Identification, Phylogeny, and Comparative Expression of the Lipoxygenase Gene Family of the Aquatic Duckweed, Spirodela polyrhiza, during Growth and in Response to Methyl Jasmonate and Salt
Abstract
1. Introduction
2. Results
2.1. Identification of the LOX Gene Family in the Greater Duckweed, Spirodela polyrhiza
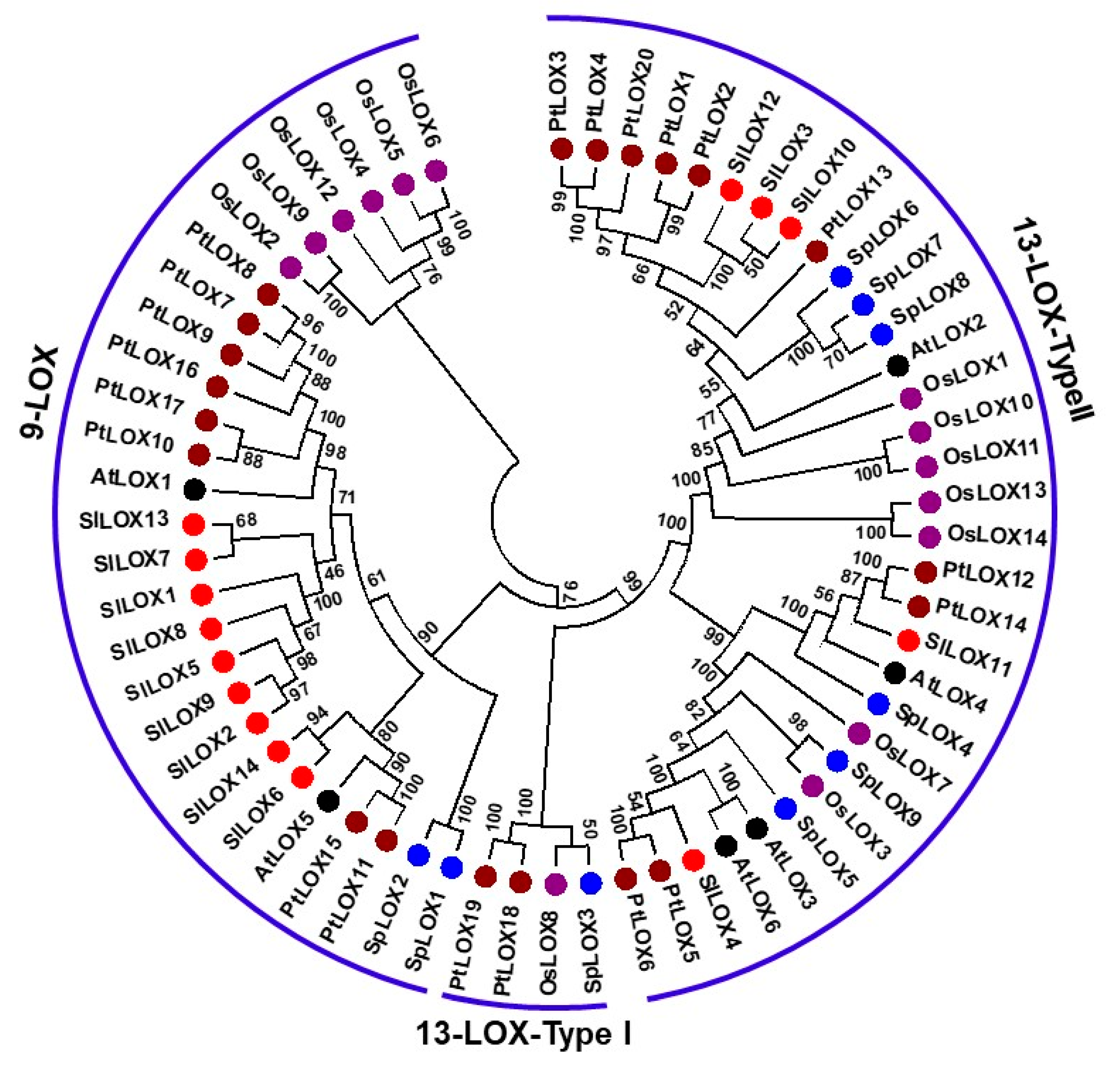
2.2. Comparative Phylogeny of the S. polyrhiza LOX Gene Family Reveals Dominance of 13-LOX Sub-Family Genes
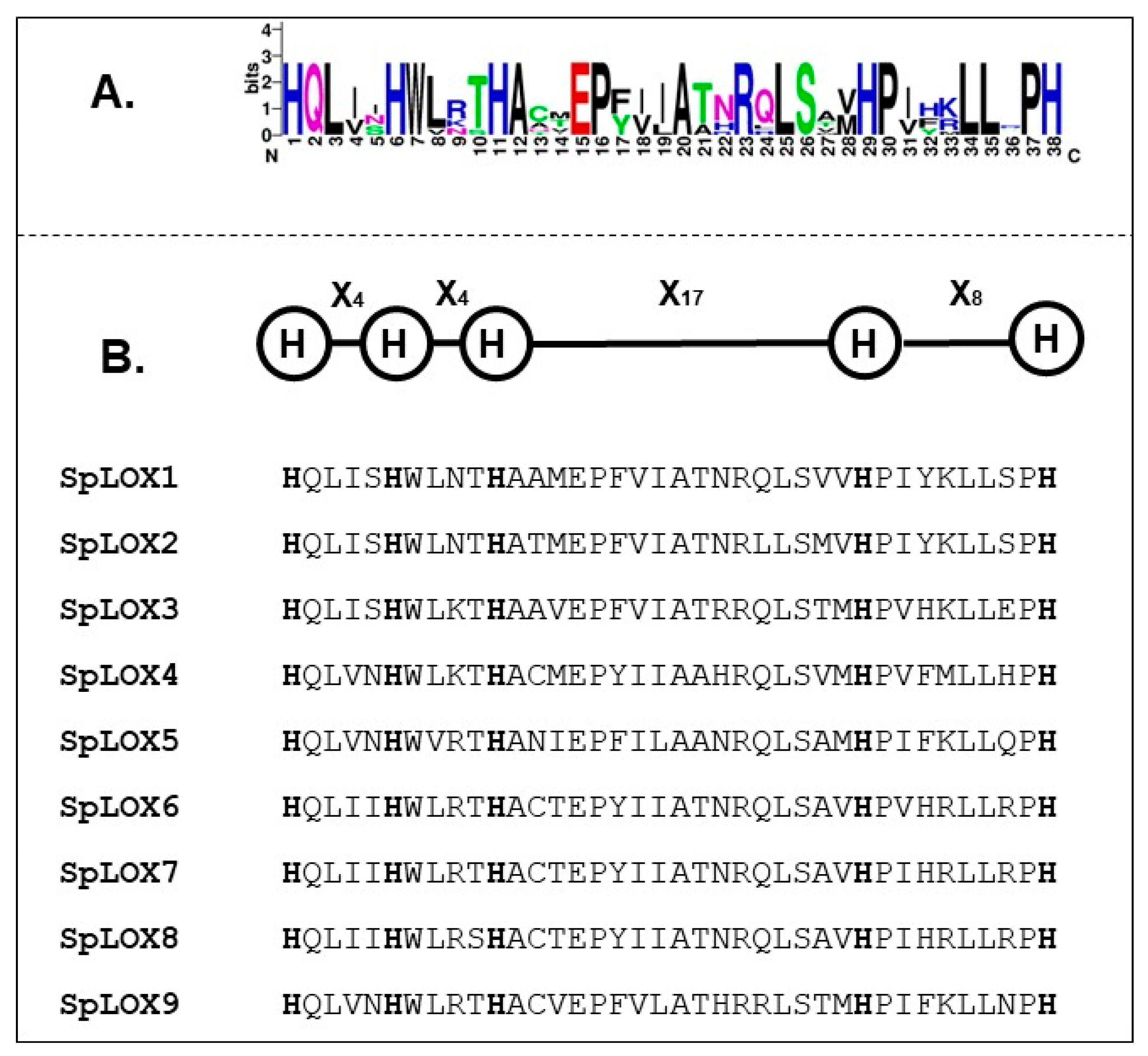
2.3. Analysis of 5-Histidine Conserved Motifs in Spirodela LOX Protein Sequences
2.4. Structural Analysis of the Spirodela LOX Genes
2.5. Tandem Duplications among Spirodela LOXs
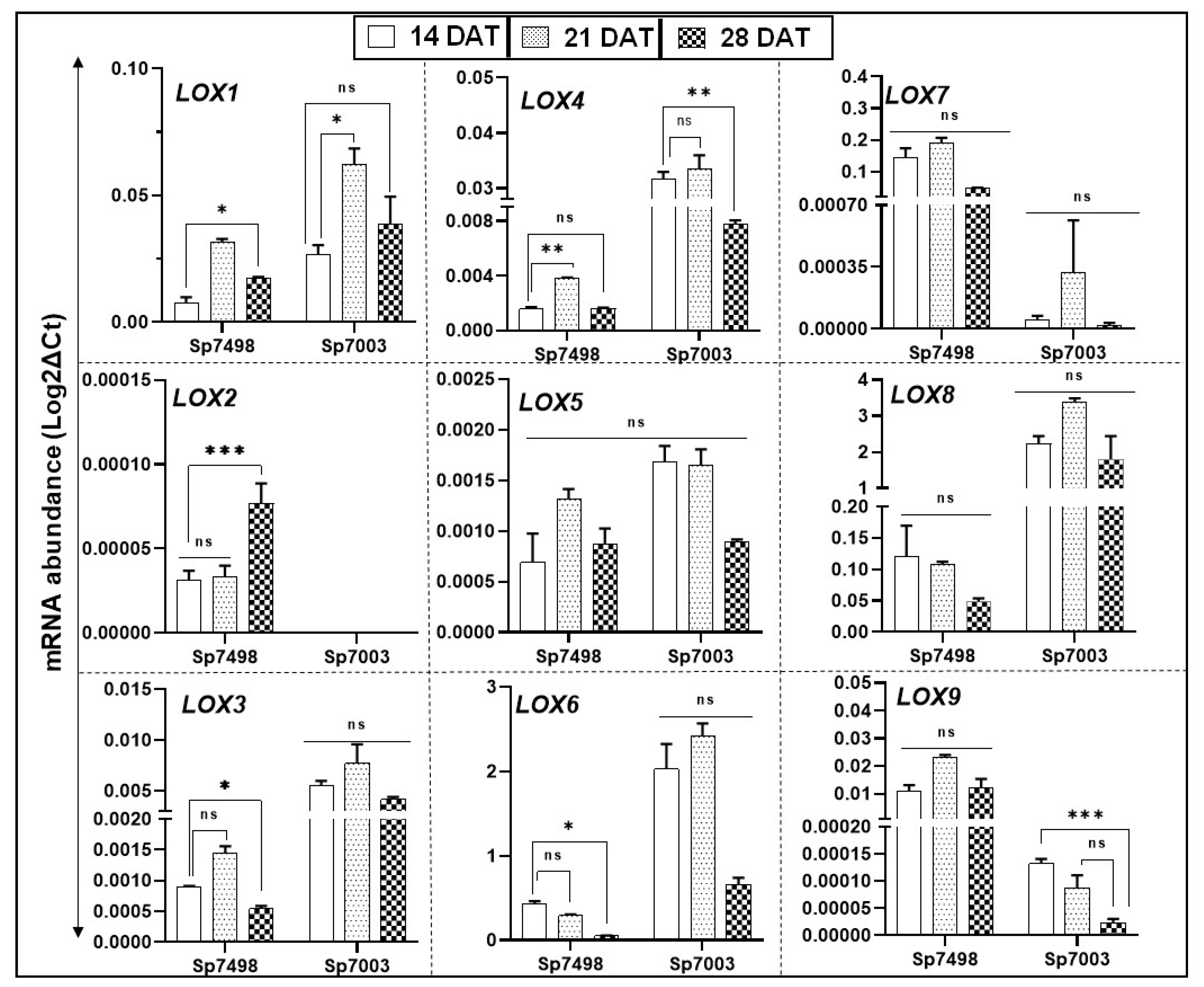
2.6. Spirodela LOX Gene Family Transcript Accumulation Changes with Culture Age
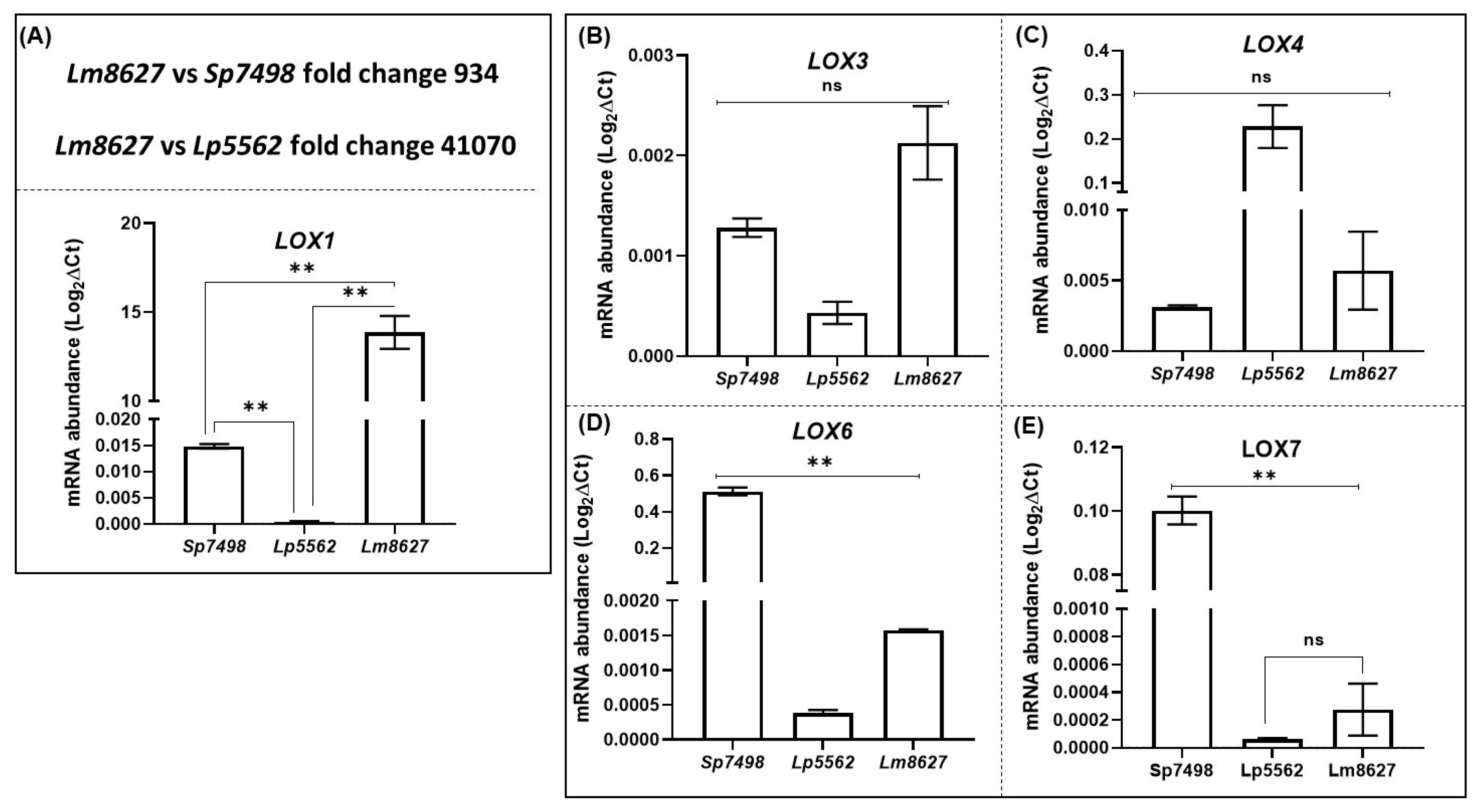
2.7. Expression of Most LOX Genes in Lemna minor and Landoltia punctata is Significantly Lower Compared to Spirodela polyrhiza
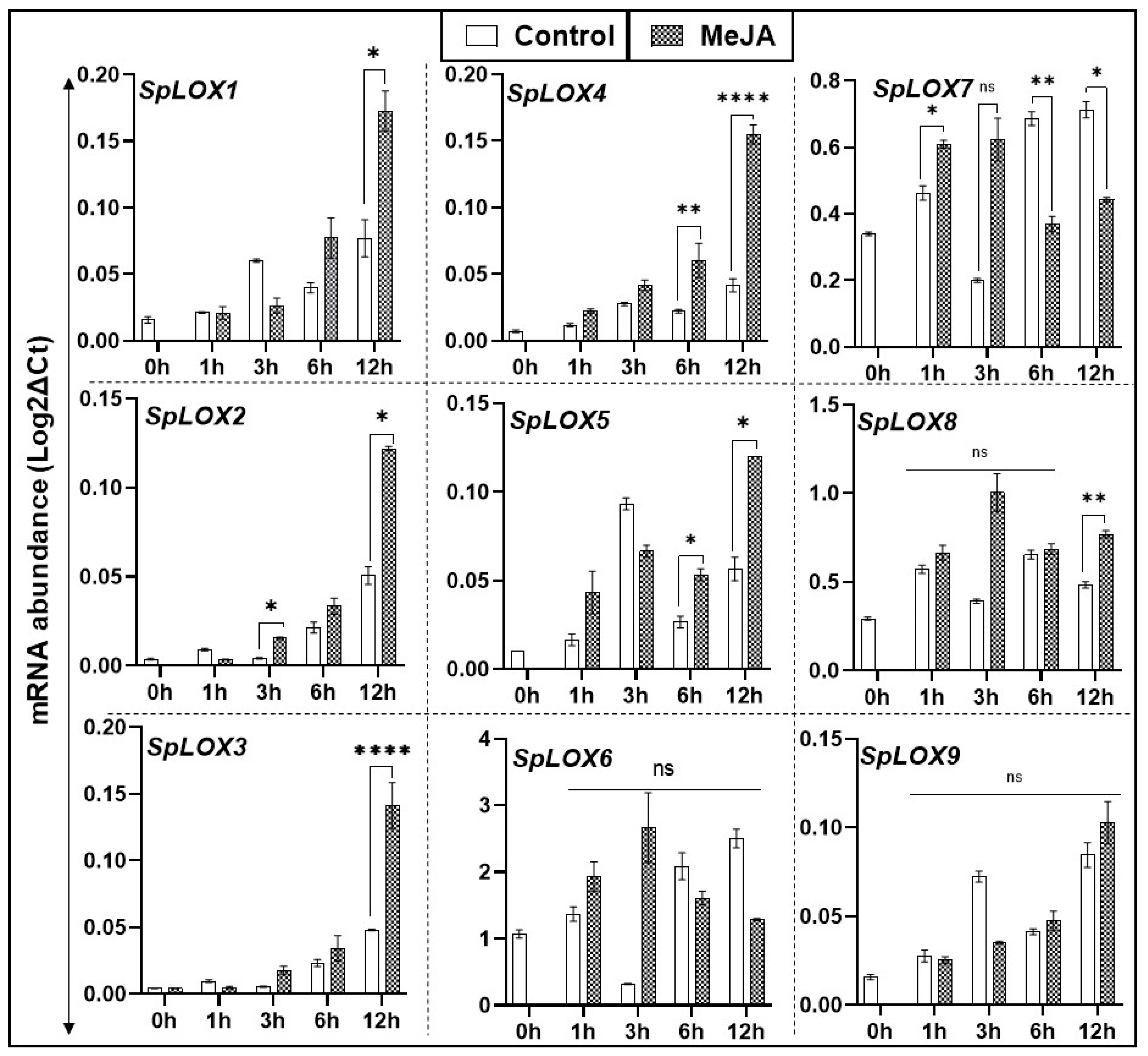
2.8. Methyl Jasmonate Treated Fronds and Transcript Abundance in Spirodela LOX Genes
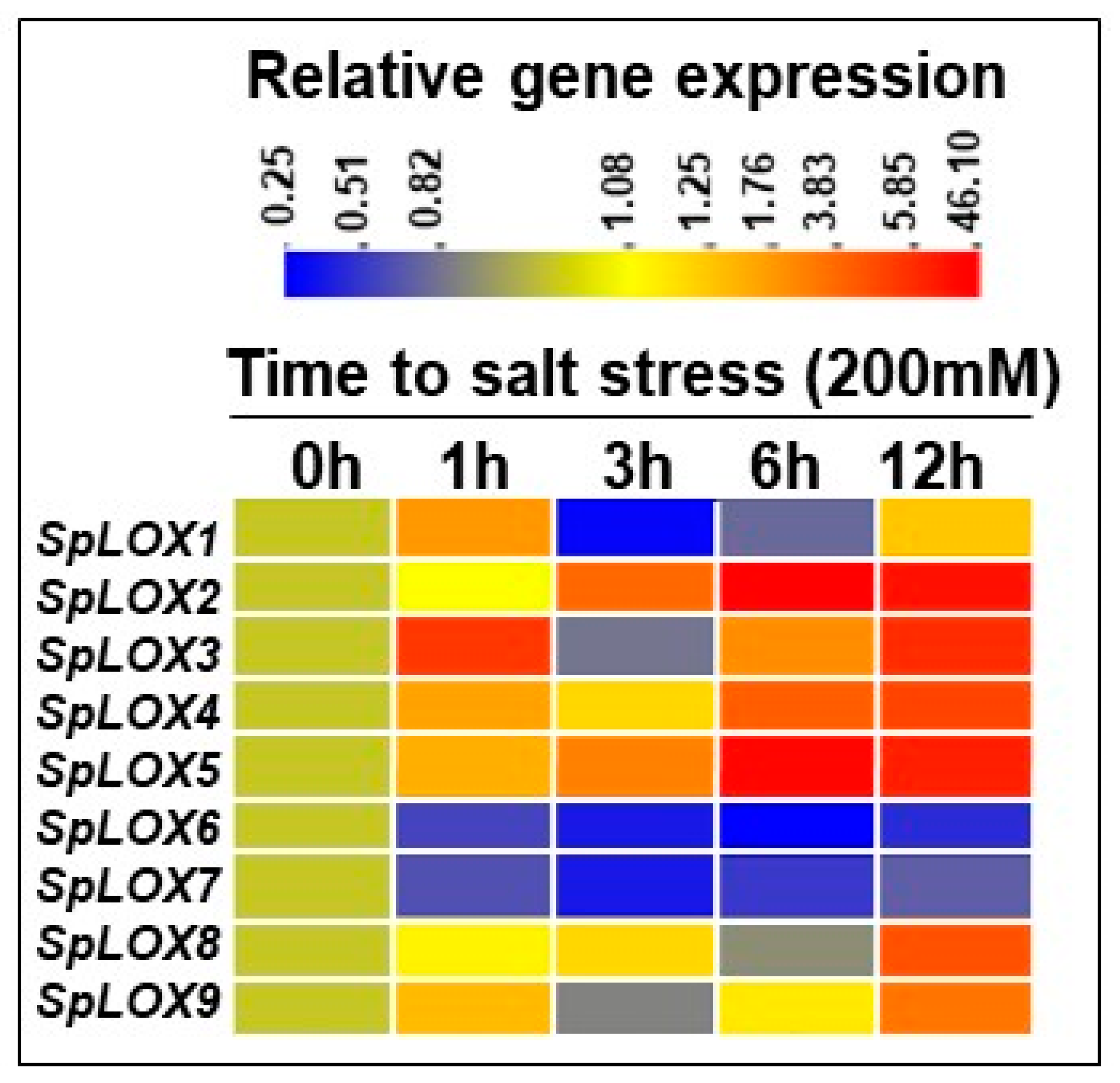
2.9. Salt Stress and Response of Spirodela LOXs
3. Discussion
4. Materials and Methods
4.1. Plant Material, Growth Conditions, and Salt Stress Treatments
4.2. Sequence Retrieval of the LOX Gene Family from Spirodela polyrhiza
4.3. Phylogenetic Analysis of the S. polyrhiza LOX Family with Previously Characterized LOX Gene Families
4.4. Conserved Motifs in LOX Protein Sequences and Subcellular Localizations
4.5. Gene Structure and Gene Duplication Analyses
4.6. Total RNA Extraction, cDNA Preparation, and Quantitative Real Time PCR (qRT-PCR)
4.7. Methyl Jasmonate Treatment
4.8. Salt Treatment
4.9. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| LOXs | Lipoxygenases |
| PLAT | Polycystin-1, lipoxygenase, alpha-toxin |
| His | Histidine |
| 9-HPOD | 9s-hydroperoxyoctadecadienoic acid |
| 13-HPOD | 13s-hydroperoxyoctadecadienoic acid |
| JA | Jasmonic acid |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| MeJA | Methyl jasmonate |
References
- Hillman, W.S. The Lemnaceae, or duckweeds. Bot. Rev. 1961, 27, 221–287. [Google Scholar] [CrossRef]
- Bog, M.; Appenroth, K.-J.; Sree, K.S. Duckweed (Lemnaceae): Its Molecular Taxonomy. Front. Sustain. Food Syst. 2019, 3, 117. [Google Scholar] [CrossRef]
- Landolt, E. The Family of Lemnaceae—A Monographic Study (Vol. 1); Geobotanische Institut ETH, Stiftung Rübel: Zurich, Switzerland, 1986; Volume 71, p. 566. [Google Scholar]
