Expression, Regulation and Function of microRNA as Important Players in the Transition of MDS to Secondary AML and Their Cross Talk to RNA-Binding Proteins
Abstract
1. The Pathology of Myelodysplastic Syndromes
2. Characteristics of miRNAs
3. Features of RBPs
4. Methodical Strategies and Specific Challenges Regarding miRNA and RBP Analyses in MDS and sAML
5. miRNA Expression in MDS and sAML
6. Functional Relevance of Differentially Expressed miRNAs in MDS and AML
7. Clinical Relevance of miRNAs in MDS and AML
8. Predictive Value of miRNA Expression from Patient’s Plasma
9. Correlation of microRNAs with Cytogenetic Features in MDS and SAML
10. Crosstalk of miRNAs and RBPs in MDS and SAML
11. Treatment Options of mRNA Binding Modulators in MDS and SAML
12. Therapeutic Possibilities of miRNAs in MDS and SAML
13. Treatment Approaches Targeting the Spliceosome in MDS and SAML
14. Targeting RNA Methylation in MDS and SAML Treatment
15. Treatment Options of mRNA Binding Modulators in MDS and SAML
16. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ALKBHS | Alk B homolog 5 |
| AML | acute myeloid leukemia |
| ASXL1 | additional sex combs-like 1 |
| BCOR | BCL6 corepressor |
| BM | bone marrow |
| CN | cytogenetically normal |
| DGCR8 | DiGeorge syndrome chromosomal region 8 |
| DNMT3A | DNA (cytosin-5) -methyltransferase 3A |
| EV | extra cellular vesicle |
| EZH2 | enhancer of zeste 2 polycomb repressive complex 2 subunit |
| FTO | fat mass- and obesity-associated |
| HC | healthy control |
| HD | healthy donor |
| HMA | hypomethylating agent |
| HSC | hematopoietic stem cell |
| ICUS | idiopathic cytopenias of undetermined significance |
| IDH1/IDH2 | isocitrate dehydrogenase (NADP(+)) 1/2 |
| IDUS | idiopathic dysplasia of undetermined significance |
| IGF2BP1 | insulin growth factor 2 binding protein 1 |
| IPSSR | Revised International Prognostic Scoring System |
| lncRNA | long noncoding RNA |
| LOH | loss of heterozygosity |
| MDS | myelodysplastic syndromes |
| MDSC | myeloid-derived suppressor cell |
| MEG3 | maternally expressed gene 3 |
| METTL | methyltransferase-like |
| miRNA | microRNA |
| MNC | mononuclear cell |
| MRD | minimal residual disease |
| MV | microvesicle |
| NF | nuclear factor |
| NGS | next generation sequencing |
| OS | overall survival |
| PD-L1 | programmed death ligand 1 |
| RBP | RNA-binding protein |
| RISC | RNA-induced silencing complex |
| RUNX1 | RUNX family transcription factor 1 |
| sAML | secondary AML |
| SF3B1 | splicing factor 3B subunit 1 |
| SNP | single nucleotide polymorphism |
| snoRNA | small nucleolar RNA |
| SNV | single nucleotide variant |
| SRSF2 | serine and arginine rich splicing factor 2 |
| TET2 | Tet methylcytosine dioxygenase 2 |
| TIM-3 | T cell immunoglobulin mucin-3 |
| TME | tumor microenvironment |
| TP53 | tumor protein 53 gene |
| TRBP | transactivation-responsive RNA-binding protein |
| U2F1 | U2 small nuclear RNA auxiliary factor 1 |
| UTR | untranslated region |
| WHO | World Health Organization |
| YTHDF1 | YTH domain-containing family protein 1 |
| ZRSR2 | zinc finger CCCH-type RNA binding motif and serine/arginine rich 2 |
References
- Ghobrial, I.; Detappe, A.; Anderson, K.C.; Steensma, D.P. The bone-marrow niche in MDS and MGUS: Implications for AML and MM. Nat. Rev. Clin. Oncol. 2018, 15, 219–233. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, P.L.; Tuechler, H.; Schanz, J.; Sanz, G.; Garcia-Manero, G.; Solé, F.; Bennett, J.M.; Bowen, D.; Fenaux, P.; Dreyfus, F.; et al. Revised International Prognostic Scoring System for Myelodysplastic Syndromes. Blood 2012, 120, 2454–2465. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef]
- Greenberg, P.L. Molecular and genetic features of myelodysplastic syndromes. Int. J. Lab. Hematol. 2011, 34, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Jhanwar, S.C. Genetic and epigenetic pathways in myelodysplastic syndromes: A brief overview. Adv. Boil. Regul. 2015, 58, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Aul, C.; Gattermann, N.; Schneider, W. Age-related incidence and other epidemiological aspects of myelodysplastic syndromes. Br. J. Haematol. 1992, 82, 358–367. [Google Scholar] [CrossRef] [PubMed]
- Bagby, G.C.; Meyers, G. Bone Marrow Failure as a Risk Factor for Clonal Evolution: Prospects for Leukemia Prevention. Hematology 2007, 2007, 40–46. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bowen, D.; Culligan, D.; Jowitt, S.; Kelsey, S.; Mufti, G.; Oscier, D.; Parker, J. Guidelines of the UK MDS Guidelines Group Guidelines for the diagnosis and therapy of adult myelodysplastic syndromes. Br. J. Haematol. 2003, 120, 187–200. [Google Scholar] [CrossRef]
- Miesner, M.; Haferlach, C.; Bacher, U.; Weiss, T.; Macijewski, K.; Kohlmann, A.; Klein, H.-U.; Dugas, M.; Kern, W.; Schnittger, S.; et al. Multilineage dysplasia (MLD) in acute myeloid leukemia (AML) correlates with MDS-related cytogenetic abnormalities and a prior history of MDS or MDS/MPN but has no independent prognostic relevance: A comparison of 408 cases classified as “AML not otherwise specified” (AML-NOS) or “AML with myelodysplasia-related changes” (AML-MRC). Blood 2010, 116, 2742–2751. [Google Scholar] [CrossRef]
- Higgins, A.; Shah, M.V. Genetic and Genomic Landscape of Secondary and Therapy-Related Acute Myeloid Leukemia. Genes 2020, 11, 749. [Google Scholar] [CrossRef]
- Visconte, V.; Tiu, R.V.; Rogers, H.J. Pathogenesis of myelodysplastic syndromes: An overview of molecular and non-molecular aspects of the disease. Blood Res. 2014, 49, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Walter, M.J.; Shen, D.; Ding, L.; Shao, J.; Koboldt, D.C.; Chen, K.; Larson, D.E.; McLellan, M.D.; Dooling, D.; Abbott, R.; et al. Clonal Architecture of Secondary Acute Myeloid Leukemia. N. Engl. J. Med. 2012, 366, 1090–1098. [Google Scholar] [CrossRef] [PubMed]
- Klaus, M.; Stavroulaki, E.; Kastrinaki, M.-C.; Fragioudaki, P.; Giannikou, K.; Psyllaki, M.; Pontikoglou, C.; Tsoukatou, D.; Mamalaki, C.; Papadaki, H. Reserves, Functional, Immunoregulatory, and Cytogenetic Properties of Bone Marrow Mesenchymal Stem Cells in Patients with Myelodysplastic Syndromes. Stem Cells Dev. 2010, 19, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Blau, O.; Hofmann, W.-K.; Baldus, C.D.; Thiel, G.; Serbent, V.; Schümann, E.; Thiel, E.; Blau, I.W. Chromosomal aberrations in bone marrow mesenchymal stroma cells from patients with myelodysplastic syndrome and acute myeloblastic leukemia. Exp. Hematol. 2007, 35, 221–229. [Google Scholar] [CrossRef]
- Pellagatti, A.; Armstrong, R.N.; Steeples, V.; Sharma, E.; Repapi, E.; Singh, S.; Sanchi, A.; Radujkovic, A.; Horn, P.; Dolatshad, H.; et al. Impact of spliceosome mutations on RNA splicing in myelodysplasia: Dysregulated genes/pathways and clinical associations. Blood 2018, 132, 1225–1240. [Google Scholar] [CrossRef]
- Milunović, V.; Rogulj, I.M.; Planinc-Peraica, A.; Bulycheva, E.; Ostojić, S.K. The role of microRNA in myelodysplastic syndromes: Beyond DNA methylation and histone modification. Eur. J. Haematol. 2016, 96, 553–563. [Google Scholar] [CrossRef]
- Genovese, G.; Kähler, A.K.; Handsaker, R.E.; Lindberg, J.; Rose, S.A.; Bakhoum, S.F.; Chambert, K.; Mick, E.; Neale, B.M.; Fromer, M.; et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 2014, 371, 2477–2487. [Google Scholar] [CrossRef]
- Papaemmanuil, E.; Gerstung, M.; Malcovati, L.; Tauro, S.; Gundem, G.; Van Loo, P.; Yoon, C.J.; Ellis, P.; Wedge, D.C.; Pellagatti, A.; et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 2013, 122, 3616–3627. [Google Scholar] [CrossRef]
- Jaiswal, S.; Fontanillas, P.; Flannick, J.; Manning, A.; Grauman, P.V.; Mar, B.G.; Lindsley, R.C.; Mermel, C.; Burtt, N.; Chavez, A.; et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014, 371, 2488–2498. [Google Scholar] [CrossRef]
- Steensma, D.P.; Bejar, R.; Jaiswal, S.; Lindsley, R.C.; Sekeres, M.A.; Hasserjian, R.P.; Ebert, B.L. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 2015, 126, 9–16. [Google Scholar] [CrossRef]
