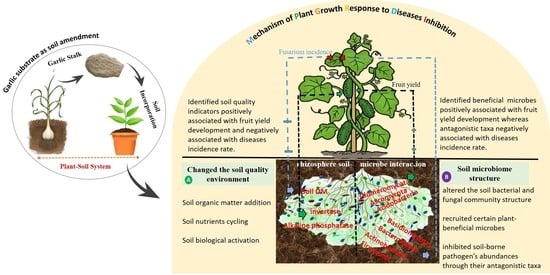
Garlic Substrate Induces Cucumber Growth Development and Decreases Fusarium Wilt through Regulation of Soil Microbial Community Structure and Diversity in Replanted Disturbed Soil
Abstract
1. Introduction
2. Results
2.1. Effect of Garlic Substrate on Plant Growth Parameters and Fusarium Incidence Rate
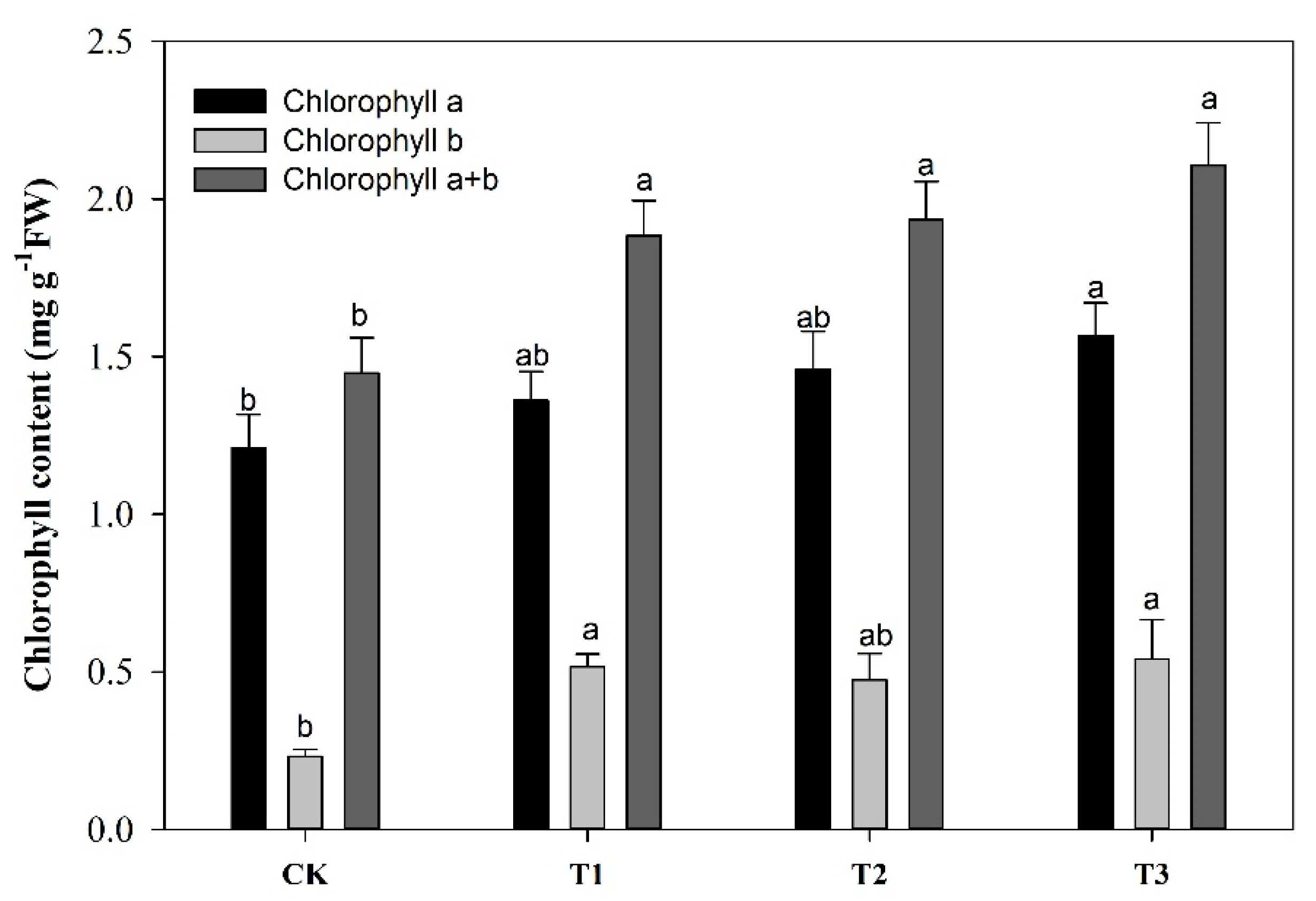
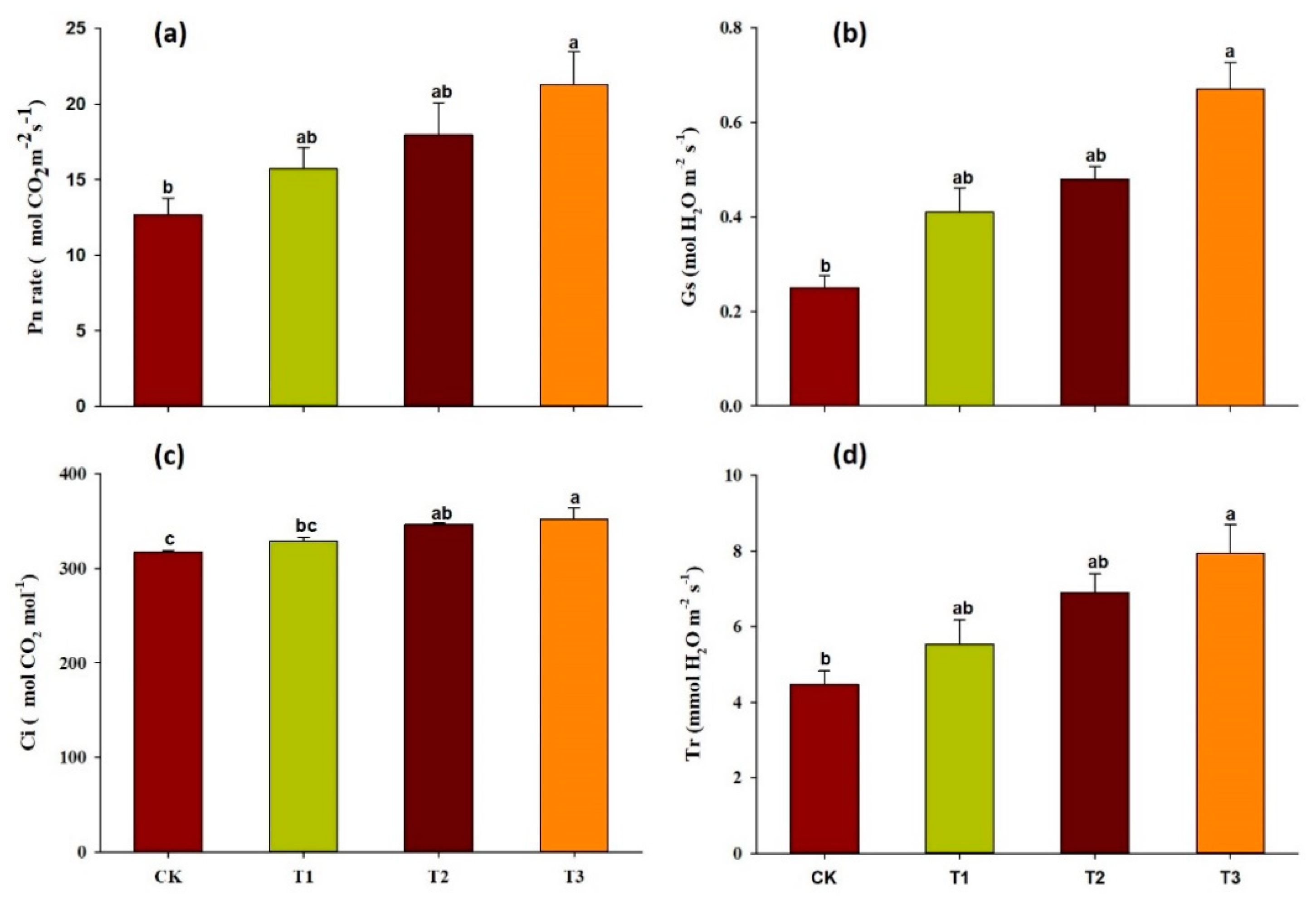
2.2. Effect of Garlic Substrate on Chlorophyll and Leaf Gas Exchange Parameters
2.3. Effects of Garlic Substrate on Changing the Soil Characteristics of Replanted Soil
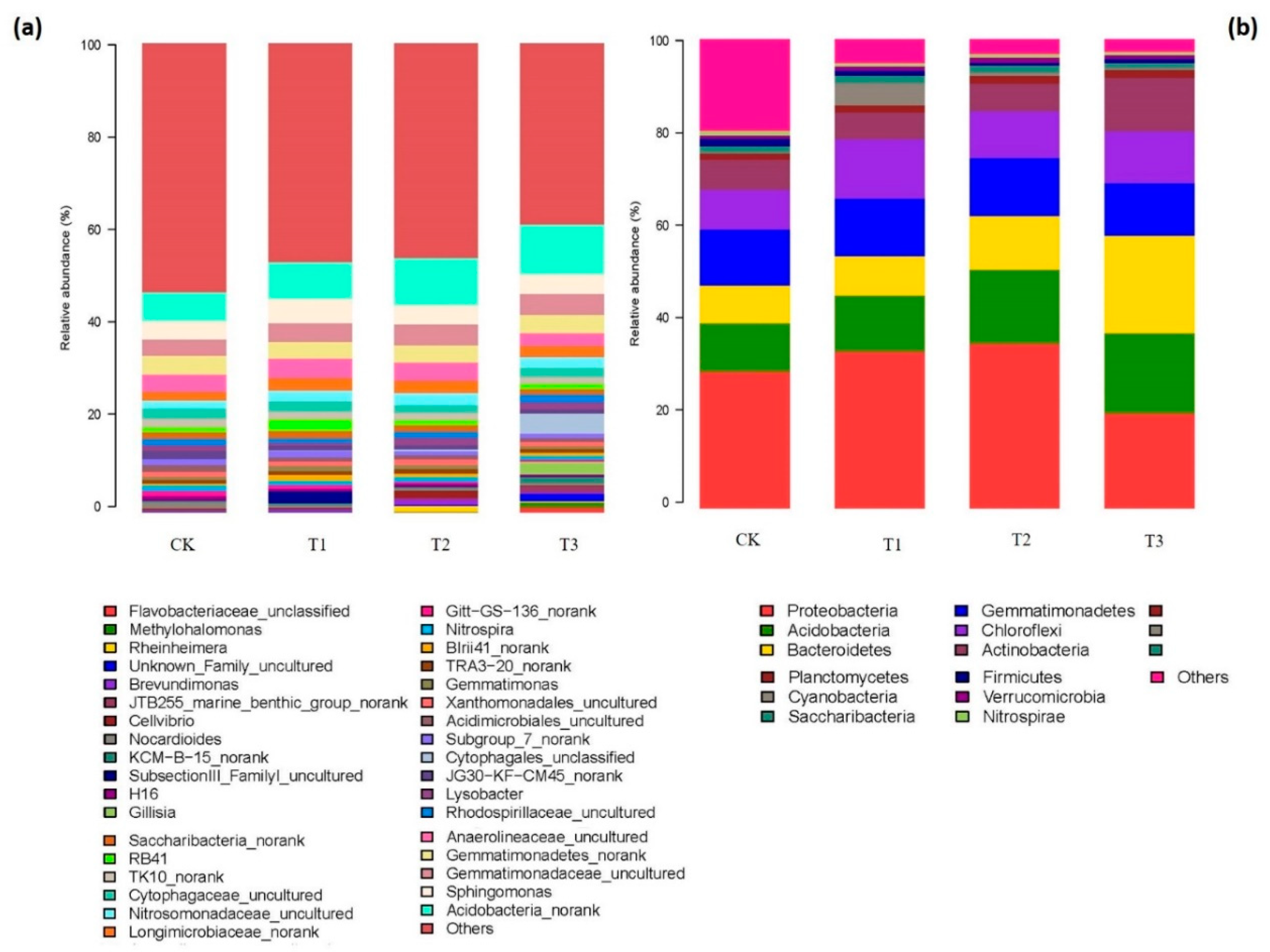
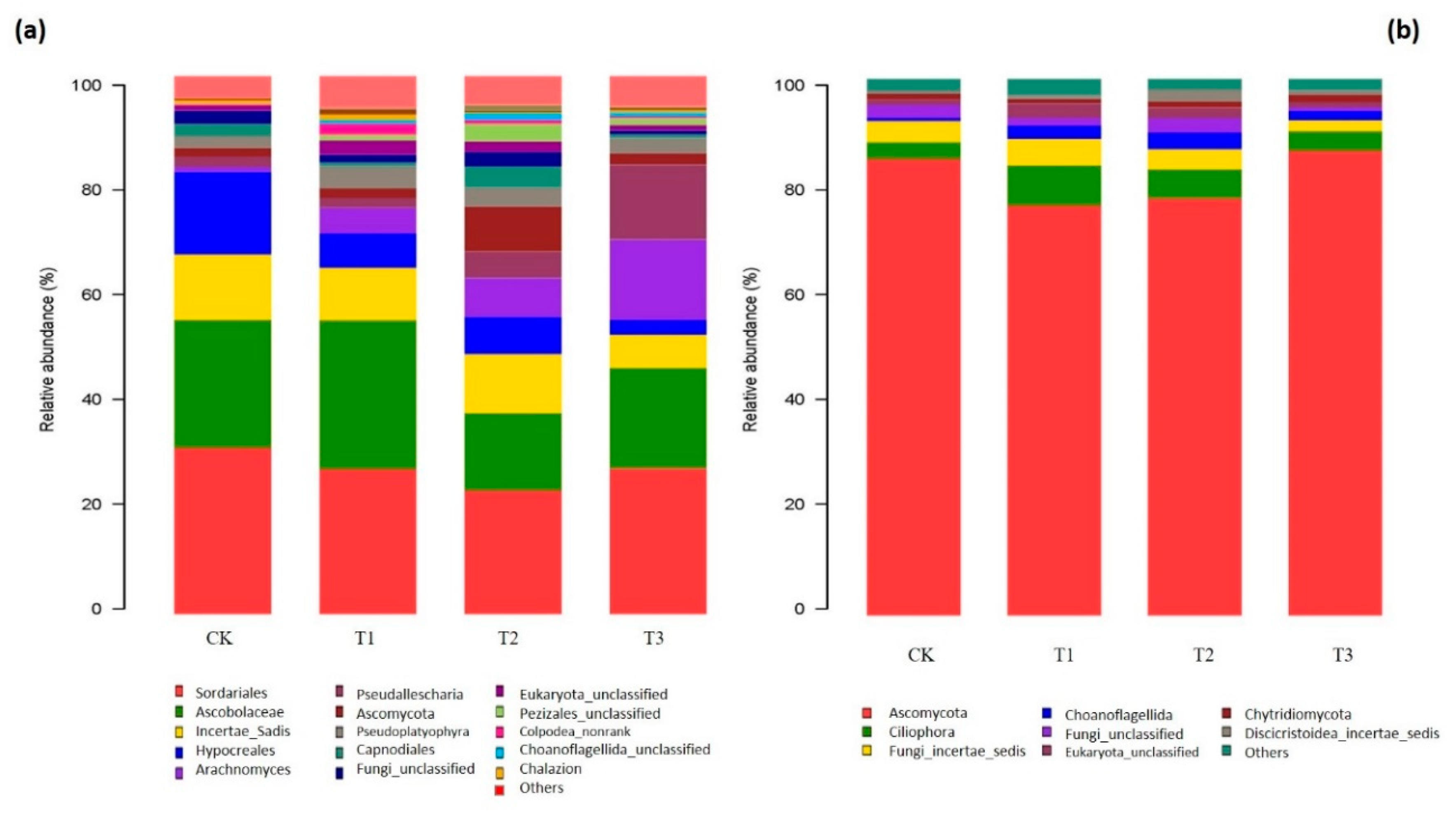
2.4. Composition of Bacterial and Fungal Communities as Influenced by Garlic Substrate
2.5. Changes in Soil Bacterial and Fungal Alpha Diversity
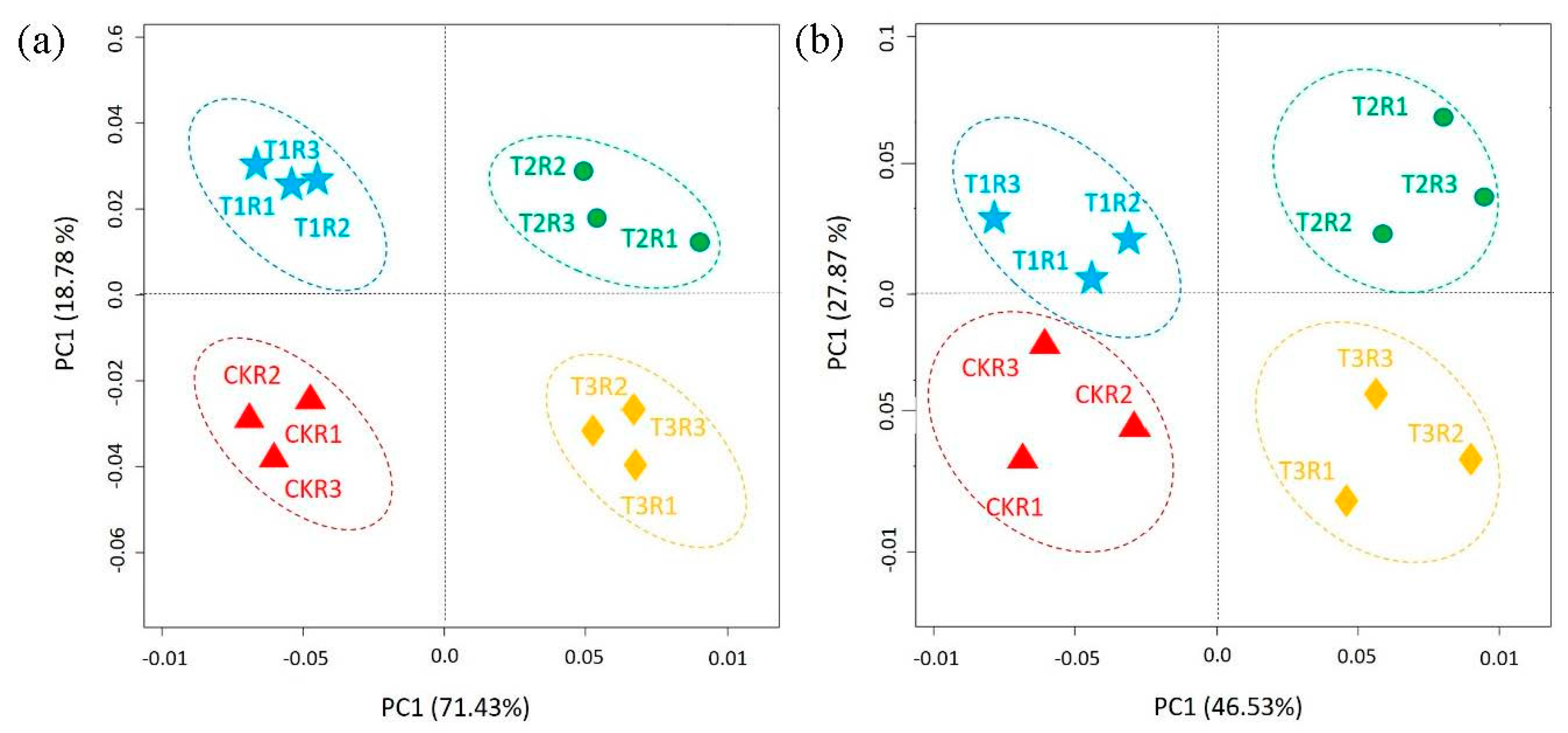
2.6. Differences in Community Composition and Diversity
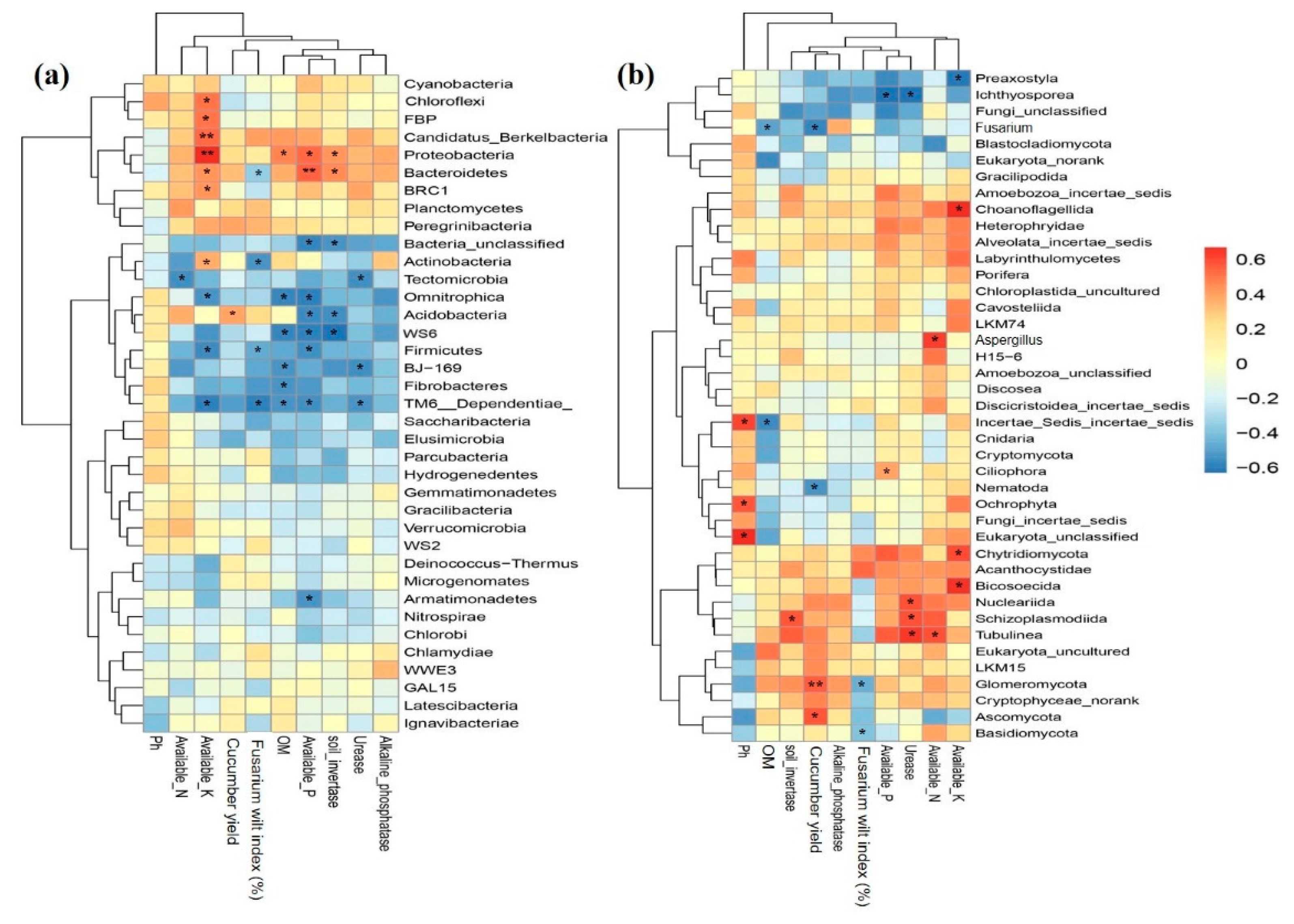
2.7. Relationship among Key Soil Properties, Crop Yield, Disease Incidence and Microbial Structure
3. Discussion
3.1. Garlic Substrate Addition Improved Cucumber Growth and Reduced Fusarium Wilt Incidence %
3.2. Garlic Substrate Addition Changed the Soil Quality Environment
3.3. Garlic Substrate Addition Alters Soil Microbial Community Composition and Diversity
3.4. Relationship Mechanism of Cucumber Yield, Fusarium Wilt Inhibition Rate and Microbial Structure
4. Materials and Methods
4.1. Site Description and Garlic Substrate Used
4.2. Experimental Set-Up
4.3. Soil Sampling and Analyses
4.4. Soil Enzyme Assays
4.5. Plant Morphological and Physiological Measurements
4.6. DNA Extraction and PCR Amplification of 16S rRNA and 18S rRNA
4.7. Sequence Processing and Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hong, Y.; Heerink, N.; Jin, S.Q.; Berentsen, P.; Zhang, L.Z.; Werf, W.V.D. Intercropping and agroforestry in China-Current state and trends. Agric. Ecosyst. Environ. 2017, 244, 52–61. [Google Scholar] [CrossRef]
- Song, Y.; Li, X.; Yao, S.; Yang, X.; Jiang, X. Correlations between soil metabolomics and bacterial community structures in the pepper rhizosphere under plastic greenhouse cultivation. Sci. Total Environ. 2020, 728, 138439. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Wu, X.; Wang, Y.; Meyerson, L.A.; Gu, B.J.; Min, Y.; Xue, H.; Peng, C.H.; Ge, Y. Does growing vegetables in plastic greenhouses enhance regional ecosystem services beyond the food supply? Front. Ecol. Environ. 2013, 11, 43–49. [Google Scholar] [CrossRef]
- Hu, W.; Zhang, Y.; Huang, B.; Teng, Y. Soil environmental quality in greenhouse vegetable production systems in eastern China: Current status and management strategies. Chemosphere 2017, 170, 183–195. [Google Scholar] [CrossRef]
- Chang, J.; Wu, X.; Liu, A.; Wang, Y.; Xu, B.; Yang, W. Assessment of net ecosystem services of plastic greenhouse vegetable cultivation in China. Ecol. Econ. 2011, 70, 740–748. [Google Scholar] [CrossRef]