- Wang, W.; Kerstetter, R.; Michael, T. Evolution of genome size in duckweeds (Lemnaceae). J. Bot. 2011, 2, 1. [Google Scholar] [CrossRef]
- Mattoo, A.K.; Pick, U.; Hoffman-Falk, H.; Edelman, M. The rapidly metabolized 32,000-dalton polypeptide of the chloroplast is the “proteinaceous shield” regulating photosystem II electron transport and mediating diuron herbicide sensitivity. Proc. Natl. Acad. Sci. USA 1981, 78, 1572–1576. [Google Scholar] [CrossRef] [PubMed]
- Mattoo, A.K.; Hoffman-Falk, H.; Marder, J.B.; Edelman, M. Regulation of protein metabolism: Coupling of photosynthetic electron transport to in vivo degradation of the rapidly metabolized 32-kilodalton protein of the chloroplast membranes. Proc. Natl. Acad. Sci. USA 1984, 81, 1380–1384. [Google Scholar] [CrossRef]
- Mattoo, A.K.; Edelman, M. Intramembrane translocation and posttranslational palmitoylation of the chloroplast 32-kDa herbicide-binding protein. Proc. Natl. Acad. Sci. USA 1987, 84, 1497–1501. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, P.; Sree, K.S.; Appenroth, K.-J. Duckweeds for water remediation and toxicity testing. Toxicol. Environ. Chem. 2016, 98, 1127–1154. [Google Scholar] [CrossRef]
- Rusoff, L.L.; Blakeney, E.W.; Culley, D.D. Duckweeds (Lemnaceae family): A potential source of protein and amino acids. J. Agric. Food Chem. 1980, 28, 848–850. [Google Scholar] [CrossRef]
- Stomp, A.-M. The duckweeds: A valuable plant for biomanufacturing. In Biotechnology Annual Review; Elsevier: Amsterdam, The Netherlands, 2005; Volume 11, pp. 69–99. [Google Scholar]
- Appenroth, K.-J.; Sree, K.S.; Fakhoorian, T.; Lam, E. Resurgence of duckweed research and applications: Report from the 3rd International Duckweed Conference. Plant Mol. Biol. 2015, 89, 647–654. [Google Scholar] [CrossRef]
- Edelman, M.; Colt, M. Nutrient value of leaf vs. seed. Front. Chem. 2016, 4, 32. [Google Scholar] [CrossRef]
- Wang, W.; Haberer, G.; Gundlach, H.; Gläßer, C.; Nussbaumer, T.; Luo, M.C.; Lomsadze, A.; Borodovsky, M.; Kerstetter, R.A.; Shanklin, J.; et al. The Spirodela polyrhiza genome reveals insights into its neotenous reduction fast growth and aquatic lifestyle. Nat. Commun. 2014, 5, 3311. [Google Scholar] [CrossRef] [PubMed]
- Van Hoeck, A.; Horemans, N.; Monsieurs, P.; Cao, H.X.; Vandenhove, H.; Blust, R. The first draft genome of the aquatic model plant Lemna minor opens the route for future stress physiology research and biotechnological applications. Biotechnol. Biofuels 2015, 8, 188. [Google Scholar] [CrossRef] [PubMed]
