Structural Complexity and Plasticity of Signaling Regulation at the Melanocortin-4 Receptor
Abstract
1. The Melanocortin-4 Receptor
2. Specific Features in the MC4R Sequence and Structure Linked with Functionalities
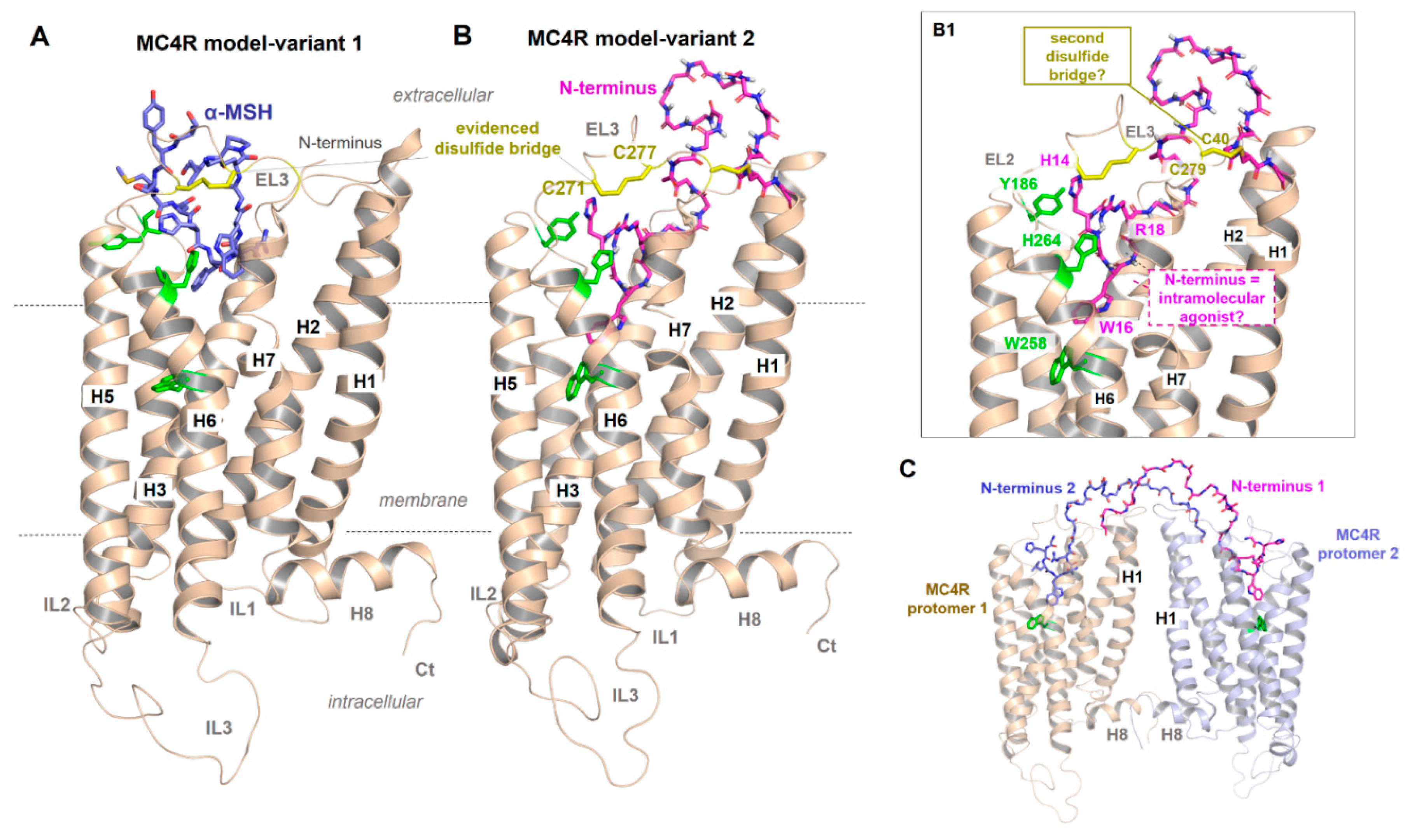
2.1. A Presumed Role of the MC4R N-terminus in Regulating Permanent Basal Activity and the Inhibitory Effect of AgRP
2.2. Basal (Constitutive, Ligand-Independent) Signaling Activity
2.3. The Short Second Extracellular Loop Has A Strong Impact on MC4R Functional Properties
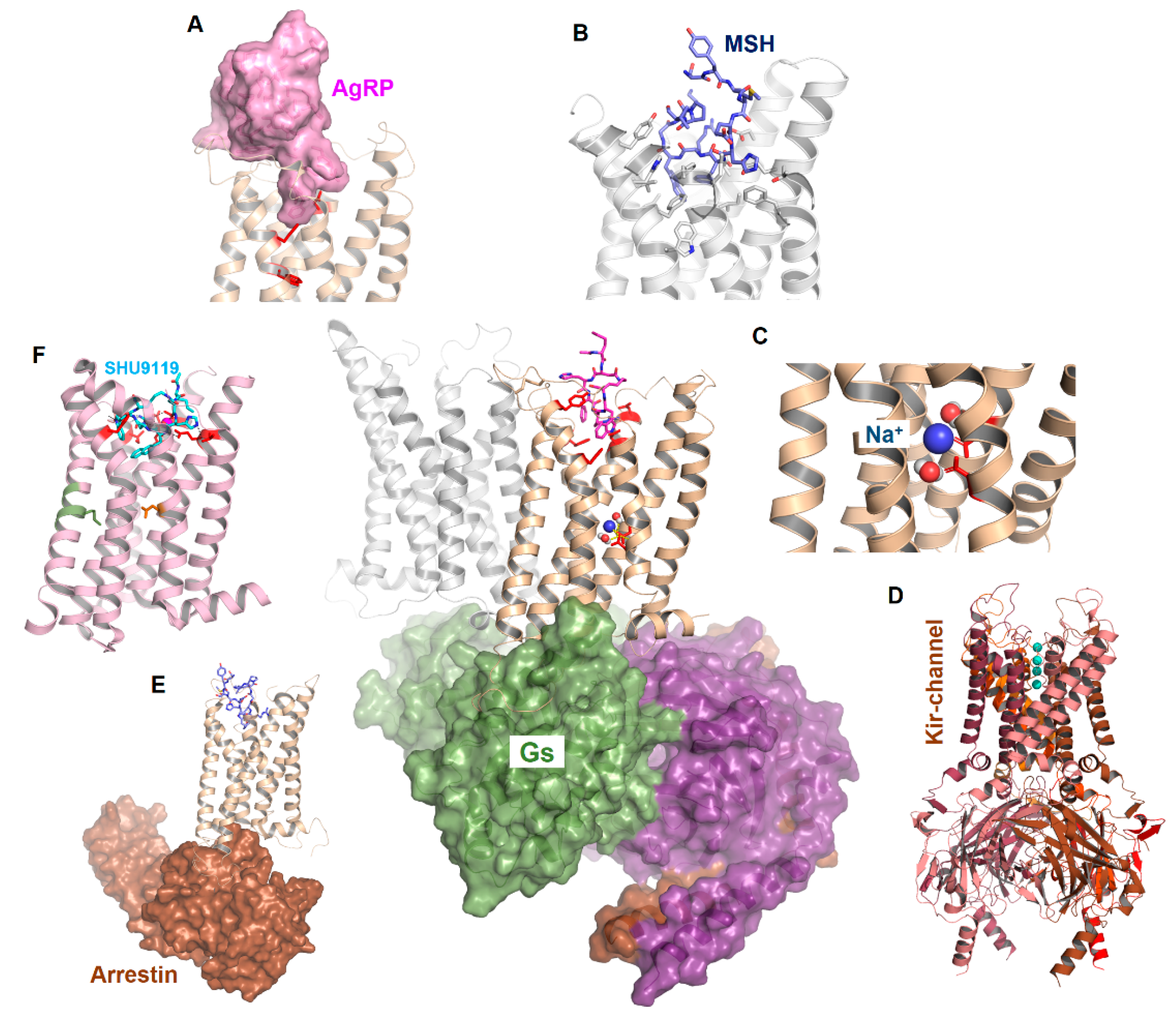
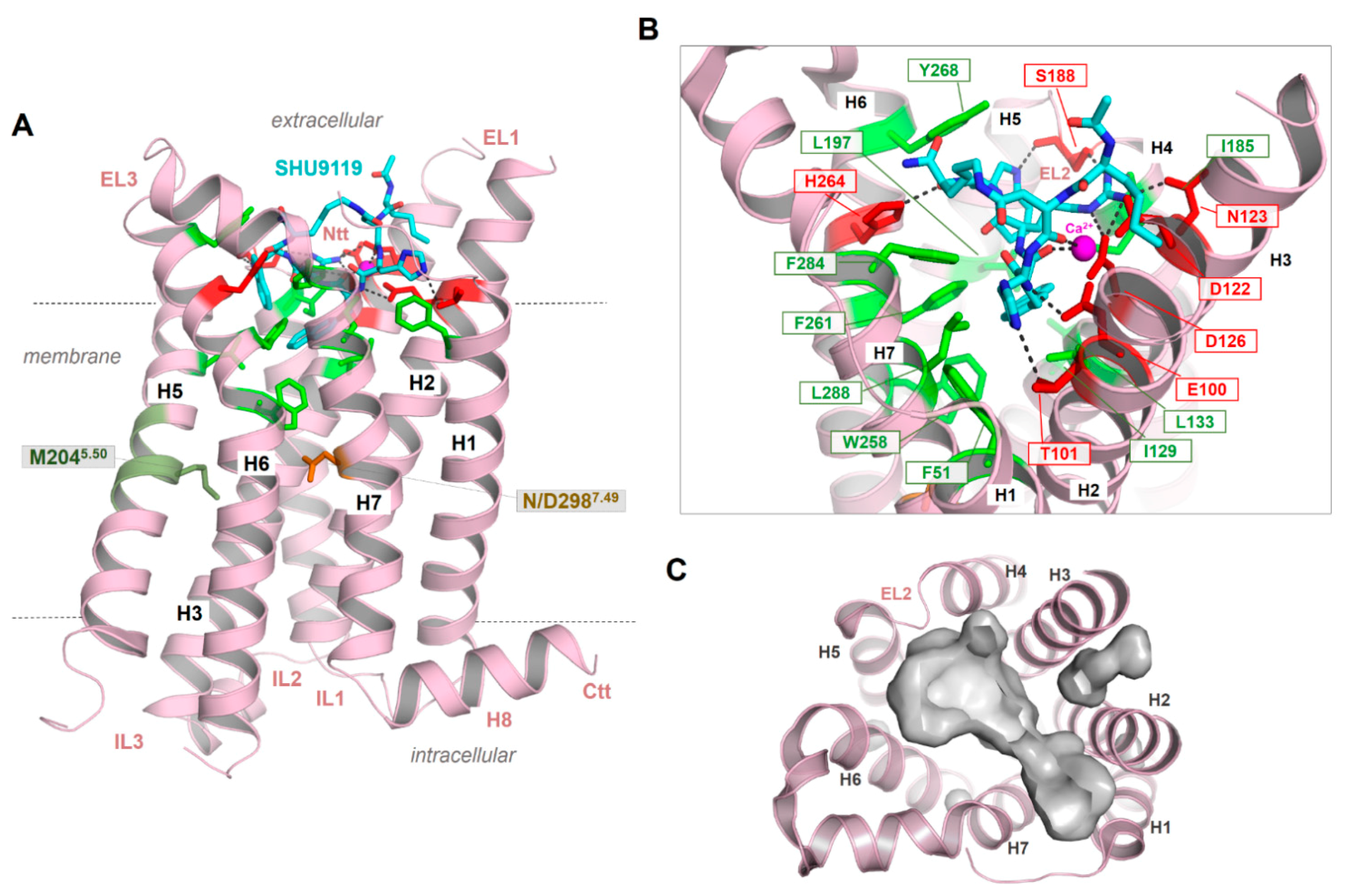
2.4. Structural Specificities of the MCRs in the Transmembrane Region
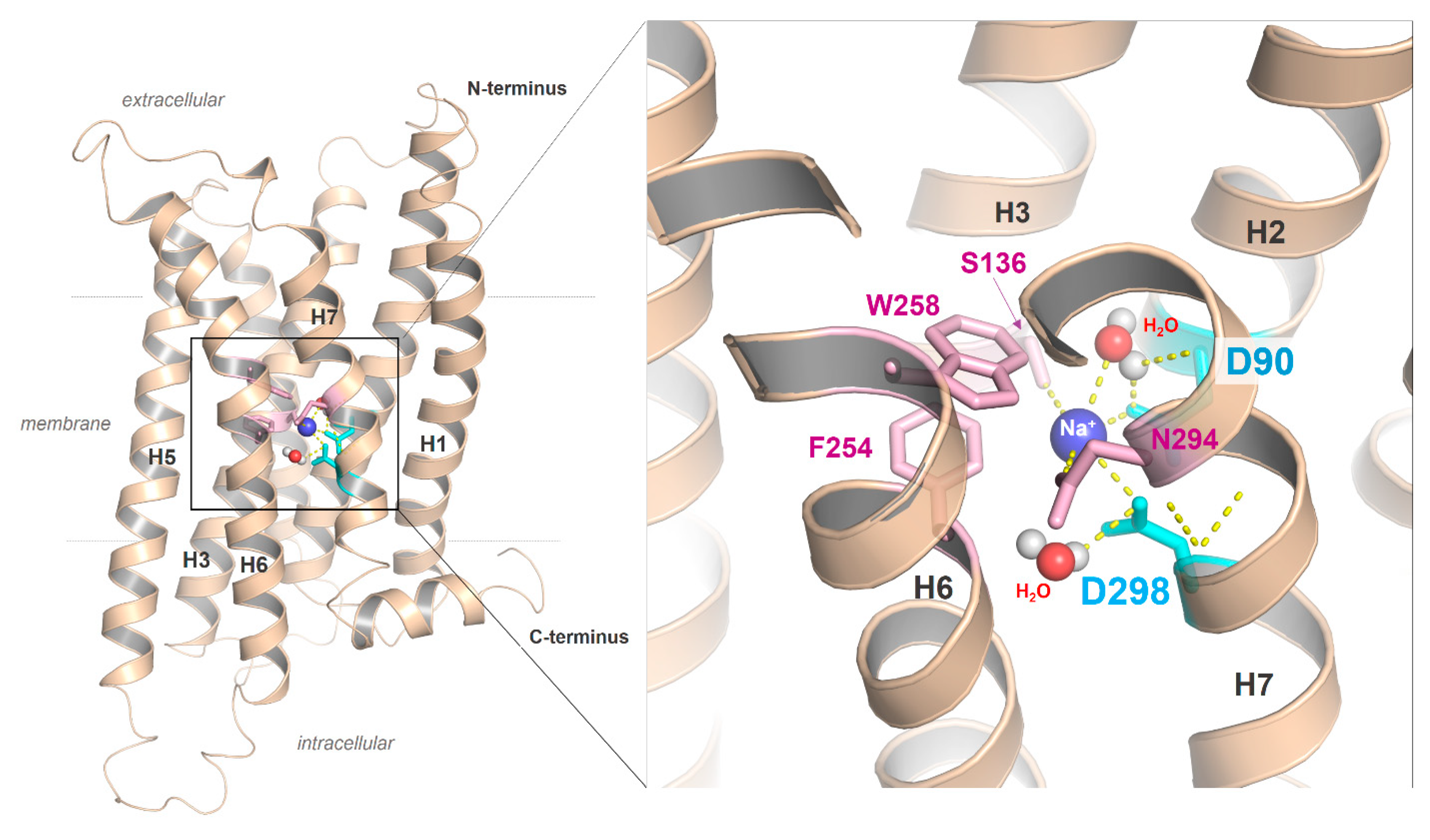
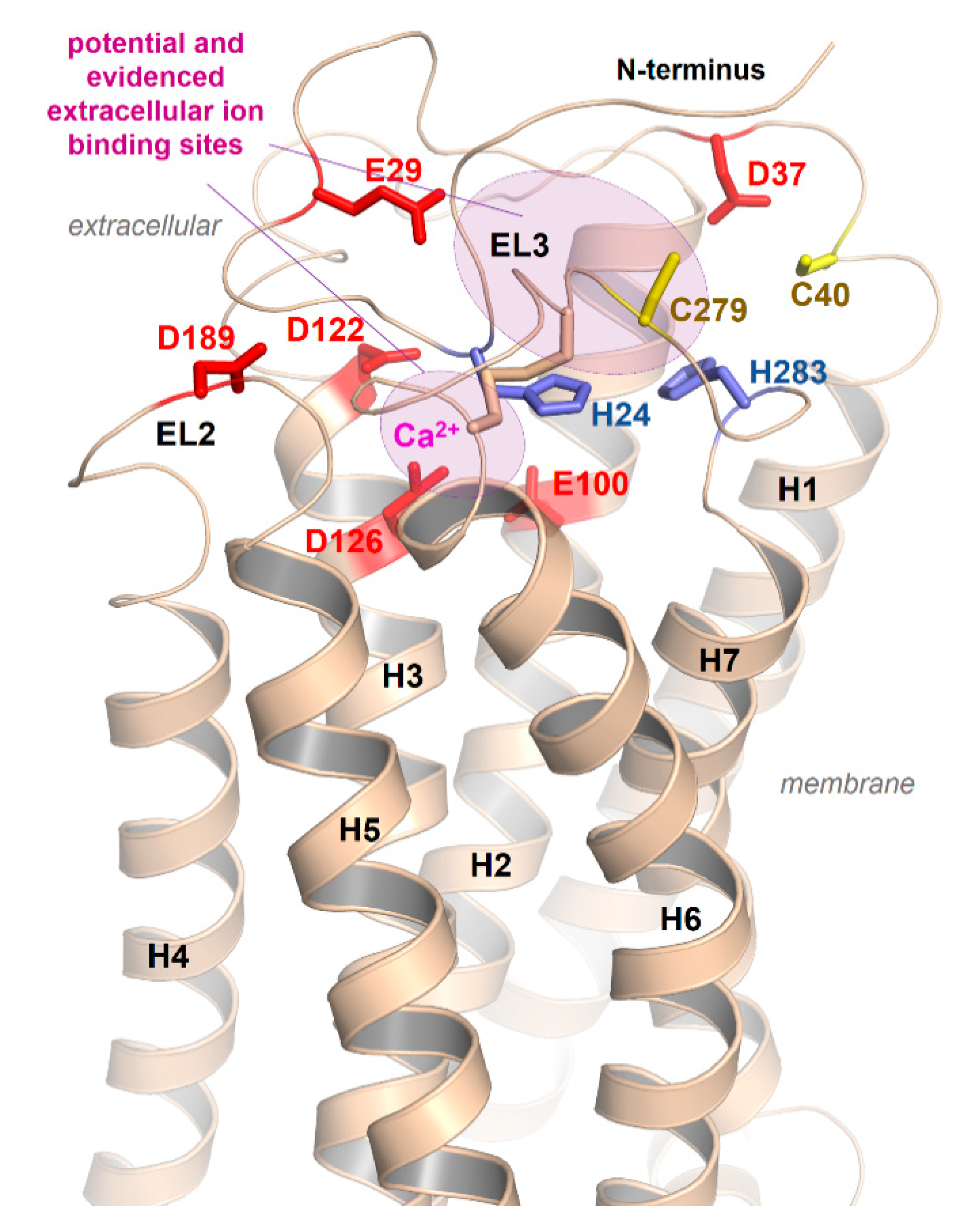
2.5. Ion Binding Sites in the MC4R
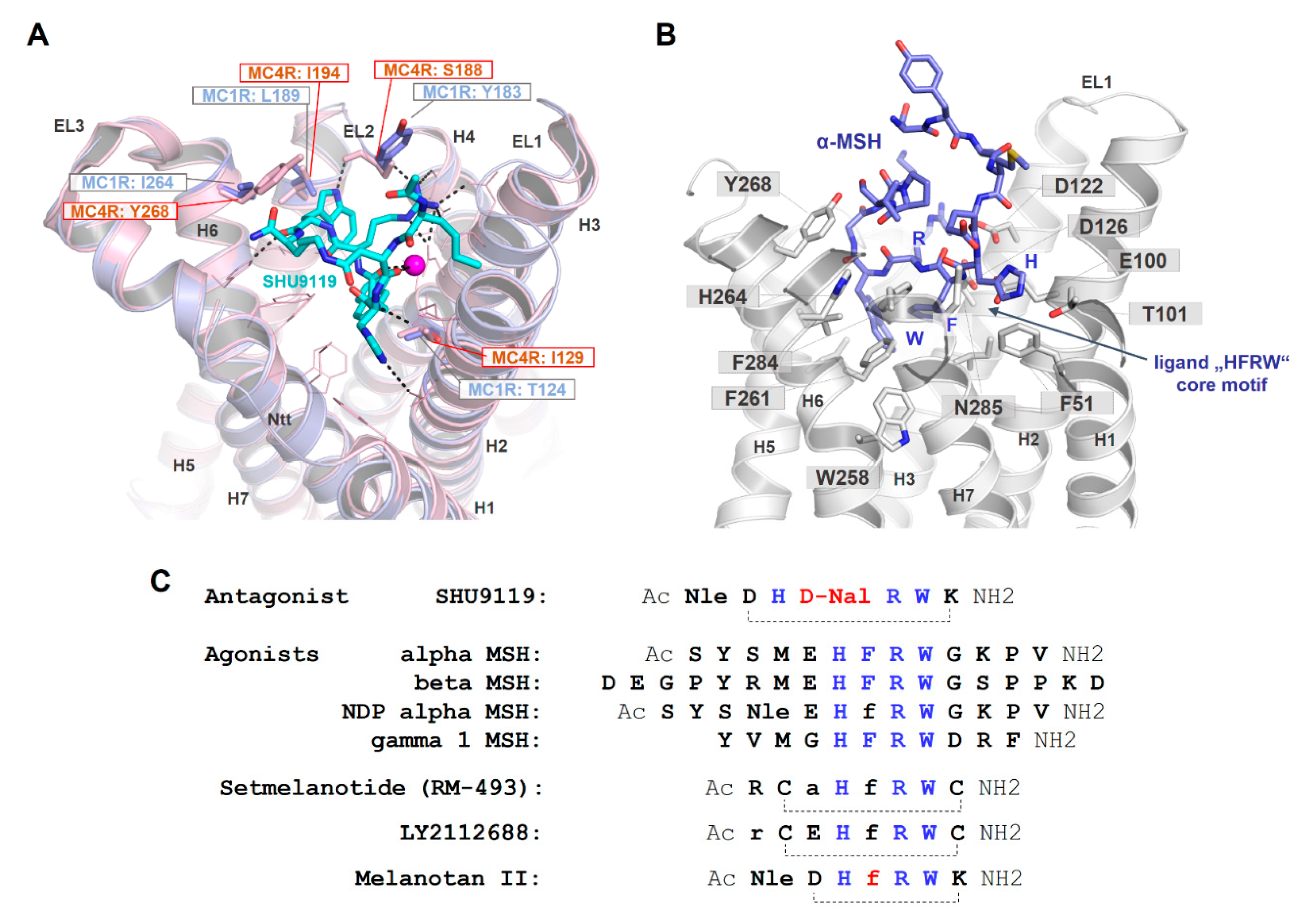
2.6. Peptidic Ligand Binding at the MC4R
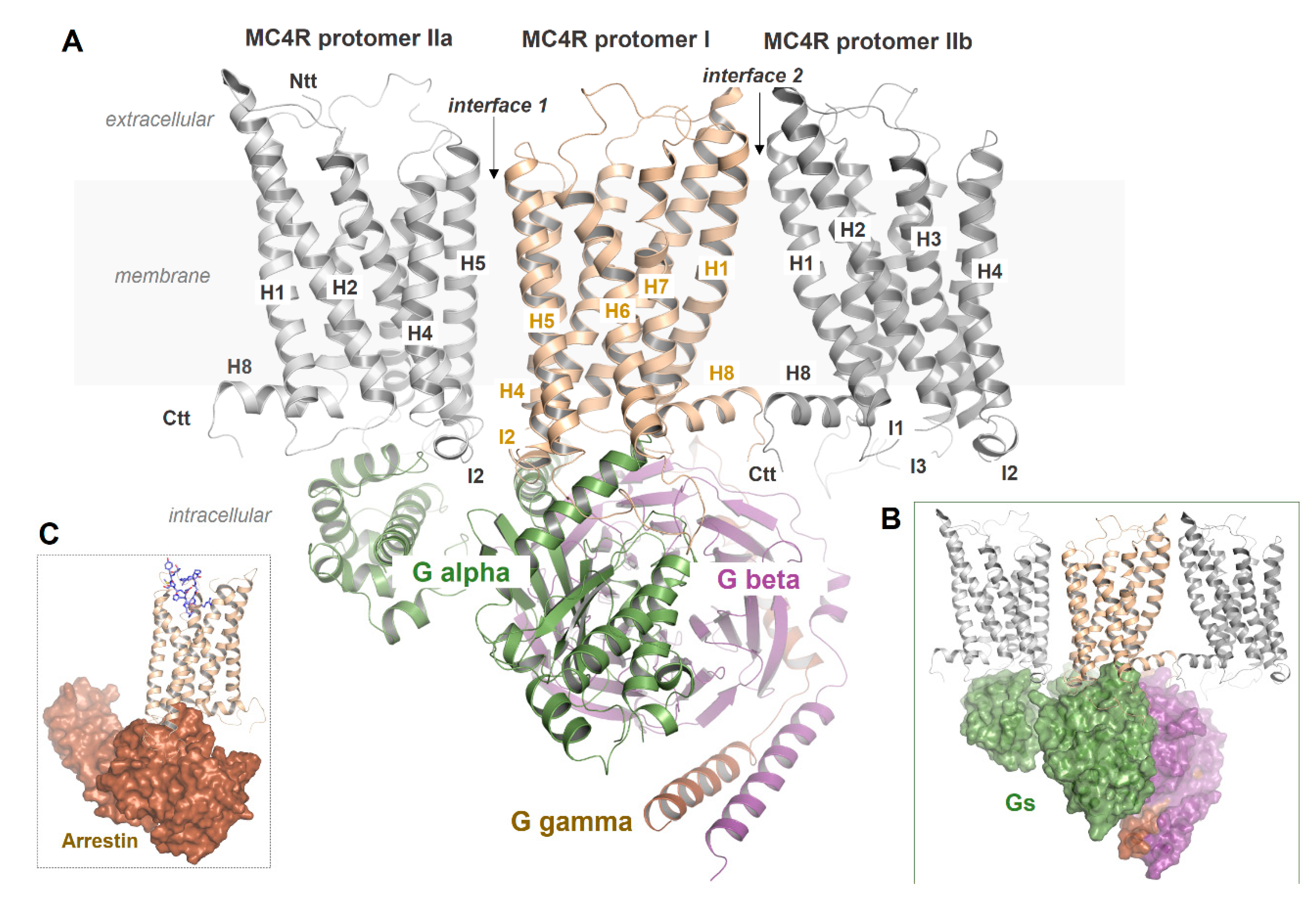
2.7. MC4R Assembly in Multimeric Constellations
3. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- De Mendoza, A.; Sebe-Pedros, A.; Ruiz-Trillo, I. The evolution of the GPCR signaling system in eukaryotes: Modularity, conservation, and the transition to metazoan multicellularity. Genome Biol. Evol. 2014, 6, 606–619. [Google Scholar] [CrossRef] [PubMed]
- Schoneberg, T.; Hofreiter, M.; Schulz, A.; Rompler, H. Learning from the past: Evolution of GPCR functions. Trends Pharmacol. Sci. 2007, 28, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Limbird, L.E. The receptor concept: A continuing evolution. Mol. Interv. 2004, 4, 326–336. [Google Scholar] [CrossRef]
- Alexander, S.P.; Christopoulos, A.; Davenport, A.P.; Kelly, E.; Mathie, A.; Peters, J.A.; Veale, E.L.; Armstrong, J.F.; Faccenda, E.; Harding, S.D.; et al. The Concise Guide to Pharmacology 2019/20: G protein-coupled receptors. Br. J. Pharmacol. 2019, 176, S21–S141. [Google Scholar] [CrossRef] [PubMed]
- Kobilka, B.K.; Deupi, X. Conformational complexity of G-protein-coupled receptors. Trends Pharmacol. Sci. 2007, 28, 397–406. [Google Scholar] [CrossRef]
- Kristiansen, K. Molecular mechanisms of ligand binding, signaling, and regulation within the superfamily of G-protein-coupled receptors: Molecular modeling and mutagenesis approaches to receptor structure and function. Pharmacol. Ther. 2004, 103, 21–80. [Google Scholar] [CrossRef]
- Weis, W.I.; Kobilka, B.K. The Molecular basis of G protein-coupled receptor activation. Annu. Rev. Biochem. 2018, 87, 897–919. [Google Scholar] [CrossRef]
- Ho, M.K.; Su, Y.; Yeung, W.W.; Wong, Y.H. Regulation of transcription factors by heterotrimeric G proteins. Curr. Mol. Pharmacol. 2009, 2, 19–31. [Google Scholar] [CrossRef]
- Veldhuis, N.A.; Poole, D.P.; Grace, M.; McIntyre, P.; Bunnett, N.W. The G protein-coupled receptor-transient receptor potential channel axis: Molecular insights for targeting disorders of sensation and inflammation. Pharmacol. Rev. 2015, 67, 36–73. [Google Scholar] [CrossRef]
- Hutchings, C.J.; Koglin, M.; Marshall, F.H. Therapeutic antibodies directed at G protein-coupled receptors. mAbs 2010, 2, 594–606. [Google Scholar] [CrossRef]
- Dorsam, R.T.; Gutkind, J.S. G-protein-coupled receptors and cancer. Nat. Rev. Cancer 2007, 7, 79–94. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Jimenez, C.; Santisteban, P. TSH signalling and cancer. Arq. Bras. Endocrinol. Metab. 2007, 51, 654–671. [Google Scholar] [CrossRef] [PubMed]
- Schoneberg, T.; Schulz, A.; Biebermann, H.; Hermsdorf, T.; Rompler, H.; Sangkuhl, K. Mutant G-protein-coupled receptors as a cause of human diseases. Pharmacol. Ther. 2004, 104, 173–206. [Google Scholar] [CrossRef] [PubMed]
- Seifert, R.; Wenzel-Seifert, K. Constitutive activity of G-protein-coupled receptors: Cause of disease and common property of wild-type receptors. Naunyn Schmiedebergs Arch. Pharmacol. 2002, 366, 381–416. [Google Scholar] [CrossRef]
- Tao, Y.X. Inactivating mutations of G protein-coupled receptors and diseases: Structure-function insights and therapeutic implications. Pharmacol. Ther. 2006, 111, 949–973. [Google Scholar] [CrossRef]
- Vassart, G.; Costagliola, S. G protein-coupled receptors: Mutations and endocrine diseases. Nat. Rev. Endocrinol. 2011, 7, 362–372. [Google Scholar] [CrossRef]
- Hauser, A.S.; Attwood, M.M.; Rask-Andersen, M.; Schioth, H.B.; Gloriam, D.E. Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 2017, 16, 829–842. [Google Scholar] [CrossRef]
- Munk, C.; Mutt, E.; Isberg, V.; Nikolajsen, L.F.; Bibbe, J.M.; Flock, T.; Hanson, M.A.; Stevens, R.C.; Deupi, X.; Gloriam, D.E. An online resource for GPCR structure determination and analysis. Nat. Methods 2019, 16, 151–162. [Google Scholar] [CrossRef]
- Thal, D.M.; Vuckovic, Z.; Draper-Joyce, C.J.; Liang, Y.L.; Glukhova, A.; Christopoulos, A.; Sexton, P.M. Recent advances in the determination of G protein-coupled receptor structures. Curr. Opin. Struct. Biol. 2018, 51, 28–34. [Google Scholar] [CrossRef]
- Congreve, M.; de Graaf, C.; Swain, N.A.; Tate, C.G. Impact of GPCR structures on drug discovery. Cell 2020, 181, 81–91. [Google Scholar] [CrossRef]
- Yu, J.; Gimenez, L.E.; Hernandez, C.C.; Wu, Y.; Wein, A.H.; Han, G.W.; McClary, K.; Mittal, S.R.; Burdsall, K.; Stauch, B.; et al. Determination of the melanocortin-4 receptor structure identifies Ca2+ as a cofactor for ligand binding. Science 2020, 368, 428–433. [Google Scholar] [PubMed]
