Lithium Chloride Sensitivity in Yeast and Regulation of Translation
Abstract
1. Introduction
2. Results
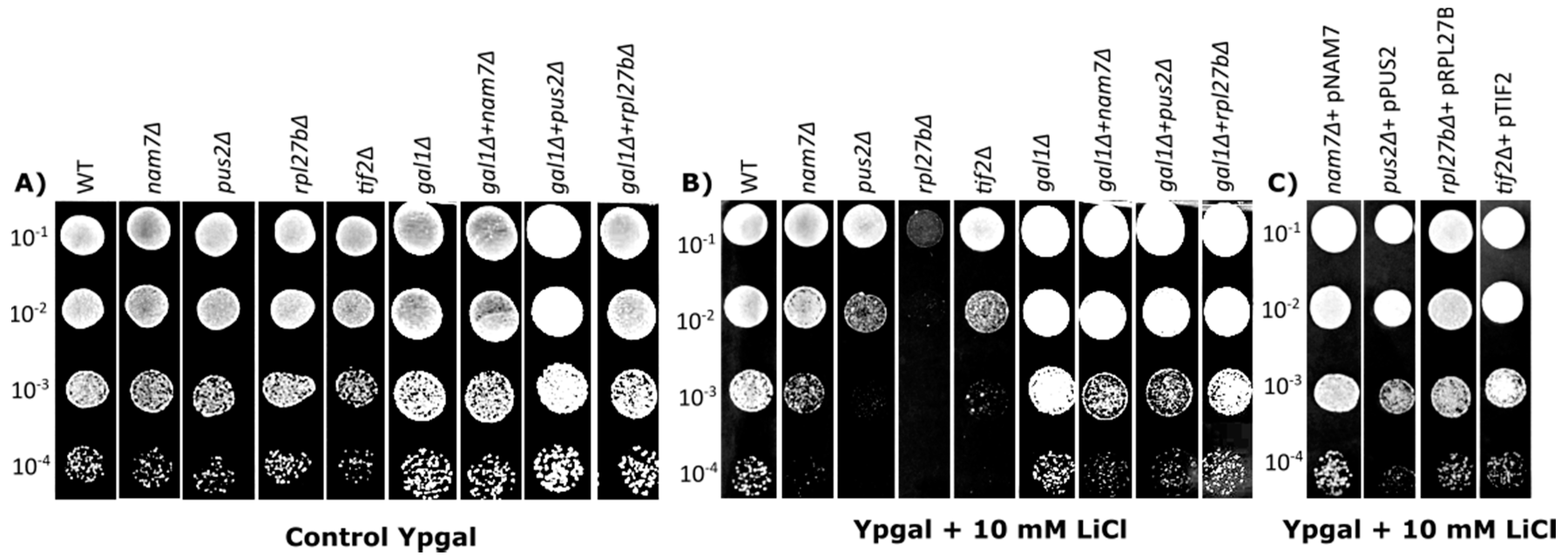
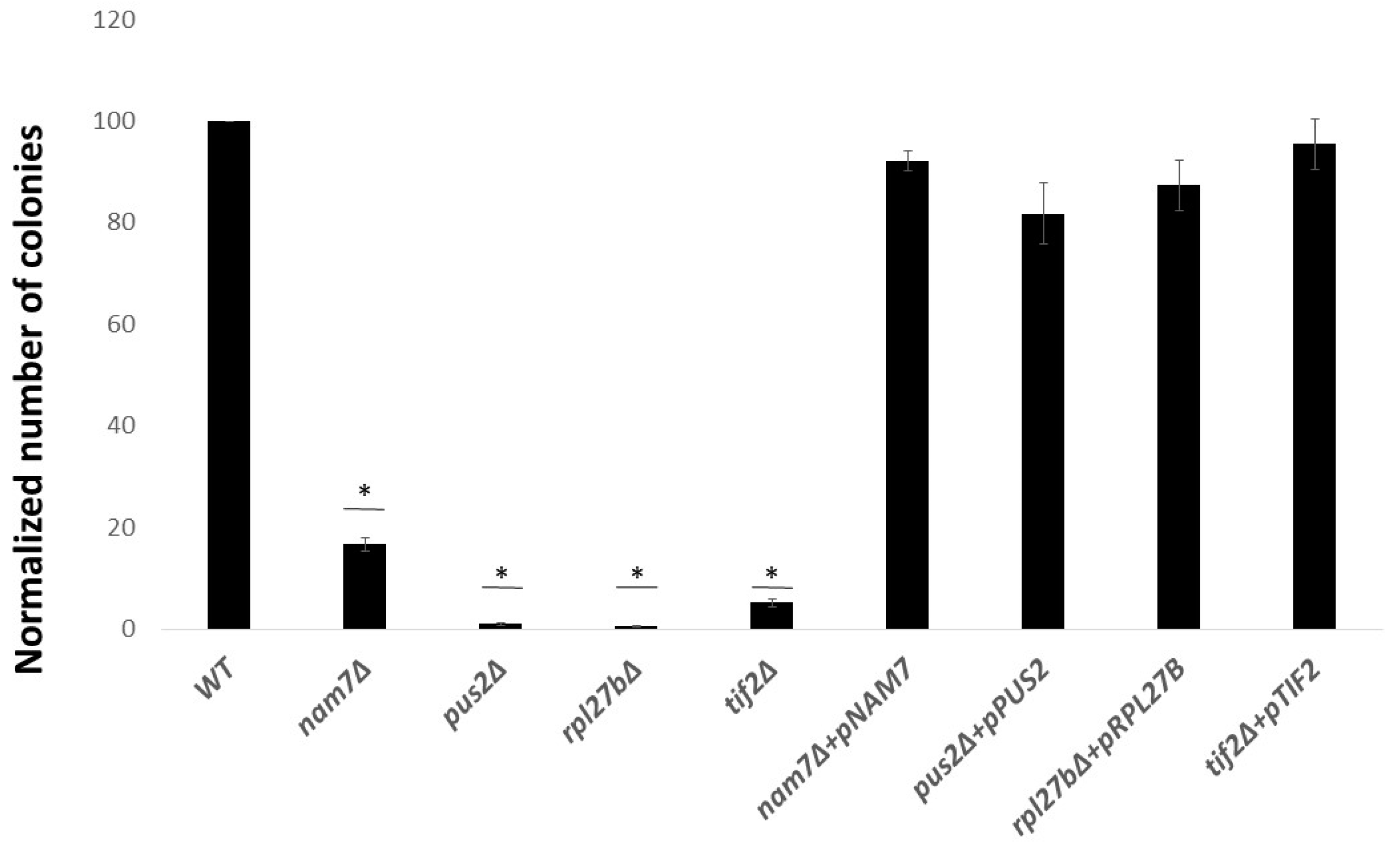
2.1. Deletion of NAM7, PUS2, and RPL27B Increases Yeast Sensitivity to Lithium Chloride
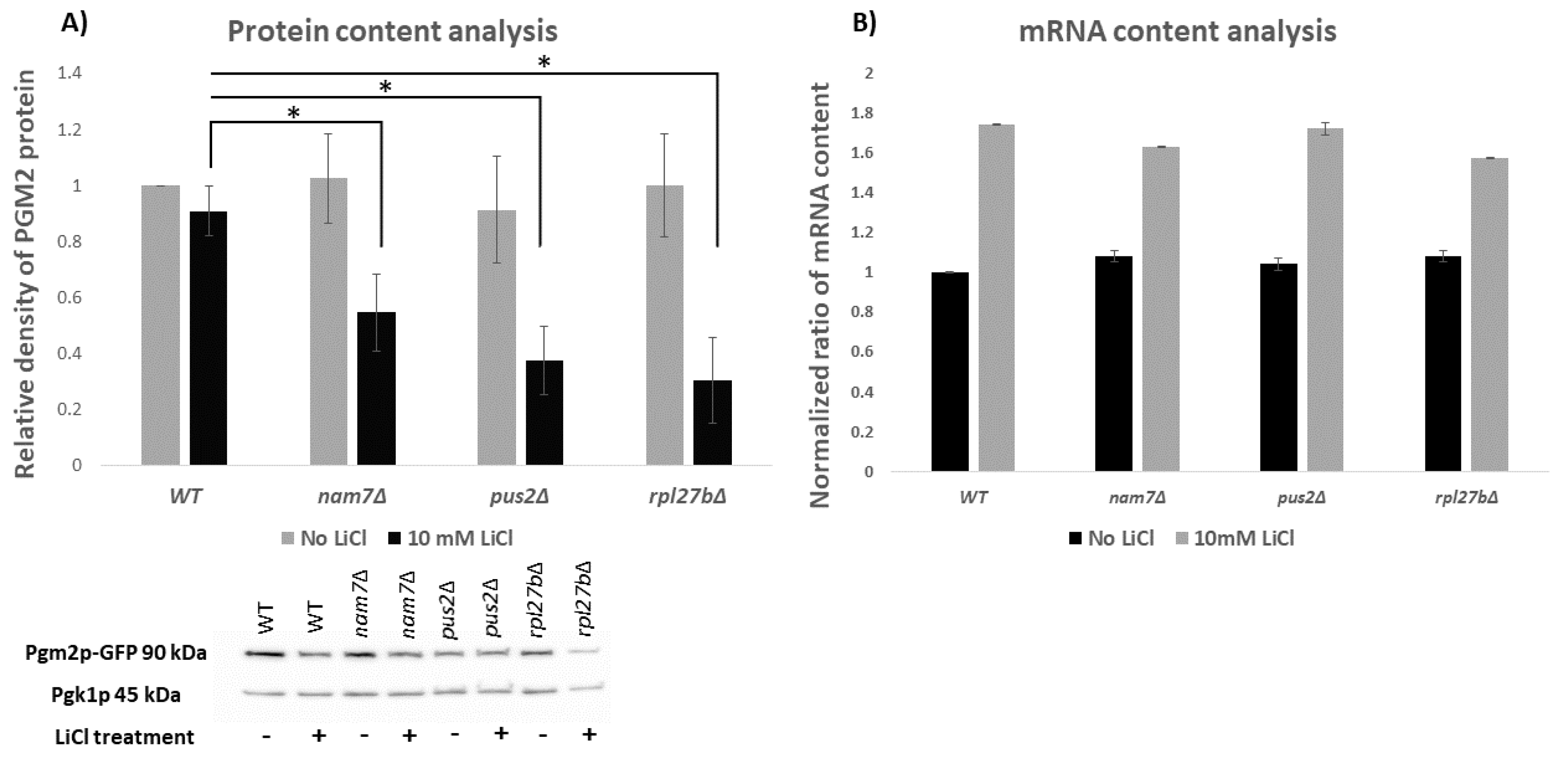
2.2. NAM7, PUS2, and RPL27B Regulate the Expression of PGM2 at the Level of Translation
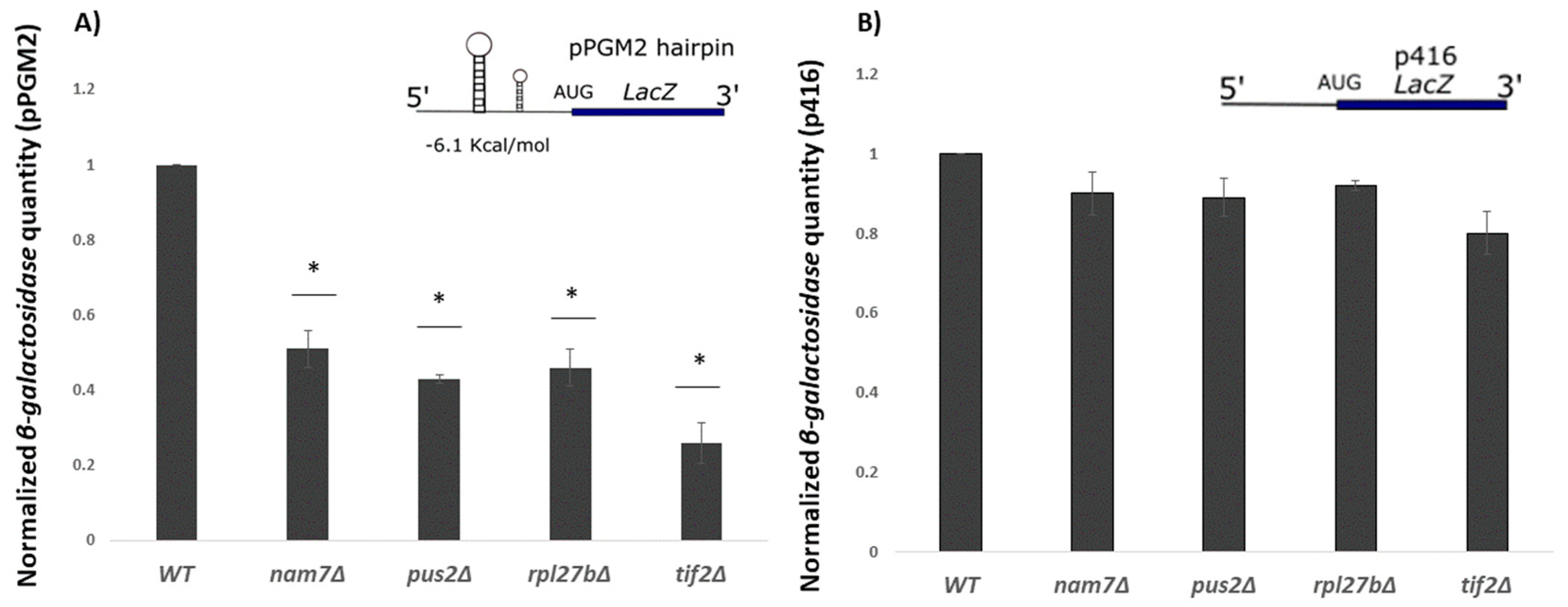
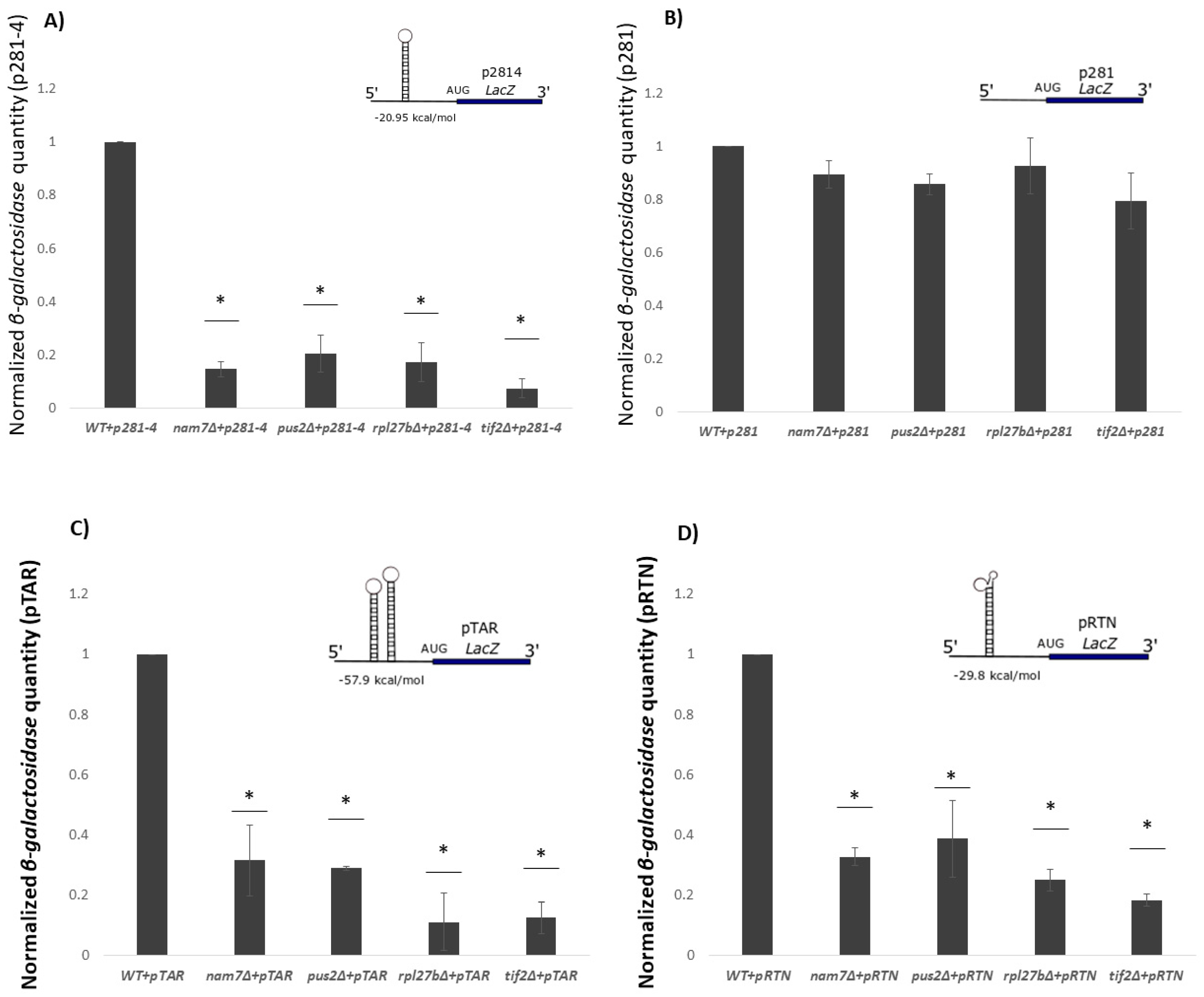
2.3. Translation of β-Galactosidase Reporter mRNA with a Hairpin Structure Is Altered by Deletion of NAM7, PUS2, and RPL27B
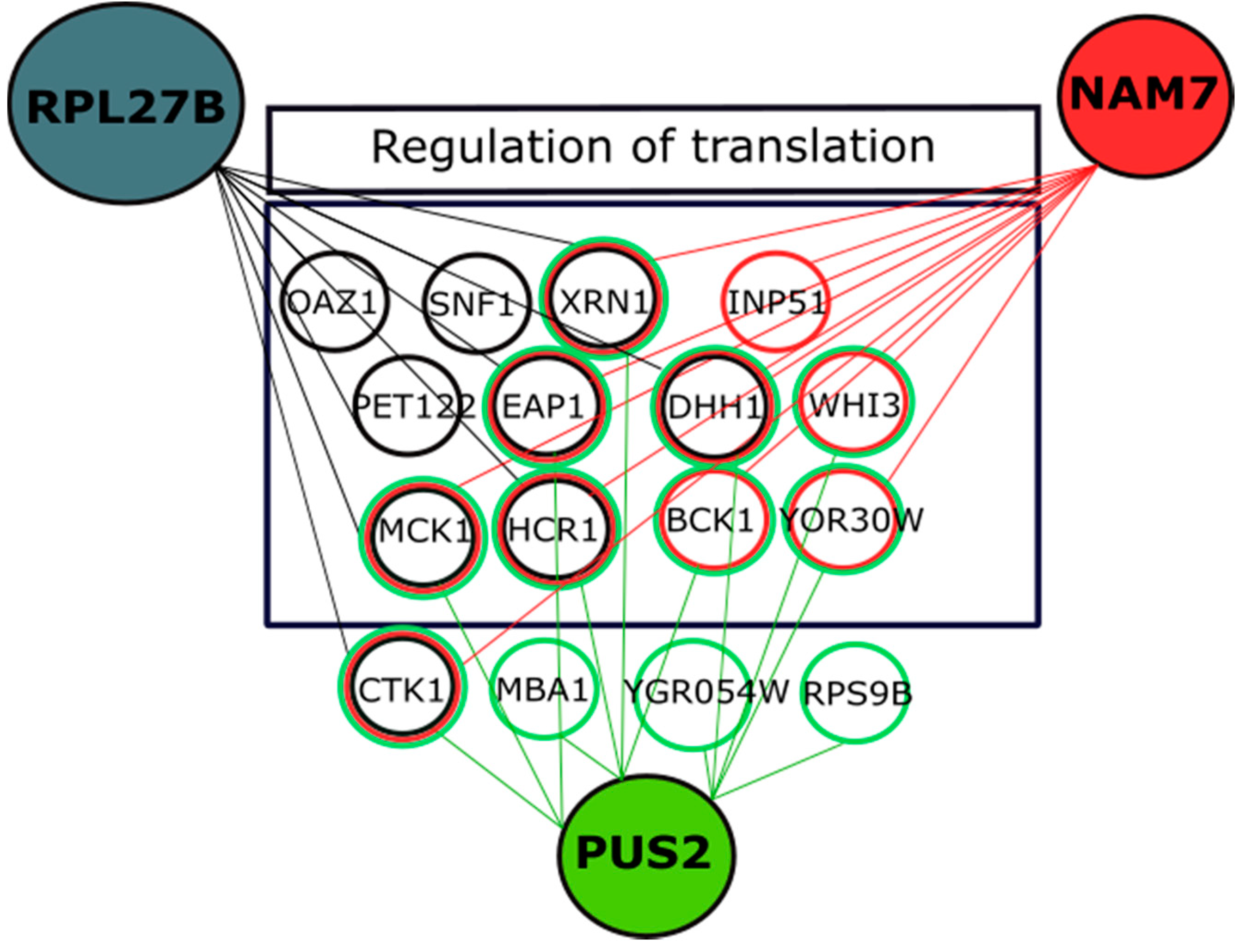
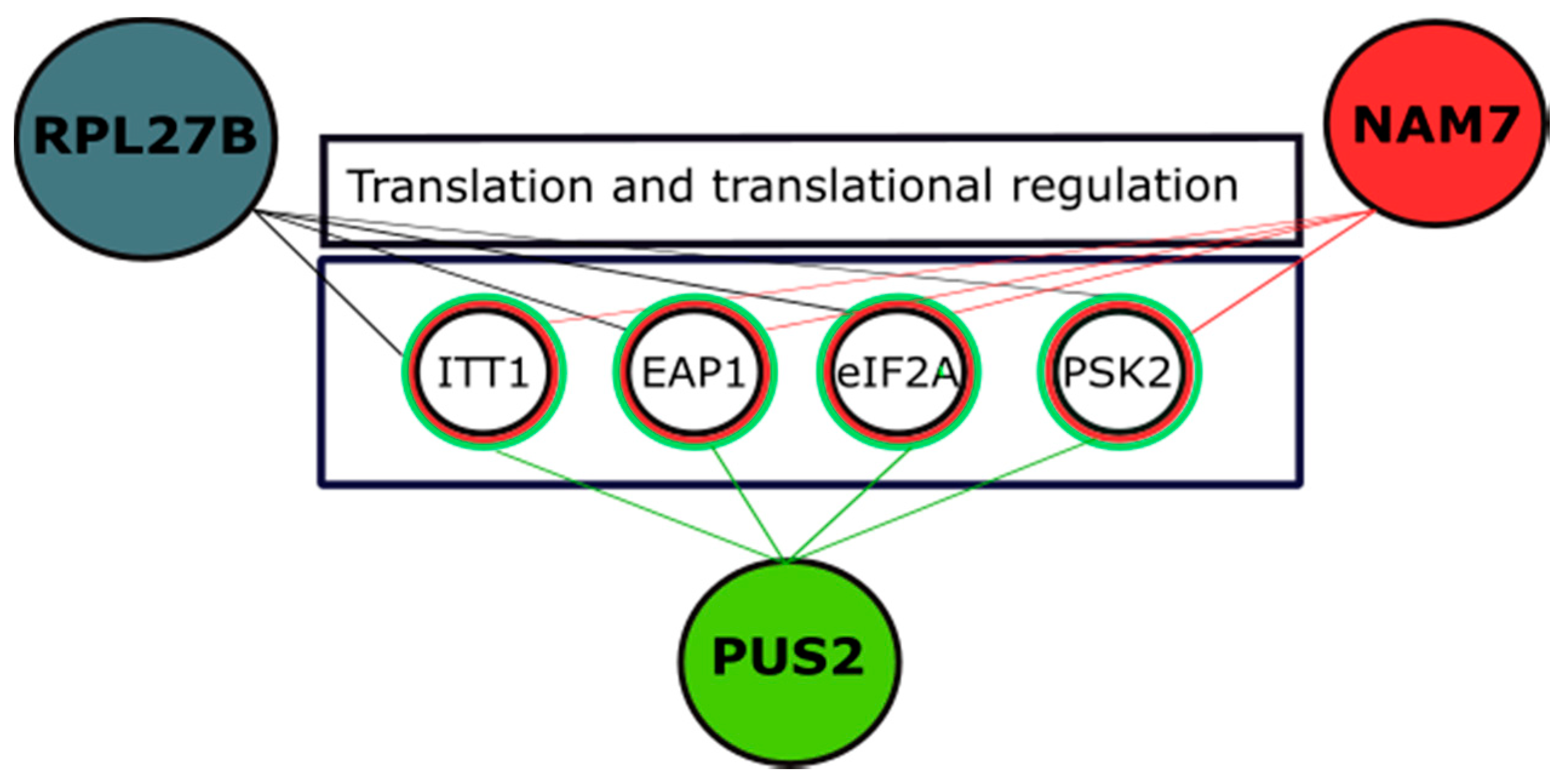
2.4. Genetic Interaction Analysis Further Connects the Activity of NAM7, PUS2, and RPL27B to Protein Biosynthesis
3. Discussion
4. Materials and Methods
4.1. Strains, Plasmids, Gene Collections, and Cell and DNA Manipulations
4.2. Chemical Sensitivity Analysis
4.3. Quantitative β-Galactosidase Assay
4.4. Quantitative Real Time PCR (qRT-PCR)
4.5. Western Blot Analysis
4.6. Genetic Interaction Analysis
4.7. Genetic Interaction Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s Disease |
| BD | Bipolar Disorder |
| LiCl | Lithium Chloride |
| WT | Wild Type |
| ORF | Open Reading Frame |
| ONPG | O-nitrophenyl-α-d-galactopyranoside |
| NAT | Nourseothricin Sulfate |
| SGA | Synthetic Genetic Array |
| PSA | Phenotypic Suppression Array |
| GO | Gene Ontology |
| YKO | yeast non-essential gene knockout mutant library |
| PI/PKC | phosphoinositide/Protein Kinase C signaling |
| UDP-Glucose | Uridine diphosphate glucose |
| Ypgal | Yeast extract, Peptone, and galactose |
| 5′-UTR | 5′-Untranslated Region |
References
- Yan, P.; Xu, D.; Ji, Y.; Yin, F.; Cui, J.; Su, R.; Wang, Y.; Zhu, Y.; Wei, S.; Lai, J. LiCl Pretreatment Ameliorates Adolescent Methamphetamine Exposure-Induced Long-Term Alterations in Behavior and Hippocampal Ultrastructure in Adulthood in Mice. Int. J. Neuropsychopharmacol. 2019, 22, 303–316. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.-T.; Chuang, D.-M. Molecular actions and therapeutic potential of lithium in preclinical and clinical studies of CNS disorders. Pharmacol Ther. 2011, 128, 281–304. [Google Scholar] [CrossRef] [PubMed]