- Ali, A.; Ghani, M.I.; Ding, H.; Fan, Y.; Cheng, Z.; Iqbal, M. Co-amended synergistic interactions between arbuscular mycorrhizal fungi and the organic substrate-induced cucumber yield and fruit quality associated with the regulation of the am-fungal community structure under anthropogenic cultivated soil. Int. J. Mol. Sci. 2019, 20, 1539. [Google Scholar] [CrossRef]
- Ding, H.; Ali, A.; Cheng, Z. Dynamics of a soil fungal community in a three-year green garlic/cucumber crop rotation system in northwest china. Sustainability 2018, 10, 1391. [Google Scholar] [CrossRef]
- Zhou, X.; Wu, F. Changes in soil chemical characters and enzyme activities during continuous monocropping of cucumber (Cucumis sativus L). Pak. J. Bot. 2015, 47, 691–697. [Google Scholar]
- Zhou, X.; Yu, G.; Wu, F. Soil phenolics in a continuously mono-cropped cucumber (Cucumis sativus L.) system and their effects on cucumber seedling growth and soil microbial communities. Eur. J. Soil Sci. 2012, 63, 332–340. [Google Scholar] [CrossRef]
- Yao, H.; Jiao, X.; Wu, F. Effects of continuous cucumber cropping and alternative rotations under protected cultivation on soil microbial community diversity. Plant Soil 2006, 284, 195–203. [Google Scholar] [CrossRef]
- Bu, R.; Xie, J.; Yu, J.; Liao, W.; Xiao, X.; Lv, J.; Calderón-Urrea, A. Autotoxicity in cucumber (Cucumis sativus L.) seedlings is alleviated by silicon through an increase in the activity of antioxidant enzymes and by mitigating lipid peroxidation. J. Plant Biol. 2016, 59, 247–259. [Google Scholar] [CrossRef]
- Fu, H.; Zhang, G.; Zhang, F.; Sun, Z.; Geng, G.; Li, T. Effects of continuous tomato monoculture on soil microbial properties and enzyme activities in a solar greenhouse. Sustainability 2017, 9, 317. [Google Scholar] [CrossRef]
- Ali, A.; Imran, G.M.; Li, Y.; Ding, H.; Meng, H.; Cheng, Z. Hiseq base molecular characterization of soil microbial community, diversity structure, and predictive functional profiling in continuous cucumber planted soil affected by diverse cropping systems in an intensive greenhouse region of northern China. Int. J. Mol. Sci. 2019, 20, 2619. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.M.; Yao, J.; Yan, F. Vegetable cultivation under greenhouse conditions leads to rapid accumulation of nutrients, acidification and salinity of soils and groundwater contamination in South-Eastern China. Nutr. Cycl. Agroecosyst. 2009, 83, 73–84. [Google Scholar] [CrossRef]
- Liang, Y.; Lin, X.; Yamada, S.; Inoue, M.; Inosako, K. Soil degradation and prevention in greenhouse production. Springer Plus. 2013, 2 (Suppl. 1), S10. [Google Scholar] [CrossRef]
- Ding, H.; Ali, A.; Cheng, Z. Effect of green garlic/cucumber crop rotation for 3 years on the dynamics of soil properties and cucumber yield in Chinese anthrosol. J. Sci. Food Agric. 2020, 100, 362–370. [Google Scholar] [CrossRef]