- Hruby, V.J.; Lu, D.; Sharma, S.D.; Castrucci, A.L.; Kesterson, R.A.; Al-Obeidi, F.A.; Hadley, M.E.; Cone, R.D. Cyclic lactam α-melanotropin analogues of Ac-Nle4-cyclo[Asp5, D-Phe7, Lys10] α-melanocyte-stimulating hormone-(4–10)-NH2 with bulky aromatic amino acids at position 7 show high antagonist potency and selectivity at specific melanocortin receptors. J. Med. Chem. 1995, 38, 3454–3461. [Google Scholar] [CrossRef] [PubMed]
- Novoselova, T.V.; Chan, L.F.; Clark, A.J.L. Pathophysiology of melanocortin receptors and their accessory proteins. Best Pract. Res. Clin. Endocrinol. Metab. 2018, 32, 93–106. [Google Scholar] [CrossRef]
- Tao, Y.X. Melanocortin receptors. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 2411–2413. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.X. The melanocortin-4 receptor: Physiology, pharmacology, and pathophysiology. Endocr. Rev. 2010, 31, 506–543. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yang, Z.; Zhang, Y.P.; He, S.; Liang, X.F.; Tao, Y.X. Molecular cloning, tissue distribution, and pharmacological characterization of melanocortin-4 receptor in grass carp (Ctenopharyngodon idella). Domest. Anim. Endocrinol. 2017, 59, 140–151. [Google Scholar] [CrossRef]
- Anderson, E.J.; Cakir, I.; Carrington, S.J.; Cone, R.D.; Ghamari-Langroudi, M.; Gillyard, T.; Gimenez, L.E.; Litt, M.J. 60 Years of POMC: Regulation of feeding and energy homeostasis by α-MSH. J. Mol. Endocrinol. 2016, 56, T157–T174. [Google Scholar] [CrossRef]
- Litt, M.J.; Okoye, G.D.; Lark, D.; Cakir, I.; Moore, C.; Barber, M.C.; Atkinson, J.; Fessel, J.; Moslehi, J.; Cone, R.D. Loss of the melanocortin-4 receptor in mice causes dilated cardiomyopathy. eLife 2017, 6, e28118. [Google Scholar] [CrossRef]
- Dhillon, S.; Keam, S.J. Bremelanotide: First approval. Drugs 2019, 79, 1599–1606. [Google Scholar] [CrossRef]
- Panaro, B.L.; Tough, I.R.; Engelstoft, M.S.; Matthews, R.T.; Digby, G.J.; Moller, C.L.; Svendsen, B.; Gribble, F.; Reimann, F.; Holst, J.J.; et al. The melanocortin-4 receptor is expressed in enteroendocrine L cells and regulates the release of peptide YY and glucagon-like peptide 1 in vivo. Cell Metab. 2014, 20, 1018–1029. [Google Scholar] [CrossRef]
- Mansour, M.; White, D.; Wernette, C.; Dennis, J.; Tao, Y.X.; Collins, R.; Parker, L.; Morrison, E. Pancreatic neuronal melanocortin-4 receptor modulates serum insulin levels independent of leptin receptor. Endocrine 2010, 37, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Alwahsh, S.M.; Ramadori, G.; Kollmar, O.; Slotta, J.E. Upregulation of hepatic melanocortin 4 receptor during rat liver regeneration. J. Surg. Res. 2016, 203, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Chen, Y.; Xu, H.; Yang, L.; Yuan, F.; Li, L.; Xu, Y.; Chen, Y.; Zhang, C.; Lin, G. Melanocortin receptor 4 signaling regulates vertebrate limb regeneration. Dev. Cell 2018, 46, 397–409. [Google Scholar] [CrossRef] [PubMed]
- Schaub, J.W.; Bruce, E.B.; Haskell-Luevano, C. Drugs, exercise, and the melanocortin-4 receptor—Different means, same ends: Treating obesity. Adv. Exp. Med. Biol. 2010, 681, 49–60. [Google Scholar]
- Yang, L.K.; Tao, Y.X. Biased signaling at neural melanocortin receptors in regulation of energy homeostasis. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 2486–2495. [Google Scholar] [CrossRef]
- Kuhnen, P.; Krude, H.; Biebermann, H. Melanocortin-4 receptor signalling: Importance for weight regulation and obesity treatment. Trends Mol. Med. 2019, 25, 136–148. [Google Scholar] [CrossRef]
- Shichida, Y.; Matsuyama, T. Evolution of opsins and phototransduction. Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 2881–2895. [Google Scholar] [CrossRef]
- Tao, Y.X. Constitutive activity in melanocortin-4 receptor: Biased signaling of inverse agonists. Adv. Pharmacol. 2014, 70, 135–154. [Google Scholar]
- Clement, K.; Biebermann, H.; Farooqi, I.S.; Van der Ploeg, L.; Wolters, B.; Poitou, C.; Puder, L.; Fiedorek, F.; Gottesdiener, K.; Kleinau, G.; et al. MC4R agonism promotes durable weight loss in patients with leptin receptor deficiency. Nat. Med. 2018, 24, 551–555. [Google Scholar] [CrossRef]
- Biebermann, H.; Krude, H.; Elsner, A.; Chubanov, V.; Gudermann, T.; Gruters, A. Autosomal-dominant mode of inheritance of a melanocortin-4 receptor mutation in a patient with severe early-onset obesity is due to a dominant-negative effect caused by receptor dimerization. Diabetes 2003, 52, 2984–2988. [Google Scholar] [CrossRef]
- Nickolls, S.A.; Maki, R.A. Dimerization of the melanocortin 4 receptor: A study using bioluminescence resonance energy transfer. Peptides 2006, 27, 380–387. [Google Scholar] [CrossRef] [PubMed]
- Rouault, A.A.J.; Srinivasan, D.K.; Yin, T.C.; Lee, A.A.; Sebag, J.A. Melanocortin receptor accessory proteins (MRAPs): Functions in the melanocortin system and beyond. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863 Pt A, 2462–2467. [Google Scholar] [CrossRef]
- Schonnop, L.; Kleinau, G.; Herrfurth, N.; Volckmar, A.L.; Cetindag, C.; Muller, A.; Peters, T.; Herpertz, S.; Antel, J.; Hebebrand, J.; et al. Decreased melanocortin-4 receptor function conferred by an infrequent variant at the human melanocortin receptor accessory protein 2 gene. Obesity 2016, 24, 1976–1982. [Google Scholar] [CrossRef]
- Soletto, L.; Hernandez-Balfago, S.; Rocha, A.; Scheerer, P.; Kleinau, G.; Cerda-Reverter, J.M. Melanocortin receptor accessory protein 2-induced adrenocorticotropic hormone response of human melanocortin 4 receptor. J. Endocr. Soc. 2019, 3, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Mosialou, I.; Shikhel, S.; Liu, J.M.; Maurizi, A.; Luo, N.; He, Z.; Huang, Y.; Zong, H.; Friedman, R.A.; Barasch, J.; et al. MC4R-dependent suppression of appetite by bone-derived lipocalin 2. Nature 2017, 543, 385–390. [Google Scholar] [CrossRef] [PubMed]