- Bejar, R.; Stevenson, K.E.; Caughey, B.A.; Abdel-Wahab, O.; Steensma, D.P.; Galili, N.; Raza, A.; Kantarjian, H.; Levine, R.L.; Neuberg, D.; et al. Validation of a Prognostic Model and the Impact of Mutations in Patients with Lower-Risk Myelodysplastic Syndromes. J. Clin. Oncol. 2012, 30, 3376–3382. [Google Scholar] [CrossRef] [PubMed]
- Bejar, R. Clinical and genetic predictors of prognosis in myelodysplastic syndromes. Haematology 2014, 99, 956–964. [Google Scholar] [CrossRef] [PubMed]
- Bejar, R.; Stevenson, K.; Abdel-Wahab, O.; Galili, N.; Nilsson, B.; Garcia-Manero, G.; Kantarjian, H.; Raza, A.; Levine, R.L.; Neuberg, D.; et al. Clinical Effect of Point Mutations in Myelodysplastic Syndromes. N. Engl. J. Med. 2011, 364, 2496–2506. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, P.; Cox, C.; Lebeau, M.M.; Fenaux, P.; Morel, P.; Sanz, G.; Sanz, M.A.; Vallespi, T.; Hamblin, T.; Oscier, D.; et al. International Scoring System for Evaluating Prognosis in Myelodysplastic Syndromes. Blood 1997, 89, 2079–2088. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Calin, G.A.; Lopez-Berestein, G.; Sood, A.K. miRNA Deregulation in Cancer Cells and the Tumor Microenvironment. Cancer Discov. 2016, 6, 235–246. [Google Scholar] [CrossRef]
- Ozkazanc, D.; Yoyen-Ermis, D.; Tavukcuoglu, E.; Buyukasik, Y.; Esendagli, G. Functional exhaustion of CD4+T cells induced by co-stimulatory signals from myeloid leukaemia cells. Immunology 2016, 149, 460–471. [Google Scholar] [CrossRef]
- Montes, P.; Bernal, M.; Campo, L.N.; González-Ramírez, A.R.; Jiménez, P.; Garrido, P.; Jurado, M.; Garrido, F.; Ruiz-Cabello, F.; Hernández, F. Tumor genetic alterations and features of the immune microenvironment drive myelodysplastic syndrome escape and progression. Cancer Immunol. Immunother. 2019, 68, 2015–2027. [Google Scholar] [CrossRef]
- Meng, F.; Li, L.; Lu, F.; Yue, J.; Liu, Z.; Zhang, W.; Fu, R. Overexpression of TIGIT in NK and T Cells Contributes to Tumor Immune Escape in Myelodysplastic Syndromes. Front. Oncol. 2020, 10, 1595. [Google Scholar] [CrossRef]
- Tcvetkov, N.; Gusak, A.; Morozova, E.; Moiseev, I.; Baykov, V.; Barabanshikova, M.; Lepik, K.; Bakin, E.; Vlasova, J.; Osipova, A.; et al. Immune checkpoints bone marrow expression as the predictor of clinical outcome in myelodysplastic syndrome. Leuk. Res. Rep. 2020, 14, 100215. [Google Scholar] [CrossRef]
- Jiang, H.; Fu, R.; Wang, H.; Li, L.; Liu, H.; Shao, Z. CD47 is expressed abnormally on hematopoietic cells in myelodysplastic syndrome. Leuk. Res. 2013, 37, 907–910. [Google Scholar] [CrossRef]
- Asayama, T.; Tamura, H.; Ishibashi, M.; Kuribayashi-Hamada, Y.; Onodera-Kondo, A.; Okuyama, N.; Yamada, A.; Shimizu, M.; Moriya, K.; Takahashi, H.; et al. Functional expression of Tim-3 on blasts and clinical impact of its ligand galectin-9 in myelodysplastic syndromes. Oncotarget 2017, 8, 88904–88917. [Google Scholar] [CrossRef] [PubMed]
- Roela, R.A.; Carraro, D.M.; Brentani, H.; Kaiano, J.H.; Simao, D.F.; Guarnieiro, R.; Lopes, L.F.; Borojevic, R.; Brentani, M.M. Gene stage-specific expression in the microenvironment of pediatric myelodysplastic syndromes. Leuk. Res. 2007, 31, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Handschuh, L. Not Only Mutations Matter: Molecular Picture of Acute Myeloid Leukemia Emerging from Transcriptome Studies. J. Oncol. 2019, 2019, 1–36. [Google Scholar] [CrossRef] [PubMed]
- Eichmüller, S.B.; Osen, W.; Mandelboim, O.; Seliger, B. Immune Modulatory microRNAs Involved in Tumor Attack and Tumor Immune Escape. J. Natl. Cancer Inst. 2017, 109. [Google Scholar] [CrossRef] [PubMed]
- Rhyasen, G.W.; Starczynowski, D.T. Deregulation of microRNAs in myelodysplastic syndrome. Leukemia 2011, 26, 13–22. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Marcucci, G.; Radmacher, M.D.; Maharry, K.; Mrozek, K.; Ruppert, A.S.; Paschka, P.; Vukosavljevic, T.; Whitman, S.P.; Baldus, C.D.; Langer, C.; et al. MicroRNA Expression in Cytogenetically Normal Acute Myeloid Leukemia. N. Engl. J. Med. 2008, 358, 1919–1928. [Google Scholar] [CrossRef]
- Sokol, L.; Caceres, G.; Volinia, S.; Alder, H.; Nuovo, G.J.; Liu, C.-G.; McGraw, K.; Clark, J.A.; Sigua, C.A.; Chen, D.-T.; et al. Identification of a risk dependent microRNA expression signature in myelodysplastic syndromes. Br. J. Haematol. 2011, 153, 24–32. [Google Scholar] [CrossRef]
- Merkerova, M.D.; Krejcik, Z.; Votavová, H.; Belickova, M.; Vasikova, A.; Cermák, J. Distinctive microRNA expression profiles in CD34+ bone marrow cells from patients with myelodysplastic syndrome. Eur. J. Hum. Genet. 2010, 19, 313–319. [Google Scholar] [CrossRef]
- Kim, H.H.; Kuwano, Y.; Srikantan, S.; Lee, E.K.; Martindale, J.L.; Gorospe, M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009, 23, 1743–1748. [Google Scholar] [CrossRef]
- Scheeres, D.E.; Scholten, D.J. DRGs and outliers in surgical critical care. Am. Surg. 1989, 55, 511–515. [Google Scholar] [PubMed]
- Wightman, B.; Burglin, T.R.; Gatto, J.; Arasu, P.; Ruvkun, G. Negative regulatory sequences in the lin-14 3′-untranslated region are necessary to generate a temporal switch during Caenorhabditis elegans development. Genes Dev. 1991, 5, 1813–1824. [Google Scholar] [CrossRef] [PubMed]
- Michlewski, G.; Cáceres, J.F. Post-transcriptional control of miRNA biogenesis. RNA 2018, 25, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Piovan, C.; Croce, C.M. Interplay between microRNAs and the epigenetic machinery: An intricate network. Biochim. Biophys. Acta 2010, 1799, 694–701. [Google Scholar] [CrossRef]
- Raaijmakers, M.H.G.P.; Mukherjee, S.; Guo, S.; Zhang, S.; Kobayashi, T.; Schoonmaker, J.A.; Ebert, B.L.; Al-Shahrour, F.; Hasserjian, R.P.; Scadden, E.O.; et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature 2010, 464, 852–857. [Google Scholar] [CrossRef]
- Ozdogan, H.; Gur-Dedeoglu, B.; Islakoglu, Y.O.; Aydos, A.; Kose, S.; Atalay, A.; Yegin, Z.A.; Avcu, F.; Uckan-Cetinkaya, D.; Ilhan, O. DICER1 gene and miRNA dysregulation in mesenchymal stem cells of patients with myelodysplastic syndrome and acute myeloblastic leukemia. Leuk. Res. 2017, 63, 62–71. [Google Scholar] [CrossRef]
- De Sousa, M.C.; Gjorgjieva, M.; Dolicka, D.; Sobolewski, C.; Foti, M. Deciphering miRNAs’ Action through miRNA Editing. Int. J. Mol. Sci. 2019, 20, 6249. [Google Scholar] [CrossRef]
- Jopling, C.; Yi, M.; Lancaster, A.M.; Lemon, S.M.; Sarnow, P. Modulation of Hepatitis C Virus RNA Abundance by a Liver-Specific MicroRNA. Science 2005, 309, 1577–1581. [Google Scholar] [CrossRef]
- Sarma, N.J.; Tiriveedhi, V.; Crippin, J.S.; Chapman, W.C.; Mohanakumar, T. Hepatitis C Virus-Induced Changes in MicroRNA 107 (miRNA-107) and miRNA-449a Modulate CCL2 by Targeting the Interleukin-6 Receptor Complex in Hepatitis. J. Virol. 2014, 88, 3733–3743. [Google Scholar] [CrossRef]
- Singaravelu, R.; Russell, R.S.; Tyrrell, D.L.; Pezacki, J.P. Hepatitis C virus and microRNAs: miRed in a host of possibilities. Curr. Opin. Virol. 2014, 7, 1–10. [Google Scholar] [CrossRef]
- Guay, C.; Regazzi, R. Circulating microRNAs as novel biomarkers for diabetes mellitus. Nat. Rev. Endocrinol. 2013, 9, 513–521. [Google Scholar] [CrossRef]
- Condorelli, G.; Latronico, M.V.; Cavarretta, E. microRNAs in Cardiovascular Diseases. J. Am. Coll. Cardiol. 2014, 63, 2177–2187. [Google Scholar] [CrossRef] [PubMed]
- Wallace, J.A.; O’Connell, R.M. MicroRNAs and acute myeloid leukemia: Therapeutic implications and emerging concepts. Blood 2017, 130, 1290–1301. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Sevignani, C.; Dumitru, C.D.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, R.; Bullrich, F.; Negrini, M.; et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 2004, 101, 2999–3004. [Google Scholar] [CrossRef] [PubMed]
- Starczynowski, D.T.; Morin, R.; McPherson, A.; Lam, J.; Chari, R.; Wegrzyn, J.; Kuchenbauer, F.; Hirst, M.; Tohyama, K.; Humphries, R.K.; et al. Genome-wide identification of human microRNAs located in leukemia-associated genomic alterations. Blood 2011, 117, 595–607. [Google Scholar] [CrossRef] [PubMed]
