Promiscuous Enzyme Activity as a Driver of Allo and Iso Convergent Evolution, Lessons from the β-Lactamases
Abstract
1. Introduction: The Concepts: Iso and Allo-Convergent Evolution, Evolutionary Shift, and Maintenance of Apomorphies
2. Enzyme Convergent Evolution via Allo-Convergent and Iso-Convergent Evolution: The Case of β-Lactamases
2.1. Allo-Convergent Evolution of Enzyme Function: The Case of β-Lactamases
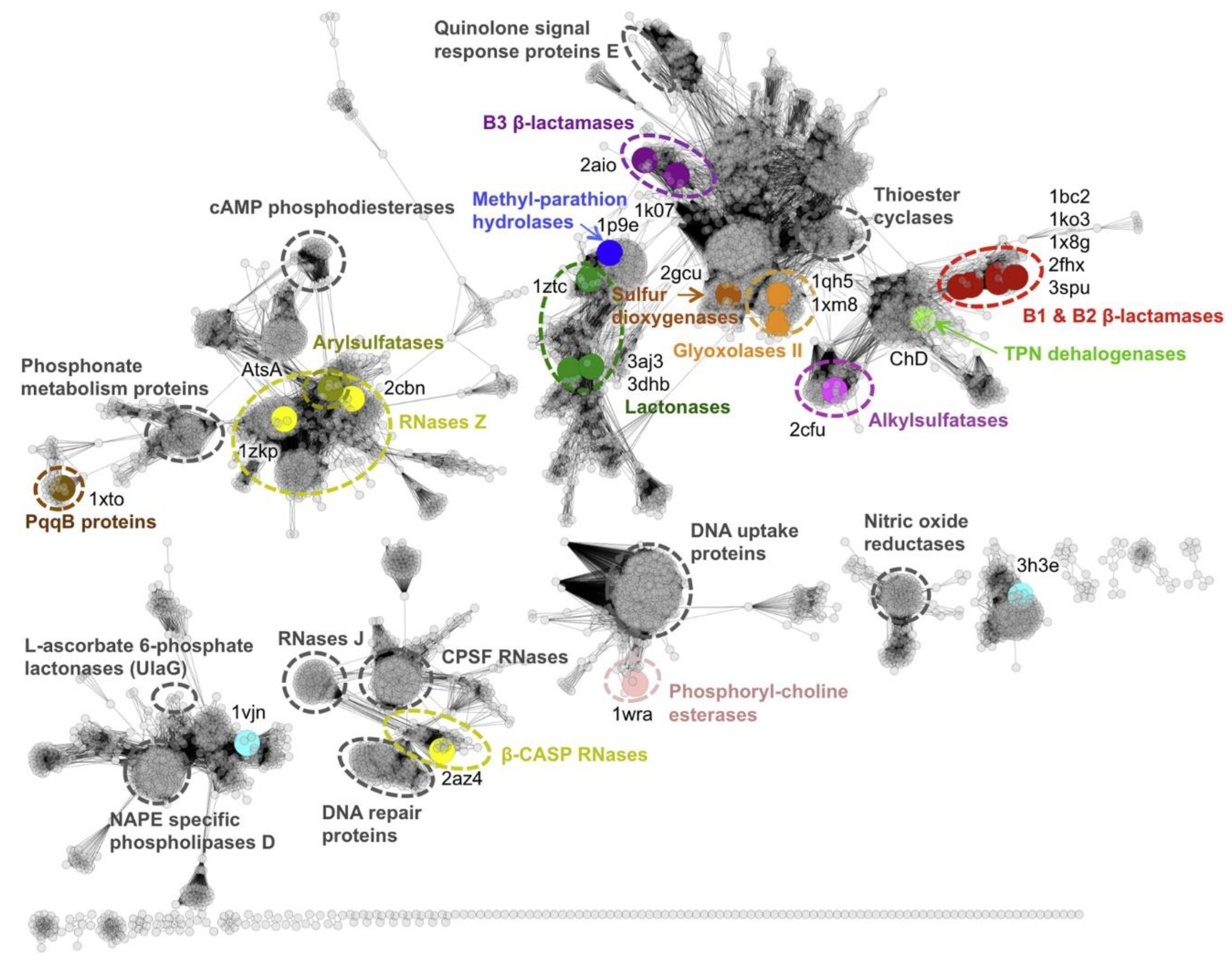
2.2. Iso-Convergent Evolution of the Metallo-β-Lactamases Functional Family
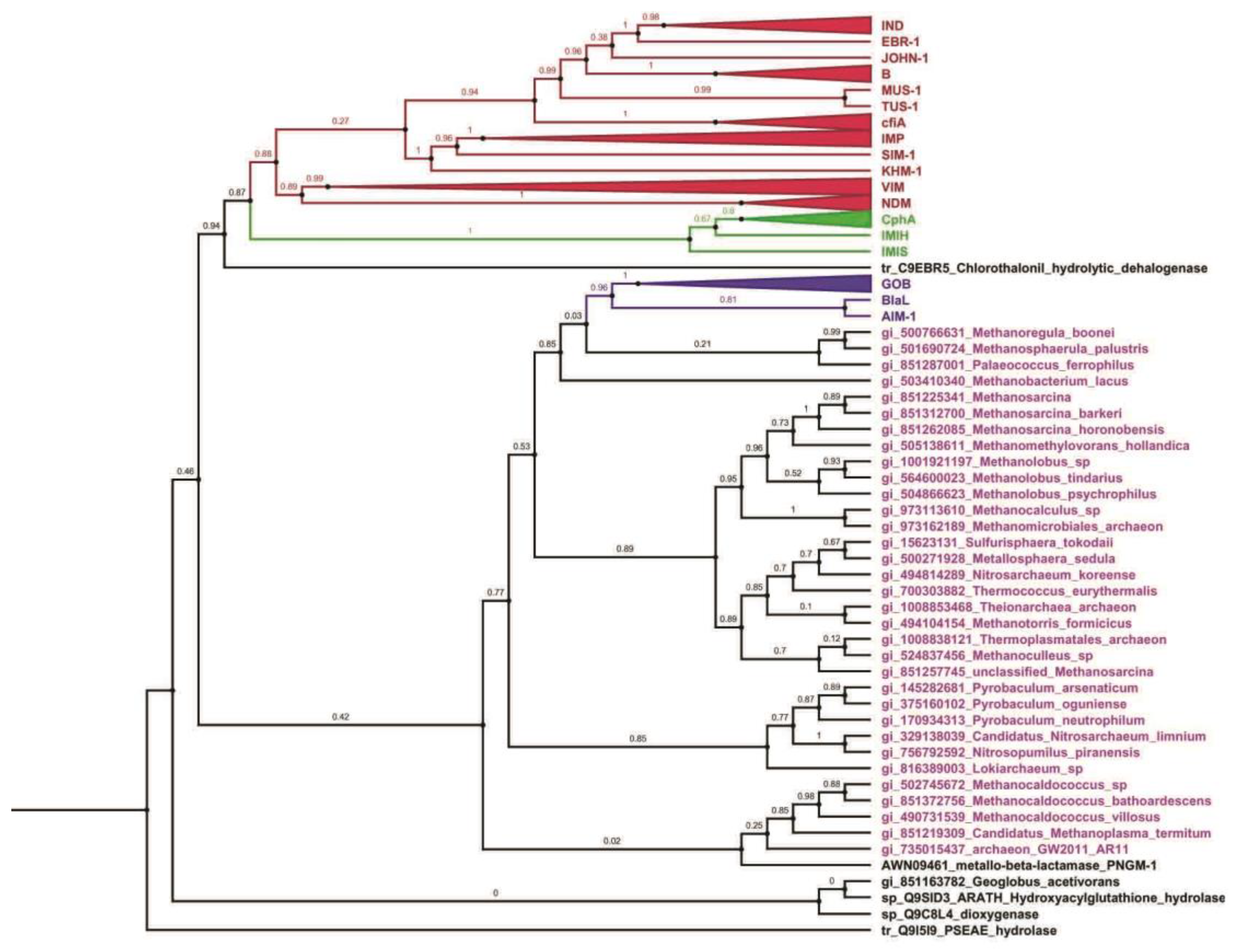
Evolutionary Analyses of the Metallo-β-Lactamases Family
2.3. Evolution of the Metallo-β-Lactamase Activity via an Increase in the Probability of Shift and Fixation of the New Character
2.4. The Metallo-β-Lactamases Evolved via Iso-Convergent Evolution Because Possibly Also of Positive Selection
2.5. Iso-Convergent Evolution of the Serine β-Lactamase Family
2.6. Hypothesis: Allo-Convergent Evolution Should Be Linked to Iso-Convergent Evolution
3. Materials and Methods
3.1. Sequence Selection
3.2. Functional Domain Identifications
3.3. Phylogenetic Tree Construction
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Pontarotti, P.; Hue, I. Road map to study convergent evolution: A proposition for evolutionary systems biology approaches. In Evolutionary Biology: Convergent Evolution, Evolution of Complex Traits, Concepts and Methods; Springer International Publishing: Cham, Switzerland, 2016; pp. 3–21. ISBN 9783319413242. [Google Scholar]
- Yampolsky, L.Y.; Stoltzfus, A. Bias in the introduction of variation as an orienting factor in evolution. Evol. Dev. 2001, 3, 73–83. [Google Scholar] [CrossRef]
