The Cotton BEL1-Like Transcription Factor GhBLH7-D06 Negatively Regulates the Defense Response against Verticillium dahliae
Abstract
1. Introduction
2. Results
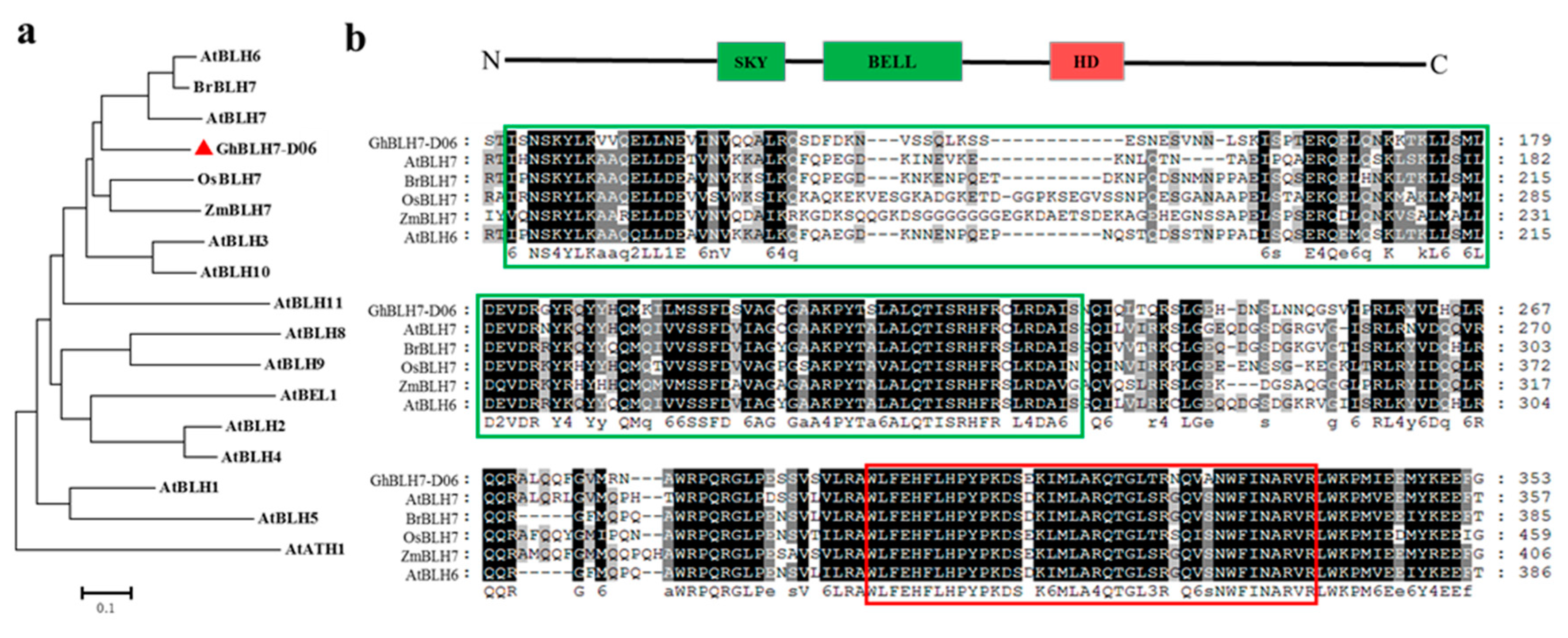
2.1. Identification and Characterization of a BEL1-Like Transcription Factor GhBLH7-D06
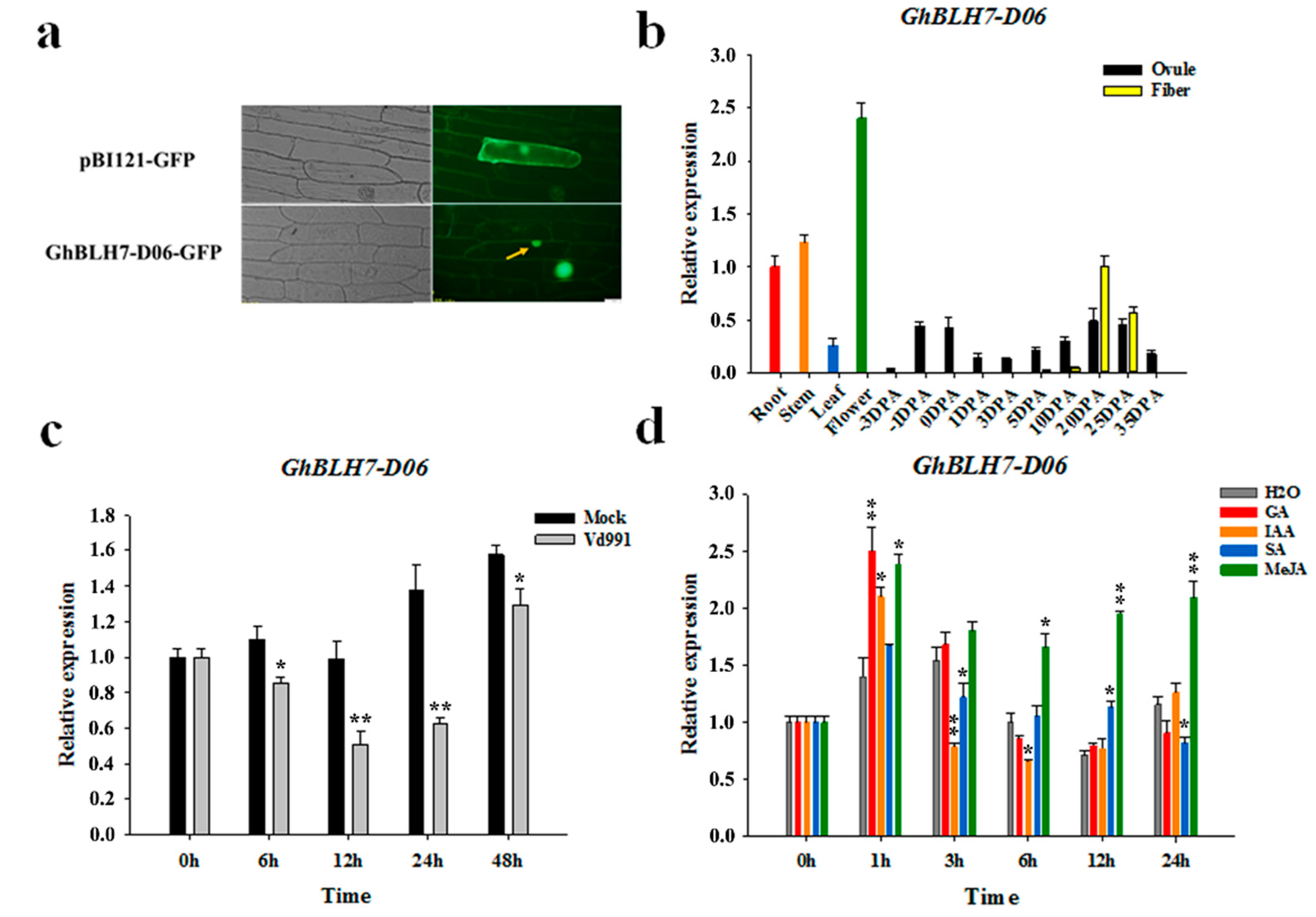
2.2. Expression Pattern Analysis of Cotton GhBLH7-D06
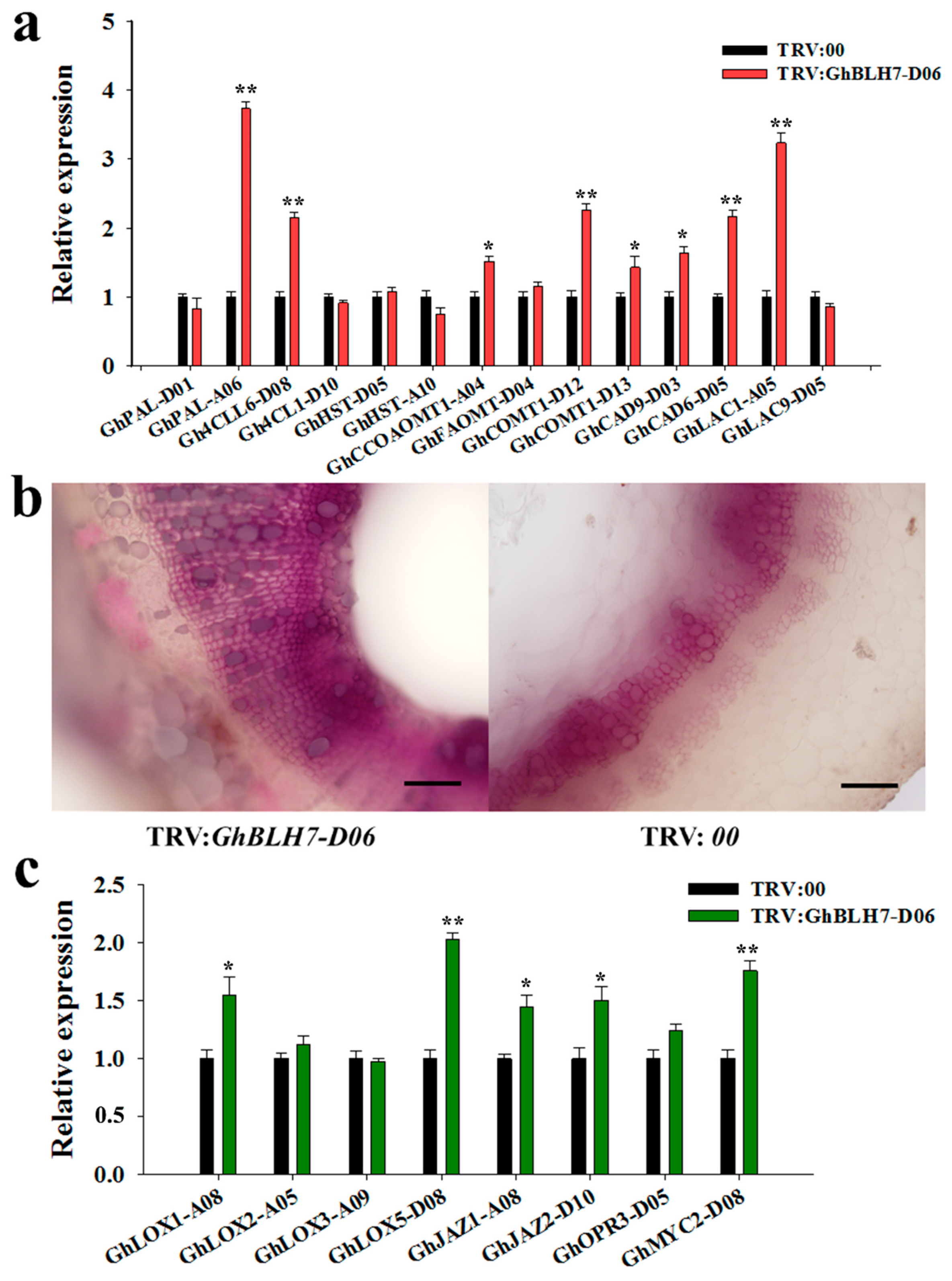
2.3. GhBLH7-D06 Silenced Cotton Displays Enhanced Resistance to V. dahliae Infection
2.4. Silencing of GhBLH7-D06 in Cotton Activates the Biosynthesis of Lignin and JA Associated Genes
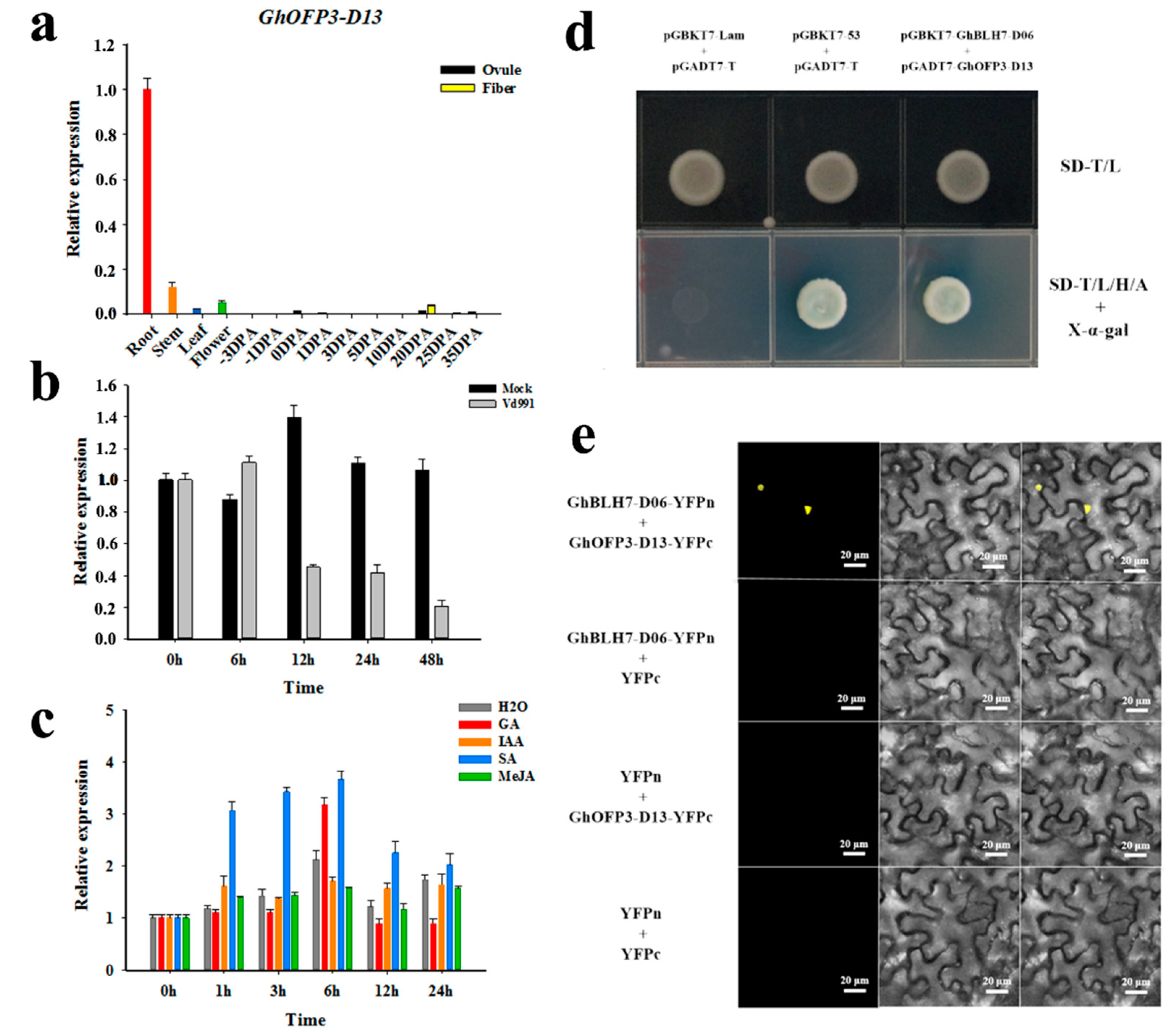
2.5. GhBLH7-D06 Interacts with GhOFP3-D13 in Response to V. dahliae Infection
2.6. GhBLH7-D06 Could Inhibit the Expression of GhPAL-A06
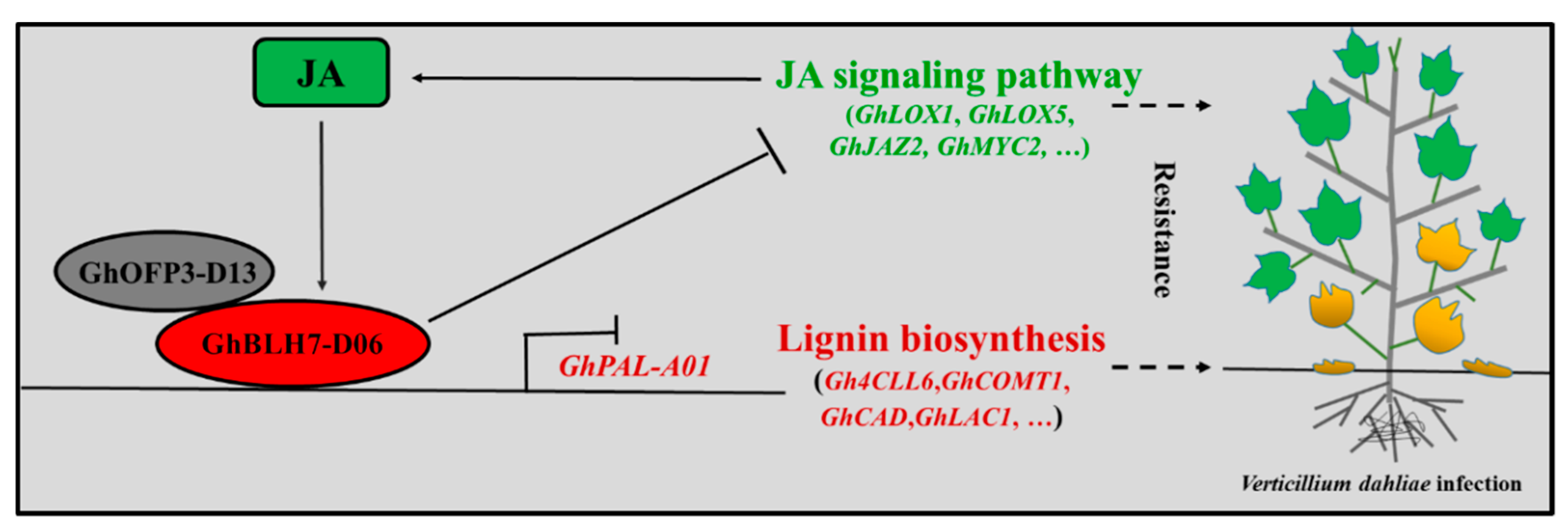
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Conditions and Stress Treatments
4.2. Gene Cloning, Multiple-Sequence Alignment and Phylogenetic Analysis
4.3. Subcellular Localization
4.4. Expression Analysis
4.5. Virus-Induced Gene Silencing (VIGS) of GhBLH7-D06
4.6. Fungal Pathogen Cultivation and Inoculation
4.7. Histochemical Test
4.8. Yeast Two-Hybrid and Library Screening Assay
4.9. BiFC Assay
4.10. Dual-Luciferase Reporter Gene Assay
4.11. Yeast One-Hybrid Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| TALE | Three-amino-acid-loop-extension |
| SCW | Secondary cell wall |
| VIGS | Virus-induced gene silencing |
| Y2H | Yeast two-hybrid |
| BiFC | Bimolecular fluorescence complementary |
| OFP | Ovate family protein |
| Y1H | Yeast one-hybrid |
| IAA | Auxin |
| SA | Salicylic acid |
| GA | Gibberellin acid |
| JA | Jasmonic acid |
| MeJA | Methyl jasmonate |
| ROS | Reactive oxygen species |
| TF | Transcription factor |
| ABA | Abscisic acid |
| BR | Brassinosteroid |
| CK | Cytokinin |
| AA | Amino acid |
References