- Stavast, C.J.; Leenen, P.J.M.; Erkeland, S.J. The interplay between critical transcription factors and microRNAs in the control of normal and malignant myelopoiesis. Cancer Lett. 2018, 427, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Ghaedi, H.; Bastami, M.; Zare-Abdollahi, D.; Alipoor, B.; Movafagh, A.; Mirfakhraie, R.; Omrani, M.D.; Masotti, A. Bioinformatics prioritization of SNPs perturbing microRNA regulation of hematological malignancy-implicated genes. Genomics 2015, 106, 360–366. [Google Scholar] [CrossRef]
- Kunze, K.; Gamerdinger, U.; Leßig-Owlanj, J.; Sorokina, M.; Brobeil, A.; Tur, M.K.; Blau, W.; Burchardt, A.; Kabisch, A.; Schliesser, G.; et al. Detection of an activated JAK3 variant and a Xq26.3 microdeletion causing loss of PHF6 and miR-424 expression in myelodysplastic syndromes by combined targeted next generation sequencing and SNP array analysis. Pathol. Res. Pract. 2014, 210, 369–376. [Google Scholar] [CrossRef]
- Nicoloso, M.S.; Sun, H.; Spizzo, R.; Kim, H.; Wickramasinghe, P.; Shimizu, M.; Wojcik, S.E.; Ferdin, J.; Kunej, T.; Xiao, L.; et al. Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility. Cancer Res. 2010, 70, 2789–2798. [Google Scholar] [CrossRef]
- Pereira, B.; Billaud, M.; Almeida, R. RNA-Binding Proteins in Cancer: Old Players and New Actors. Trends Cancer 2017, 3, 506–528. [Google Scholar] [CrossRef]
- Corley, M.; Burns, M.C.; Yeo, G.W. How RNA-Binding Proteins Interact with RNA: Molecules and Mechanisms. Mol. Cell 2020, 78, 9–29. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Ni, H.; Liu, Y.; Yuan, Y.; Xi, T.; Li, X.; Zheng, L. RNA-binding proteins in tumor progression. J. Hematol. Oncol. 2020, 13, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-L.; Li, B.; Luo, Y.-X.; Lin, Q.; Liu, S.-R.; Zhang, X.-Q.; Zhou, H.; Yang, J.; Qu, L.-H. Comprehensive Genomic Characterization of RNA-Binding Proteins across Human Cancers. Cell Rep. 2018, 22, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Hodson, D.J.; Screen, M.; Turner, M. RNA-binding proteins in hematopoiesis and hematological malignancy. Blood 2019, 133, 2365–2373. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Murmu, K.C.; Biswas, M.; Chakraborty, S.; Basu, J.; Madhulika, S.; Kolapalli, S.P.; Chauhan, S.; Sengupta, A.; Prasad, P. Transcriptomic Analysis Identifies RNA Binding Proteins as Putative Regulators of Myelopoiesis and Leukemia. Front. Oncol. 2019, 9, 692. [Google Scholar] [CrossRef] [PubMed]
- Schuschel, K.; Helwig, M.; Hüttelmaier, S.; Heckl, D.; Klusmann, J.-H.; Hoell, J.I. RNA-Binding Proteins in Acute Leukemias. Int. J. Mol. Sci. 2020, 21, 3409. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Plessmann, U.; Harlander, S.; Daiß, J.-L.; Eichner, N.; Lehmann, G.; Schall, K.; Urlaub, H.; Meister, G. A Compendium of RNA-Binding Proteins that Regulate MicroRNA Biogenesis. Mol. Cell 2017, 66, 270–284.e13. [Google Scholar] [CrossRef]
- Keene, J.D. RNA regulons: Coordination of post-transcriptional events. Nat. Rev. Genet. 2007, 8, 533–543. [Google Scholar] [CrossRef]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef]
- Haferlach, T.; Nagata, Y.; Grossmann, V.; Okuno, Y.; Bacher, U.; Nagae, G.; Schnittger, S.; Sanada, M.; Kon, A.; Alpermann, T.; et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 2013, 28, 241–247. [Google Scholar] [CrossRef]
- Papaemmanuil, E.; Cazzola, M.; Boultwood, J.; Malcovati, L.; Vyas, P.; Bowen, D.; Pellagatti, A.; Wainscoat, J.; Hellström-Lindberg, E.; Gambacorti-Passerini, C.; et al. SomaticSF3B1Mutation in Myelodysplasia with Ring Sideroblasts. N. Engl. J. Med. 2011, 365, 1384–1395. [Google Scholar] [CrossRef]
- Wang, E.; Aifantis, I. RNA Splicing and Cancer. Trends Cancer 2020, 6, 631–644. [Google Scholar] [CrossRef] [PubMed]
- Aldridge, S.; Hadfield, J. Introduction to miRNA Profiling Technologies and Cross-Platform Comparison. Adv. Struct. Saf. Stud. 2011, 822, 19–31. [Google Scholar] [CrossRef]
- Lu, T.X.; Rothenberg, M.E. MicroRNA. J. Allergy Clin. Immunol. 2018, 141, 1202–1207. [Google Scholar] [CrossRef]
- Kalogianni, D.P.; Kalligosfyri, P.M.; Kyriakou, I.K.; Christopoulos, T.K. Advances in microRNA analysis. Anal. Bioanal. Chem. 2017, 410, 695–713. [Google Scholar] [CrossRef]
- Pradervand, S.; Weber, J.; Lemoine, F.; Consales, F.; Paillusson, A.; Dupasquier, M.; Thomas, J.; Richter, H.; Kaessmann, H.; Beaudoing, E.; et al. Concordance among digital gene expression, microarrays, and qPCR when measuring differential expression of microRNAs. Biotechnology 2010, 48, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Faridani, O.R.; Abdullayev, I.; Hagemann-Jensen, M.; Schell, J.P.; Lanner, F.; Sandberg, R. Single-cell sequencing of the small-RNA transcriptome. Nat. Biotechnol. 2016, 34, 1264–1266. [Google Scholar] [CrossRef]
- Pandita, A.; Ramadas, P.; Poudel, A.; Saad, N.; Anand, A.; Basnet, A.; Wang, D.; Middleton, F.A.; Gilligan, D.M. Differential expression of miRNAs in acute myeloid leukemia quantified by Nextgen sequencing of whole blood samples. PLoS ONE 2019, 14, e0213078. [Google Scholar] [CrossRef]
- Arnold, C.P.; Tan, R.; Zhou, B.; Yue, S.-B.; Schaffert, S.; Biggs, J.R.; Doyonnas, R.; Lo, M.-C.; Perry, J.M.; Renault, V.M.; et al. MicroRNA programs in normal and aberrant stem and progenitor cells. Genome Res. 2011, 21, 798–810. [Google Scholar] [CrossRef]
- Siebolts, U.; Varnholt, H.; Drebber, U.; Dienes, H.-P.; Wickenhauser, C.; Odenthal, M. Tissues from routine pathology archives are suitable for microRNA analyses by quantitative PCR. J. Clin. Pathol. 2008, 62, 84–88. [Google Scholar] [CrossRef]
- Wang, N.; Zheng, J.; Chen, Z.; Liu, Y.; Dura, B.; Kwak, M.; Xavier-Ferrucio, J.; Lu, Y.-C.; Zhang, M.; Roden, C.; et al. Single-cell microRNA-mRNA co-sequencing reveals non-genetic heterogeneity and mechanisms of microRNA regulation. Nat. Commun. 2019, 10, 95. [Google Scholar] [CrossRef] [PubMed]
- Braun, J.; Misiak, D.; Busch, B.; Krohn, K.; Hüttelmaier, S. Rapid identification of regulatory microRNAs by miTRAP (miRNA trapping by RNA in vitro affinity purification). Nucleic Acids Res. 2014, 42, e66. [Google Scholar] [CrossRef] [PubMed]
- Jasinski-Bergner, S.; Stoehr, C.; Bukur, J.; Massa, C.; Braun, J.; Hüttelmaier, S.; Spath, V.; Wartenberg, R.; Legal, W.; Täubert, H.; et al. Clinical relevance of miR-mediated HLA-G regulation and the associated immune cell infiltration in renal cell carcinoma. Oncoimmunology 2015, 4, e1008805. [Google Scholar] [CrossRef]
- Hrustincova, A.; Krejcik, Z.; Kundrat, D.; Szikszai, K.; Belickova, M.; Pecherkova, P.; Klema, J.; Vesela, J.; Hruba, M.; Cermak, J.; et al. Circulating Small Noncoding RNAs Have Specific Expression Patterns in Plasma and Extracellular Vesicles in Myelodysplastic Syndromes and Are Predictive of Patient Outcome. Cells 2020, 9, 794. [Google Scholar] [CrossRef]
- Krause, D.S.; Fackler, M.J.; Civin, C.I.; May, W.S. CD34: Structure, biology, and clinical utility. Blood 1996, 87, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Madadi, S.; Schwarzenbach, H.; Lorenzen, J.; Soleimani, M. MicroRNA expression studies: Challenge of selecting reliable reference controls for data normalization. Cell. Mol. Life Sci. 2019, 76, 3497–3514. [Google Scholar] [CrossRef]
- Pan, X.; Yang, Y.; Xia, C.; Mirza, A.H.; Shen, H. Recent methodology progress of deep learning for RNA–protein interaction prediction. Wiley Interdiscip. Rev. RNA 2019, 10, e1544. [Google Scholar] [CrossRef]
- Xu, J.; Wang, Z.; Jin, X.; Li, L.; Pan, T. Methods for Identification of Protein-RNA Interaction. Adv. Exp. Med. Biol. 2018, 117–126. [Google Scholar] [CrossRef]
- Wang, H.-C.; Chan, L.-P.; Cho, S.-F. Targeting the Immune Microenvironment in the Treatment of Head and Neck Squamous Cell Carcinoma. Front. Oncol. 2019, 9, 1084. [Google Scholar] [CrossRef]
- Lee, F.; Ule, J. Advances in CLIP Technologies for Studies of Protein-RNA Interactions. Mol. Cell 2018, 69, 354–369. [Google Scholar] [CrossRef]
- Van Nues, R.; Schweikert, G.; De Leau, E.; Selega, A.; Langford, A.; Franklin, R.; Iosub, I.; Wadsworth, P.; Sanguinetti, G.; Granneman, S. Kinetic CRAC uncovers a role for Nab3 in determining gene expression profiles during stress. Nat. Commun. 2017, 8, 12. [Google Scholar] [CrossRef] [PubMed]