- Ariyasinghe, D.; Perera, S.R. The role of lithium in the treatment of bipolar disorder, Sri Lanka. J. Psychiatry 2018, 28–30. [Google Scholar] [CrossRef][Green Version]
- Hull, M.; Lee, E.; Lee, T.; Anand, N.; Lalone, V.; Parameswaran, N. Lithium chloride induces TNFα in mouse macrophages Via MEK-ERK-dependent pathway. J. Cell. Biochem. 2014, 115, 71–80. [Google Scholar] [CrossRef]
- Mu, F.; Huang, J.; Xing, T.; Jing, Y.; Cui, T.; Guo, Y.; Yan, X.; Li, H.; Wang, N. The Wnt/β-catenin/Lef1 pathway promotes cell proliferation at least in part through direct upregulation of miR-17-92 cluster. Front. Genet. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Won, E.; Kim, Y. An Oldie but Goodie: Lithium in the Treatment of Bipolar Disorder through Neuroprotective and Neurotrophic Mechanisms. Int. J. Mol. Sci. 2017, 18, 2679. [Google Scholar] [CrossRef]
- Sofola-adesakin, O.; Castillo-quan, J.; Rallis, C.; Tain, L.; Bjedov, I.; Rogers, I.; Li, L.; Martinez, P.; Khericha, M.; Cabecinha, M.; et al. Lithium suppresses A β pathology by inhibiting translation in an adult Drosophila model of Alzheimer ’ s disease. Front. Aging Neurosci. 2014, 6, 1–10. [Google Scholar] [CrossRef]
- Zhang, J. Lithium chloride promotes proliferation of neural stem cells in vitro, possibly by triggering the Wnt signaling pathway. Taylor Fr. Gr. 2019, 23, 32–41. [Google Scholar] [CrossRef]
- Williams, R.S.; Harwood, A. Lithium therapy and signal transduction. Trends Pharmacol. Sci. 2000, 21, 61–64. [Google Scholar] [CrossRef]
- Hampel, H.; Ewers, M.; Bürger, K.; Annas, P.; Mörtberg, A.; Bogstedt, A.; Frölich, L.; Schröder, J.; Schönknecht, P.; Riepe, M.W.; et al. Lithium trial in Alzheimer’s disease: A randomized, single-blind, placebo-controlled, multicenter 10-week study. J. Clin. Psychiatry. 2009, 70, 922–931. [Google Scholar] [CrossRef]
- Castillo-Quan, J.; Li, L.; Kinghorn, K.; Hardy, J.; Bjedov, I.; Partridge, L. Lithium Promotes Longevity through GSK3/NRF2- Dependent Hormesis Lithium Promotes Longevity. Cell Rep. 2016, 15, 638–650. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Song, L.; Hu, J.; Yin, W.; Li, Z.; Chen, Y.; Su, X.; Wang, R.; Hu, Z. Biochemical and Biophysical Research Communications Enhanced tolerance to NaCl and LiCl stresses by over-expressing Caragana korshinskii sodium/proton exchanger 1 (CkNHX1) and the hydrophilic C terminus is required for the activity of CkNHX1 in Atsos3. Biochem. Biophys. Res. Commun. 2012, 417, 732–737. [Google Scholar] [CrossRef] [PubMed]