- Li, F.; Fan, G.; Lu, C.; Xiao, G.; Zou, C.; Kohel, R.J.; Ma, Z.; Shang, H.; Ma, X.; Wu, J. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 2015, 33, 524. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, M.A.C.; Duarte, J.B.; Morello, C.L.; Suassuna, N.D. Mixed inheritance in the genetic control of ramulosis (Colletotrichum gossypii var. cephalosporioides) resistance in cotton. Genet. Mol. Res. 2016, 15, gmr.15038667. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.J.; Negrāo, S.; Tester, M. Salt resistant crop plants. Curr. Opin. Biotechnol. 2014, 26, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Xiaohong, H.; Mo, J.; Sun, Q.; Liu, J. Molecular research and genetic engineering of resistance to Verticillium wilt in cotton: A review. Afr. J. Biotechnol. 2009, 8, 7363–7372. [Google Scholar]
- Fradin, E.F.; Thomma, B.P.H.J. Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant Pathol. 2006, 7, 71–86. [Google Scholar] [CrossRef]
- Yang, C.; Liang, S.; Wang, H.; Han, L.; Wang, F.; Cheng, H.; Wu, X.; Qu, Z.; Wu, J.; Xia, G. Cotton major latex protein 28 functions as a positive regulator of the ethylene responsive factor 6 in defense against verticillium dahliae. Mol. Plant 2015, 8, 399–411. [Google Scholar] [CrossRef]
- Dubery, I.A.; Meyer, R. Specific binding of a Verticillium dahliae phytotoxin to protoplasts of cotton, Gossypium hirsutum. Plant Cell Rep. 1996, 15, 777–780. [Google Scholar] [CrossRef]
- Xu, L.; Zhu, L.; Tu, L.; Liu, L.; Yuan, D.; Jin, L.; Long, L.; Zhang, X. Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J. Exp. Bot. 2011, 62, 5607–5621. [Google Scholar] [CrossRef]
- Sink, K.C.; Grey, W.E. A root-injection method to assess verticillium wilt resistance of peppermint (Mentha × piperita L.) and its use in identifying resistant somaclones of cv. Black Mitcham. Euphytica 1999, 106, 223–230. [Google Scholar] [CrossRef]
- Zhang, D.; Wang, J.; Wang, D.; Kong, Z.; Zhou, L.; Zhang, G.; Gui, Y.; Li, J.; Huang, J.; Wang, B. Population genomics demystifies the defoliation phenotype in the plant pathogen Verticillium dahliae. New Phytol. 2019, 222, 1012–1029. [Google Scholar] [CrossRef]