- An, D.; Li, C.; Zhou, Y.; Wu, Y.; Wang, W. Genomes and Transcriptomes of Duckweeds. Front. Chem. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Hoang, P.N.T.; Michael, T.P.; Gilbert, S.; Chu, P.; Motley, S.T.; Appenroth, K.J.; Schubert, I.; Lam, E. Generating a high-confidence reference genome map of the Greater Duckweed by integration of cytogenomic, optical mapping, and Oxford Nanopore technologies. Plant J. 2018, 96, 670–684. [Google Scholar] [CrossRef] [PubMed]
- An, D.; Zhou, Y.; Li, C.; Xiao, Q.; Wang, T.; Zhang, Y.; Wu, Y.; Li, Y.; Chao, D.-Y.; Messing, J.; et al. Plant evolution and environmental adaptation unveiled by long-read whole-genome sequencing of Spirodela. Proc. Natl. Acad. Sci. USA 2019, 116, 18893–18899. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C. Jasmonates: An Update on Biosynthesis, Signal Transduction and Action in Plant Stress Response, Growth and Development. Ann. Bot. 2007, 100, 681–697. [Google Scholar] [CrossRef]
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 2017, 68, 1303–1321. [Google Scholar] [CrossRef]
- Wasternack, C.; Feussner, I. The oxylipin pathways: Biochemistry and function. Annu. Rev. Plant Biol. 2018, 69, 363–386. [Google Scholar] [CrossRef]
- Andre, E.; Hou, K. The lipoxydases of seeds of Glycine soja (Sieb) and Phaseolus vulgaris (L.). CR Hebd. Seances. Acad. Sci. 1932, 195, 172–174. [Google Scholar]
- Siedow, J.N. Plant lipoxygenase: Structure and function. Annu. Rev. Plant Biol. 1991, 42, 145–188. [Google Scholar] [CrossRef]
- Porta, H.; Rocha-Sosa, M. Plant lipoxygenases. Physiological and molecular features. Plant Physiol. 2002, 130, 15–21. [Google Scholar] [CrossRef]
- Liavonchanka, A.; Feussner, I. Lipoxygenases: Occurrence, functions and catalysis. J. Plant Physiol. 2006, 163, 348–357. [Google Scholar] [CrossRef] [PubMed]
- Rance, I.; Fournier, J.; Esquerre-Tugaye, M.-T. The incompatible interaction between Phytophthora parasitica var. nicotianae race 0 and tobacco is suppressed in transgenic plants expressing antisense lipoxygenase sequences. Proc. Natl. Acad. Sci. USA 1998, 95, 6554–6559. [Google Scholar] [CrossRef] [PubMed]
- Baysal, T.; Demirdöven, A. Lipoxygenase in fruits and vegetables: A review. Enzym. Microb. Technol. 2007, 40, 491–496. [Google Scholar] [CrossRef]
- Hayward, S.; Cilliers, T.; Swart, P. Lipoxygenases: From Isolation to Application. Compr. Rev. Food Sci. Food Saf. 2017, 16, 199–211. [Google Scholar] [CrossRef]
- Bell, E.; Creelman, R.A.; Mullet, J.E. A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 1995, 92, 8675–8679. [Google Scholar] [CrossRef]
