D.L. Preclinical development of moxidectin as a novel therapeutic for alcohol use disorder. Neuropharmacology 2017, 113, 60–70. [Google Scholar] [CrossRef]
- Bertin, E.; Deluc, T.; Pilch, K.S.; Martinez, A.; Pougnet, J.-T.; Doudnikoff, E.; Allain, A.-E.; Bergmann, P.; Russeau, M.; Toulmé, E.; et al. Increased surface P2X4 receptor regulates anxiety and memory in P2X4 internalization-defective knock-in mice. Mol. Psychiatry 2020, 1–16. [Google Scholar] [CrossRef]
- Stokes, L.; Layhadi, J.A.; Bibic, L.; Dhuna, K.; Fountain, S.J. P2X4 Receptor Function in the Nervous System and Current Breakthroughs in Pharmacology. Front. Pharmacol. 2017, 8, 291. [Google Scholar] [CrossRef]
- Boué-Grabot, E.; Toulme, E.; Emerit, M.B.; Garret, M. Subunit-specific Coupling between γ-Aminobutyric Acid Type A and P2X2Receptor Channels. J. Boil. Chem. 2004, 279, 52517–52525. [Google Scholar] [CrossRef]
- Boué-Grabot, É.; Émerit, M.B.; Toulmé, E.; Séguéla, P.; Garret, M. Cross-talk and Co-trafficking between ρ1/GABA Receptors and ATP-gated Channels. J. Biol. Chem. 2004. 279, 6967–6975. [CrossRef]
- Toulme, E.; Blais, D.; Léger, C.; Landry, M.; Garret, M.; Seguela, P.; Boué-Grabot, E. An intracellular motif of P2X3receptors is required for functional cross-talk with GABAAreceptors in nociceptive DRG neurons. J. Neurochem. 2007, 102, 1357–1368. [Google Scholar] [CrossRef]
- Jo, Y.-H.; Donier, E.; Martinez, A.; Garret, M.; Toulmé, E.; Boué-Grabot, E. Cross-talk between P2X4 and γ-Aminobutyric Acid, Type A Receptors Determines Synaptic Efficacy at a Central Synapse. J. Boil. Chem. 2011, 286, 19993–20004. [Google Scholar] [CrossRef]
- Khakh, B.S.; Zhou, X.; Sydes, J.; Galligan, J.J.; Lester, H.A. State-dependent cross-inhibition between transmitter-gated cation channels. Nature 2000, 406, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Boué-Grabot, E.; Barajas-López, C.; Chakfe, Y.; Blais, D.; Bélanger, D.; Emerit, M.B.; Seguela, P. Intracellular Cross Talk and Physical Interaction between Two Classes of Neurotransmitter-Gated Channels. J. Neurosci. 2003, 23, 1246–1253. [Google Scholar] [CrossRef] [PubMed]
- Emerit, M.B.; Baranowski, C.; Díaz, J.; Martinez, A.; Areias, J.; Alterio, J.; Masson, J.; Boué-Grabot, E.; Darmon, M.C. A New Mechanism of Receptor Targeting by Interaction between Two Classes of Ligand-Gated Ion Channels. J. Neurosci. 2016, 36, 1456–1470. [Google Scholar] [CrossRef] [PubMed]
- Soto, P.; Gaete, P.S.; Fuentes, C.; Lozano, B.; Naulin, P.A.; Figueroa, X.F.; Barrera, N.P. Function of P2X4 Receptors Is Directly Modulated by a 1:1 Stoichiometric Interaction With 5-HT3A Receptors. Front. Cell. Neurosci. 2020, 14, 106. [Google Scholar] [CrossRef]
- Gordon, G.R.; Baimoukhametova, D.V.; A Hewitt, S.; Rajapaksha, W.R.A.K.J.S.; Fisher, T.E.; Bains, J.S. Norepinephrine triggers release of glial ATP to increase postsynaptic efficacy. Nat. Neurosci. 2005, 8, 1078–1086. [Google Scholar] [CrossRef]
- Pougnet, J.-T.; Toulme, E.; Martinez, A.; Choquet, D.; Hosy, E.; Boué-Grabot, E. ATP P2X Receptors Downregulate AMPA Receptor Trafficking and Postsynaptic Efficacy in Hippocampal Neurons. Neuron 2014, 83, 417–430. [Google Scholar] [CrossRef]
- Pougnet, J.-T.; Compans, B.; Martinez, A.; Choquet, D.; Hosy, E.; Boué-Grabot, E. P2X-mediated AMPA receptor internalization and synaptic depression is controlled by two CaMKII phosphorylation sites on GluA1 in hippocampal neurons. Sci. Rep. 2016, 6, 31836. [Google Scholar] [CrossRef]
- Pankratov, Y.; Lalo, U.; Krishtal, O.; Verkhratsky, A. P2X receptors and synaptic plasticity. Neuroscience 2009, 158, 137–148. [Google Scholar] [CrossRef]
- Khakh, B.S.; Bao, X.R.; Labarca, C.; Lester, H.A. Neuronal P2X transmitter-gated cation channels change their ion selectivity in seconds. Nat. Neurosci. 1999, 2, 322–330. [Google Scholar] [CrossRef]
- Zheng, X.; Zhang, L.; Wang, A.P.; Bennett, M.V.; Zukin, R.S. Ca2+ Influx Amplifies Protein Kinase C Potentiation of Recombinant NMDA Receptors. J. Neurosci. 1997, 17, 8676–8686. [Google Scholar] [CrossRef]