- Lacombe, S.; Rougon-Cardoso, A.; Sherwood, E.; Peeters, N.; Zipfel, C. Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat. Biotechnol. 2010, 28, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X.; Rong, W.; Yang, J.; Li, Z.; Wu, L.; Zhang, G.; Ma, Z. Histochemical analyses reveal that stronger intrinsic defenses in gossypium barbadense than in G. hirsutum are associated with resistance to Verticillium dahliae. Mol. Plant-Microbe Interact. (MPMI) 2017, 30, 984. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Long, L.; Zhu, L.F.; Xu, L.; Gao, W.H.; Sun, L.Q.; Liu, L.L.; Zhang, X.L. Proteomic and virus-induced gene silencing (VIGS) analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae. Mol. Cell. Proteom. 2013, 12, 3690–3703. [Google Scholar] [CrossRef] [PubMed]
- Fradin, E.F.; Abdelhaliem, A.; Masini, L.; Den Berg, G.V.; Joosten, M.H.A.J.; Thomma, B.P.H.J. Interfamily transfer of tomato Ve1 mediates Verticillium resistance in arabidopsis. Plant Physiol. 2011, 156, 2255–2265. [Google Scholar] [CrossRef] [PubMed]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef] [PubMed]
- Thaler, J.S. The role of the jasmonate response in plant susceptibility to diverse pathogens with a range of lifestyles. Plant Physiol. 2004, 135, 530–538. [Google Scholar] [CrossRef]
- He, X.; Zhu, L.; Wassan, G.M.; Wang, Y.; Miao, Y.; Shaban, M.; Hu, H.; Sun, H.; Zhang, X. GhJAZ2 attenuates cotton resistance to biotic stresses via inhibiting the transcriptional activity of GhbHLH171. Mol. Plant Pathol. 2018, 19, 896–908. [Google Scholar] [CrossRef]
- Li, C.; He, X.; Luo, X.; Xu, L.; Liu, L.; Min, L.; Jin, L.; Zhu, L.; Zhang, X. Cotton WRKY1 mediates the plant defense-to-development transition during infection of cotton by Verticillium dahliae by activating JASMONATE ZIM-DOMAIN1 expression. Plant Physiol. 2014, 166, 2179–2194. [Google Scholar] [CrossRef]
- He, X.; Wang, T.; Zhu, W.; Wang, Y.; Zhu, L. GhHB12, a HD-ZIP I transcription factor, negatively regulates the cotton resistance to Verticillium dahliae. Int. J. Mol. Sci. 2018, 19, 3997. [Google Scholar] [CrossRef]
- Gao, W.; Long, L.; Xu, L.; Lindsey, K.; Zhang, X.; Zhu, L. Suppression of the homeobox gene HDTF1 enhances resistance to Verticillium dahliae and Botrytis cinerea in cotton. J. Integr. Plant Biol. 2016, 58, 503–513. [Google Scholar] [CrossRef]