- Chauvin, A.; Caldelari, D.; Wolfender, J.-L.; Farmer, E.E. Four 13-lipoxygenases contribute to rapid jasmonate synthesis in wounded Arabidopsis thaliana leaves: A role for lipoxygenase 6 in responses to long-distance wound signals. New Phytol. 2013, 197, 566–575. [Google Scholar] [CrossRef]
- Griffiths, A.; Barry, C.; Alpuche-Solis, A.G.; Grierson, D. Ethylene and developmental signals regulate expression of lipoxygenase genes during tomato fruit ripening. J. Exp. Bot. 1999, 50, 793–798. [Google Scholar] [CrossRef]
- Kausch, K.D.; Handa, A.K. Molecular Cloning of a Ripening-Specific Lipoxygenase and Its Expression during Wild-Type and Mutant Tomato Fruit Development. Plant Physiol. 1997, 113, 1041–1050. [Google Scholar] [CrossRef]
- Kausch, K.D.; Sobolev, A.P.; Goyal, R.K.; Fatima, T.; Laila-Beevi, R.; Saftner, R.A.; Handa, A.K.; Mattoo, A.K. Methyl jasmonate deficiency alters cellular metabolome, including the aminome of tomato (Solanum lycopersicum L.) fruit. Amino Acids 2012, 42, 843–856. [Google Scholar] [CrossRef]
- Yan, L.; Zhai, Q.; Wei, J.; Li, S.; Wang, B.; Huang, T.; Du, M.; Sun, J.; Kang, L.; Li, C.-B.; et al. Role of Tomato Lipoxygenase D in Wound-Induced Jasmonate Biosynthesis and Plant Immunity to Insect Herbivores. PLoS Genet. 2013, 9, e1003964. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Tieman, D.; Jones, J.B.; Taylor, M.G.; Schmelz, E.; Huffaker, A.; Bies, D.; Chen, K.; Klee, H.J. A 13-lipoxygenase, TomloxC, is essential for synthesis of C5 flavour volatiles in tomato. J. Exp. Bot. 2014, 65, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, R.K.; Mattoo, A.K. Genome-wide identification of tomato (Solanum lycopersicum L.) lipoxygenases coupled with expression profiles during plant development and in response to methyl-jasmonate and wounding. J. Plant Physiol. 2018, 231, 318–328. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, R.K.; Handa, A.K.; Mattoo, A.K. Transcript Abundance Patterns of 9- and 13-Lipoxygenase Subfamily Gene Members in Response to Abiotic Stresses (Heat, Cold, Drought or Salt) in Tomato (Solanum lycopersicum L.) Highlights Member-Specific Dynamics Relevant to Each Stress. Genes 2019, 10, 683. [Google Scholar] [CrossRef]
- Shin, J.H.; Van, K.; Kim, D.H.; Kim, K.D.; Jang, Y.E.; Choi, B.-S.; Kim, M.Y.; Lee, S.-H. The lipoxygenase gene family: A genomic fossil of shared polyploidy between Glycine max and Medicago truncatula. BMC Plant Biol. 2008, 8, 133. [Google Scholar] [CrossRef]