- Streisfeld, M.A.; Rausher, M.D. Population genetics, pleiotropy, and the preferential fixation of mutations during adaptive evolution. Evolution (N. Y.) 2011, 65, 629–642. [Google Scholar] [CrossRef]
- Losos, J.B. Convergence, adaptation, and constraint. Evolution (N. Y.) 2011, 65, 1827–1840. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cheng, T.C.; Huang, G.; Lu, Q.; Surleac, M.D.; Mandell, J.D.; Pontarotti, P.; Petrescu, A.J.; Xu, A.; Xiong, Y.; et al. Transposon molecular domestication and the evolution of the RAG recombinase. Nature 2019, 569, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Doolittle, R.F. Convergent evolution: The need to be explicit. Trends Biochem. Sci. 1994, 19, 15–18. [Google Scholar] [CrossRef]
- Gherardini, P.F.; Wass, M.N.; Helmer-Citterich, M.; Sternberg, M.J.E. Convergent Evolution of Enzyme Active Sites Is not a Rare Phenomenon. J. Mol. Biol. 2007, 372, 817–845. [Google Scholar] [CrossRef]
- Khersonsky, O.; Tawfik, D.S. Enzyme Promiscuity: A Mechanistic and Evolutionary Perspective. Annu. Rev. Biochem. 2010, 79, 471–505. [Google Scholar] [CrossRef]
- Spratt, B.G. Penicillin-binding proteins and the future of β-lactam antibiotics. The seventh Fleming lecture. J. Gen. Microbiol. 1983, 129, 1247–1260. [Google Scholar] [CrossRef][Green Version]
- Burroughs, A.M.; Allen, K.N.; Dunaway-Mariano, D.; Aravind, L. Evolutionary Genomics of the HAD Superfamily: Understanding the Structural Adaptations and Catalytic Diversity in a Superfamily of Phosphoesterases and Allied Enzymes. J. Mol. Biol. 2006, 361, 1003–1034. [Google Scholar] [CrossRef]
- Keshri, V.; Arbuckle, K.; Chabrol, O.; Rolain, J.M.; Raoult, D.; Pontarotti, P. The functional convergence of antibiotic resistance in β-lactamases is not conferred by a simple convergent substitution of amino acid. Evol. Appl. 2019, 12, 1812–1822. [Google Scholar] [CrossRef]