- Krupp, J.J.; Vissel, B.; Heinemann, S.F.; Westbrook, G.L. Calcium-dependent inactivation of recombinant N-methyl-D-aspartate receptors is NR2 subunit specific. Mol. Pharmacol. 1996, 50, 1680–1688. [Google Scholar]
- Michailidis, I.E.; Helton, T.D.; Petrou, V.I.; Mirshahi, T.; Ehlers, M.D.; Logothetis, D.E. Phosphatidylinositol-4,5-Bisphosphate Regulates NMDA Receptor Activity through α-Actinin. J. Neurosci. 2007, 27, 5523–5532. [Google Scholar] [CrossRef] [PubMed]
- Royle, S.J.; Bobanović, L.K.; Murrell-Lagnado, R.D. Identification of a Non-canonical Tyrosine-based Endocytic Motif in an Ionotropic Receptor. J. Boil. Chem. 2002, 277, 35378–35385. [Google Scholar] [CrossRef] [PubMed]
- Royle, S.J.; Qureshi, O.S.; Bobanović, L.K.; Evans, P.R.; Owen, D.J.; Murrell-Lagnado, R.D. Non-canonical YXXG endocytic motifs: Recognition by AP2 and preferential utilization in P2X4 receptors. J. Cell Sci. 2005, 118, 3073–3080. [Google Scholar] [CrossRef] [PubMed]
- Pankratov, Y.V.; Lalo, U.V.; Krishtal, O. Role for P2X Receptors in Long-Term Potentiation. J. Neurosci. 2002, 22, 8363–8369. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, T.; Kanazawa, I.; Sakurai, M. Differential induction of LTP and LTD is not determined solely by instantaneous calcium concentration: An essential involvement of a temporal factor. Eur. J. Neurosci. 2001, 14, 701–708. [Google Scholar] [CrossRef]
- Huganir, R.L.; A Nicoll, R. AMPARs and synaptic plasticity: The last 25 years. Neuron 2013, 80, 704–717. [Google Scholar] [CrossRef]
- Toulme, E.; Soto, F.; Garret, M.; Boué-Grabot, E. Functional Properties of Internalization-Deficient P2X4 Receptors Reveal a Novel Mechanism of Ligand-Gated Channel Facilitation by Ivermectin. Mol. Pharmacol. 2005, 69, 576–587. [Google Scholar] [CrossRef]
- Richler, E.; Shigetomi, E.; Khakh, B.S. Neuronal P2X2 receptors are mobile ATP sensors that explore the plasma membrane when activated. J. Neurosci. 2011, 31, 16716–16730. [Google Scholar] [CrossRef]
- Bobanovic, L.K.; Royle, S.J.; Murrell-Lagnado, R.D. P2X Receptor Trafficking in Neurons Is Subunit Specific. J. Neurosci. 2002, 22, 4814–4824. [Google Scholar] [CrossRef]
- Popova, M.; Rodriguez, L.; Trudell, J.R.; Nguyen, S.; Bloomfield, M.; Davies, D.L.; Asatryan, L. Residues in Transmembrane Segments of the P2X4 Receptor Contribute to Channel Function and Ethanol Sensitivity. Int. J. Mol. Sci. 2020, 21, 2471. [Google Scholar] [CrossRef] [PubMed]
- Davies, D.L.; Kochegarov, A.A.; Kuo, S.T.; Kulkarni, A.A.; Woodward, J.J.; King, B.F.; Alkana, R.L. Ethanol differentially affects ATP-gated P2X and P2X receptor subtypes expressed in oocytes. Neuropharmacology 2005, 49, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Ogata, J.; Shiraishi, M.; Namba, T.; Smothers, C.T.; Woodward, J.J.; Harris, R.A. Effects of Anesthetics on Mutant N-Methyl-d-Aspartate Receptors Expressed in Xenopus Oocytes. J. Pharmacol. Exp. Ther. 2006, 318, 434–443. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodriguez, L.; Yi, C.; Chu, C.; Duriez, Q.; Watanabe, S.; Ryu, M.; Reyes, B.; Asatryan, L.; Boué-Grabot, E.; Davies, D. Cross-Talk between P2X and NMDA Receptors. Int. J. Mol. Sci. 2020, 21, 7187. https://doi.org/10.3390/ijms21197187
Rodriguez L, Yi C, Chu C, Duriez Q, Watanabe S, Ryu M, Reyes B, Asatryan L, Boué-Grabot E, Davies D. Cross-Talk between P2X and NMDA Receptors. International Journal of Molecular Sciences. 2020; 21(19):7187. https://doi.org/10.3390/ijms21197187
Chicago/Turabian StyleRodriguez, Larry, Catherine Yi, Cameron Chu, Quentin Duriez, Sharyse Watanabe, Megan Ryu, Brandon Reyes, Liana Asatryan, Eric Boué-Grabot, and Daryl Davies. 2020. "Cross-Talk between P2X and NMDA Receptors" International Journal of Molecular Sciences 21, no. 19: 7187. https://doi.org/10.3390/ijms21197187
APA StyleRodriguez, L., Yi, C., Chu, C., Duriez, Q., Watanabe, S., Ryu, M., Reyes, B., Asatryan, L., Boué-Grabot, E., & Davies, D. (2020). Cross-Talk between P2X and NMDA Receptors. International Journal of Molecular Sciences, 21(19), 7187. https://doi.org/10.3390/ijms21197187