- Xiong, W.; Li, Z.; Liu, H.; Xue, C.; Zhang, R.; Wu, H.; Shen, Q. The effect of long-term continuous cropping of black pepper on soil bacterial communities as determined by 454 pyrosequencing. PLoS ONE 2015, 10, e0136946. [Google Scholar] [CrossRef]
- Wang, Z.; Li, T.; Wen, X.; Liu, Y.; Han, J.; Liao, Y.; DeBruyn, J.M. Fungal communities in rhizosphere soil under conservation tillage shift in response to plant growth. Front. Microbiol. 2017, 8, 1301. [Google Scholar] [CrossRef]
- Xiao, X.; Cheng, Z.; Lv, J.; Xie, J.; Ma, N.; Yu, J. A green garlic (Allium sativum L.) based intercropping system reduces the strain of continuous monocropping in cucumber (Cucumis sativus L.) by adjusting the micro-ecological environment of soil. PeerJ 2019, 7, e7267. [Google Scholar] [CrossRef]
- Xiao, X.; Cheng, Z.; Meng, H.; Liu, L.; Li, H.; Dong, Y. Intercropping of green garlic (Allium sativum L.) induces nutrient concentration changes in the soil and plants in continuously cropped cucumber (Cucumis sativus L.) in a plastic tunnel. PLoS ONE 2013, 8, e62173. [Google Scholar] [CrossRef]
- Wang, M.; Wu, C.; Cheng, Z.; Meng, H. Growth and physiological changes in continuously cropped eggplant (Solanum melongena L.) upon relay intercropping with garlic (Allium sativum L.). Front. Plant Sci. 2015, 6, 262. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Cheng, Z.; Liu, M.; Hayat, S.; Feng, H. Garlic exerts allelopathic effects on pepper physiology in a hydroponic co-culture system. Biol. Open 2016, 5, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Ali, A.; Cheng, Z. An Allelopathic Role for garlic root exudates in the regulation of carbohydrate metabolism in cucumber in a hydroponic co-culture system. Plants 2020, 9, 45. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Cheng, Z.; Abdul-rehman, K.; Shah, J.R.; Bushra, G. Pepper-Garlic intercropping system improves soil biology and nutrient status in plastic tunnel. Int. J. Agric. Biol. 2015, 17, 869–880. [Google Scholar] [CrossRef]
- Ahmad, I.; Cheng, Z.; Meng, H.; Liu, T.; Wang, M.; Ejaz, M.; Wasila, H. Effect of pepper-garlic intercropping system on soil microbial and bio-chemical properties. Pak. J. Bot. 2013, 45, 695–702. [Google Scholar]
- Ghani, M.I.; Ali, A.; Atif, M.J.; Ali, M.; Amin, B.; Anees, M.; Cheng, Z. Soil amendment with raw garlic stalk: A novel strategy to stimulate growth and the antioxidative defense system in monocropped eggplant in the north of China. Agronomy 2019, 9, 89. [Google Scholar] [CrossRef]
- Ghani, M.I.; Ali, A.; Atif, M.J.; Ali, M.; Amin, B.; Anees, M.; Cheng, Z. Changes in the soil microbiome in eggplant monoculture revealed by high-throughput Illumina MiSeq sequencing as influenced by raw garlic stalk amendment. Int. J. Mol. Sci. 2019, 20, 2125. [Google Scholar] [CrossRef]
- Beare, M.H.; Wilson, P.E.; Fraser, P.M.; Butler, R.C. Management effect of barley straw decomposition, nitrogen release, and crop production. Soil Sci. Soc. Am. J. 2002, 66, 848–856. [Google Scholar] [CrossRef]