- Bebrone, C. Metallo-β-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem. Pharmacol. 2007, 74, 1686–1701. [Google Scholar] [CrossRef]
- Baier, F.; Tokuriki, N. Connectivity between catalytic landscapes of the metallo-β-lactamase superfamily. J. Mol. Biol. 2014, 426, 2442–2456. [Google Scholar] [CrossRef]
- Atkinson, H.J.; Morris, J.H.; Ferrin, T.E.; Babbitt, P.C. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS ONE 2009, 4, e4345. [Google Scholar] [CrossRef]
- Alderson, R.G.; Barker, D.; Mitchell, J.B.O. One origin for metallo-β-lactamase activity, or two? An investigation assessing a diverse set of reconstructed ancestral sequences based on a sample of phylogenetic trees. J. Mol. Evol. 2014, 79, 117–129. [Google Scholar] [CrossRef]
- Keshri, V.; Panda, A.; Levasseur, A.; Rolain, J.M.; Pontarotti, P.; Raoult, D. Phylogenomic analysis of b-lactamase in Archaea and bacteria enables the identification of putative new members. Genome Biol. Evol. 2018, 10, 1106–1114. [Google Scholar] [CrossRef]
- Diene, S.M.; Pinault, L.; Armstrong, N.; Keshri, V.; Khelaifia, S.; Chabrière, E.; Caetano-Anolles, G.; Rolain, J.-M.; Pontarotti, P.; Raoult, D. Paradoxical β-lactamase activity of archaeal encoding enzymes. bioRxiv 2019, 667907. [Google Scholar] [CrossRef]
- Colson, P.; Pinault, L.; Azza, S.; Armstrong, N.; Chabriere, E.; La Scola, B.; Pontarotti, P.; Raoult, D. A metallo-beta-lactamase with both beta-lactamase and ribonuclease activity is linked with traduction in giant viruses. bioRxiv 2019, 819797. [Google Scholar] [CrossRef]
- CATH. CATH Superfamily 3.40.449.10. Available online: http://www.cathdb.info/version/latest/superfamily/3.60.15.10 (accessed on 15 February 2020).