- Hu, Q.; Zhu, L.; Zhang, X.; Guan, Q.; Zhang, X. GhCPK33 negatively regulates defense against Verticillium dahliae by phosphorylating GhOPR3. Plant Physiol. 2018, 178, 876–889. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Sun, L.; Wassan, G.M.; He, X.; Muhammad, S. GbSOBIR1 confers Verticillium wilt resistance by phosphorylating the transcriptional factor GbbHLH171 in Gossypium barbadense. Plant Biotechnol. J. 2019, 17, 152–163. [Google Scholar] [CrossRef]
- Loake, G.; Grant, M. Salicylic acid in plant defence—The players and protagonists. Curr. Opin. Plant Biol. 2007, 10, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Xu, L.; He, X.; Zhang, L.; Shaban, M.; Zhang, X.; Zhu, L. Suppression of tryptophan synthase activates cotton immunity by triggering cell death via promoting SA synthesis. Plant J. 2019, 98, 329–345. [Google Scholar] [CrossRef] [PubMed]
- Gong, Q.; Yang, Z.; Wang, X.; Butt, H.I.; Chen, E.; He, S.; Zhang, C.; Zhang, X.; Li, F. Salicylic acid-related cotton (Gossypium arboreum) ribosomal protein GaRPL18 contributes to resistance to Verticillium dahliae. BMC Plant Biol. 2017, 17, 59. [Google Scholar] [CrossRef]
- Derksen, H.; Rampitsch, C.; Daayf, F. Signaling cross-talk in plant disease resistance. Plant Sci. 2013, 207, 79–87. [Google Scholar] [CrossRef]
- Xiong, X.; Sun, S.; Li, Y.; Zhang, X.; Sun, J.; Xue, F. The cotton WRKY transcription factor GhWRKY70 negatively regulates the defense response against Verticillium dahliae. Crop J. 2019, 7, 393–402. [Google Scholar] [CrossRef]
- Xiong, X.P.; Sun, S.C.; Zhang, X.Y.; Li, Y.J.; Liu, F.; Zhu, Q.H.; Xue, F.; Sun, J. GhWRKY70D13 regulates resistance to Verticillium dahliae in cotton through the ethylene and jasmonic acid signaling pathways. Front. Plant Sci. 2020, 11, 69. [Google Scholar] [CrossRef]
- He, X.; Zhu, L.; Xu, L.; Guo, W.W.; Zhang, X. GhATAF1, a NAC transcription factor, confers abiotic and biotic stress responses by regulating phytohormonal signaling networks. Plant Cell Rep. 2016, 35, 2167–2179. [Google Scholar] [CrossRef]
- Hamann, T. Plant cell wall integrity maintenance as an essential component of biotic stress response mechanisms. Front. Plant Sci. 2012, 3, 77. [Google Scholar] [CrossRef]