- Kuang, X.; Chi, J.; Wang, L. Deregulated microRNA expression and its pathogenetic implications for myelodysplastic syndromes. Hematology 2016, 21, 593–602. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Enjeti, A.K.; Ariyarajah, A.; D’Crus, A.; Riveros, C.; Seldon, M.; Lincz, L.F. Circulating microvesicles are less procoagulant and carry different miRNA cargo in myelodysplasia. Blood Cells Mol. Dis. 2019, 74, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Hur, E.-H.; Moon, J.H.; Goo, B.-K.; Choi, D.R.; Lee, J.-H. Expression and prognostic significance of microRNAs in Korean patients with myelodysplastic syndrome. Korean J. Intern. Med. 2019, 34, 390–400. [Google Scholar] [CrossRef] [PubMed]
- Merkerova, M.D.; Hrustincova, A.; Krejcik, Z.; Votavová, H.; Ratajova, E.; Cermak, J.; Belickova, M. Microarray profiling defines circulating microRNAs associated with myelodysplastic syndromes. Neoplasma 2017, 64, 571–578. [Google Scholar] [CrossRef]
- Wen, J.; Huang, Y.; Li, H.; Zhang, X.; Cheng, P.; Deng, D.; Peng, Z.; Luo, J.; Zhao, W.; Lai, Y.; et al. Over-expression of miR-196b-5p is significantly associated with the progression of myelodysplastic syndrome. Int. J. Hematol. 2017, 105, 777–783. [Google Scholar] [CrossRef]
- Jang, S.J.; Choi, I.-S.; Park, G.; Moon, D.-S.; Choi, J.-S.; Nam, M.-H.; Yoon, S.-Y.; Choi, C.H.; Kang, S.H. MicroRNA-205-5p is upregulated in myelodysplastic syndromes and induces cell proliferation via PTEN suppression. Leuk. Res. 2016, 47, 172–177. [Google Scholar] [CrossRef]
- Alkhatabi, H.A.; McLornan, D.P.; Kulasekararaj, A.G.; Malik, F.; Seidl, T.; Darling, D.; Gaken, J.; Mufti, G.J. RPL27A is a target of miR-595 and may contribute to the myelodysplastic phenotype through ribosomal dysgenesis. Oncotarget 2016, 7, 47875–47890. [Google Scholar] [CrossRef]
- Cull, A.H.; Rauh, M.J. Success in bone marrow failure? Novel therapeutic directions based on the immune environment of myelodysplastic syndromes. J. Leukoc. Biol. 2017, 102, 209–219. [Google Scholar] [CrossRef]
- Guo, Y.; Strickland, S.A.; Mohan, S.; Li, S.; Bosompem, A.; Vickers, K.C.; Zhao, S.; Sheng, Q.; Kim, A.S. MicroRNAs and tRNA-derived fragments predict the transformation of myelodysplastic syndromes to acute myeloid leukemia. Leuk. Lymphoma 2017, 58, 2144–2155. [Google Scholar] [CrossRef]
- Haase, D. Cytogenetic features in myelodysplastic syndromes. Ann. Hematol. 2008, 87, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Su, R.; Lin, H.-S.; Zhang, X.-H.; Yin, X.-L.; Ning, H.-M.; Liu, B.; Zhai, P.-F.; Gong, J.-N.; Shen, C.; Song, L.; et al. MiR-181 family: Regulators of myeloid differentiation and acute myeloid leukemia as well as potential therapeutic targets. Oncogene 2014, 34, 3226–3239. [Google Scholar] [CrossRef] [PubMed]
- Marcucci, G.; Maharry, K.; Radmacher, M.D.; Mrózek, K.; Vukosavljevic, T.; Paschka, P.; Whitman, S.P.; Langer, C.; Baldus, C.D.; Liu, C.-G.; et al. Prognostic Significance of, and Gene and MicroRNA Expression Signatures Associated with, CEBPAMutations in Cytogenetically Normal Acute Myeloid Leukemia with High-Risk Molecular Features: A Cancer and Leukemia Group B Study. J. Clin. Oncol. 2008, 26, 5078–5087. [Google Scholar] [CrossRef]
- Lovat, F.; Fassan, M.; Sacchi, D.; Ranganathan, P.; Palamarchuk, A.; Bill, M.; Karunasiri, M.; Gasparini, P.; Nigita, G.; Distefano, R.; et al. Knockout of both miR-15/16 loci induces acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 2018, 115, 13069–13074. [Google Scholar] [CrossRef]
- Lovat, F.; Nigita, G.; Distefano, R.; Nakamura, T.; Gasparini, P.; Tomasello, L.; Fadda, P.; Ibrahimova, N.; Catricala, S.; Palamarchuk, A.; et al. Combined loss of function of two different loci of miR-15/16 drives the pathogenesis of acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 2020, 117, 12332–12340. [Google Scholar] [CrossRef]
- Tsitsiou, E.; Lindsay, M.A. microRNAs and the immune response. Curr. Opin. Pharmacol. 2009, 9, 514–520. [Google Scholar] [CrossRef] [PubMed]
- Pellagatti, A.; Boultwood, J. Splicing factor mutant myelodysplastic syndromes: Recent advances. Adv. Biol. Regul. 2020, 75, 100655. [Google Scholar] [CrossRef]
- Kim, E.H.; Ilagan, J.O.; Liang, Y.; Daubner, G.M.; Lee, S.C.-W.; Ramakrishnan, A.; Li, Y.; Chung, Y.R.; Micol, J.-B.; Murphy, M.E.; et al. SRSF2 Mutations Contribute to Myelodysplasia by Mutant-Specific Effects on Exon Recognition. Cancer Cell 2015, 27, 617–630. [Google Scholar] [CrossRef]
- Yoshida, K.; Sanada, M.; Shiraishi, Y.; Nowak, D.; Nagata, Y.; Yamamoto, R.; Sato, Y.; Sato-Otsubo, A.; Kon, A.; Nagasaki, M.; et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011, 478, 64–69. [Google Scholar] [CrossRef]
- Van Kouwenhove, M.; Kedde, M.; Agami, R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat. Rev. Cancer 2011, 11, 644–656. [Google Scholar] [CrossRef]
- Fan, Y.; Xia, J. miRNet—Functional Analysis and Visual Exploration of miRNA–Target Interactions in a Network Context. Adv. Struct. Saf. Stud. 2018, 215–233. [Google Scholar] [CrossRef]
- Shastri, A.; Will, B.; Steidl, U.; Verma, A. Stem and progenitor cell alterations in myelodysplastic syndromes. Blood 2017, 129, 1586–1594. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Zhu, Y.; He, Q.; Wu, L.-Y.; Zhang, Z.; Shi, W.-H.; Liu, L.; Chang, C.-K.; Li, X. Identification of microRNA-regulated pathways using an integration of microRNA-mRNA microarray and bioinformatics analysis in CD34+ cells of myelodysplastic syndromes. Sci. Rep. 2016, 6, 32232. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Jin, F.; Zhao, Y.; Chen, Y.; Ge, L.; Liu, L.; Yang, M. Downregulation of microRNA-144 inhibits proliferation and promotes the apoptosis of myelodysplastic syndrome cells through the activation of the AKAP12-dependent ERK1/2 signaling pathway. Cell. Signal. 2020, 68, 109493. [Google Scholar] [CrossRef]
- Ganan-Gomez, I.; Wei, Y.; Yang, H.; Pierce, S.; Bueso-Ramos, C.; Calin, G.; del Carmen Boyano-Adanez, M.; Garcia-Manero, G. Overexpression of miR-125a in myelodysplastic syndrome CD34+ cells modulates NF-kappaB activation and enhances erythroid differentiation arrest. PLoS ONE 2014, 9, e93404. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yin, Y.; Liu, X.; Xi, X.; Xue, W.; Qu, Y.-Q. Non-small cell lung cancer associated microRNA expression signature: Integrated bioinformatics analysis, validation and clinical significance. Oncotarget 2017, 8, 24564–24578. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, M.S. Combining Chemotherapy with Biological Response Modifiers in Treatment of Cancer. J. Natl. Cancer Inst. 1988, 80, 1445–1450. [Google Scholar] [CrossRef]
- Li, G.; Song, Y.; Li, G.; Ren, J.; Xie, J.; Zhang, Y.; Gao, F.; Mu, J.; Dai, J. Downregulation of microRNA-21 expression inhibits proliferation, and induces G1 arrest and apoptosis via the PTEN/AKT pathway in SKM-1 cells. Mol. Med. Rep. 2018, 18, 2771–2779. [Google Scholar] [CrossRef]
- Zhao, J.L.; Rao, D.S.; Boldin, M.P.; Taganov, K.D.; O’Connell, R.M.; Baltimore, D. NF-kappaB dysregulation in microRNA-146a-deficient mice drives the development of myeloid malignancies. Proc. Natl. Acad. Sci. USA 2011, 108, 9184–9189. [Google Scholar] [CrossRef]
- Fang, J.; Barker, B.; Bolanos, L.; Liu, X.; Jerez, A.; Makishima, H.; Christie, S.; Chen, X.; Rao, D.S.; Grimes, H.L.; et al. Myeloid malignancies with chromosome 5q deletions acquire a dependency on an intrachromosomal NF-kappaB gene network. Cell Rep. 2014, 8, 1328–1338. [Google Scholar] [CrossRef]
- Zhang, Y.C.; Ye, H.; Zeng, Z.; Chin, Y.E.; Huang, Y.N.; Fu, G.H. The NF-kappaB p65/miR-23a-27a-24 cluster is a target for leukemia treatment. Oncotarget 2015, 6, 33554–33567. [Google Scholar] [CrossRef] [PubMed]
- Rhyasen, G.W.; Bolanos, L.; Fang, J.; Jerez, A.; Wunderlich, M.; Rigolino, C.; Mathews, L.; Ferrer, M.; Southall, N.; Guha, R.; et al. Targeting IRAK1 as a Therapeutic Approach for Myelodysplastic Syndrome. Cancer Cell 2013, 24, 90–104. [Google Scholar] [CrossRef] [PubMed]