- Farooqi, I.S.; Yeo, G.S.; O’Rahilly, S. Binge eating as a phenotype of melanocortin 4 receptor gene mutations. N. Engl. J. Med. 2003, 349, 606–609. [Google Scholar] [PubMed]
- Dandona, P.; Aljada, A.; Bandyopadhyay, A. Inflammation: The link between insulin resistance, obesity and diabetes. Trends Immunol. 2004, 25, 4–7. [Google Scholar] [CrossRef] [PubMed]
- Heyder, N.; Kleinau, G.; Szczepek, M.; Kwiatkowski, D.; Speck, D.; Soletto, L.; Cerda-Reverter, J.M.; Krude, H.; Kuhnen, P.; Biebermann, H.; et al. Signal transduction and pathogenic modifications at the melanocortin-4 receptor: A structural perspective. Front. Endocrinol. 2019, 10, 515. [Google Scholar] [CrossRef]
- Gillyard, T.; Fowler, K.; Williams, S.Y.; Cone, R.D. Obesity-associated mutant melanocortin-4 receptors with normal Gαs coupling frequently exhibit other discoverable pharmacological and biochemical defects. J. Neuroendocrinol. 2019, 31, e12795. [Google Scholar] [CrossRef]
- He, S.; Tao, Y.X. Defect in MAPK signaling as a cause for monogenic obesity caused by inactivating mutations in the melanocortin-4 receptor gene. Int. J. Boil. Sci. 2014, 10, 1128–1137. [Google Scholar] [CrossRef]
- Lotta, L.A.; Mokrosinski, J.; De Oliveira, E.M.; Li, C.; Sharp, S.J.; Luan, J.; Brouwers, B.; Ayinampudi, V.; Bowker, N.; Kerrison, N.; et al. Human gain-of-function MC4R variants show signaling bias and protect against obesity. Cell 2019, 177, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Paisdzior, S.; Dimitriou, I.M.; Schope, P.C.; Annibale, P.; Scheerer, P.; Krude, H.; Lohse, M.J.; Biebermann, H.; Kuhnen, P. Differential signaling profiles of MC4R mutations with three different ligands. Int J. Mol. Sci. 2020, 21, 1224. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.Y.; Muniyappa, R.; Abel, B.S.; Mullins, K.P.; Staker, P.; Brychta, R.J.; Zhao, X.; Ring, M.; Psota, T.L.; Cone, R.D.; et al. RM-493, a melanocortin-4 receptor (MC4R) agonist, increases resting energy expenditure in obese individuals. J. Clin. Endocrinol. Metab. 2015, 100, 1639–1645. [Google Scholar] [CrossRef]
- Collet, T.H.; Dubern, B.; Mokrosinski, J.; Connors, H.; Keogh, J.M.; De Oliveira, E.M.; Henning, E.; Poitou-Bernert, C.; Oppert, J.M.; Tounian, P.; et al. Evaluation of a melanocortin-4 receptor (MC4R) agonist (Setmelanotide) in MC4R deficiency. Mol. Metab. 2017, 6, 1321–1329. [Google Scholar] [CrossRef]
- Kievit, P.; Halem, H.; Marks, D.L.; Dong, J.Z.; Glavas, M.M.; Sinnayah, P.; Pranger, L.; Cowley, M.A.; Grove, K.L.; Culler, M.D. Chronic treatment with a melanocortin-4 receptor agonist causes weight loss, reduces insulin resistance, and improves cardiovascular function in diet-induced obese rhesus macaques. Diabetes 2013, 62, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Kuhnen, P.; Clement, K.; Wiegand, S.; Blankenstein, O.; Gottesdiener, K.; Martini, L.L.; Mai, K.; Blume-Peytavi, U.; Gruters, A.; Krude, H. Proopiomelanocortin deficiency treated with a melanocortin-4 receptor agonist. N. Engl. J. Med. 2016, 375, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, S.M.; Lee, Y.S.; Gillies, R.J.; Hruby, V.J. Synthesis and evaluation of bivalent ligands for binding to the human melanocortin-4 receptor. Bioorg. Med. Chem. 2014, 22, 6360–6365. [Google Scholar] [CrossRef]
- Lensing, C.J.; Adank, D.N.; Doering, S.R.; Wilber, S.L.; Andreasen, A.; Schaub, J.W.; Xiang, Z.; Haskell-Luevano, C. Ac-Trp-DPhe(p-I)-Arg-Trp-NH2, a 250-fold selective melanocortin-4 receptor (MC4R) antagonist over the melanocortin-3 receptor (MC3R), affects energy homeostasis in male and female mice differently. ACS Chem. Neurosci. 2016, 7, 1283–1291. [Google Scholar] [CrossRef]
- Lensing, C.J.; Adank, D.N.; Wilber, S.L.; Freeman, K.T.; Schnell, S.M.; Speth, R.C.; Zarth, A.T.; Haskell-Luevano, C. A direct in vivo comparison of the melanocortin monovalent agonist Ac-His-DPhe-Arg-Trp-NH2 versus the bivalent agonist Ac-His-DPhe-Arg-Trp-PEDG20-His-DPhe-Arg-Trp-NH2: A bivalent advantage. ACS Chem. Neurosci. 2017, 8, 1262–1278. [Google Scholar] [CrossRef]
- Lensing, C.J.; Freeman, K.T.; Schnell, S.M.; Adank, D.N.; Speth, R.C.; Haskell-Luevano, C. An in vitro and in vivo investigation of bivalent ligands that display preferential binding and functional activity for different melanocortin receptor homodimers. J. Med. Chem. 2016, 59, 3112–3128. [Google Scholar]
- Fani, L.; Bak, S.; Delhanty, P.; Van Rossum, E.F.; Van den Akker, E.L. The melanocortin-4 receptor as target for obesity treatment: A systematic review of emerging pharmacological therapeutic options. Int. J. Obes. 2014, 38, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Wikberg, J.E.; Mutulis, F. Targeting melanocortin receptors: An approach to treat weight disorders and sexual dysfunction. Nat. Rev. Drug Discov. 2008, 7, 307–323. [Google Scholar] [CrossRef] [PubMed]
- Ericson, M.D.; Lensing, C.J.; Fleming, K.A.; Schlasner, K.N.; Doering, S.R.; Haskell-Luevano, C. Bench-top to clinical therapies: A review of melanocortin ligands from 1954 to 2016. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 2414–2435. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.M.; Nawaz, A.; Evans, M. Drug therapy in obesity: A review of current and emerging treatments. Diabetes Ther. 2020, 11, 1199–1216. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Lubrano-Berthelier, C.; Govaerts, C.; Picard, F.; Santiago, P.; Conklin, B.R.; Vaisse, C. Constitutive activity of the melanocortin-4 receptor is maintained by its N-terminal domain and plays a role in energy homeostasis in humans. J. Clin. Invest. 2004, 114, 1158–1164. [Google Scholar] [CrossRef]