- Hidayatullah, A.; Jan, A.; Shah, Z. Residual effect of organic nitrogen sources applied to rice on the subsequent wheat crop. Int. J. Agron. Plant. Prod. 2013, 4, 620–631. [Google Scholar]
- Yadvinder, S.; Bijay, S.; Timsina, J. Plant residues management for nutrient cycling and improving soil productivity in rice-based cropping systems in the tropics. Adv. Agron. 2005, 85, 269–407. [Google Scholar]
- Amanullah, S.U.T.; Iqbal, A.; Fahad, S. Growth and productivity response of hybrid rice to application of animal manures, plant residues and phosphorus. Front. Plant Sci. 2016, 7, 1440. [Google Scholar] [CrossRef] [PubMed]
- Opala, P.A.; Othieno, C.O.; Okalebo, J.R.; Kisinyo, P.O. Effects of combining organic materials with inorganic phosphorus sources on maize yield and financial benefits in western Kenya. Exper. Agric. 2010, 46, 23–34. [Google Scholar] [CrossRef]
- Ding, H.; Ahmad, I.; Ali, A.; Khan, A.R.; Cheng, Z. Bioassay evaluation of the potential allelopathic effects of garlic (Allium sativum L.) root exudates on lettuce and cucumber. Pak. J. Bot. 2018, 50, 371–380. [Google Scholar]
- Hayat, S.; Cheng, Z.; Ahmad, H.; Ali, M.; Chen, X.; Wang, M. Garlic, from remedy to stimulant: Evaluation of antifungal potential reveals diversity in phytoalexin allicin content among garlic cultivars; allicin containing aqueous garlic extracts trigger antioxidants in Cucumber. Front. Plant Sci. 2016, 7, 1235. [Google Scholar] [CrossRef]
- Bertoli, A.; Tozzi, S.; Pistelli, L.; Angelini, L.G. Fibre hemp inflorescences: From crop-residues to essential oil production. Ind. Crops Prod. 2010, 32, 329–337. [Google Scholar] [CrossRef]
- Xuan, T.D.; Tawata, S.; Khanh, T.D.; Chung, I.M. Decomposition of allelopathic plants in soil. J. Agron. Crop. Sci. 2005, 191, 162–171. [Google Scholar] [CrossRef]
- Kobayashi, K. Factors affecting phytotoxic activity of allelochemicals in soil. Weed Biol. Manag. 2004, 4, 1–7. [Google Scholar] [CrossRef]
- Gonzalez, M.; Gomez, E.; Comese, R.; Quesada, M.; Conti, M. Influence of organic amendments on soil quality potential indicators in an urban horticultural system. Bioresour. Technol. 2010, 101, 8897–8901. [Google Scholar] [CrossRef]
- Butterly, C.; Baldock, J.; Tang, C. The contribution of crop residues to changes in soil pH under field conditions. Plant Soil 2013, 366, 185–198. [Google Scholar] [CrossRef]
- Shaw, J.N.; Mask, P.L. Crop residue effects on electrical conductivity of tennessee valley soils. Commun. Soil Sci. Plant Anal. 2003, 34, 747–763. [Google Scholar] [CrossRef]
- Clay, D.E.; Mishra, U. The importance of crop residues in maintaining soil organic carbon in agroecosystems. Bioenergy Land Use Chang. 2017, 4, 115–124. [Google Scholar]
- Mi, W.; Wu, L.; Brookes, P.C.; Liu, Y.; Zhang, X.; Yang, X. Changes in soil organic carbon fractions under integrated management systems in a low-productivity paddy soil given different organic amendments and chemical fertilizers. Soil Till. Res. 2016, 163, 64–70. [Google Scholar] [CrossRef]