- Lenox, R.H.; Wang, L. Molecular basis of lithium action: Integration of lithium-responsive signaling and gene expression networks. Mol. Psychiatry. 2003, 8, 135–144. [Google Scholar] [CrossRef]
- Montero-Lomelí, M.; Morais, B.L.B.; Figueiredo, D.L.; Neto, D.C.S.; Martins, J.R.P.; Masuda, C.A. The initiation factor eIF4A is involved in the response to lithium stress in Saccharomyces cerevisiae. J. Biol. Chem. 2002, 277, 21542–21548. [Google Scholar] [CrossRef]
- Masuda, C.A.; Xavier, M.A.; Mattos, K.A.; Galina, A.; Montero-Lomeli, M. Phosphoglucomutase Is an in Vivo Lithium Target in Yeast. Biol. Chem. 2001, 276, 37794–37801. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, G.C.; Kong, I.; Yun, E.; Zheng, J.; Kweon, D.; Jin, Y. A mutation in PGM2 causing inefficient Galactose metabolism in the Probiotic Yeast Saccharomyces boulardii. Appl. Environ. Microbiol. 2018, 84, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bergkessel, M.; Whitworth, G.; Guthrie, C. Diverse environmental stresses elicit distinct responses at the level of pre-mRNA processing in yeast. RNA 2011, 17, 1461–1478. [Google Scholar] [CrossRef]
- Dichtl, B.; Stevens, A.; Tollervey, D. Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J. 1997, 16, 7184–7195. [Google Scholar] [CrossRef]
- Asensio, J.; Ruiz-Argüeso, T. Rodríguez-Navarro, A. Sensitivity of yeasts to lithium. Antonie Van Leeuwenhoek. 1976, 42, 1–8. [Google Scholar] [CrossRef]
- Zhao, J.; Lin, W.; Ma, X.; Lu, Q.; Ma, X.; Bian, G.; Jiang, L. The protein kinase Hal5p is the high-copy suppressor of lithium-sensitive mutations of genes involved in the sporulation and meiosis as well as the ergosterolbiosynthesis in Saccharomyces cerevisiae. Genomics 2010, 95, 290–298. [Google Scholar] [CrossRef]