- Velez Rueda, A.J.; Palopoli, N.; Zacarías, M.; Sommese, L.M.; Parisi, G. ProtMiscuity: A database of promiscuous proteins. Database (Oxford) 2019, 2019. [Google Scholar] [CrossRef]
- Copley, S.D. Shining a light on enzyme promiscuity. Curr. Opin. Struct. Biol. 2017, 47, 167–175. [Google Scholar] [CrossRef]
- Jensen, R.A. Enzyme Recruitment in Evolution of New Function. Annu. Rev. Microbiol. 1976, 30, 409–425. [Google Scholar] [CrossRef]
- Diene, S.M.; Pinault, L.; Keshri, V.; Armstrong, N.; Khelaifia, S.; Chabrière, E.; Caetano-Anolles, G.; Colson, P.; La Scola, B.; Rolain, J.M.; et al. Human metallo-β-lactamase enzymes degrade penicillin. Sci. Rep. 2019, 9, 12173. [Google Scholar] [CrossRef] [PubMed]
- Lisa, M.N.; Morán-Barrio, J.; Guindón, M.F.; Vila, A.J. Probing the role of met221 in the unusual metallo-β-lactamase gob-18. Inorg. Chem. 2012, 51, 12419–12425. [Google Scholar] [CrossRef]
- Keshri, V.; Diene, S.M.; Estienne, A.; Dardaillon, J.; Chabrol, O.; Tichit, L.; Rolain, J.-M.; Raoult, D.; Pontarotti, P. An Integrative Database of β-Lactamase Enzymes: Sequences, Structures, Functions, and Phylogenetic Trees. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef]
- Morán-Barrio, J.; González, J.M.; Lisa, M.N.; Costello, A.L.; Peraro, M.D.; Carloni, P.; Bennett, B.; Tierney, D.L.; Limansky, A.S.; Viale, A.M.; et al. The metallo-beta-lactamase GOB is a mono-Zn(II) enzyme with a novel active site. J. Biol. Chem. 2007, 282, 18286–18293. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, M.M.; Herman, R.; Ghiglione, B.; Kerff, F.; D’Amico González, G.; Bouillenne, F.; Galleni, M.; Handelsman, J.; Charlier, P.; Gutkind, G.; et al. Crystal structure and kinetic analysis of the class B3 di-zinc metallo-β-lactamase LRA-12 from an Alaskan soil metagenome. PLoS ONE 2017, 12, e0182043. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Takahashi, M.; Jeon, J.H.; Kang, L.W.; Seki, M.; Park, K.S.; Hong, M.K.; Park, Y.S.; Kim, T.Y.; Karim, A.M.; et al. Dual activity of PNGM-1 pinpoints the evolutionary origin of subclass B3 metallo-β-lactamases: A molecular and evolutionary study. Emerg. Microbes Infect. 2019, 8, 1688–1700. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.G.; Barlow, M. Evolution of the serine β-lactamases: Past, present and future. Drug Resist. Updat. 2004, 7, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Dcosta, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.L.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R.; et al. Antibiotic resistance is ancient. Nature 2011, 477, 457–461. [Google Scholar] [CrossRef]
- Townsend, C.A. Convergent biosynthetic pathways to β-lactam antibiotics. Curr. Opin. Chem. Biol. 2016, 35, 97–108. [Google Scholar] [CrossRef]
- Levasseur, A.; Pontarotti, P. The role of duplications in the evolution of genomes highlights the need for evolutionary-based approaches in comparative genomics. Boil. Direct 2011, 6, 11. [Google Scholar] [CrossRef]
- Shapiro, A.B.; Gao, N.; Gu, R.F.; Thresher, J. Fluorescence anisotropy-based measurement of Pseudomonas aeruginosa penicillin-binding protein 2 transpeptidase inhibitor acylation rate constants. Anal. Biochem. 2014, 463, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, A.B.; Gu, R.F.; Gao, N.; Livchak, S.; Thresher, J. Continuous fluorescence anisotropy-based assay of BOCILLIN FL penicillin reaction with penicillin binding protein 3. Anal. Biochem. 2013, 439, 37–43. [Google Scholar] [CrossRef]