- Ortiz-Cornejo, N.L.; Romero-Salas, E.A.; Navarro-Noya, Y.E.; González-Zúñiga, J.C.; Ramirez-Villanueva, D.A.; Vásquez-Murrieta, M.S.; Verhulst, N.; Govaerts, B.; Dendooven, L.; Luna-Guido, M. Incorporation of bean plant residue in soil with different agricultural practices and its effect on the soil bacteria. Appl. Soil Ecol. 2017, 119, 417–427. [Google Scholar] [CrossRef]
- Malhi, S.S.; Lemke, R.; Wang, Z.H.; Chhabra, B.S. Tillage, nitrogen and crop residue effects on crop yield, nutrient uptake, soil quality, and greenhouse gas emissions. Soil Tillage Res. 2006, 90, 171–183. [Google Scholar] [CrossRef]
- Wei, T.; Zhang, P.; Wang, K.; Ding, R.; Yang, B.; Nie, J.; Jia, Z.; Han, Q. Effects of wheat straw incorporation on the availability of soil nutrients and enzyme activities in semiarid areas. PLoS ONE 2015, 10, e0120994. [Google Scholar] [CrossRef]
- Vanzolini, J.I.; Juan, A.; Galantini, B.; Juan, M.M.; Liliana, S. Changes in soil pH and phosphorus availability during decomposition of cover crop residues. Arch. Agron. Soil Sci. 2017, 60, 457–470. [Google Scholar] [CrossRef]
- Bhattacharjya, S.; Chandra, R.; Pareek, N.; Raverkar, K.P. Biochar and crop residue application to soil: Effect on soil biochemical properties, nutrient availability and yield of rice (Oryza sativa L.) and wheat (Triticum aestivum L.). Arch. Agron. Soil Sci. 2016, 62, 1095–1108. [Google Scholar] [CrossRef]
- Truong, T.H.H.; Marschner, P. Respiration, available N and microbial biomass N in soil amended with mixes of organic materials differing in C/N ratio and decomposition stage. Geoderma 2018, 319, 167–174. [Google Scholar] [CrossRef]
- Bending, G.D.; Turner, M.K.; Jones, J.E. Interactions between crop residue and soil organic matter quality and the functional diversity of soil microbial communities. Soil Biol. Biochem. 2002, 34, 1073–1082. [Google Scholar] [CrossRef]
- He, M.; Zhang, K.; Tan, H.; Hu, R.; Su, J.; Wang, J.; Huang, L.; Zhang, Y.; Li, X. Nutrient levels within leaves, stems, and roots of the xeric species Reaumuria soongorica in relation to geographical, climatic, and soil conditions. Ecol. Evol. 2015, 5, 1494–1503. [Google Scholar] [CrossRef]
- Moreno-Cornejo, J.; Zornoza, R.; Faz, A. Carbon and nitrogen mineralization during decomposition of crop residues in a calcareous soil. Geoderma 2014, 230–231, 58–63. [Google Scholar] [CrossRef]
- Muhammad, W.; Vaughan, S.M.; Dalal, R.C.; Menzies, N.W. Crop residues and fertilizer nitrogen influence residue decomposition and nitrous oxide emission from a Vertisol. Biol. Fertil. Soils 2011, 47, 15–23. [Google Scholar] [CrossRef]
- Cesarano, G.; De Filippis, F.; La Storia, A.; Scala, F.; Bonanomi, G. Organic amendment type and application frequency affect crop yields, soil fertility and microbiome composition. Appl. Soil Ecol. 2017, 120, 254–264. [Google Scholar] [CrossRef]