- Shaban, M.; Miao, Y.; Ullah, A.; Khan, A.Q.; Menghwar, H.; Khan, A.H.; Ahmed, M.M.; Tabassum, M.A.; Zhu, L. Physiological and molecular mechanism of defense in cotton against Verticillium dahliae. Plant Physiol. Biochem. 2018, 125, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Mottiar, Y.; Vanholme, R.; Boerjan, W.; Ralph, J.; Mansfield, S.D. Designer lignins: Harnessing the plasticity of lignification. Curr. Opin. Biotechnol. 2016, 37, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Reusche, M.; Thole, K.; Janz, D.; Truskina, J.; Rindfleisch, S.; Drubert, C.; Polle, A.; Lipka, V.; Teichmann, T. Verticillium infection triggers VASCULAR-RELATED NAC DOMAIN7-dependent de novo xylem formation and enhances drought tolerance in Arabidopsis. Plant Cell 2012, 24, 3823–3837. [Google Scholar] [CrossRef] [PubMed]
- Gayoso, C.; Pomar, F.; Novo-Uzal, E.; Merino, F.; Ilárduya, ó.M.d. The Ve-mediated resistance response of the tomato to Verticillium dahliae involves H2O2, peroxidase and lignins and drives PAL gene expression. BMC Plant Biol. 2010, 10, 232. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, W.; He, X.; Liu, M.; Zhang, K.; Shaban, M.; Sun, L.; Zhu, J.; Luo, Y.; Yuan, D. Functional characterization of cotton genes responsive to Verticillium dahliae through bioinformatics and reverse genetics strategies. J. Exp. Bot. 2014, 65, 6679–6692. [Google Scholar] [CrossRef]
- Shi, H.; Liu, Z.; Zhu, L.; Zhang, C.; Chen, Y.; Zhou, Y.; Li, F.; Li, X. Overexpression of cotton (Gossypium hirsutum) dirigent1 gene enhances lignification that blocks the spread of Verticillium dahliae. Acta Biochim. Biophys. Sin. 2012, 44, 555–564. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, L.; Wang, X.; Chen, B.; Zhao, J. The cotton laccase gene GhLAC15 enhanced Verticillium wilt resistance via increasing defense-induced lignification and lignin components in the cell wall of plants. Mol. Plant Pathol. 2019, 20, 309–322. [Google Scholar] [CrossRef]
- Hu, Q.; Min, L.; Yang, X.; Jin, S.; Zhang, L.; Li, Y.; Ma, Y.; Qi, X.; Li, D.; Liu, H. Laccase GhLac1 modulates broad-spectrum biotic stress tolerance via manipulating phenylpropanoid pathway and jasmonic acid synthesis. Plant Physiol. 2017, 176, 1808–1823. [Google Scholar] [CrossRef]