- Asghar, F.; Yan, H.; Jiang, L. The putative transcription factor CaMaf1 controls the sensitivity to lithium and rapamycin and represses RNA polymerase III transcription in Candida albicans. FEMS Yeast Res. 2018, 18, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lopez, F.; Leube, M.; Gil-Mascarell, R.; Navarro-Aviñó, J.P.; Serrano, R. The yeast inositol monophosphatase is a lithium- and sodium-sensitive enzyme encoded by a non-essential gene pair. Mol. Microbiol. 1999, 31, 1255–1264. [Google Scholar] [CrossRef] [PubMed]
- Hajikarimlou, M.; Moteshareie, H.; Omidi, K.; Hooshyar, M.; Shaikho, S.; Kazmirchuk, T.; Burnside, D.; Takallou, S.; Zare, N.; Jagadeesan, S.K.; et al. Sensitivity of yeast to lithium chloride connects the activity of YTA6 and YPR096C to translation of structured mRNAs. PLoS ONE 2020, 15, e0235033. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Cao, C.; Jiang, L. Genome-scale genetic screen of lead ion-sensitive gene deletion mutations in Saccharomyces cerevisiae. Gene 2015, 563, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Cao, C.; Liu, Y.; Wang, J.; Li, S.; Li, J.; Deng, Y. Genetic analysis of oxidative and endoplasmic reticulum stress responses induced by cobalt toxicity in budding yeast. Biochim. Biophys. Acta Gen. Subj. 2020, 1864, 129516. [Google Scholar] [CrossRef]
- Moteshareie, H.; Hajikarimlou, M.; Indrayanti, A.M.; Burnside, D.; Paula, A.; Id, D.; Lettl, C.; Ahmed, D.; Omidi, K.; Id, T.K.; et al. Heavy metal sensitivities of gene deletion strains for ITT1 and RPS1A connect their activities to the expression of URE2, a key gene involved in metal detoxification in yeast. PLoS ONE 2018, 13, e0198704. [Google Scholar] [CrossRef]
- Alamgir, M.; Jessulat, M.; Azizi, A.; Golshani, A. Chemical-genetic profile analysis of five inhibitory compounds in yeast. BMC Chem. Biol. 2010, 10. [Google Scholar] [CrossRef]
- Omidi, K.; Jessulat, M.; Hooshyar, M.; Burnside, D.; Schoenrock, A.; Kazmirchuk, T.; Hajikarimlou, M.; Daniel, M.; Moteshareie, H.; Bhojoo, U.; et al. Uncharacterized ORF HUR1 influences the efficiency of non-homologous end-joining repair in Saccharomyces cerevisiae. Gene 2018, 639, 128–136. [Google Scholar] [CrossRef]
- Bro, C.; Regenberg, B.; Lagniel, G.; Labarre, J.; Montero-Lomelí, M.; Nielsen, J. Transcriptional, proteomic, and metabolic responses to lithium in galactose-grown yeast cells. J. Biol. Chem. 2003, 278, 32141–32149. [Google Scholar] [CrossRef]