- Hussein, K.; Theophile, K.; Büsche, G.; Schlegelberger, B.; Göhring, G.; Kreipe, H.; Bock, O. Aberrant microRNA expression pattern in myelodysplastic bone marrow cells. Leuk. Res. 2010, 34, 1169–1174. [Google Scholar] [CrossRef] [PubMed]
- Erdogan, B.; Facey, C.; Qualtieri, J.; Tedesco, J.; Rinker, E.; Isett, R.B.; Tobias, J.; Baldwin, D.A.; Thompson, J.E.; Carroll, M.; et al. Diagnostic microRNAs in myelodysplastic syndrome. Exp. Hematol. 2011, 39, 915–926.e2. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Z.; Maiti, S.; Hu, S.; Loghavi, S.; Calin, G.A.; Garcia-Manero, G.; Kantarjian, H.M.; Medeiros, L.J.; Cooper, L.J.; Bueso-Ramos, C.E. Plasma circulating-microRNA profiles are useful for assessing prognosis in patients with cytogenetically normal myelodysplastic syndromes. Mod. Pathol. 2014, 28, 373–382. [Google Scholar] [CrossRef]
- Daschkey, S.; Röttgers, S.; Giri, A.; Bradtke, J.; Teigler-Schlegel, A.; Meister, G.; Borkhardt, A.; Landgraf, P. MicroRNAs Distinguish Cytogenetic Subgroups in Pediatric AML and Contribute to Complex Regulatory Networks in AML-Relevant Pathways. PLoS ONE 2013, 8, e56334. [Google Scholar] [CrossRef]
- Díaz-Beyá, M.; Brunet, S.; Nomdedeu, J.; Tejero, R.; Díaz-Campelo, M.; Pratcorona, M.; Tormo, M.; Ribera, J.M.; Escoda, L.; Duarte, L.; et al. MicroRNA expression at diagnosis adds relevant prognostic information to molecular categorization in patients with intermediate-risk cytogenetic acute myeloid leukemia. Leukemia 2013, 28, 804–812. [Google Scholar] [CrossRef]
- Gerloff, D.; Grundler, R.; Wurm, A.A.; Bräuer-Hartmann, D.; Katzerke, C.; Hartmann, J.-U.; Madan, V.; Müller-Tidow, C.; Duyster, J.; Tenen, D.G.; et al. NF-κB/STAT5/miR-155 network targets PU.1 in FLT3-ITD-driven acute myeloid leukemia. Leukemia 2014, 29, 535–547. [Google Scholar] [CrossRef]
- Wallace, J.A.; Kagele, D.A.; Eiring, A.M.; Kim, C.N.; Hu, R.; Runtsch, M.C.; Alexander, M.; Huffaker, T.B.; Lee, S.-H.; Patel, A.B.; et al. miR-155 promotes FLT3-ITD–induced myeloproliferative disease through inhibition of the interferon response. Blood 2017, 129, 3074–3086. [Google Scholar] [CrossRef]
- Quandt, D.; Zucht, H.D.; Amann, A.; Wulf-Goldenberg, A.; Borrebaeck, C.; Cannarile, M.; Lambrechts, D.; Oberacher, H.; Garrett, J.; Nayak, T.; et al. Implementing liquid biopsies into clinical decision making for cancer immunotherapy. Oncotarget 2017, 8, 48507–48520. [Google Scholar] [CrossRef]
- Zuo, Z.; Calin, G.A.; De Paula, H.M.; Medeiros, L.J.; Fernandez, M.H.; Shimizu, M.; Garcia-Manero, G.; Bueso-Ramos, C.E. Circulating microRNAs let-7a and miR-16 predict progression-free survival and overall survival in patients with myelodysplastic syndrome. Blood 2011, 118, 413–415. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Volinia, S.; Liu, C.-G.; Fernandez-Cymering, C.; Palumbo, T.; Pichiorri, F.; Fabbri, M.; Coombes, K.R.; Alder, H.; Nakamura, T.; et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood 2008, 111, 3183–3189. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Melo, S.A.; Sugimoto, H.; O’Connell, J.T.; Kato, N.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A.; et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014, 26, 707–721. [Google Scholar] [CrossRef] [PubMed]
- Huan, J.; Hornick, N.; Shurtleff, M.J.; Skinner, A.M.; Goloviznina, N.A.; Roberts, J.C.T.; Kurre, P. RNA Trafficking by Acute Myelogenous Leukemia Exosomes. Cancer Res. 2012, 73, 918–929. [Google Scholar] [CrossRef]
- Muntión, S.; Ramos, T.L.; Diez-Campelo, M.; Rosón, B.; Sánchez-Abarca, L.I.; Misiewicz-Krzemińska, I.; Preciado, S.; Sarasquete, M.E.; Rivas, J.D.L.; Gonzalez, M.; et al. Microvesicles from Mesenchymal Stromal Cells Are Involved in HPC-Microenvironment Crosstalk in Myelodysplastic Patients. PLoS ONE 2016, 11, e0146722. [Google Scholar] [CrossRef]
- Hornick, N.I.; Huan, J.; Doron, B.; Goloviznina, N.A.; Lapidus, J.; Chang, B.H.; Kurre, P. Serum Exosome MicroRNA as a Minimally-Invasive Early Biomarker of AML. Sci. Rep. 2015, 5, 11295. [Google Scholar] [CrossRef]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Nonlinear partial differential equations and applications: Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Pellagatti, A.; Boultwood, J. The molecular pathogenesis of the myelodysplastic syndromes. Eur. J. Haematol. 2015, 95, 3–15. [Google Scholar] [CrossRef]
- Ebert, B.L. Genetic deletions in AML and MDS. Best Pr. Res. Clin. Haematol. 2010, 23, 457–461. [Google Scholar] [CrossRef][Green Version]
- Starczynowski, D.T.; Kuchenbauer, F.; Argiropoulos, B.; Sung, S.; Morin, R.D.; Muranyi, A.; Hirst, M.; Hogge, D.; Marra, M.; Wells, R.A.; et al. Identification of miR-145 and miR-146a as mediators of the 5q– syndrome phenotype. Nat. Med. 2009, 16, 49–58. [Google Scholar] [CrossRef]
- Yin, X.; Huang, S.; Xu, A.; Fan, F.; Chen, L.; Sun, C.; Hu, Y. Identification of distinctive long noncoding RNA competitive interactions and a six-methylated-gene prognostic signature in acute myeloid leukemia with -5/del(5q) or -7/del(7q). J. Cell Biochem. 2020, 121, 1563–1574. [Google Scholar] [CrossRef] [PubMed]
- Pellagatti, A.; Boultwood, J. RECENT ADVANCES IN THE 5Q- SYNDROME. Mediterr. J. Hematol. Infect. Dis. 2015, 7, e2015037. [Google Scholar] [CrossRef] [PubMed]
- Harada, N.; Watanabe, Y.; Yoshimura, Y.; Sakumoto, H.; Makishima, F.; Tsuchiya, M.; Nakanishi, K.; Nakanishi, M.; Aoki, Y. Identification of a checkpoint modulator with synthetic lethality to p53 mutants. Anti-Cancer Drugs 2011, 22, 986–994. [Google Scholar] [CrossRef]
- Vos, P.D.; Leedman, P.J.; Filipovska, A.; Rackham, O. Modulation of miRNA function by natural and synthetic RNA-binding proteins in cancer. Cell. Mol. Life Sci. 2019, 76, 3745–3752. [Google Scholar] [CrossRef]
- Wang, E.; Lu, S.X.; Pastore, A.; Chen, X.; Imig, J.; Lee, S.C.-W.; Hockemeyer, K.; Ghebrechristos, Y.E.; Yoshimi, A.; Inoue, D.; et al. Targeting an RNA-Binding Protein Network in Acute Myeloid Leukemia. Cancer Cell 2019, 35, 369–384.e7. [Google Scholar] [CrossRef] [PubMed]
- Aslan, D.; Garde, C.; Nygaard, M.K.; Helbo, A.S.; Dimopoulos, K.; Hansen, J.W.; Severinsen, M.T.; Treppendahl, M.B.; Sjø, L.D.; Grønbæk, K.; et al. Tumor suppressor microRNAs are downregulated in myelodysplastic syndrome with spliceosome mutations. Oncotarget 2016, 7, 9951–9963. [Google Scholar] [CrossRef]
- Coutinho, D.F.; Monte-Mór, B.C.; Vianna, D.T.; Rouxinol, S.T.; Batalha, A.B.W.; Bueno, A.P.S.; Boulhosa, A.M.; Fernandez, T.S.; Pombo-De-Oliveira, M.S.; Gutiyama, L.M.; et al. TET2 expression level and 5-hydroxymethylcytosine are decreased in refractory cytopenia of childhood. Leuk. Res. 2015, 39, 1103–1108. [Google Scholar] [CrossRef]
- Jiang, S.; Yan, W.; Wang, S.E.; Baltimore, D. Dual mechanisms of posttranscriptional regulation of Tet2 by Let-7 microRNA in macrophages. Proc. Natl. Acad. Sci. USA 2019, 116, 12416–12421. [Google Scholar] [CrossRef]
- Song, S.J.; Ito, K.; Ala, U.; Kats, L.; Webster, K.; Sun, S.M.; Jongen-Lavrencic, M.; Manova-Todorova, K.; Teruya-Feldstein, J.; Avigan, D.E.; et al. The oncogenic microRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell Stem Cell 2013, 13, 87–101. [Google Scholar] [CrossRef]
- Macedo, L.C.; Silvestre, A.P.A.; Rodrigues, C.; De Alencar, J.B.; Zacarias, J.M.V.; Ambrosio-Albuquerque, E.P.; Sell, A.M.; Visentainer, J.E.L. Genetics factors associated with myelodysplastic syndromes. Blood Cells Mol. Dis. 2015, 55, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Maki, K.; Sasaki, K.; Nagata, Y.; Nagasawa, F.; Nakamura, Y.; Ogawa, S.; Mitani, K. Expressional changes of genes and miRNA in common megakaryocyte-erythroid progenitors from lower-risk myelodysplastic syndrome. Int. J. Hematol. 2014, 100, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.H.; Choi, J.-S. MicroRNA-661 upregulation in myelodysplastic syndromes induces apoptosis through p53 activation and associates with decreased overall survival. Leuk. Lymphoma 2019, 60, 2779–2786. [Google Scholar] [CrossRef] [PubMed]