- Schoneberg, T.; Kleinau, G.; Bruser, A. What are they waiting for? Tethered agonism in G protein-coupled receptors. Pharmacol. Res. 2016, 108, 9–15. [Google Scholar] [CrossRef]
- Coleman, J.L.; Ngo, T.; Smith, N.J. The G protein-coupled receptor N-terminus and receptor signalling: N-tering a new era. Cell Signal. 2017, 33, 1–9. [Google Scholar] [CrossRef]
- Muller, A.; Berkmann, J.C.; Scheerer, P.; Biebermann, H.; Kleinau, G. Insights into basal signaling regulation, oligomerization, and structural organization of the human G-protein coupled receptor 83. PLoS ONE 2016, 11, e0168260. [Google Scholar] [CrossRef]
- Bruser, A.; Schulz, A.; Rothemund, S.; Ricken, A.; Calebiro, D.; Kleinau, G.; Schoneberg, T. The activation mechanism of glycoprotein hormone receptors with implications in the cause and therapy of endocrine diseases. J. Biol. Chem. 2016, 291, 508–520. [Google Scholar] [CrossRef]
- Vlaeminck-Guillem, V.; Ho, S.C.; Rodien, P.; Vassart, G.; Costagliola, S. Activation of the cAMP pathway by the TSH receptor involves switching of the ectodomain from a tethered inverse agonist to an agonist. Mol. Endocrinol. 2002, 16, 736–746. [Google Scholar] [CrossRef]
- Krause, G.; Kreuchwig, A.; Kleinau, G. Extended and structurally supported insights into extracellular hormone binding, signal transduction and organization of the thyrotropin receptor. PLoS ONE 2012, 7, e52920. [Google Scholar] [CrossRef] [PubMed]
- El Buri, A.; Adams, D.R.; Smith, D.; Tate, R.J.; Mullin, M.; Pyne, S.; Pyne, N.J. The sphingosine 1-phosphate receptor 2 is shed in exosomes from breast cancer cells and is N-terminally processed to a short constitutively active form that promotes extracellular signal regulated kinase activation and DNA synthesis in fibroblasts. Oncotarget 2018, 9, 29453–29467. [Google Scholar] [CrossRef] [PubMed]
- Sethi, A.; Bruell, S.; Patil, N.; Hossain, M.A.; Scott, D.J.; Petrie, E.J.; Bathgate, R.A.; Gooley, P.R. The complex binding mode of the peptide hormone H2 relaxin to its receptor RXFP1. Nat. Commun. 2016, 7, 11344. [Google Scholar] [CrossRef] [PubMed]
- Kountz, T.S.; Lee, K.S.; Aggarwal-Howarth, S.; Curran, E.; Park, J.M.; Harris, D.A.; Stewart, A.; Hendrickson, J.; Camp, N.D.; Wolf-Yadlin, A.; et al. Endogenous N-terminal domain cleavage modulates α1D-adrenergic receptor pharmacodynamics. J. Biol. Chem. 2016, 291, 18210–18221. [Google Scholar] [CrossRef] [PubMed]
- Liebscher, I.; Schon, J.; Petersen, S.C.; Fischer, L.; Auerbach, N.; Demberg, L.M.; Mogha, A.; Coster, M.; Simon, K.U.; Rothemund, S.; et al. A tethered agonist within the ectodomain activates the adhesion G protein-coupled receptors GPR126 and GPR133. Cell Rep. 2014, 9, 2018–2026. [Google Scholar] [CrossRef] [PubMed]
- Coughlin, S.R. Protease-activated receptors in hemostasis, thrombosis and vascular biology. J. Thromb. Haemost. 2005, 3, 1800–1814. [Google Scholar] [CrossRef]
- Zhang, M.; Tong, K.P.; Fremont, V.; Chen, J.; Narayan, P.; Puett, D.; Weintraub, B.D.; Szkudlinski, M.W. The extracellular domain suppresses constitutive activity of the transmembrane domain of the human TSH receptor: Implications for hormone-receptor interaction and antagonist design. Endocrinology 2000, 141, 3514–3517. [Google Scholar] [CrossRef]
- Lin, X.; Li, M.; Wang, N.; Wu, Y.; Luo, Z.; Guo, S.; Han, G.W.; Li, S.; Yue, Y.; Wei, X.; et al. Structural basis of ligand recognition and self-activation of orphan GPR52. Nature 2020, 579, 152–157. [Google Scholar] [CrossRef]
- Ersoy, B.A.; Pardo, L.; Zhang, S.; Thompson, D.A.; Millhauser, G.; Govaerts, C.; Vaisse, C. Mechanism of N-terminal modulation of activity at the melanocortin-4 receptor GPCR. Nat. Chem. Biol. 2012, 8, 725–730. [Google Scholar] [CrossRef]
- Li, J.T.; Yang, Z.; Chen, H.P.; Zhu, C.H.; Deng, S.P.; Li, G.L.; Tao, Y.X. Molecular cloning, tissue distribution, and pharmacological characterization of melanocortin-4 receptor in spotted scat, Scatophagus argus. Gen. Comp. Endocrinol. 2016, 230, 143–152. [Google Scholar] [CrossRef]
- Rao, Y.Z.; Chen, R.; Zhang, Y.; Tao, Y.X. Orange-spotted grouper melanocortin-4 receptor: Modulation of signaling by MRAP2. Gen. Comp. Endocrinol. 2019, 284, 113234. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.Q.; Hou, Z.S.; Wen, H.S.; Li, Y.; Qi, X.; Li, W.J.; Tao, Y.X. Melanocortin-4 receptor in spotted sea bass, Lateolabrax maculatus: Cloning, tissue distribution, physiology, and pharmacology. Front. Endocrinol. 2019, 10, 705. [Google Scholar] [CrossRef] [PubMed]
- Ballesteros, J.A.; Weinstein, H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relationships in G-protein coupled receptors. Methods Neurosci. 1995, 25, 366–428. [Google Scholar]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Yang, Y.; Chen, M.; Kesterson, R.A., Jr.; Harmon, C.M. Structural insights into the role of the ACTH receptor cysteine residues on receptor function. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007, 293, R1126. [Google Scholar] [CrossRef][Green Version]
- Tarnow, P.; Schoneberg, T.; Krude, H.; Gruters, A.; Biebermann, H. Mutationally induced disulfide bond formation within the third extracellular loop causes melanocortin 4 receptor inactivation in patients with obesity. J. Biol. Chem. 2003, 278, 48666–48673. [Google Scholar] [CrossRef]