- Nagalakshmi, U.; Wang, Z.; Waern, K.; Shou, C.; Raha, D.; Gerstein, M.; Synder, M. The Transcriptional Landscape of the Yeast Genome Defined by RNA Sequencing. Science 2008, 320, 1344–1350. [Google Scholar] [CrossRef]
- Altmann, M.; Müller, P.; Wittmer, B.; Ruchti, F.; Lanker, S.; Trachsel, H. A Saccharomyces cerevisiae homologue of mammalian translation initiation factor 4B contributes to RNA helicase activity. EMBO J. 1993, 12, 3997–4003. [Google Scholar] [CrossRef] [PubMed]
- Bolinger, C.; Sharma, A.; Singh, D.; Yu, L. RNA helicase A modulates translation of HIV-1 and infectivity of progeny virions. Nucleic Acids Res. 2010, 38, 1686–1696. [Google Scholar] [CrossRef]
- Washietl, S.; Hofacker, I.L.; Lukasser, M.; Hüttenhofer, A.; Stadler, P.F. Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat. Biotechnol. 2005, 23, 1383–1390. [Google Scholar] [CrossRef] [PubMed]
- Wagih, O.; Usaj, M.; Baryshnikova, A.; Vandersluis, B.; Kuzmin, E.; Costanzo, M.; Myers, C.L.; Andrews, B.J.; Boone, C.M.; Parts, L. SGAtools: One-stop analysis and visualization of array-based genetic interaction screens. Nucleic Acids Res. 2013, 41, 591–596. [Google Scholar] [CrossRef] [PubMed]
- Jessulat, M.; Malty, R.H.; Nguyen-tran, D.; Deineko, V.; Aoki, H.; Vlasblom, J.; Omidi, K. Spindle Checkpoint Factors Bub1 and Bub2 Promote DNA DoubleStrand Break Repair by Nonhomologous End Joining. Mcb 2015, 35, 2448–2463. [Google Scholar] [CrossRef]
- Samanfar, B.; Shostak, K.; Moteshareie, H.; Hajikarimlou, M.; Shaikho, S.; Omidi, K.; Hooshyar, M.; Burnside, D.; Márquez, I.G.; Kazmirchuk, T.; et al. The sensitivity of the yeast, Saccharomyces cerevisiae, to acetic acid is influenced by DOM34 and RPL36A. PeerJ 2017, 2017. [Google Scholar] [CrossRef][Green Version]
- Tong, A.; Evangelista, M.; Parsons, A.B.; Xu, H.; Bader, G.D.; Page, N.; Robinson, M.; Raghibizadeh, S.; Hogue, C.W.V.; Bussey, H.; et al. Systematic Genetic Analysis with Ordered Arrays of Yeast Deletion Mutants. Science 2001, 294, 2364–2369. [Google Scholar] [CrossRef]
- Toufighi, K.; Youn, J.; Ou, J.; Luis, B.S.; Hibbs, M.; Hess, D.; Gingras, A.; Bader, G.D.; Troyanskaya, O.G.; Brown, G.W.; et al. Quantitative analysis of fitness and genetic interactions in yeast on a genome scale. Nat. Methods 2011, 7, 1017–1024. [Google Scholar] [CrossRef]