- Guo, W.; Jin, L.; Miao, Y.; He, X.; Hu, Q.; Guo, K.; Zhu, L.; Zhang, X. An ethylene response-related factor, GbERF1-like, from Gossypium barbadense improves resistance to Verticillium dahliae via activating lignin synthesis. Plant Mol. Biol. 2016, 91, 305–318. [Google Scholar] [CrossRef]
- Liu, Y.; You, S.; Taylor-Teeples, M.; Wenhua, L.L.; Schuetz, M. BEL1-LIKE HOMEODOMAIN6 and KNOTTED ARABIDOPSIS THALIANA7 interact and regulate secondary cell wall formation via repression of REVOLUTA. Plant Cell 2014, 26, 4843. [Google Scholar] [CrossRef]
- Yoon, J.; Cho, L.H.; Antt, H.W.; Koh, H.J.; Gynheung, A. KNOX protein OSH15 induces grain shattering by repressing lignin biosynthesis genes. Plant Physiol. 2017, 174, 312–325. [Google Scholar] [CrossRef]
- Yoon, J.; Cho, L.; Lim, S.; Heebak, K.; Hee-Jong, C. The BEL1-type homeobox gene SH5 induces seed shattering by enhancing abscission-zone development and inhibiting lignin biosynthesis. Plant J. 2014, 79, 717–728. [Google Scholar] [CrossRef]
- Luo, H.; Song, F.; Goodman, R.M.; Zheng, Z. Up-regulation of OsBIHD1, a rice gene encoding BELL homeodomain transcriptional factor, in disease resistance responses. Plant Biol. 2005, 7, 459–468. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, P.; Luo, X.; Yang, C.; Wu, J. Cotton plant defence against a fungal pathogen is enhanced by expanding BLADE-ON-PETIOLE1 expression beyond lateral-organ boundaries. Commun. Biol. 2019, 2, 238. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Wang, N.; Hao, P.; Sun, H.; Yu, S. Genome-wide identification and characterization of TALE superfamily genes in cotton reveals their functions in regulating secondary cell wall biosynthesis. BMC Plant Biol. 2019, 19, 432. [Google Scholar] [CrossRef]
- Hamant, O.; Pautot, V. Plant development: A tale story. Comptes Rendus Biol. 2010, 333, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Pooja, S.; Tian, L.; Carolina, G.; Mei, Y.; Hannapel, D.J. The BEL1-like family of transcription factors in potato. J. Exp. Bot. 2014, 65, 709–723. [Google Scholar]