- Bannenberg, G.; Martínez, M.; Hamberg, M.; Castresana, C. Diversity of the Enzymatic Activity in the Lipoxygenase Gene Family of Arabidopsis thaliana. Lipids 2009, 44, 85. [Google Scholar] [CrossRef]
- Podolyan, A.; White, J.; Jordan, B.; Winefield, C. Identification of the lipoxygenase gene family from Vitis vinifera and biochemical characterisation of two 13-lipoxygenases expressed in grape berries of Sauvignon Blanc. Funct. Plant Biol. 2010, 37, 767–784. [Google Scholar] [CrossRef]
- Liu, S.Q.; Liu, X.H.; Jiang, L.W. Genome-wide identification, phylogeny and expression analysis of the lipoxygenase gene family in cucumber. Genet. Mol. Res. 2011, 10, 2613–2636. [Google Scholar] [CrossRef]
- Umate, P. Genome-wide analysis of lipoxygenase gene family in Arabidopsis and rice. Plant Signal. Behav. 2011, 6, 335–338. [Google Scholar] [CrossRef]
- Vogt, J.; Schiller, D.; Ulrich, D.; Schwab, W.; Dunemann, F. Identification of lipoxygenase (LOX) genes putatively involved in fruit flavour formation in apple (Malus × domestica). Tree Genet. Genomes 2013, 9, 1493–1511. [Google Scholar] [CrossRef]
- Li, M.; Li, L.; Dunwell, J.M.; Qiao, X.; Liu, X.; Zhang, S. Characterization of the lipoxygenase (LOX) gene family in the Chinese white pear (Pyrus bretschneideri) and comparison with other members of the Rosaceae. BMC Genom. 2014, 15, 444. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Chen, X.; Yan, H.; Li, W.; Li, Y.; Cai, R.; Xiang, Y. The Lipoxygenase Gene Family in Poplar: Identification, Classification, and Expression in Response to MeJA Treatment. PLoS ONE 2015, 10, e0125526. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Wang, P.; Li, C.; Han, S.; Lopez-Baltazar, J.; Zhang, X.; Wang, X. Identification of lipoxygenase (LOX) genes from legumes and their responses in wild type and cultivated peanut upon Aspergillus flavus infection. Sci. Rep. 2016, 6, 35245. [Google Scholar] [CrossRef] [PubMed]
- Sarde, S.J.; Kumar, A.; Remme, R.N.; Dicke, M. Genome-wide identification, classification and expression of lipoxygenase gene family in pepper. Plant Mol. Biol. 2018, 98, 375–387. [Google Scholar] [CrossRef]
- Shaban, M.; Ahmed, M.M.; Sun, H.; Ullah, A.; Zhu, L. Genome-wide identification of lipoxygenase gene family in cotton and functional characterization in response to abiotic stresses. BMC Genom. 2018, 19, 599. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, X.; Guo, L.; Xu, Q.; Zhao, S.; Li, F.; Yan, X.; Liu, S.; Wei, C. Characterization and Alternative Splicing Profiles of the Lipoxygenase Gene Family in Tea Plant (Camellia sinensis). Plant Cell Physiol. 2018, 59, 1765–1781. [Google Scholar] [CrossRef]