- Karásková, M.; Gunis, S.; Herrmannová, A.; Wagner, S.; Munzarová, V. Functional Characterization of the Role of the N-terminal Domain of the c/Nip1 Subunit of Eukaryotic Initiation Factor 3 (eIF3) in AUG Recognition. JBC 2012, 287, 28420–28434. [Google Scholar] [CrossRef]
- Firczuk, H.; Kannambath, S.; Pahle, J.; Claydon, A.; Beynon, R.; Duncan, J.; Westerhoff, H.; Mendes, P.; McCarthy, J.E. An in vivo control map for the eukaryotic mRNA translation machinery. Mol. Syst. Biol. 2013, 9, 1–13. [Google Scholar] [CrossRef]
- Pisareva, V.P.; Pisarev, A.V. DHX29 and eIF3 cooperate in ribosomal scanning on structured mRNAs during translation initiation. RNA 2016, 22, 1859–1870. [Google Scholar] [CrossRef] [PubMed]
- Cuchalova, L.; Kouba, T.; Herrmannova, A.; Danyi, I.; Chiu, W.-L.; Valasek, L. The RRM of eIF3g is required for resumption of scanning of post-termination ribosomes for reinitiation on GCN4 and together with eIF3i stimulates linear scanning. Mol. Cell. Biol. 2010, 30, 4671–4686. [Google Scholar] [CrossRef] [PubMed]
- Valášek, L.; Phan, L.; Schoenfeld, L.W.; Valášková, V.; Hinnebusch, A.G. Related eIF3 subunits TIF32 and HCR1 interact with an RNA recognition motif in PRT1 required for eIF3 integrity and ribosome binding. EMBO J. 2001, 20, 891–904. [Google Scholar] [CrossRef] [PubMed]
- Blasco-moreno, B.; De Campos-mata, L.; Böttcher, R.; García-martínez, J.; Jung, J.; Nedialkova, D.D.; Chattopadhyay, S.; Gas, M.; Oliva, B.; Pérez-ortín, J.E.; et al. The exonuclease Xrn1 activates transcription and translation of mRNAs encoding membrane proteins. Nat. Commun. 2019, 1–15. [Google Scholar] [CrossRef]
- Hinnebusch, A.G. Molecular Mechanism of Scanning and Start Codon Selection in Eukaryotes. Microbiol. Mol. Biol. Rev. 2011, 75, 434–467. [Google Scholar] [CrossRef]
- Sen, N.D.; Zhou, F.; Ingolia, N.T.; Hinnebusch, A.G. Genome-wide analysis of translational efficiency reveals distinct but overlapping functions of yeast DEAD-box RNA helicases Ded1 and eIF4A. Genome Res. 2015, 25, 1196–1205. [Google Scholar] [CrossRef] [PubMed]