- Kiefer, L.L.; Veal, J.M.; Mountjoy, K.G.; Wilkison, W.O. Melanocortin receptor binding determinants in the agouti protein. Biochemistry 1998, 37, 991–997. [Google Scholar] [CrossRef]
- Tota, M.R.; Smith, T.S.; Mao, C.; MacNeil, T.; Mosley, R.T.; Van der Ploeg, L.H.; Fong, T.M. Molecular interaction of Agouti protein and Agouti-related protein with human melanocortin receptors. Biochemistry 1999, 38, 897–904. [Google Scholar] [CrossRef]
- Elsner, A.; Tarnow, P.; Schaefer, M.; Ambrugger, P.; Krude, H.; Gruters, A.; Biebermann, H. MC4R oligomerizes independently of extracellular cysteine residues. Peptides 2006, 27, 372–379. [Google Scholar] [CrossRef]
- Piechowski, C.L.; Rediger, A.; Lagemann, C.; Muhlhaus, J.; Muller, A.; Pratzka, J.; Tarnow, P.; Gruters, A.; Krude, H.; Kleinau, G.; et al. Inhibition of melanocortin-4 receptor dimerization by substitutions in intracellular loop 2. J. Mol. Endocrinol. 2013, 51, 109–118. [Google Scholar] [CrossRef]
- Chapman, K.L.; Findlay, J.B. The melanocortin 4 receptor: Oligomer formation, interaction sites and functional significance. Biochim. Biophys. Acta 2013, 1828, 535–542. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Saleh, N.; Kleinau, G.; Heyder, N.; Clark, T.; Hildebrand, P.W.; Scheerer, P. Binding, thermodynamics, and selectivity of a non-peptide antagonist to the melanocortin-4 receptor. Front. Pharmacol. 2018, 9, 560. [Google Scholar] [CrossRef] [PubMed]
- Chavkin, C.; McLaughlin, J.P.; Celver, J.P. Regulation of opioid receptor function by chronic agonist exposure: Constitutive activity and desensitization. Mol. Pharmacol. 2001, 60, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Kenakin, T. The physiological significance of constitutive receptor activity. Trends Pharmacol. Sci. 2005, 26, 603–605. [Google Scholar] [CrossRef]
- Smit, M.J.; Vischer, H.F.; Bakker, R.A.; Jongejan, A.; Timmerman, H.; Pardo, L.; Leurs, R. Pharmacogenomic and structural analysis of constitutive G protein-coupled receptor activity. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 53–87. [Google Scholar] [CrossRef]
- Link, R.; Veiksina, S.; Tahk, M.J.; Laasfeld, T.; Paiste, P.; Kopanchuk, S.; Rinken, A. The constitutive activity of melanocortin-4 receptors in cAMP pathway is allosterically modulated by zinc and copper ions. J. Neurochem. 2020, 153, 346–361. [Google Scholar] [CrossRef]
- Holst, B.; Elling, C.E.; Schwartz, T.W. Metal ion-mediated agonism and agonist enhancement in melanocortin MC1 and MC4 receptors. J. Biol. Chem. 2002, 277, 47662–47670. [Google Scholar] [CrossRef]
- Zhang, J.; Li, X.; Zhou, Y.; Cui, L.; Li, J.; Wu, C.; Wan, Y.; Li, J.; Wang, Y. The interaction of MC3R and MC4R with MRAP2, ACTH, α-MSH and AgRP in chickens. J. Endocrinol. 2017, 234, 155–174. [Google Scholar] [CrossRef]
- Tao, Y.X. Molecular chaperones and G protein-coupled receptor maturation and pharmacology. Mol. Cell Endocrinol. 2020, 511, 110862. [Google Scholar] [CrossRef]
- Kay, E.I.; Botha, R.; Montgomery, J.M.; Mountjoy, K.G. hMRAPα specifically alters hMC4R molecular mass and N-linked complex glycosylation in HEK293 cells. J. Mol. Endocrinol. 2013, 50, 217–227. [Google Scholar] [CrossRef]
- Kay, E.I.; Botha, R.; Montgomery, J.M.; Mountjoy, K.G. hMRAPα increases αMSH-induced hMC1R and hMC3R functional coupling and hMC4R constitutive activity. J. Mol. Endocrinol. 2013, 50, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.X.; Conn, P.M. Chaperoning G protein-coupled receptors: From cell biology to therapeutics. Endocr. Rev. 2014, 35, 602–647. [Google Scholar] [CrossRef] [PubMed]
- Hay, D.L.; Walker, C.S.; Gingell, J.J.; Ladds, G.; Reynolds, C.A.; Poyner, D.R. Receptor activity-modifying proteins; multifunctional G protein-coupled receptor accessory proteins. Biochem. Soc. Trans. 2016, 44, 568–573. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.L.; Khoshouei, M.; Deganutti, G.; Glukhova, A.; Koole, C.; Peat, T.S.; Radjainia, M.; Plitzko, J.M.; Baumeister, W.; Miller, L.J.; et al. Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor. Nature 2018, 561, 492–497. [Google Scholar] [CrossRef]
- McLatchie, L.M.; Fraser, N.J.; Main, M.J.; Wise, A.; Brown, J.; Thompson, N.; Solari, R.; Lee, M.G.; Foord, S.M. RAMPs regulate the transport and ligand specificity of the calcitonin-receptor-like receptor. Nature 1998, 393, 333–339. [Google Scholar] [CrossRef]
- Klein, K.R.; Matson, B.C.; Caron, K.M. The expanding repertoire of receptor activity modifying protein (RAMP) function. Crit. Rev. Biochem. Mol. Biol. 2016, 51, 65–71. [Google Scholar] [CrossRef]
- Pioszak, A.A.; Hay, D.L. RAMPs as allosteric modulators of the calcitonin and calcitonin-like class B G protein-coupled receptors. Adv. Pharmacol. 2020, 88, 115–141. [Google Scholar]
- Tao, Y.X. Constitutive activation of G protein-coupled receptors and diseases: Insights into mechanism of activation and therapeutics. Pharmacol. Ther. 2008, 120, 129–148. [Google Scholar] [CrossRef]
- Kleinau, G.; Biebermann, H. Constitutive activities in the thyrotropin receptor: Regulation and significance. Adv. Pharmacol. 2014, 70, 81–119. [Google Scholar]
- Zhang, M.; Tao, Y.X.; Ryan, G.L.; Feng, X.; Fanelli, F.; Segaloff, D.L. Intrinsic differences in the response of the human lutropin receptor versus the human follitropin receptor to activating mutations. J. Boil. Chem. 2007, 282, 25527–25539. [Google Scholar] [CrossRef]
- Rosenkilde, M.M.; Smit, M.J.; Waldhoer, M. Structure, function and physiological consequences of virally encoded chemokine seven transmembrane receptors. Br. J. Pharmacol. 2008, 153, S154–S166. [Google Scholar] [CrossRef] [PubMed]