- Bedi, R.K.; Huang, D.; Eberle, S.A.; Wiedmer, L.; Śledź, P.; Caflisch, A. Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer. ChemMedChem 2020, 15, 744–748. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.-G.; et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Methods 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Lan, Q.; Liu, P.Y.; Haase, J.; Bell, J.L.; Hüttelmaier, S.; Liu, T. The Critical Role of RNA m6A Methylation in Cancer. Cancer Res. 2019, 79, 1285–1292. [Google Scholar] [CrossRef]
- Rottman, F.; Bokar, J.; Narayan, P.; Shambaugh, M.; Ludwiczak, R. N6-Adenosine methylation in mRNA: Substrate specificity and enzyme complexity. Biochimie 1994, 76, 1109–1114. [Google Scholar] [CrossRef]
- Bajan, S.; Hutvagner, G. RNA-Based Therapeutics: From Antisense Oligonucleotides to miRNAs. Cells 2020, 9, 137. [Google Scholar] [CrossRef]
- Innao, V.; Allegra, A.; Pulvirenti, N.; Allegra, A.G.; Musolino, C. Therapeutic potential of antagomiRs in haematological and oncological neoplasms. Eur. J. Cancer Care 2020, 29, e13208. [Google Scholar] [CrossRef]
- Vautrin, A.; Manchon, L.; Garcel, A.; Campos, N.; Lapasset, L.; Laaref, A.M.; Bruno, R.; Gislard, M.; Dubois, E.; Scherrer, D.; et al. Both anti-inflammatory and antiviral properties of novel drug candidate ABX464 are mediated by modulation of RNA splicing. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Seto, A.G.; Beatty, X.; Lynch, J.M.; Hermreck, M.; Tetzlaff, M.; Duvic, M.; Jackson, A.L. Cobomarsen, an oligonucleotide inhibitor of miR-155, co-ordinately regulates multiple survival pathways to reduce cellular proliferation and survival in cutaneous T-cell lymphoma. Br. J. Haematol. 2018, 183, 428–444. [Google Scholar] [CrossRef] [PubMed]
- Ratti, M.; Lampis, A.; Ghidini, M.; Salati, M.; Mirchev, M.B.; Valeri, N.; Hahne, J.C. MicroRNAs (miRNAs) and Long Non-Coding RNAs (lncRNAs) as New Tools for Cancer Therapy: First Steps from Bench to Bedside. Target. Oncol. 2020, 15, 261–278. [Google Scholar] [CrossRef] [PubMed]
- Gomez, I.G.; MacKenna, D.A.; Johnson, B.G.; Kaimal, V.; Roach, A.; Ren, S.; Nakagawa, N.; Xin, C.; Newitt, R.; Pandya, S.; et al. Anti-microRNA-21 oligonucleotides prevent Alport nephropathy progression by stimulating metabolic pathways. J. Clin. Investig. 2014, 125, 141–156. [Google Scholar] [CrossRef] [PubMed]
- Reid, G.; Kao, S.C.; Pavlakis, N.; Brahmbhatt, H.; MacDiarmid, J.; Clarke, S.; Boyer, M.; Van Zandwijk, N. Clinical development of TargomiRs, a miRNA mimic-based treatment for patients with recurrent thoracic cancer. Epigenomics 2016, 8, 1079–1085. [Google Scholar] [CrossRef]
- Seiler, M.; Yoshimi, A.; Darman, R.; Chan, B.; Keaney, G.; Thomas, M.; Agrawal, A.A.; Caleb, B.; Csibi, A.; Sean, E.; et al. H3B-8800, an orally available small-molecule splicing modulator, induces lethality in spliceosome-mutant cancers. Nat. Med. 2018, 24, 497–504. [Google Scholar] [CrossRef]
- Lee, S.C.; Dvinge, H.; Kim, E.; Cho, H.; Micol, J.-B.; Chung, Y.R.; Durham, B.H.; Yoshimi, A.; Kim, Y.J.; Thomas, M.; et al. Modulation of splicing catalysis for therapeutic targeting of leukemia with mutations in genes encoding spliceosomal proteins. Nat. Med. 2016, 22, 672–678, Erratum in 2016, 22, 692. [Google Scholar] [CrossRef]
- Xiao, L.; Li, X.; Mu, Z.; Zhou, J.; Zhou, P.; Xie, C.; Jiang, S. FTO inhibition enhances the anti-tumor effect of temozolomide by targeting MYC-miR-155/23a cluster-MXI1 feedback circuit in glioma. Cancer Res. 2020, 80, 3945–3958. [Google Scholar] [CrossRef]
- Chen, B.; Ye, F.; Yu, L.; Jia, G.-F.; Huang, X.; Zhang, X.; Peng, S.; Chen, K.; Wang, M.; Gong, S.; et al. Development of Cell-ActiveN6-Methyladenosine RNA Demethylase FTO Inhibitor. J. Am. Chem. Soc. 2012, 134, 17963–17971. [Google Scholar] [CrossRef]
- Huang, Y.; Su, R.; Sheng, Y.; Dong, L.; Dong, Z.; Xu, H.; Ni, T.; Zhang, Z.; Zhang, T.; Li, C.; et al. Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell 2019, 35, 677–691.e10. [Google Scholar] [CrossRef]
- Shibayama, Y.; Kondo, T.; Ohya, H.; Fujisawa, S.-I.; Teshima, T.; Iseki, K. Upregulation of microRNA-126-5p is associated with drug resistance to cytarabine and poor prognosis in AML patients. Oncol. Rep. 2015, 33, 2176–2182. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, J.; Dong, K.; Lin, F.; Long, M.; Ouyang, Y.; Wei, J.; Chen, X.; Weng, Y.; He, T.; et al. Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell. Signal. 2015, 27, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Butrym, A.; Rybka, J.; Baczynska, D.; Poręba, R.; Mazur, G.; Kuliczkowski, K. Expression of microRNA-181 determines response to treatment with azacitidine and predicts survival in elderly patients with acute myeloid leukaemia. Oncol. Lett. 2016, 12, 2296–2300. [Google Scholar] [CrossRef] [PubMed]
- Czech, M.P. MicroRNAs as Therapeutic Targets. N. Engl. J. Med. 2006, 354, 1194–1195. [Google Scholar] [CrossRef] [PubMed]
- Bansal, P.; Christopher, A.F.; Kaur, R.P.; Kaur, G.; Kaur, A.; Gupta, V. MicroRNA therapeutics: Discovering novel targets and developing specific therapy. Perspect. Clin. Res. 2016, 7, 68. [Google Scholar] [CrossRef]
- Yamamoto, H.; Lu, J.; Oba, S.; Kawamata, T.; Yoshimi, A.; Kurosaki, N.; Yokoyama, K.; Matsushita, H.; Kurokawa, M.; Tojo, A.; et al. miR-133 regulates Evi1 expression in AML cells as a potential therapeutic target. Sci. Rep. 2016, 6, 19204. [Google Scholar] [CrossRef]
- Jiang, X.; Hu, C.; Arnovitz, S.; Bugno, J.; Yu, M.; Zuo, Z.; Chen, P.; Huang, H.; Ulrich, B.; Gurbuxani, S.; et al. miR-22 has a potent anti-tumour role with therapeutic potential in acute myeloid leukaemia. Nat. Commun. 2016, 7, 11452. [Google Scholar] [CrossRef]
- Chen, K.; Chen, Y.; Chen, Z.; Shi, Y.; He, Z.; Ding, B.; Wang, C.; Yu, L. miR-134 increases the antitumor effects of cytarabine by targeting Mnks in acute myeloid leukemia cells. Oncol. Targets Ther. 2018, 11, 3141–3147. [Google Scholar] [CrossRef]
- Huang, X.; Schwind, S.; Santhanam, R.; Eisfeld, A.-K.; Chiang, C.-L.; Lankenau, M.; Yu, B.; Hoellerbauer, P.; Jin, Y.; Tarighat, S.S.; et al. Targeting the RAS/MAPK pathway with miR-181a in acute myeloid leukemia. Oncotarget 2016, 7, 59273–59286. [Google Scholar] [CrossRef]
- Sun, J.; Huang, Q.; Li, S.; Meng, F.; Li, X.; Gong, X. miR-330-5p/Tim-3 axis regulates macrophage M2 polarization and insulin resistance in diabetes mice. Mol. Immunol. 2018, 95, 107–113. [Google Scholar] [CrossRef]
- Moghaddam, Y.; Andalib, A.; Ganji, M.M.; Homayouni, V.; Sharifi, M.; Ganjalikhani-Hakemi, M. Evaluation of the effect of TIM-3 suppression by miR-498 and its effect on apoptosis and proliferation rate of HL-60 cell line. Pathol. Res. Pract. 2018, 214, 1482–1488. [Google Scholar] [CrossRef] [PubMed]
- Hojati, Z.; Ganjalikhani-Hakemi, M.; Ameri, M.; Alimohammadi-Jelodar, S.F.; Dehbashi, M.; Ganji, M.M.; Homayouni, V.; Khanahmad, H. Evaluation of Silencing Effect of miR-133a-5p Mimic on TIM-3 Expression in AML (HL-60) Cell Line. Indian J. Clin. Biochem. 2019, 35, 359–366. [Google Scholar] [CrossRef]
- Wang, Y.; Cheng, Q.; Liu, J.; Dong, M. Leukemia Stem Cell-Released Microvesicles Promote the Survival and Migration of Myeloid Leukemia Cells and These Effects Can Be Inhibited by MicroRNA34a Overexpression. Stem Cells Int. 2016, 2016, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Jiang, Y.; Zhang, H.; Chen, L.; Luo, P.; Li, L.; Zhao, J.; Lv, F.; Zou, D.; Zhang, Y.; et al. Clinicopathological implications of TIM3+ tumor-infiltrating lymphocytes and the miR-455-5p/Galectin-9 axis in skull base chordoma patients. Cancer Immunol. Immunother. 2019, 68, 1157–1169. [Google Scholar] [CrossRef] [PubMed]
- Khalife, J.; Radomska, H.S.; Santhanam, R.; Huang, X.; Neviani, P.; Saultz, J.; Wang, H.; Wu, Y.-Z.; Alachkar, H.; Anghelina, M.; et al. Pharmacological targeting of miR-155 via the NEDD8-activating enzyme inhibitor MLN4924 (Pevonedistat) in FLT3-ITD acute myeloid leukemia. Leukemia 2015, 29, 1981–1992. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, R.; Deng, Y.; Qian, J.; Lu, Z.; Wang, Y.; Zhang, D.; Luo, F.; Chu, Y. MiR-15a/16 deficiency enhances anti-tumor immunity of glioma-infiltrating CD8+ T cells through targeting mTOR. Int. J. Cancer 2017, 141, 2082–2092. [Google Scholar] [CrossRef]