- Murvin, M.; Banruo, Y.; Alec, D.; Christian, F.; Angie, F. Methylation of Ded1 Affects Its Role in Translation. FASEB J. 2019. Available online: https://www.fasebj.org/doi/abs/10.1096/fasebj.2019.33.1_supplement.629.5 (accessed on 20 January 2020). [CrossRef]
- Minshall, N.; Kress, M.; Weil, D.; Standart, N. Role of p54 RNA Helicase Activity and Its C-terminal Domain in Translational Repression, P-body Localization and Assembly. Mol. Biol. Cell. 2009, 20, 2464–2472. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Minami, M.; Suzuki, M.; Abe, K.; Zenno, S.; Tsujimoto, M.; Matsumoto, K.; Minami, Y. The Hsp90 Inhibitor Geldanamycin Abrogates Colocalization of eIF4E and eIF4E-Transporter into Stress Granules and Association of eIF4E with eIF4G. J. Biol. Chem. 2009, 284, 35597–35604. [Google Scholar] [CrossRef] [PubMed]
- Omidi, K.; Hooshyar, M.; Jessulat, M.; Samanfar, B.; Sanders, M.; Burnside, D.; Pitre, S.; Schoenrock, A.; Xu, J.; Babu, M.; et al. Phosphatase Complex Pph3/Psy2 Is Involved in Regulation of Efficient Non-Homologous End-Joining Pathway in the Yeast Saccharomyces cerevisiae. PLoS ONE 2014, 9, e87248. [Google Scholar] [CrossRef][Green Version]
- Fischer, N.; Weis, K. The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J. 2002, 21, 2788–2797. [Google Scholar] [CrossRef] [PubMed]
- Carroll, J.S.; Munchel, S.E.; Weis, K. The DExD/H box ATPase Dhh1 functions in translational repression, mRNA decay, and processing body dynamics. Cell Biol. 2011, 194, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Blewett, N.H.; Goldstrohm, A.C. A Eukaryotic Translation Initiation Factor 4E-Binding Protein Promotes mRNA Decapping and Is Required for PUF Repression. Mcb 2012, 32, 4181–4194. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ohmori, T.; Irie, K.; Kimura, Y.; Suda, Y.; Mizuno, T.; Ire, K. Different Regulations of ROM2 and LRG1 Expression by Ccr4, Pop2, and Dhh1 in the Saccharomyces cerevisiae Cell Wall Integrity Pathway. MSphere 2016, 1. [Google Scholar] [CrossRef] [PubMed]
- Coordes, B. Ctk1 Function is Ccrucial for Efficient Translation Initiation and Interacts with the mRNP Processing Factor Npl3. Ph.D. Thesis, University of Munich, Munich, Germany, 2011. Available online: https://edoc.ub.uni-muenchen.de/13203/1/Coordes_Britta.pdf (accessed on 20 January 2020).