- Mo, X.L.; Tao, Y.X. Activation of MAPK by inverse agonists in six naturally occurring constitutively active mutant human melanocortin-4 receptors. Biochim. Biophys. Acta 2013, 1832, 1939–1948. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Tao, Y.X. Biased signaling initiated by agouti-related peptide through human melanocortin-3 and -4 receptors. Biochim. Biophys. Acta 2016, 1862, 1485–1494. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Liang, X.F.; Li, G.L.; Tao, Y.X. Biased signaling in fish melanocortin-4 receptors (MC4Rs): Divergent pharmacology of four ligands on spotted scat (Scatophagus argus) and grass carp (Ctenopharyngodon idella) MC4Rs. Mol. Cell Endocrinol. 2020, 515, 110929. [Google Scholar] [CrossRef] [PubMed]
- Agosti, F.; Cordisco Gonzalez, S.; Martinez Damonte, V.; Tolosa, M.J.; Di Siervi, N.; Schioth, H.B.; Davio, C.; Perello, M.; Raingo, J. Melanocortin 4 receptor constitutive activity inhibits L-type voltage-gated calcium channels in neurons. Neuroscience 2017, 346, 102–112. [Google Scholar] [CrossRef]
- Ghamari-Langroudi, M.; Digby, G.J.; Sebag, J.A.; Millhauser, G.L.; Palomino, R.; Matthews, R.; Gillyard, T.; Panaro, B.L.; Tough, I.R.; Cox, H.M.; et al. G-protein-independent coupling of MC4R to Kir7.1 in hypothalamic neurons. Nature 2015, 520, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Doronin, S.V.; Potapova, I.A.; Lu, Z.; Cohen, I.S. Angiotensin receptor type 1 forms a complex with the transient outward potassium channel Kv4.3 and regulates its gating properties and intracellular localization. J. Biol. Chem. 2004, 279, 48231–48237. [Google Scholar] [CrossRef]
- Fischer, J.; Kleinau, G.; Rutz, C.; Zwanziger, D.; Khajavi, N.; Muller, A.; Rehders, M.; Brix, K.; Worth, C.L.; Fuhrer, D.; et al. Evidence of G-protein-coupled receptor and substrate transporter heteromerization at a single molecule level. Cell Mol. Life Sci. 2018, 75, 2227–2239. [Google Scholar] [CrossRef]
- Ahuja, S.; Smith, S.O. Multiple switches in G protein-coupled receptor activation. Trends Pharmacol. Sci. 2009, 30, 494–502. [Google Scholar] [CrossRef]
- Bokoch, M.P.; Zou, Y.; Rasmussen, S.G.; Liu, C.W.; Nygaard, R.; Rosenbaum, D.M.; Fung, J.J.; Choi, H.J.; Thian, F.S.; Kobilka, T.S.; et al. Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor. Nature 2010, 463, 108–112. [Google Scholar] [CrossRef]
- Massotte, D.; Kieffer, B.L. The second extracellular loop: A damper for G protein-coupled receptors? Nat. Struct. Mol. Biol. 2005, 12, 287–288. [Google Scholar] [CrossRef] [PubMed]
- Peeters, M.C.; Van Westen, G.J.; Li, Q.; AP, I.J. Importance of the extracellular loops in G protein-coupled receptors for ligand recognition and receptor activation. Trends Pharmacol. Sci. 2011, 32, 35–42. [Google Scholar] [CrossRef] [PubMed]
- De Graaf, C.; Song, G.; Cao, C.; Zhao, Q.; Wang, M.W.; Wu, B.; Stevens, R.C. Extending the structural view of Class B GPCRs. Trends Biochem. Sci. 2017, 42, 946–960. [Google Scholar] [CrossRef] [PubMed]
- Wheatley, M.; Wootten, D.; Conner, M.T.; Simms, J.; Kendrick, R.; Logan, R.T.; Poyner, D.R.; Barwell, J. Lifting the lid on GPCRs: The role of extracellular loops. Br. J. Pharmacol. 2012, 165, 1688–1703. [Google Scholar] [CrossRef]
- Woolley, M.J.; Conner, A.C. Understanding the common themes and diverse roles of the second extracellular loop (ECL2) of the GPCR super-family. Mol. Cell Endocrinol. 2017, 449, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Karnik, S.S.; Khorana, H.G. Assembly of functional rhodopsin requires a disulfide bond between cysteine residues 110 and 187. J. Biol. Chem. 1990, 265, 17520–17524. [Google Scholar] [PubMed]
- Ryu, K.; Lee, H.; Kim, S.; Beauchamp, J.; Tung, C.S.; Isaacs, N.W.; Ji, I.; Ji, T.H. Modulation of high affinity hormone binding. Human choriogonadotropin binding to the exodomain of the receptor is influenced by exoloop 2 of the receptor. J. Biol. Chem. 1998, 273, 6285–6291. [Google Scholar] [CrossRef]
- Wichard, J.D.; Ter Laak, A.; Krause, G.; Heinrich, N.; Kuhne, R.; Kleinau, G. Chemogenomic analysis of G-protein coupled receptors and their ligands deciphers locks and keys governing diverse aspects of signalling. PLoS ONE 2011, 6, e16811. [Google Scholar] [CrossRef]
- Sebhat, I.K.; Martin, W.J.; Ye, Z.; Barakat, K.; Mosley, R.T.; Johnston, D.B.; Bakshi, R.; Palucki, B.; Weinberg, D.H.; MacNeil, T.; et al. Design and pharmacology of N-[(3R)-1,2,3,4-tetrahydroisoquinolinium- 3-ylcarbonyl]-(1R)-1-(4-chlorobenzyl)- 2-[4-cyclohexyl-4-(1H-1,2,4-triazol- 1-ylmethyl)piperidin-1-yl]-2-oxoethylamine (1), a potent, selective, melanocortin subtype-4 receptor agonist. J. Med. Chem. 2002, 45, 4589–4593. [Google Scholar] [CrossRef] [PubMed]
- Vos, T.J.; Caracoti, A.; Che, J.L.; Dai, M.; Farrer, C.A.; Forsyth, N.E.; Drabic, S.V.; Horlick, R.A.; Lamppu, D.; Yowe, D.L.; et al. Identification of 2-[2-[2-(5-bromo-2- methoxyphenyl)-ethyl]-3-fluorophenyl]-4,5-dihydro-1H-imidazole (ML00253764), a small molecule melanocortin 4 receptor antagonist that effectively reduces tumor-induced weight loss in a mouse model. J. Med. Chem. 2004, 47, 1602–1604. [Google Scholar] [CrossRef] [PubMed]
- Yi, T.L.; Yang, L.K.; Ruan, G.L.; Yang, D.Q.; Tao, Y.X. Melanocortin-4 receptor in swamp eel (Monopterus albus): Cloning, tissue distribution, and pharmacology. Gene 2018, 678, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, S.; Hornak, V.; Yan, E.C.; Syrett, N.; Goncalves, J.A.; Hirshfeld, A.; Ziliox, M.; Sakmar, T.P.; Sheves, M.; Reeves, P.J.; et al. Helix movement is coupled to displacement of the second extracellular loop in rhodopsin activation. Nat. Struct. Mol. Biol. 2009, 16, 168–175. [Google Scholar] [CrossRef]