- Wang, J.; Hu, T.; Wang, W.; Hu, H.; Wei, Q.; Wei, X.; Bao, C. Bioinformatics Analysis of the Lipoxygenase Gene Family in Radish (Raphanus sativus) and Functional Characterization in Response to Abiotic and Biotic Stresses. Int. J. Mol. Sci. 2019, 20, 6095. [Google Scholar] [CrossRef]
- Michael, T.P.; Bryant, D.; Gutierrez, R.; Borisjuk, N.; Chu, P.; Zhang, H.; Xia, J.; Zhou, J.; Peng, H.; Baidouri, M.E.; et al. Comprehensive definition of genome features in Spirodela polyrhiza by high-depth physical mapping and short-read DNA sequencing strategies. Plant J. 2017, 89, 617–635. [Google Scholar] [CrossRef]
- Sharma, M.; Laxmi, A. Jasmonates: Emerging Players in Controlling Temperature Stress Tolerance. Front. Plant Sci. 2016, 6. [Google Scholar] [CrossRef]
- Edelman, M.; Mattoo, A. The D1 protein: Past and future. In Photoprotection, Photoinhibition, Gene Regulation and Environment; Demmig-Adams, B., Adams, W., Mattoo, A.K., Eds.; Springer: Dordecht, The Netherlands, 2006; pp. 23–38. [Google Scholar]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Krajnčič, B.; Kristl, J.; Janžekovič, I. Possible role of jasmonic acid in the regulation of floral induction, evocation and floral differentiation in Lemna minor L. Plant Physiol. Biochem. 2006, 44, 752–758. [Google Scholar] [CrossRef] [PubMed]
- Appenroth, K.-J.; Dathe, W.; Hertel, W.; Augsten, H. Photophysiology of Turion Germination in Spirodela polyrhiza (L.) SCHLEIDEN. VII. Action of Jasmonic acid. J. Plant Physiol. 1991, 138, 345–349. [Google Scholar] [CrossRef]
- Lu, Q.; Zhang, T.; Zhang, W.; Su, C.; Yang, Y.; Hu, D.; Xu, Q. Alleviation of cadmium toxicity in Lemna minor by exogenous salicylic acid. Ecotoxicol. Environ. Saf. 2018, 147, 500–508. [Google Scholar] [CrossRef] [PubMed]
- De Domenico, S.; Taurino, M.; Gallo, A.; Poltronieri, P.; Pastor, V.; Flors, V.; Santino, A. Oxylipin dynamics in Medicago truncatula in response to salt and wounding stresses. Physiol. Plant. 2019, 165, 198–208. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, Q.; Zhai, H.; Li, Y.; Wang, X.; Liu, Q.; He, S. Transcript profile analysis reveals important roles of jasmonic acid signalling pathway in the response of sweet potato to salt stress. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Jiang, M.; Xu, F.; Peng, M.; Huang, F.; Meng, F. Methyl jasmonate regulated diploid and tetraploid black locust (Robinia pseudoacacia L.) tolerance to salt stress. Acta Physiol. Plant. 2016, 38, 106. [Google Scholar] [CrossRef]