- Zhang, P.; Chen, X.; Wei, T.; Yang, Z.; Jia, Z.; Yang, B.; Han, Q.; Ren, X. Effects of straw incorporation on the soil nutrient contents enzyme activities, and crop yield in a semiarid region of China. Soil Tillage Res. 2016, 160, 65–72. [Google Scholar] [CrossRef]
- Bakht, J.; Shafi, M.; Jan, M.T.; Shah, Z. Influence of crop residue management: Cropping system and N fertilizer on soil N and C dynamics and sustainable wheat (Triticum aestivum L.) production. Soil Tillage Res. 2009, 104, 233–240. [Google Scholar] [CrossRef]
- Innangi, M.; Niro, E.; D’Ascoli, R.; Danise, T.; Proietti, P.; Nasini, L.; Regni, L.; Castaldi, S.; Fioretto, A. Effects of olive pomace amendment on soil enzyme activities. Appl. Soil Ecol. 2009, 119, 242–249. [Google Scholar] [CrossRef]
- Zhao, J.; Ni, T.; Li, J.; Lu, Q.; Fang, Z.; Huang, Q.; Zhang, R.; Li, R.; Shen, B.; Shen, Q. Effects of organic–inorganic compound fertilizer with reduced chemical fertilizer application on crop yields, soil biological activity and bacterial community structure in a rice–Wheat cropping system. Appl. Soil Ecol. 2016, 99, 1–12. [Google Scholar] [CrossRef]
- Marschner, P.; Umar, S.; Baumann, K. The microbial community composition changes rapidly in the early stages of decomposition of wheat residue. Soil Biol. Biochem. 2011, 43, 445–451. [Google Scholar] [CrossRef]
- Rui, J.; Peng, J.; Lu, Y. Succession of bacterial populations during plant residue decomposition in rice field soil. Appl. Environ. Microbiol. 2009, 75, 4879–4886. [Google Scholar] [CrossRef]
- Su, P.; Lou, J.; Brookes, P.C.; Luo, Y.; He, Y.; Xu, J. Taxon-specific responses of soil microbial communities to different soil priming effects induced by addition of plant residues and their biochars. J. Soils Sediments 2017, 17, 674–684. [Google Scholar] [CrossRef]
- Jiang, C.; Wu, Y.P.; Cheng, Y.X. Bacterial and fungal communities and contribution of physicochemical factors during cattle farm waste composting. Microbiol. Open 2017, 6, 6–14. [Google Scholar]
- Ma, A.; Zhuang, X.; Wu, J.; Cui, M.; Lv, D.; Liu, C.; Zhuang, G. Ascomycota members dominate fungal communities during straw residue decomposition in arable soil. PLoS ONE 2013, 6, e66146. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Mercado, D.I.; Lynn, D.H. Soil ciliate species richness and abundance associated with the rhizosphere of different subtropical plant species. J. Eukaryot. Microbiol. 2004, 51, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Bastian, F.; Bouziri, L.; Nicolardot, B.; Ranjard, L. Impact of wheat straw decomposition on successional patterns of soil microbial community structure. Soil Biol. Biochem. 2009, 41, 262–275. [Google Scholar] [CrossRef]
- Liu, J.; Yu, Z.; Yao, Q.; Hu, X.; Zhang, W.; Mi, G.; Chen, X.; Wang, G. Distinct soil bacterial communities in response to the cropping system in a Mollisol of northeast China. Appl. Soil Ecol. 2017, 119, 407–416. [Google Scholar] [CrossRef]