- Coordes, B.; Brünger, K.M.; Burger, K.; Soufi, B.; Horenk, J.; Eick, D. Ctk1 Function Is Necessary for Full Translation Initiation Activity in Saccharomyces cerevisiae. Am. Soc. Microbiol. 2015, 14, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Boone, C.; Bussey, H.; Andrews, B.J. Exploring genetic interactions and networks with yeast. Nat. Rev. Genet. 2007, 8, 437–449. [Google Scholar] [CrossRef]
- Douglas, A.C.; Smith, A.M.; Sharifpoor, S.; Yan, Z.; Durbic, T.; Heisler, L.E.; Lee, A.Y.; Ryan, O.; Göttert, H.; Surendra, A.; et al. Functional Analysis With a Barcoder Yeast Gene Overexpression System. G3 Genes Genomes Genet. 2012, 2, 1279–1289. [Google Scholar] [CrossRef]
- Kodama, H.; Ito, K.; Nakamura, Y. The role of N-terminal domain of translational release factor eRF3 for the control of functionality and stability in S. cerevisiae. Genes Cells 2007, 12, 639–650. [Google Scholar] [CrossRef]
- Xiao, R.; Gao, Y.; Shen, Q.; Li, C.; Chang, W.; Chai, B. Polypeptide chain release factor eRF3 is a novel molecular partner Of survivin. Cell Biol. Int. 2013, 37, 359–369. [Google Scholar] [CrossRef]
- Reineke, L.C.; Cao, Y.; Baus, D.; Hossain, N.M.; Merrick, W.C. Insights into the role of yeast eiF2A in Ires-Mediated translation. PLoS ONE 2011, 6. [Google Scholar] [CrossRef]
- Rutter, J.; Probst, B.L.; Mcknight, S.L. Coordinate Regulation of Sugar Flux and Translation by PAS Kinase. Cell 2002, 111, 17–28. [Google Scholar] [CrossRef]
- Jeon, S.; Lim, S.; Ha, J.; Kim, J. Identification of Psk2, Skp1, and Tub4 as trans-acting factors for uORF-containing ROK1 mRNA in Saccharomyces cerevisiae. Microbiology 2015, 53, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Alamgir, M.; Eroukova, V.; Jessulat, M.; Xu, J.; Golshani, A. Chemical-genetic profile analysis in yeast suggests that a previously uncharacterized open reading frame, YBR261C, affects protein synthesis. BMC Genom. 2008, 9, 583. [Google Scholar] [CrossRef] [PubMed]
- Samanfar, B.; Omidi, K.; Hooshyar, M.; Laliberte, B.; Alamgir, M.; Seal, A.J.; Ahmed-Muhsin, E.; Viteri, D.F.; Said, K.; Chalabian, F.; et al. Large-scale investigation of oxygen response mutants in Saccharomyces cerevisiae. Mol. Biosyst. 2013, 9, 1351–1359. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Costanzo, M.; Heisler, L.E.; Paw, J.; Kaper, F.; Andrews, B.J.; Boone, C.; Giaever, G.; Nislow, C. Yeast Barcoders: A chemogenomic application of a universal donor-strain collection carrying bar-code identifiers. Nat. Methods 2008, 5, 719–725. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Berbenetz, N.M.; Giaever, G.; Nislow, C. Precise gene-dose alleles for chemical genetics. Genetics 2009, 182, 623–626. [Google Scholar] [CrossRef] [PubMed]
- Giaever, G.; Nislow, C. The Yeast Deletion Collection: A Decade of Functional Genomics. Genetics 2014, 197, 451–465. [Google Scholar] [CrossRef]
- Gelperin, D.M.; White, M.A.; Wilkinson, M.L.; Kon, Y.; Kung, L.A.; Wise, K.J.; Lopez-hoyo, N.; Jiang, L.; Piccirillo, S.; Yu, H.; et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 2005, 2816–2826. [Google Scholar] [CrossRef]
- Karlsson-rosenthal, C.; Millar, J.B.A. Cdc25: Mechanisms of checkpoint inhibition and recovery. Trends Cell Biol. 2006, 16. [Google Scholar] [CrossRef]
- Tong, A.H.; Lesage, G.; Bader, G.D.; Ding, H.; Xu, H.; Xin, X.; Young, J.; Berriz, G.F.; Brost, R.L.; Chang, M.; et al. Global mapping of the yeast genetic interaction network. Science 2004, 303, 808–813. [Google Scholar] [CrossRef]
- Krogan, N.J.; Kim, M.; Tong, A.; Golshani, A.; Cagney, G.; Canadien, V.; Richards, D.P.; Beattie, B.K.; Emili, A.; Boone, C.; et al. Methylation of Histone H3 by Set2 in Saccharomyces cerevisiae Is Linked to Transcriptional Elongation by RNA Polymerase II. Mcb 2003, 23, 4207–4218. [Google Scholar] [CrossRef]
- Stansfield, I.; Akhmaloka, M.; Tuite, F. A mutant allele of the SUP45 (SAL4) gene of Saccharomyces cerevisiae shows temperature-dependent aUosuppressor and omnipotent suppressor phenotypes. Curr. Genet. 1995, 27, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; . Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Szymanski, E.P.; Kerscher, O. Budding Yeast Protein Extraction and Purification for the Study of Function, Interactions, and Post-translational Modifications. J. Vis. Exp. 2013, 80, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Baryshnikova, A.; Costanzo, M.; Dixon, S.; Vizeacoumar, F.J.; Myers, C.L.; Andrews, B.; Boone, C. Synthetic Genetic Array (SGA) Analysis in Saccharomyces Cerevisiae and Schizosaccharomyces Pombe, 2nd ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2010. [Google Scholar] [CrossRef]
- Memarian, N.; Jessulat, M.; Alirezaie, J.; Mir-Rashed, N.; Xu, J.; Zareie, M.; Smith, M.; Golshani, A. Colony size measurement of the yeast gene deletion strains for functional genomics. BMC Bioinform. 2007, 8. [Google Scholar] [CrossRef]
- Samanfar, B.; Tan, L.H.; Shostak, K.; Chalabian, F.; Wu, Z.; Alamgir, M.; Sunba, N.; Burnside, D.; Omidi, K.; Hooshyar, M.; et al. A global investigation of gene deletion strains that affect premature stop codon bypass in yeast, Saccharomyces cerevisiae. Mol. Biosyst. 2014, 10, 916–924. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hajikarimlou, M.; Hunt, K.; Kirby, G.; Takallou, S.; Jagadeesan, S.K.; Omidi, K.; Hooshyar, M.; Burnside, D.; Moteshareie, H.; Babu, M.; et al. Lithium Chloride Sensitivity in Yeast and Regulation of Translation. Int. J. Mol. Sci. 2020, 21, 5730. https://doi.org/10.3390/ijms21165730
Hajikarimlou M, Hunt K, Kirby G, Takallou S, Jagadeesan SK, Omidi K, Hooshyar M, Burnside D, Moteshareie H, Babu M, et al. Lithium Chloride Sensitivity in Yeast and Regulation of Translation. International Journal of Molecular Sciences. 2020; 21(16):5730. https://doi.org/10.3390/ijms21165730
Chicago/Turabian StyleHajikarimlou, Maryam, Kathryn Hunt, Grace Kirby, Sarah Takallou, Sasi Kumar Jagadeesan, Katayoun Omidi, Mohsen Hooshyar, Daniel Burnside, Houman Moteshareie, Mohan Babu, and et al. 2020. "Lithium Chloride Sensitivity in Yeast and Regulation of Translation" International Journal of Molecular Sciences 21, no. 16: 5730. https://doi.org/10.3390/ijms21165730
APA StyleHajikarimlou, M., Hunt, K., Kirby, G., Takallou, S., Jagadeesan, S. K., Omidi, K., Hooshyar, M., Burnside, D., Moteshareie, H., Babu, M., Smith, M., Holcik, M., Samanfar, B., & Golshani, A. (2020). Lithium Chloride Sensitivity in Yeast and Regulation of Translation. International Journal of Molecular Sciences, 21(16), 5730. https://doi.org/10.3390/ijms21165730