- Faghih, S.; Ghobadi, C.; Zarei, A. Response of strawberry plant cv. ‘Camarosa’ to salicylic acid and methyl jasmonate application under salt stress condition. J. Plant Growth Regul. 2017, 36, 651–659. [Google Scholar] [CrossRef]
- Fu, L.; Ding, Z.; Sun, X.; Zhang, J. Physiological and Transcriptomic Analysis Reveals Distorted Ion Homeostasis and Responses in the Freshwater Plant Spirodela polyrhiza L. under Salt Stress. Genes 2019, 10, 743. [Google Scholar] [CrossRef]
- Mattoo, A.K.; Mehta, R.A.; Baker, J.E. Copper-induced ethylene biosynthesis in terrestrial (Nicotiana tabacum) and aquatic (Spirodela oligorrhiza) higher plants. Phytochemistry 1992, 31, 405–409. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Zuckerkandl, E.; Pauling, L. Evolutionary Divergence and Convergence in Proteins. In Evolving Genes and Proteins; Bryson, V., Vogel, H.J., Eds.; Academic Press: Cambridge, MA, USA, 1965; pp. 97–166. ISBN 978-1-4832-2734-4. [Google Scholar]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Yang, X.; Kalluri, U.C.; Jawdy, S.; Gunter, L.E.; Yin, T.; Tschaplinski, T.J.; Weston, D.J.; Ranjan, P.; Tuskan, G.A. The F-Box Gene Family Is Expanded in Herbaceous Annual Plants Relative to Woody Perennial Plants. Plant Physiol. 2008, 148, 1189–1200. [Google Scholar] [CrossRef]
- Sharp, A.J.; Locke, D.P.; McGrath, S.D.; Cheng, Z.; Bailey, J.A.; Vallente, R.U.; Pertz, L.M.; Clark, R.A.; Schwartz, S.; Segraves, R.; et al. Segmental Duplications and Copy-Number Variation in the Human Genome. Am. J. Hum. Genet. 2005, 77, 78–88. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


| Gene Name | Sequence ID | Sequence Coordinates | Genomic (bp *) | ORF (bp *) | Protein (aa *) | Mol. Wt. (kDa *) | pI | Predicted Subfamily # |
|---|---|---|---|---|---|---|---|---|
| SpLOX1 | Spipo2G0068200 | 5245164:5249140 (−) | 3977 | 2607 | 868 | 97.22 | 5.58 | 9-LOX |
| SpLOX2 | Spipo2G0068500 | 5257861:5263185 (−) | 5325 | 2535 | 844 | 95.40 | 6.12 | 9-LOX |
| SpLOX3 | Spipo4G0070100 | 5947369:5951085 (+) | 3717 | 2421 | 806 | 90.60 | 6.24 | 13-LOX |
| SpLOX4 | Spipo7G0050500 | 4554161:4559214 (+) | 5054 | 2751 | 916 | 102.81 | 7.10 | 13-LOX |
| SpLOX5 | Spipo15G0045200 | 4070955:4073755 (−) | 2801 | 2379 | 792 | 88.63 | 6.04 | 13-LOX |
| SpLOX6 | Spipo28G0005500 | 470166:475175 (−) | 5010 | 2715 | 904 | 102.13 | 6.29 | 13-LOX |
| SpLOX7 | Spipo28G0005600 | 487129:491614 (+) | 4486 | 2715 | 904 | 103.52 | 7.16 | 13-LOX |
| SpLOX8 | Spipo28G0005700 | 497951:504441 (+) | 6491 | 1914 | 637 | 71.66 | 6.99 | 13-LOX |