- Bai, W.-Q.; Xiao, Y.-H.; Zhao, J.; Song, S.-Q.; Hu, L.; Zeng, J.-Y.; Li, X.-B.; Hou, L.; Luo, M.; Li, D.-M. Gibberellin overproduction promotes sucrose synthase expression and secondary cell wall deposition in cotton fibers. PLoS ONE 2014, 9, e96537. [Google Scholar] [CrossRef] [PubMed]
- Bednarek, P. Chemical warfare or modulators of defence responses -the function of secondary metabolites in plant immunity. Curr. Opin. Plant Biol. 2012, 15, 407–414. [Google Scholar] [CrossRef]
- Liu, Y.; Douglas, C.J. A role for OVATE FAMILY PROTEIN1 (OFP1) and OFP4 in a BLH6-KNAT7 multi-protein complex regulating secondary cell wall formation in Arabidopsis thaliana. Plant Signal. Behav. 2015, 10, e1033126. [Google Scholar] [CrossRef] [PubMed]
- Ragni, L.; Belles-Boix, E.; Gunl, M.; Pautot, V. Interaction of KNAT6 and KNAT2 with BREVIPEDICELLUS and PENNYWISE in Arabidopsis inflorescences. Plant Cell 2008, 20, 888–900. [Google Scholar] [CrossRef]
- Kim, D.; Cho, Y.H.; Ryu, H.; Kim, Y.; Kim, T.H.; Hwang, I. BLH1 and KNAT3 modulate ABA responses during germination and early seedling development in Arabidopsis. Plant J. 2013, 75, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Wang, S.; Liu, Y.; Chen, J.G.; Douglas, C.J. OVATE FAMILY PROTEIN4 (OFP4) interaction with KNAT7 regulates secondary cell wall formation in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 2011, 67, 328–341. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, X.; Ju, H.; Chen, J.; Wang, S.; Wang, H.; Zhao, Y.; Chang, Y. Ovate family protein1 interaction with BLH3 regulates transition timing from vegetative to reproductive phase in Arabidopsis. Biochem. Biophys. Res. Commun. 2016, 470, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Hackbusch, J.; Richter, K.; Müller, J.; Salamini, F.; Uhrig, J.F. A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loon extension homeodomain proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 4908–4912. [Google Scholar] [CrossRef] [PubMed]