- Lemaire, S.; Fuda, C.; Van Bambeke, F.; Tulkens, P.M.; Mobashery, S. Restoration of susceptibility of methicillin-resistant Staphylococcus aureus to beta-lactam antibiotics by acidic pH: Role of penicillin-binding protein PBP 2a. J. Biol. Chem. 2008, 283, 12769–12776. [Google Scholar] [CrossRef] [PubMed]
- Massova, I.; Mobashery, S. Kinship and diversification of bacterial penicillin-binding proteins and β-lactamases. Antimicrob. Agents Chemother. 1998, 42, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.P.; Kincaid, E.; Sun, Y.; Bauer, M.D. Kinetics of β-lactam interactions with penicillin-susceptible and -resistant penicillin-binding protein 2x proteins from Streptococcus pneumoniae. Involvement of acylation and deacylation β-lactam resistance. J. Biol. Chem. 2001, 276, 31494–31501. [Google Scholar] [CrossRef] [PubMed]
- Elias, M.; Tawfik, D.S. Divergence and convergence in enzyme evolution: Parallel evolution of paraoxonases from quorum-quenching lactonases. J. Biol. Chem. 2012, 287, 11–20. [Google Scholar] [CrossRef]
- Gould, S.J.; Vrba, E.S. Exaptation—A Missing Term in the Science of Form. Paleobiology 1982, 1, 4–15. [Google Scholar] [CrossRef]
- Fuchs, G. Alternative Pathways of Carbon Dioxide Fixation: Insights into the Early Evolution of Life? Annu. Rev. Microbiol. 2011, 65, 631–658. [Google Scholar] [CrossRef]
- Bada, J.L. ORIGIN OF LIFE: Some Like It Hot, But Not the First Biomolecules. Science 2002, 296, 1982–1983. [Google Scholar] [CrossRef]
- Schönheit, P.; Buckel, W.; Martin, W.F. On the Origin of Heterotrophy. Trends Microbiol. 2016, 24, 12–25. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J.; Finn, R. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2016, 44, D343–D350. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.M. ARG-annot, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Keshri, V.; Chabrière, E.; Pinault, L.; Colson, P.; Diene, S.M.; Rolain, J.-M.; Raoult, D.; Pontarotti, P. Promiscuous Enzyme Activity as a Driver of Allo and Iso Convergent Evolution, Lessons from the β-Lactamases. Int. J. Mol. Sci. 2020, 21, 6260. https://doi.org/10.3390/ijms21176260
Keshri V, Chabrière E, Pinault L, Colson P, Diene SM, Rolain J-M, Raoult D, Pontarotti P. Promiscuous Enzyme Activity as a Driver of Allo and Iso Convergent Evolution, Lessons from the β-Lactamases. International Journal of Molecular Sciences. 2020; 21(17):6260. https://doi.org/10.3390/ijms21176260
Chicago/Turabian StyleKeshri, Vivek, Eric Chabrière, Lucile Pinault, Philippe Colson, Seydina M Diene, Jean-Marc Rolain, Didier Raoult, and Pierre Pontarotti. 2020. "Promiscuous Enzyme Activity as a Driver of Allo and Iso Convergent Evolution, Lessons from the β-Lactamases" International Journal of Molecular Sciences 21, no. 17: 6260. https://doi.org/10.3390/ijms21176260
APA StyleKeshri, V., Chabrière, E., Pinault, L., Colson, P., Diene, S. M., Rolain, J.-M., Raoult, D., & Pontarotti, P. (2020). Promiscuous Enzyme Activity as a Driver of Allo and Iso Convergent Evolution, Lessons from the β-Lactamases. International Journal of Molecular Sciences, 21(17), 6260. https://doi.org/10.3390/ijms21176260