| SpLOX9 | Spipo0G0030100 | 2519505:2523103 (+) | 3599 | 2763 | 920 | 103.52 | 7.16 | 13-LOX |
| Gene Name | Sequence ID | PLAT Domain 1, 2 | LOX Domain | 5-Histidine Domain |
|---|---|---|---|---|
| SpLOX1 | Spipo2G0068200 | 55–162HMMER | 175–846HMMER | 519–556 |
| SpLOX2 | Spipo2G0068500 | 71–165HMMER | 178–822HMMER | 522–559 |
| SpLOX3 | Spipo4G0070100 | 16–131CDART | 135–784HMMER | 479–516 |
| SpLOX4 | Spipo7G0050500 | 115–216HMMER | 229–899HMMER | 567–604 |
| SpLOX5 | Spipo15G0045200 | 28–99HMMER | 112–775HMMER | 447–484 |
| SpLOX6 | Spipo28G0005500 | 123–207HMMER | 220–887HMMER | 557–594 |
| SpLOX7 | Spipo28G0005600 | 121–207HMMER | 222–887HMMER | 557–594 |
| SpLOX8 | Spipo28G0005700 | 23–120HMMER | 129–620HMMER | 466–503 |
| SpLOX9 | Spipo0G0030100 | 138–223 HMMER | 236–903 HMMER | 574–611 |
| Growth Stages * | Culture Stage *(Days After Inoculation) | Endogenous JA Levels (ng−1 FW) * | 13-LOX Genes with Similar Patterns (Figure 7—This Study) # |
|---|---|---|---|
| Vegetative stage | Day 14 | 389 ± 8 | LOX3, 4, 6, 7, 8, 9 |
| Apical floral induction | Day 21 | 217 ± 13 | |
| Flowering plants | Day 28 | 37.5 ± 1.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Upadhyay, R.K.; Edelman, M.; Mattoo, A.K. Identification, Phylogeny, and Comparative Expression of the Lipoxygenase Gene Family of the Aquatic Duckweed, Spirodela polyrhiza, during Growth and in Response to Methyl Jasmonate and Salt. Int. J. Mol. Sci. 2020, 21, 9527. https://doi.org/10.3390/ijms21249527
Upadhyay RK, Edelman M, Mattoo AK. Identification, Phylogeny, and Comparative Expression of the Lipoxygenase Gene Family of the Aquatic Duckweed, Spirodela polyrhiza, during Growth and in Response to Methyl Jasmonate and Salt. International Journal of Molecular Sciences. 2020; 21(24):9527. https://doi.org/10.3390/ijms21249527
Chicago/Turabian StyleUpadhyay, Rakesh K., Marvin Edelman, and Autar K. Mattoo. 2020. "Identification, Phylogeny, and Comparative Expression of the Lipoxygenase Gene Family of the Aquatic Duckweed, Spirodela polyrhiza, during Growth and in Response to Methyl Jasmonate and Salt" International Journal of Molecular Sciences 21, no. 24: 9527. https://doi.org/10.3390/ijms21249527
APA StyleUpadhyay, R. K., Edelman, M., & Mattoo, A. K. (2020). Identification, Phylogeny, and Comparative Expression of the Lipoxygenase Gene Family of the Aquatic Duckweed, Spirodela polyrhiza, during Growth and in Response to Methyl Jasmonate and Salt. International Journal of Molecular Sciences, 21(24), 9527. https://doi.org/10.3390/ijms21249527