- Venkatakrishnan, A.J.; Deupi, X.; Lebon, G.; Heydenreich, F.M.; Flock, T.; Miljus, T.; Balaji, S.; Bouvier, M.; Veprintsev, D.B.; Tate, C.G.; et al. Diverse activation pathways in class A GPCRs converge near the G-protein-coupling region. Nature 2016, 536, 484–487. [Google Scholar] [CrossRef]
- Hofmann, K.P.; Scheerer, P.; Hildebrand, P.W.; Choe, H.W.; Park, J.H.; Heck, M.; Ernst, O.P. A G protein-coupled receptor at work: The rhodopsin model. Trends Biochem. Sci. 2009, 34, 540–552. [Google Scholar] [CrossRef]
- Nygaard, R.; Frimurer, T.M.; Holst, B.; Rosenkilde, M.M.; Schwartz, T.W. Ligand binding and micro-switches in 7TM receptor structures. Trends Pharmacol. Sci. 2009, 30, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Katritch, V.; Fenalti, G.; Abola, E.E.; Roth, B.L.; Cherezov, V.; Stevens, R.C. Allosteric sodium in class A GPCR signaling. Trends Biochem. Sci. 2014, 39, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Venkatakrishnan, A.J.; Ma, A.K.; Fonseca, R.; Latorraca, N.R.; Kelly, B.; Betz, R.M.; Asawa, C.; Kobilka, B.K.; Dror, R.O. Diverse GPCRs exhibit conserved water networks for stabilization and activation. Proc. Natl. Acad. Sci. USA 2019, 116, 3288–3293. [Google Scholar] [CrossRef]
- Zarzycka, B.; Zaidi, S.A.; Roth, B.L.; Katritch, V. Harnessing ion-binding sites for GPCR pharmacology. Pharmacol. Rev. 2019, 71, 571–595. [Google Scholar] [CrossRef]
- Reiersen, H.; Rees, A.R. The hunchback and its neighbours: Proline as an environmental modulator. Trends Biochem. Sci. 2001, 26, 679–684. [Google Scholar] [CrossRef]
- Yohannan, S.; Faham, S.; Yang, D.; Whitelegge, J.P.; Bowie, J.U. The evolution of transmembrane helix kinks and the structural diversity of G protein-coupled receptors. Proc. Natl. Acad. Sci. USA 2004, 101, 959–963. [Google Scholar] [CrossRef]
- Sansuk, K.; Deupi, X.; Torrecillas, I.R.; Jongejan, A.; Nijmeijer, S.; Bakker, R.A.; Pardo, L.; Leurs, R. A structural insight into the reorientation of transmembrane domains 3 and 5 during family A G protein-coupled receptor activation. Mol. Pharmacol. 2011, 79, 262–269. [Google Scholar] [CrossRef]
- Hanson, M.A.; Roth, C.B.; Jo, E.; Griffith, M.T.; Scott, F.L.; Reinhart, G.; Desale, H.; Clemons, B.; Cahalan, S.M.; Schuerer, S.C.; et al. Crystal structure of a lipid G protein-coupled receptor. Science 2012, 335, 851–855. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zhang, J.; Gao, Z.G.; Zhang, D.; Zhu, L.; Han, G.W.; Moss, S.M.; Paoletta, S.; Kiselev, E.; Lu, W.; et al. Structure of the human P2Y12 receptor in complex with an antithrombotic drug. Nature 2014, 509, 115–118. [Google Scholar] [CrossRef]
- Hua, T.; Vemuri, K.; Pu, M.; Qu, L.; Han, G.W.; Wu, Y.; Zhao, S.; Shui, W.; Li, S.; Korde, A.; et al. Crystal structure of the human cannabinoid receptor CB1. Cell 2016, 167, 750–762. [Google Scholar] [CrossRef]
- Li, X.; Hua, T.; Vemuri, K.; Ho, J.H.; Wu, Y.; Wu, L.; Popov, P.; Benchama, O.; Zvonok, N.; Locke, K.; et al. Crystal structure of the human cannabinoid receptor CB2. Cell 2019, 176, 459–467. [Google Scholar] [CrossRef]
- Chrencik, J.E.; Roth, C.B.; Terakado, M.; Kurata, H.; Omi, R.; Kihara, Y.; Warshaviak, D.; Nakade, S.; Asmar-Rovira, G.; Mileni, M.; et al. Crystal structure of antagonist bound human lysophosphatidic acid receptor 1. Cell 2015, 161, 1633–1643. [Google Scholar] [CrossRef]
- Manivet, P.; Schneider, B.; Smith, J.C.; Choi, D.S.; Maroteaux, L.; Kellermann, O.; Launay, J.M. The serotonin binding site of human and murine 5-HT2B receptors: Molecular modeling and site-directed mutagenesis. J. Biol. Chem. 2002, 277, 17170–17178. [Google Scholar] [CrossRef]
- White, K.L.; Eddy, M.T.; Gao, Z.G.; Han, G.W.; Lian, T.; Deary, A.; Patel, N.; Jacobson, K.A.; Katritch, V.; Stevens, R.C. Structural connection between activation microswitch and allosteric sodium site in GPCR signaling. Structure 2018, 26, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Schiffmann, A.; Gimpl, G. Sodium functions as a negative allosteric modulator of the oxytocin receptor. Biochim. Biophys. Acta Biomembr. 2018, 1860, 1301–1308. [Google Scholar] [CrossRef]
- Cong, X.; Golebiowski, J. Allosteric Na+-binding site modulates CXCR4 activation. Phys. Chem. Chem. Phys. 2018, 20, 24915–24920. [Google Scholar] [CrossRef]
- Sun, X.; Laroche, G.; Wang, X.; Agren, H.; Bowman, G.R.; Giguere, P.M.; Tu, Y. Propagation of the allosteric modulation induced by sodium in the δ-opioid receptor. Chemistry 2017, 23, 4615–4624. [Google Scholar] [CrossRef]
- Vickery, O.N.; Carvalheda, C.A.; Zaidi, S.A.; Pisliakov, A.V.; Katritch, V.; Zachariae, U. Intracellular transfer of Na+ in an active-state G-protein-coupled receptor. Structure 2018, 26, 171–180. [Google Scholar] [CrossRef]
- Yang, Y.K.; Fong, T.M.; Dickinson, C.J.; Mao, C.; Li, J.Y.; Tota, M.R.; Mosley, R.; Van Der Ploeg, L.H.; Gantz, I. Molecular determinants of ligand binding to the human melanocortin-4 receptor. Biochemistry 2000, 39, 14900–14911. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.C.; Sartin, J.L.; Tao, Y.X. Pharmacological analyses of two naturally occurring porcine melanocortin-4 receptor mutations in domestic pigs. Domest. Anim. Endocrinol. 2008, 34, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Coster, M.; Wittkopf, D.; Kreuchwig, A.; Kleinau, G.; Thor, D.; Krause, G.; Schoneberg, T. Using ortholog sequence data to predict the functional relevance of mutations in G-protein-coupled receptors. FASEB J. 2012, 26, 3273–3281. [Google Scholar] [CrossRef] [PubMed]