- Zhang, M.; Gao, D.; Shi, Y.; Wang, Y.; Joshi, R.; Yu, Q.; Liu, D.; Alotaibi, F.; Zhang, Y.; Wang, H.; et al. miR-149-3p reverses CD8 + T-cell exhaustion by reducing inhibitory receptors and promoting cytokine secretion in breast cancer cells. Open Biol. 2019, 9, 190061. [Google Scholar] [CrossRef]
- Cheng, Y.Q.; Cheng, Y.Q.; Ren, J.P.; Zhao, J.; Wang, J.M.; Zhou, Y.; Li, G.Y.; Moorman, J.P.; Yao, Z.Q. MicroRNA-155 regulates interferon-gamma production in natural killer cells via Tim-3 signalling in chronic hepatitis C virus infection. Immunology 2015, 145, 485–497. [Google Scholar] [CrossRef]
- Jiang, X.; Zhou, T.; Xiao, Y.; Yu, J.; Dou, S.; Chen, G.; Wang, R.; Xiao, H.; Hou, C.; Wang, W.; et al. Tim-3 promotes tumor-promoting M2 macrophage polarization by binding to STAT1 and suppressing the STAT1-miR-155 signaling axis. Oncoimmunology 2016, 5, e1211219. [Google Scholar] [CrossRef]
- Fooladinezhad, H.; Khanahmad, H.; Ganjalikhani-Hakemi, M.; Doosti, A. Negative regulation of TIM-3 expression in AML cell line (HL-60) using miR-330-5p. Br. J. Biomed. Sci. 2016, 73, 129–133. [Google Scholar] [CrossRef]
- Junker, A.; Krumbholz, M.; Eisele, S.; Mohan, H.; Augstein, F.; Bittner, R.; Lassmann, H.; Wekerle, H.; Hohlfeld, R.; Meinl, E. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain 2009, 132, 3342–3352. [Google Scholar] [CrossRef] [PubMed]
- Yildirim, B.A.; Onal, C.; Dolek, Y. Is it essential to use fiducial markers during cone-beam CT-based radiotherapy for prostate cancer patients? Jpn. J. Radiol. 2016, 35, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Rastgoo, N.; Wu, J.; Liu, M.; Pourabdollah, M.; Atenafu, E.G.; Reece, D.; Chen, W.; Chang, H. Targeting CD47/TNFAIP8 by miR-155 overcomes drug resistance and inhibits tumor growth through induction of phagocytosis and apoptosis in multiple myeloma. Haematologica 2019. [Google Scholar] [CrossRef]
- Zhao, Y.; Yu, X.; Tang, H.-C.; Han, R.; Wang, X.; Wang, J.; Wang, K.; Li, G. MicroRNA-200a Promotes Phagocytosis of Macrophages and Suppresses Cell Proliferation, Migration, and Invasion in Nasopharyngeal Carcinoma by Targeting CD47. BioMed Res. Int. 2020, 2020, 3723781–13. [Google Scholar] [CrossRef]
- Tan, W.; Tang, H.; Jiang, X.; Ye, F.; Huang, L.; Shi, D.; Li, L.; Huang, X.; Li, L.; Xie, X.; et al. Metformin mediates induction of miR-708 to inhibit self-renewal and chemoresistance of breast cancer stem cells through targeting CD47. J. Cell. Mol. Med. 2019, 23, 5994–6004. [Google Scholar] [CrossRef]
- Hoy, S.M. Patisiran: First Global Approval. Drugs 2018, 78, 1625–1631. [Google Scholar] [CrossRef]
- Rizk, M.; Tuzmen, S. Patisiran for the treatment of patients with familial amyloid polyneuropathy. Drugs Today (Barc.) 2019, 55, 315–327. [Google Scholar] [CrossRef]
- Obeng, E.A.; Chappell, R.J.; Seiler, M.; Chen, M.C.; Campagna, D.R.; Schmidt, P.J.; Schneider, R.K.; Lord, A.M.; Wang, L.; Gambe, R.G.; et al. Physiologic Expression of Sf3b1(K700E) Causes Impaired Erythropoiesis, Aberrant Splicing, and Sensitivity to Therapeutic Spliceosome Modulation. Cancer Cell 2016, 30, 404–417. [Google Scholar] [CrossRef]
- Shirai, C.L.; White, B.S.; Tripathi, M.; Tapia, R.; Ley, J.N.; Ndonwi, M.; Kim, S.; Shao, J.; Carver, A.; Saez, B.; et al. Mutant U2AF1-expressing cells are sensitive to pharmacological modulation of the spliceosome. Nat. Commun. 2017, 8, 14060. [Google Scholar] [CrossRef]
- Vu, L.; Pickering, B.F.; Cheng, Y.; Zaccara, S.; Nguyen, D.; Minuesa, G.; Chou, T.; Chow, A.; Saletore, Y.; Mackay, M.; et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 2017, 23, 1369–1376. [Google Scholar] [CrossRef]
- Huang, X.; Schwind, S.; Yu, B.; Santhanam, R.; Wang, H.; Hoellerbauer, P.; Mims, A.; Klisovic, R.; Walker, A.R.; Chan, K.K.; et al. Targeted delivery of microRNA-29b by transferrin-conjugated anionic lipopolyplex nanoparticles: A novel therapeutic strategy in acute myeloid leukemia. Clin. Cancer Res. 2013, 19, 2355–2367. [Google Scholar] [CrossRef]
- Wang, H.; Li, Y.; Lv, N.; Li, Y.-H.; Wang, L.; Yu, L. Predictors of clinical responses to hypomethylating agents in acute myeloid leukemia or myelodysplastic syndromes. Ann. Hematol. 2018, 97, 2025–2038. [Google Scholar] [CrossRef]
- Kim, Y.; Cheong, J.-W.; Kim, Y.-K.; Eom, J.-I.; Jeung, H.-K.; Kim, S.-J.; Hwang, H.; Kim, J.S.; Kim, H.J.; Min, Y.H. Serum microRNA-21 as a Potential Biomarker for Response to Hypomethylating Agents in Myelodysplastic Syndromes. PLoS ONE 2014, 9, e86933. [Google Scholar] [CrossRef]
- Solly, F.; Koering, C.; Mohamed, A.M.; Maucort-Boulch, D.; Robert, G.; Auberger, P.; Flandrin-Gresta, P.; Adès, L.; Fenaux, P.; Kosmider, O.; et al. An miRNA–DNMT1 Axis Is Involved in Azacitidine Resistance and Predicts Survival in Higher-Risk Myelodysplastic Syndrome and Low Blast Count Acute Myeloid Leukemia. Clin. Cancer Res. 2016, 23, 3025–3034. [Google Scholar] [CrossRef] [PubMed]
- Blum, W.; Garzon, R.; Klisovic, R.B.; Schwind, S.; Walker, A.; Geyer, S.; Liu, S.; Havelange, V.; Becker, H.; Schaaf, L.; et al. Clinical response and miR-29b predictive significance in older AML patients treated with a 10-day schedule of decitabine. Proc. Natl. Acad. Sci. USA 2010, 107, 7473–7478. [Google Scholar] [CrossRef] [PubMed]
- Krejcik, Z.; Belickova, M.; Hrustincova, A.; Votavová, H.; Jonášová, A.; Cermák, J.; Dyr, J.E.; Merkerova, M.D. MicroRNA profiles as predictive markers of response to azacitidine therapy in myelodysplastic syndromes and acute myeloid leukemia. Cancer Biomark. 2018, 22, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Haussecker, D. Current issues of RNAi therapeutics delivery and development. J. Control. Release 2014, 195, 49–54. [Google Scholar] [CrossRef] [PubMed]
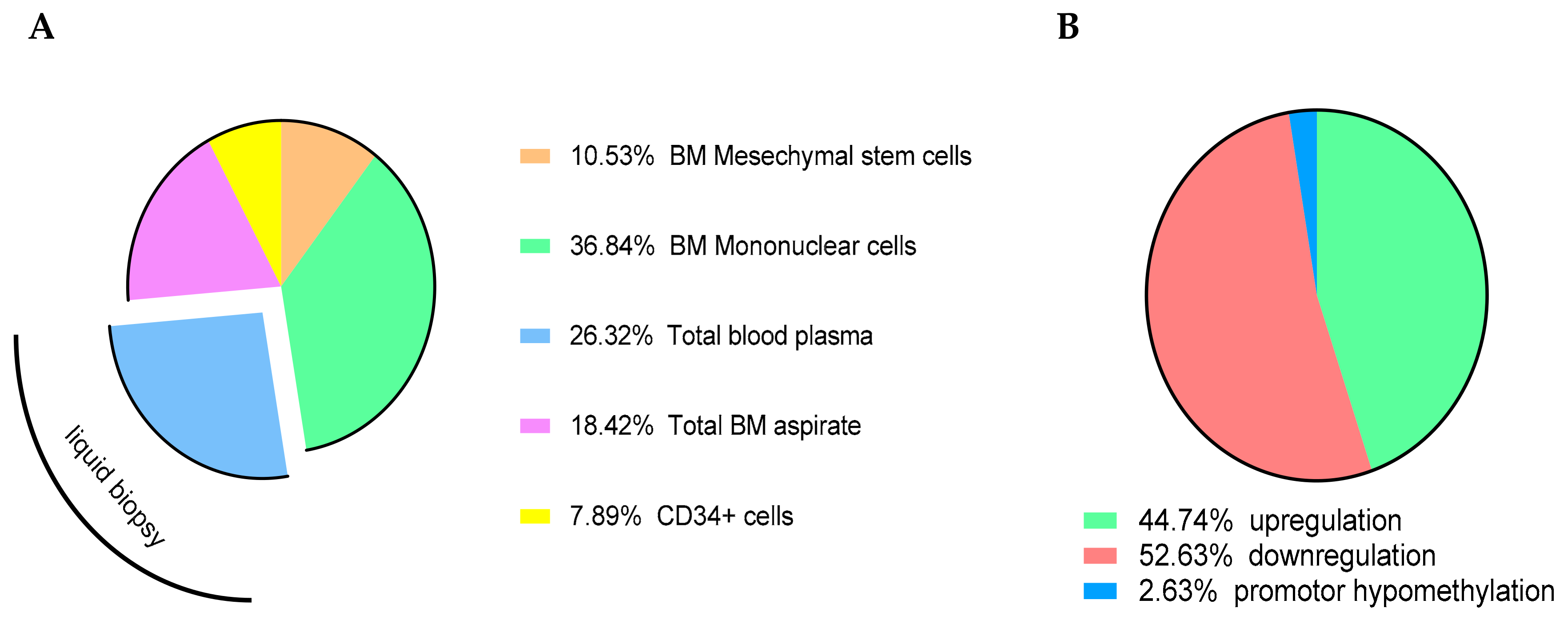
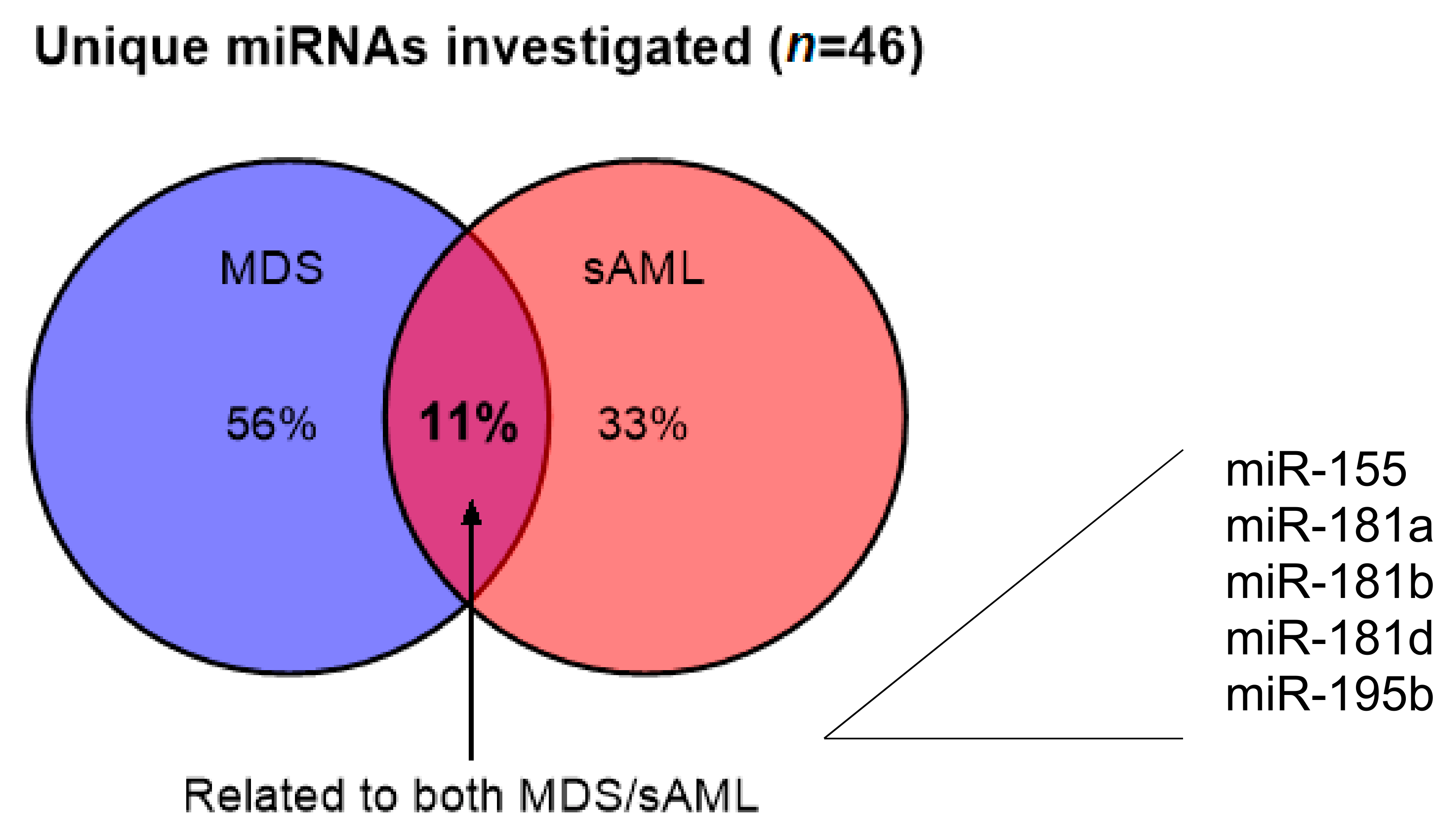
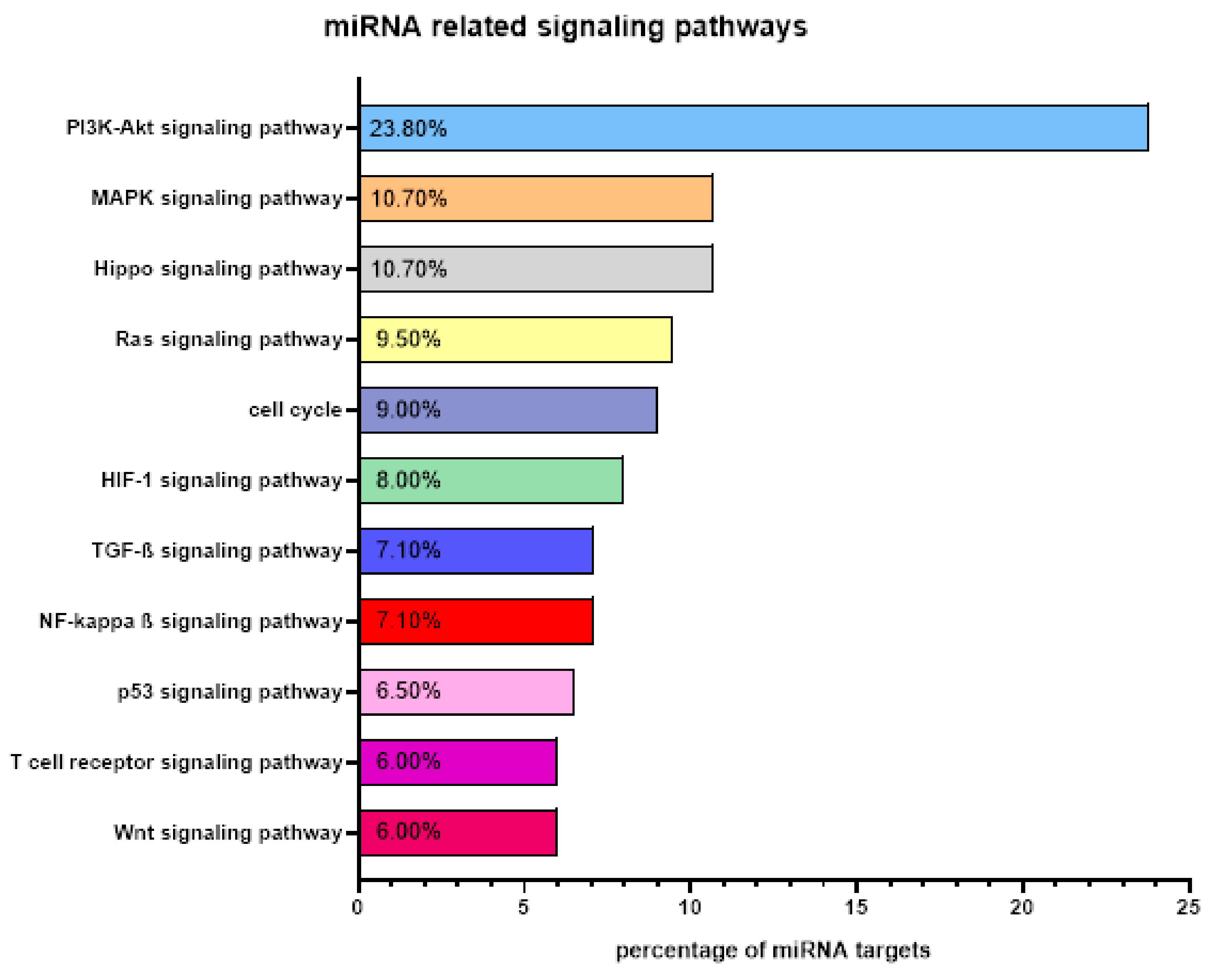
| A: miRNAs with Diagnostic Role | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| miRNA | Disease | Cellular Source | Regulation *1 | Chromosomal Location | Cytogenetic Aberration | % (n = 2072) | Target RBPs | Other Targets | Ref. |
| let-7a | MDS | BM mononuclear cells | ↓ | 9q22.32 | −9q | 1.10 | EWSR1 | HMGA2, KRAS, HRAS, HMGA1, RRM2 | [94] |
| miR-16-5p | MDS | Total blood plasma | ↓ | 3q25.33, 13q14.2 | inv/t(3q), −13/−13q | 2.0/1.9 | n.a. | BCL2, VEGFA, CCNE1, FGF2, CCND1 | [95] |
| miR-27a-3p | MDS | Total blood plasma | ↑ | 19p13.12 | No | - | n.a. | FOXO1, SP1, ZBTB10, IFNG HOXA10 | [95] |
| miR-30a-3p | MDS | BM mesenchymal stem cells | ↓ | 6q13 | No | - | n.a. | CDK6, SLC7A6, TMEM2, THBS1, CYR61 | [46] |
| miR-30d-5p | MDS | BM mesenchymal stem cells | ↓ | 8q24.22 | 8 | 8.40 | n.a. | TP53, SNAI1, EZH2, BCL9, NOTCH1 | [46] |
| miR-124a | MDS | BM mononuclear cells | ↓ | 8p23.1 | 8 | 8.40 | PTBP1, HNRNPA2B1 | CDK6, STAT3, SLC16A1, EZH2, AMOTL1 | [94] |
| miR-150-5p | MDS | Total blood plasma | ↑ | 19q13.33 | No | - | n.a. | MZB, EGR2, P2RX7, ZEB1, MUC4 | [95] |
| miR-155 | MDS | BM mononuclear cells | ↓ | 21q21.3 | +21, −21 | 2.2/1.6 | n.a. | CEBPB, TO53INP1, SOCS1, BACH1, INPP5D | [94] |
| miR-182 | MDS | BM mononuclear cells | ↓ | 7q32.2 | −7/−7q, t(7q) | 11.1, 1.1 | n.a. | RECK, MITF, FOXO1, BCL2, PDCD4 | [94] |
| miR-195b-5p | MDS/sAML | BM mononuclear cells | ↑ | 17p13.1 | −17/−17p | 2.00 | n.a. | CCND1, WEE1, BCL2, E2F3, CDK6 | [96] |
| miR-199a-5p | MDS | Total blood plasma | ↑ | 1q24.3, 19p13.2 | +1/ +1q | 1.80 | n.a. | HIF1A, SIRT1, MAP3K11, ERBB2, CAV1 | [95] |
| miR-200c | MDS | BM mononuclear cells | ↓ | 12p13.31 | −12, −12p | 1.3, 1.2 | n.a. | ZEB1, ZEB2, TUBB3, BMI1, KRAS | [94] |
| miR-205-5p | MDS | Total BM aspirate | ↑ | 1q32.2 | +1/+1q | 1.80 | n.a. | ERBB3, VEGFA, ZEB1, ZEB2, E2F1 | [97] |
| miR-222-3p | MDS | BM mesenchymal stem cells | ↓ | Xp11.3 | No | - | n.a. | CDKN1B, KIT, PTEN, TIMP3, ETS1 | [46] |
| miR-342-5p | MDS | BM mononuclear cells | ↓ | 14q32.2 | No | - | n.a. | NAA10 | [94] |
| miR-451a | MDS | Total blood plasma | ↓ | 17q11.2 | −17/−17p | 2.00 | n.a. | [95] | |