- Chen, C.; Liu, C.H.; Cai, J.; Zhang, W.; Qi, W.L.; Wang, Z.; Liu, Z.-B.; Yang, Y. Broad-spectrum antimicrobial activity, chemical composition and mechanism of action of garlic (Allium sativum) extracts. Food Control 2018, 86, 117–125. [Google Scholar] [CrossRef]
- Food and Agriculture Organization (FAO). Soils Map of the World: Revised Legend; Food and Agriculture Organization of the United Nations: Rome, Italy, 1988; p. 119. [Google Scholar]
- Han, X.; Cheng, Z.; Meng, H. Soil properties, nutrient dynamics, and soil enzyme activities associated with garlic stalk decomposition under various conditions. PLoS ONE 2012, 7, e50868. [Google Scholar] [CrossRef]
- Nelson, D.W.; Sommers, L.E. Total carbon, organic carbon, and organic matter. In Methods of Soil Analysis Part 3-Chemical Methods; John Wiley & Sons: Madison, WI, USA, 1996; pp. 961–1010. [Google Scholar]
- Jackson, M.L. Soil Chemical Analysis; Constable and Co. Ltd.: London, UK, 1962. [Google Scholar]
- Bao, S.D. Soil Agricultural Chemistry Analysis; China Agriculture Press: Beijing, China, 2000; pp. 100–109. [Google Scholar]
- Guan, S.Y. Soil Enzyme and Its Research Methods; Agricultural Press: Beijing, China, 1986. [Google Scholar]
- Margesin, R. Acid and alkaline phosphomonoesterase activity with the substrate p-nitrophenyl phosphate. In Methods in Soil Biology; Schinner, F., Öhlinger, R., Kandeler, E., Margesin, R., Eds.; Springler: Berlin, Germany, 1997; pp. 213–217. [Google Scholar]
- Gao, J.F. Experimental Guidance for Plant Physiology; Higher Education Press: Beijing, China, 2006. [Google Scholar]
| Plant Growth Indexes | CK | T1 (1:100) | T2 (3:100) | T3 (5:100) |
|---|---|---|---|---|
| Plant height (cm) | 75.16 ± 8.34 b | 81.58 ± 3.33 b | 88.27 ± 1.46 ab | 94.53 ± 4.23 a |
| Leaf area (cm−2) | 163.05 ± 7.48 b | 166.82 ± 3.73 b | 177.65 ± 3.39 b | 219.91 ± 4.69 a |
| Shoot fresh weight (g/plant) | 75.84 ± 10.92 b | 72.51 ± 8.27 b | 91.79 ± 3.44 ab | 107.85 ± 4.64 a |
| Shoot dry weight (g/plant) | 12.47 ± 2.90 b | 10.94 ± 2.74 b | 16.69 ± 0.99 a | 17.21 ± 0.98 a |
| Root fresh weight (g/plant) | 5.90 ± 0.65 bc | 5.02 ± 0.32 c | 8.97 ± 0.88 a | 7.13 ± 0.31 ab |
| Root dry weight (g/plant) | 0.86 ± 0.14 b | 0.87 ± 0.06 b | 1.87 ± 0.12 a | 1.45 ± 0.23 a |
| Fruit fresh weight (g/plant) | 171.45 ± 6.39 b | 163.74 ± 5.49 b | 213.98 ± 5.82 a | 197.30 ± 9.41 a |
| Fruit length (cm) | 31.60 ± 1.16 a | 32.75 ± 1.43 a | 34.74 ± 1.03 a | 35.01 ± 1.53 a |
| Cucumber yield (g/plant) | 725.12 ± 7.72 c | 749.50 ± 16.76 c | 822.8 ± 9.25 b | 876.92 ± 6.00 a |
| Fusarium wilt incidence % | 33.63 ± 0.52 c | 30.12 ± 1.21 b | 26.53 ± 2.52 ab | 21.53 ± 1.32 a |
| Soil Properties | CK | T1 (1:100) | T2 (3:100) | T3 (5:100) |
|---|---|---|---|---|
| Soil pH (1:5 soil: Water) | 7.75 ± 0.04 b | 7.94 ± 0.01 a | 7.80 ± 0.04 b | 7.63 ± 0.02 c |
| EC (µs·cm−1) | 588.56 ± 2.93 bc | 581.12 ± 1.81 c | 595.57 ± 0.11 b | 611.59 ± 1.09 a |
| Organic matter (g·kg−1) | 25.70 ± 2.93 c | 28.16 ± 1.81 bc | 31.94 ± 0.11 ab | 35.89 ± 1.09 a |