- Gong, S.Y.; Huang, G.Q.; Sun, X.; Qin, L.X.; Li, Y.; Zhou, L.; Li, X.B. Cotton KNL1, encoding a class II KNOX transcription factor, is involved in regulation of fibre development. J. Exp. Bot. 2014, 65, 4133–4147. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Mao, G.; Ma, Q.; Wei, H.; Su, J.; Wang, H.; Ma, Q.; Fan, S.; Song, M.; Zhang, X.; Yu, S. Fine mapping and candidate gene analysis of the virescent gene v 1 in Upland cotton (Gossypium hirsutum). Mol. Genet. Genom. 2018, 293, 249–264. [Google Scholar] [CrossRef]
- Xu, M.; Gui, Y.J.; Qi, W.Y.; Liu, S.Y.; Chen, J.Y.; Dai, X.F. Verticillium dahliae labeled with green fluorescent protein gene. Plant. Prot. 2013, 39, 128–133. [Google Scholar]
- Pomar, F.; Novo, M.; Bernal, M.A.; Merino, F.; Barcel, A.R. Changes in stem lignins (monomer composition and crosslinking) and peroxidase are related with the maintenance of leaf photosynthetic integrity during Verticillium wilt in Capsicum annuum. New Phytol. 2010, 163, 111–123. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Q.; Wang, N.; Ma, L.; Lu, J.; Wang, H.; Wang, C.; Yu, S.; Wei, H. The Cotton BEL1-Like Transcription Factor GhBLH7-D06 Negatively Regulates the Defense Response against Verticillium dahliae. Int. J. Mol. Sci. 2020, 21, 7126. https://doi.org/10.3390/ijms21197126
Ma Q, Wang N, Ma L, Lu J, Wang H, Wang C, Yu S, Wei H. The Cotton BEL1-Like Transcription Factor GhBLH7-D06 Negatively Regulates the Defense Response against Verticillium dahliae. International Journal of Molecular Sciences. 2020; 21(19):7126. https://doi.org/10.3390/ijms21197126
Chicago/Turabian StyleMa, Qiang, Nuohan Wang, Liang Ma, Jianhua Lu, Hantao Wang, Congcong Wang, Shuxun Yu, and Hengling Wei. 2020. "The Cotton BEL1-Like Transcription Factor GhBLH7-D06 Negatively Regulates the Defense Response against Verticillium dahliae" International Journal of Molecular Sciences 21, no. 19: 7126. https://doi.org/10.3390/ijms21197126
APA StyleMa, Q., Wang, N., Ma, L., Lu, J., Wang, H., Wang, C., Yu, S., & Wei, H. (2020). The Cotton BEL1-Like Transcription Factor GhBLH7-D06 Negatively Regulates the Defense Response against Verticillium dahliae. International Journal of Molecular Sciences, 21(19), 7126. https://doi.org/10.3390/ijms21197126