| miR-595 | MDS | CD34+ cells | ↓ | 7q36.3 | −7/−7q, t(7q) | 11.1, 1.1 | n.a. | [98] | |
| miR-4462 | MDS | BM mesenchymal stem cells | ↑ | 6p21.2 | No | - | n.a. | [46] | |
| B: miRNAs with Prognostic Role | |||||||||
| miRNA | Disease | Cellular source | Prognostic marker *2 | Chromosomal Location | Cytogenetic Aberration | % (n = 2072) | Target RBPs | Other targets | Ref. |
| let-7a-3 | MDS | BM mononuclear cells | Adverse *3 | 12q13.31 | −12, −12p | 1.3,1.2 | EWSR1 | HMGA2, KRAS. HRAS, HMGA1, RRM2 | [98] |
| miR-021 | MDS | BM mononuclear cells | Adverse | 17q23.1 | −17/−17p | 2.00 | n.a. | PTEN, RECK, PDCD4, BCL2, TPM1 | [94] |
| miR-27a-3p | MDS | Total blood plasma | Good | 19p13.12 | No | - | n.a. | FOXO1, SP1, ZBTB10, IFNG HOXA10 | [95] |
| miR-124a | MDS | BM mononuclear cells | Adverse | 8p23.1 | 8 | 8.40 | PTBP1, HNRNPA2B1 | CDK6, STAT3, SLC16A1, EZH2, AMOTL1 | [94] |
| miR-126 | MDS | BM mononuclear cells | Adverse | 9q34.3 | −9q | 1.10 | n.a. | VEGFA, CRK, IRS1, PIK3R2, SPRED1 | [94] |
| miR-146b-5p | MDS | BM mononuclear cells | Adverse | 10q24.32 | No | - | n.a. | MMP16, TRAF6, IRAK1, IL6, PDGFRA | [94] |
| miR-150-5p | MDS | Total blood plasma | Good | 19q13.33 | No | - | n.a. | MZB, EGR2, P2RX7, ZEB1, MUC4 | [95] |
| miR-155 | MDS | BM mononuclear cells | Adverse | 21q21.3 | +21, −21 | 2.2/1.6 | n.a. | CEBPB, SOCS1, TO53INP1, BACH1, INPP5D | [94] |
| miR-195b-5p | MDS/sAML | BM mononuclear cells | Adverse | 17p13.1 | −17/−17p | 2.00 | n.a. | CCND1, WEE1, BCL2, E2F3, CDK6 | [96] |
| miR-199a-5p | MDS | Total blood plasma | Good | 1q24.3, 19p13.2 | +1/+1q | 1.80 | n.a. | HIF1A, SIRT1, MAP3K11, ERBB2, CAV1 | [95] |
| miR-223-3p | MDS | Total blood plasma | Good | Xq12 | No | - | n.a. | LMO2, IGF1R, NFIA, RHOB, FBXW7 | [95] |
| miR-451a | MDS | Total blood plasma | Good | 17q11.2 | −17/−17p | 2.00 | n.a. | n.a. | [95] |
| C: miRNAs Indicative of Progression from MDS to sAML | |||||||||
| miRNA | Disease | Cellular Source | Regulation *4 | Chromosomal Location | Cyto-genetic Aberration | % (n = 2072) | Target RBPs | Other Targets | Ref. |
| miR-145 | MDS | CD34+ cells | ↓ | 5q33.3 | −5q, −5, t(5q) | 15.1, 3.3, 1.2 | n.a. | IRS1, POU5F1, FSCN1, KLF4, IGF1R | [99] |
| miR-146a | MDS | CD34+ cells | ↓ | 5q33.3 | −5q, −5, t(5q) | 15.1, 3.3, 1.2 | ELAVL1 | IRAK1, TRAF6, EGFR, NFKB1, STAT1 | [99] |
| miR-181a-5p | MDS | Total BM aspirate | ↑ | 1q32.1, 9q33.3 | +1/ + 1q, −9q | 1.8, 1.1 | n.a. | BCL2, ATM, DDX3X, PRKCD, CDKN1B | [100] |
| miR-181b-5p | MDS | Total BM aspirate | ↑ | 1q32.1 | +1/ + 1q | 1.80 | n.a. | TIMP3, BCL2, CYLD, TCL1A, CREB1 | [100] |
| miR-181d-5p | MDS | Total BM aspirate | ↑ | 19p13.12 | No | - | n.a. | BCL2, HRAS, MGMT, RAP1B, MEG3 | [100] |
| miR-195b-5p | MDS/sAML | BM mononuclear cells | ↑ | 17p13.1 | −17/−17p | 2.00 | n.a. | CCND1, WEE1, BCL2, E2F3, CDK6 | [96] |
| miR-199b-5p | MDS | Total BM aspirate | ↑ | 9q34.11 | −9q | 1.10 | n.a. | JAG1, HES1, PODXL, ERBB2, SET | [100] |
| miR-451a-5p | MDS | Total BM aspirate | ↓ | 17q11.2 | −17/−17p | 2.00 | n.a. | n.a. | [100] |
| miR-486-5p | MDS | Total BM aspirate | ↓ | 8p11.21 | 8 | 8.40 | n.a. | OLFM4, IGF1R, DOCK, CIT, ARHGAP5 | [100] |
| miRNA | HC | MDS with Del (5q) | MDS-RS | MDS-MLD | MDS-EB1 | MDS-EB-2 | AML-MLD |
|---|---|---|---|---|---|---|---|
| (n = 9) | (n = 6) | (n = 5) | (n = 6) | (n = 7) | (n = 10) | (n = 9) | |
| miR-10a | ↑ | ↑ | ↑ | ↓ | ↓ | ↓ | ↓ |
| miR34a | ↑ | ↑ | ↑ | ↑ | ↑ | ↓ | ↑ |
| miR322-3p | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ |
| miR-378 | ↓ | ↓ | ↓ | ↑ | ↓ | ↓ | ↓ |
| Mutated Genes | miRNAs Identified | Cells/Tissues Analyzed | References |
|---|---|---|---|
| TET 2 | miR-22 let-7 family | MDS Cytopenia Macrophages | [148,149,150] [148] [149] |
| ASXL1/IDH1 | miR-21 miR-125a miR-143/-145 miR-146a | Various cell types | [151] |
| SF3B1/SRSF2/42AF1 | let-7 family miR-103a miR-423 | MDS | [147] |
| RUNX1 | miR-9-5p | AML | [152] |
| TP53 | miR-661 | MDS | [153] |
| Context | Target | Importance in MDS Pathogenesis | Disease for Which the Drug Is Being Developed | Drug Name and Company | Mechanism of the Drug | Clinical Relevance | Clinical Trial | Phase of Study | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| miRNA | miR-124a | Downregulation | Ulcerative colitis | Abivax, ABX464 | Enhanced splicing of miR-124 | Yes | NCT03093259 | Phase 2 | [161] |
| Ulcerative colitis | Abivax, ABX464 | Enhanced splicing of miR-124 | Yes | NCT03760003 | Phase 2 | ||||
| Crohn’s disease | Abivax, ABX464 | Enhanced splicing of miR-124 | Yes | NCT03905109 | Phase 2 | ||||
| miR-155 | Upregulation | Cutaneous T-cell lymphoma (CTCL), mycosis fungoides (MF), CLL, DLBCL-ACB type, adult T-cell leukemia/lymphoma (ATLL) | Cobomarsen, MRG-106 | Inhibitor of miR-155 | Yes | NCT02580552 | Phase 1 | [162] | |
| Mycosis fungoides | Cobomarsen, MRG-106 | Inhibitor of miR-155 | Yes | NCT03837457 | Phase 1 | ||||
| miR-200c | Downregulation | Osteogenesis | pSil-miR200c plasmids | Locally delivering plasmid DNAs encoding miR-200c NAs | Yes | NCT02579187 | Withdrawn | [163] | |
| miR-21 | Downregulation | Alport syndrome | Regulus, RG-102 | Inhibitor of miR-21 | No | NCT03373786 | Phase 1 | [164] | |
| miR-16-5p | Upregulation | Malignant pleural mesothelioma, NSCLC * | MesomiR | mirR-16 based mimic | No | NCT02369198 | Phase 1 | [165] | |
| Spliceosome | SF3B1 * | Frequently mutated in MDS (>30%) | MDS, CMML *, AML | H3B-8800 | Inhibitor | Yes | NCT02841540 | Phase 1 | [166] |
| SRSF2 * | Frequently mutated in MDS (>15%) | Cancer | E7107 | Inhibitor | Yes | NCT00499499 | Phase 1 | [167] | |
| RBM39 * | Auxiliary RNA splicing factor | MDS, AML, CLL * | E7820 | Inhibitor | Yes | - | Phase 0 | [81] | |
| mRNA methylation | METTL3 * | RNA methylase | Hematologic neoplasm | Not yet identified | Inhibitor | Yes | - | Phase 0 | [154] |
| FTO * | RNA demethylase | Brain tumors | Meclofenamic acid | Inhibitor | Yes | - | Phase 0 | [168] | |
| - | Rhein | Inhibitor | Yes | - | Phase 0 | [169] | |||
| AML | FB23-2 | Inhibitor | Yes | - | Phase 0 | [170] |
| Immune Checkpoint | miRNA | Reference |
|---|---|---|
| TIM-3 | miR-15a/-16 | [186] |
| miR-34a * | [183] | |
| miR-133a-5p * | [182] | |
| miR-149-3p | [187] | |
| miR-155 | [188,189] | |
| miR-330-5p * | [180,190] | |
| miR-455-5p * | [184] | |
| miR-498 * | [181] | |
| CD47 | miR-34 | [191] |
| miR-133a * | [192] | |
| miR-155 | [193] | |
| miR-200a | [194] | |
| miR-326 | [191] | |
| miR-708 | [195] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bauer, M.; Vaxevanis, C.; Heimer, N.; Al-Ali, H.K.; Jaekel, N.; Bachmann, M.; Wickenhauser, C.; Seliger, B. Expression, Regulation and Function of microRNA as Important Players in the Transition of MDS to Secondary AML and Their Cross Talk to RNA-Binding Proteins. Int. J. Mol. Sci. 2020, 21, 7140. https://doi.org/10.3390/ijms21197140
Bauer M, Vaxevanis C, Heimer N, Al-Ali HK, Jaekel N, Bachmann M, Wickenhauser C, Seliger B. Expression, Regulation and Function of microRNA as Important Players in the Transition of MDS to Secondary AML and Their Cross Talk to RNA-Binding Proteins. International Journal of Molecular Sciences. 2020; 21(19):7140. https://doi.org/10.3390/ijms21197140
Chicago/Turabian StyleBauer, Marcus, Christoforos Vaxevanis, Nadine Heimer, Haifa Kathrin Al-Ali, Nadja Jaekel, Michael Bachmann, Claudia Wickenhauser, and Barbara Seliger. 2020. "Expression, Regulation and Function of microRNA as Important Players in the Transition of MDS to Secondary AML and Their Cross Talk to RNA-Binding Proteins" International Journal of Molecular Sciences 21, no. 19: 7140. https://doi.org/10.3390/ijms21197140
APA StyleBauer, M., Vaxevanis, C., Heimer, N., Al-Ali, H. K., Jaekel, N., Bachmann, M., Wickenhauser, C., & Seliger, B. (2020). Expression, Regulation and Function of microRNA as Important Players in the Transition of MDS to Secondary AML and Their Cross Talk to RNA-Binding Proteins. International Journal of Molecular Sciences, 21(19), 7140. https://doi.org/10.3390/ijms21197140