| Available N (mg·kg−1) | 75.16 ± 1.97 c | 94.64 ± 2.15 b | 107.83 ± 1.07 a | 113.39 ± 1.46 a |
| Available P (mg·kg−1) | 72.83 ± 3.07 c | 119.26 ± 2.34 b | 129.83 ± 1.09 a | 136.61 ± 2.91 a |
| Available K (mg·kg−1) | 333.18 ± 1.46 c | 348 ± 1.60 b | 355.83 ± 1.83 a | 346.35 ± 3.31 b |
| Soil invertase (glucose mg·g−1 soil d−1) | 27.3 ± 1.97 b | 31.08 ± 1.47 b | 32.64 ± 1.14 b | 39.31 ± 1.94 a |
| Urease (NH3-N mg·g−1 soil d−1) | 3.06 ± 0.23 c | 4.27 ± 0.39 b | 4.69 ± 0.44 b | 6.71 ± 0.23 a |
| Catalase (KMnO4 mL·g−1 20 min−1) | 10.71 ± 0.64 b | 11.08 ± 0.81 b | 12.78 ± 0.23 ab | 13.96 ± 0.97 a |
| Alkaline phosphatase (P2O5 mg 100 g−1 soil d−1) | 1.56 ± 0.10 c | 1.93 ± 0.26 bc | 2.28 ± 0.10 b | 3.35 ± 0.27 a |
| Treat-Ments | Bacterial 16S rRNA | Fungal 18S rRNA | ||||||
|---|---|---|---|---|---|---|---|---|
| OTUs | ACE | Chao1 | Shannon | OTUs | ACE | Chao1 | Shannon | |
| CK | 2190 ± 24.36 ab | 2601 ± 13.09 b | 2612 ± 13.5 b | 6.66 ± 0.05 ab | 110.9 ± 7.24 bc | 125 ± 7.0 b | 124 ± 8.08 b | 2.13 ± 0.23 ab |
| T1 | 2232 ± 23.48 ab | 2625 ± 24.42 ab | 2637 ± 23.11ab | 6.59 ± 0.08 ab | 141.3 ± 11.31 a | 153.3 ± 8.37 a | 155 ± 10.40 a | 2.45 ± 0.38 ab |
| T2 | 2268 ± 47.92 a | 2680 ± 21.36 a | 2691 ± 16.55 a | 6.98 ± 0.03 a | 141.7 ± 6.43 a | 150.3 ± 6.11 a | 152.7 ± 7.35 a | 2.89 ± 0.24 a |
| T3 | 2134 ± 26.74 b | 2627 ± 17.98 ab | 2605 ± 26.92 b | 6.32 ± 0.43 b | 128.7 ± 13.9 ab | 151 ± 4.93 a | 147 ± 7.0 ab | 2.08 ± 0.19 b |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, A.; Ghani, M.I.; Haiyan, D.; Iqbal, M.; Cheng, Z.; Cai, Z. Garlic Substrate Induces Cucumber Growth Development and Decreases Fusarium Wilt through Regulation of Soil Microbial Community Structure and Diversity in Replanted Disturbed Soil. Int. J. Mol. Sci. 2020, 21, 6008. https://doi.org/10.3390/ijms21176008
Ali A, Ghani MI, Haiyan D, Iqbal M, Cheng Z, Cai Z. Garlic Substrate Induces Cucumber Growth Development and Decreases Fusarium Wilt through Regulation of Soil Microbial Community Structure and Diversity in Replanted Disturbed Soil. International Journal of Molecular Sciences. 2020; 21(17):6008. https://doi.org/10.3390/ijms21176008
Chicago/Turabian StyleAli, Ahmad, Muhammad Imran Ghani, Ding Haiyan, Muhammad Iqbal, Zhihui Cheng, and Zucong Cai. 2020. "Garlic Substrate Induces Cucumber Growth Development and Decreases Fusarium Wilt through Regulation of Soil Microbial Community Structure and Diversity in Replanted Disturbed Soil" International Journal of Molecular Sciences 21, no. 17: 6008. https://doi.org/10.3390/ijms21176008
APA StyleAli, A., Ghani, M. I., Haiyan, D., Iqbal, M., Cheng, Z., & Cai, Z. (2020). Garlic Substrate Induces Cucumber Growth Development and Decreases Fusarium Wilt through Regulation of Soil Microbial Community Structure and Diversity in Replanted Disturbed Soil. International Journal of Molecular Sciences, 21(17), 6008. https://doi.org/10.3390/ijms21176008