- Salomon, Y. Melanocortin receptors: Targets for control by extracellular calcium. Mol. Cell Endocrinol. 1990, 70, 139–145. [Google Scholar] [CrossRef]
- Kopanchuk, S.; Veiksina, S.; Petrovska, R.; Mutule, I.; Szardenings, M.; Rinken, A.; Wikberg, J.E. Co-operative regulation of ligand binding to melanocortin receptor subtypes: Evidence for interacting binding sites. Eur. J. Pharmacol. 2005, 512, 85–95. [Google Scholar] [CrossRef]
- Rose, J.P.; Wang, B.C.; Weiss, M.S. Native SAD is maturing. IUCrJ 2015, 2 Pt 4, 431–440. [Google Scholar] [CrossRef]
- Ye, L.; Neale, C.; Sljoka, A.; Lyda, B.; Pichugin, D.; Tsuchimura, N.; Larda, S.T.; Pomes, R.; Garcia, A.E.; Ernst, O.P.; et al. Mechanistic insights into allosteric regulation of the A2A adenosine G protein-coupled receptor by physiological cations. Nat. Commun. 2018, 9, 1372. [Google Scholar] [CrossRef]
- Antoni, F.A.; Chadio, S.E. Essential role of magnesium in oxytocin-receptor affinity and ligand specificity. Biochem. J. 1989, 257, 611–614. [Google Scholar] [CrossRef]
- Huang, Y.; Zhou, Y.; Castiblanco, A.; Yang, W.; Brown, E.M.; Yang, J.J. Multiple Ca2+-binding sites in the extracellular domain of the Ca2+-sensing receptor corresponding to cooperative Ca2+ response. Biochemistry 2009, 48, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, E.J.; Bathgate, R.A.; Gooley, P.R. The human LGR7 low-density lipoprotein class A module requires calcium for structure. Ann. N. Y. Acad. Sci. 2005, 1041, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Garfield, A.S.; Shah, B.; Kleyn, P.; Ichetovkin, I.; Moeller, I.H.; Mowrey, W.R.; Van der Ploeg, L.H.T. Current mechanistic and pharmacodynamic understanding of melanocortin-4 receptor activation. Molecules 2019, 24, 1892. [Google Scholar] [CrossRef] [PubMed]
- Mountjoy, K.G.; Robbins, L.S.; Mortrud, M.T.; Cone, R.D. The cloning of a family of genes that encode the melanocortin receptors. Science 1992, 257, 1248–1251. [Google Scholar] [CrossRef]
- Gantz, I.; Fong, T.M. The melanocortin system. Am. J. Physiol. Endocronol. Metab. 2003, 284, E468–E474. [Google Scholar] [CrossRef]
- Cone, R.D. Studies on the physiological functions of the melanocortin system. Endocr. Rev. 2006, 27, 736–749. [Google Scholar] [CrossRef]
- Lu, D.; Willard, D.; Patel, I.R.; Kadwell, S.; Overton, L.; Kost, T.; Luther, M.; Chen, W.; Woychik, R.P.; Wilkison, W.O.; et al. Agouti protein is an antagonist of the melanocyte-stimulating-hormone receptor. Nature 1994, 371, 799–802. [Google Scholar] [CrossRef]
- Ollmann, M.M.; Wilson, B.D.; Yang, Y.K.; Kerns, J.A.; Chen, Y.; Gantz, I.; Barsh, G.S. Antagonism of central melanocortin receptors in vitro and in vivo by agouti-related protein. Science 1997, 278, 135–138. [Google Scholar] [CrossRef]
- Büch, T.R.; Heling, D.; Damm, E.; Gudermann, T.; Breit, A. Pertussis toxin-sensitive signaling of melanocortin-4 receptors in hypothalamic GT1–7 cells defines agouti-related protein as a biased agonist. J. Biol. Chem. 2009, 284, 26411–26420. [Google Scholar] [CrossRef]
- Inoue, A.; Raimondi, F.; Kadji, F.M.N.; Singh, G.; Kishi, T.; Uwamizu, A.; Ono, Y.; Shinjo, Y.; Ishida, S.; Arang, N.; et al. Illuminating G-protein-coupling selectivity of GPCRs. Cell 2019, 177, 1933–1947. [Google Scholar] [CrossRef]
- Gantz, I.; Miwa, H.; Konda, Y.; Shimoto, Y.; Tashiro, T.; Watson, S.J.; DelValle, J.; Yamada, T. Molecular cloning, expression, and gene localization of a fourth melanocortin receptor. J. Biol. Chem. 1993, 268, 15174–15179. [Google Scholar] [PubMed]
- Daniels, D.; Patten, C.S.; Roth, J.D.; Yee, D.K.; Fluharty, S.J. Melanocortin receptor signaling through mitogen-activated protein kinase in vitro and in rat hypothalamus. Brain Res. 2003, 986, 1–11. [Google Scholar] [CrossRef]
- Haskell-Luevano, C.; Monck, E.K. Agouti-related protein functions as an inverse agonist at a constitutively active brain melanocortin-4 receptor. Regul. Pept. 2001, 99, 1–7. [Google Scholar] [CrossRef]
- Nijenhuis, W.A.; Oosterom, J.; Adan, R.A. AgRP(83–132) acts as an inverse agonist on the human melanocortin-4 receptor. Mol. Endocrinol. 2001, 15, 164–171. [Google Scholar] [PubMed]
- Breit, A.; Wolff, K.; Kalwa, H.; Jarry, H.; Buch, T.; Gudermann, T. The natural inverse agonist agouti-related protein induces arrestin-mediated endocytosis of melanocortin-3 and -4 receptors. J. Biol. Chem. 2006, 281, 37447–37456. [Google Scholar] [CrossRef]
- Cai, M.; Hruby, V.J. Design of cyclized selective melanotropins. Biopolymers 2016, 106, 876–883. [Google Scholar] [CrossRef]
- Pfaus, J.; Giuliano, F.; Gelez, H. Bremelanotide: An overview of preclinical CNS effects on female sexual function. J. Sex. Med. 2007, 4, 269–279. [Google Scholar] [CrossRef]
- Simon, J.A.; Kingsberg, S.A.; Portman, D.; Williams, L.A.; Krop, J.; Jordan, R.; Lucas, J.; Clayton, A.H. Long-term safety and efficacy of bremelanotide for hypoactive sexual desire disorder. Obstet. Gynecol. 2019, 134, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Molinoff, P.B.; Shadiack, A.M.; Earle, D.; Diamond, L.E.; Quon, C.Y. PT-141: A melanocortin agonist for the treatment of sexual dysfunction. Ann. N. Y. Acad. Sci. 2003, 994, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, J.P.L.; Palmer, D.; Meldal, M. MC4R agonists: Structural overview on antiobesity therapeutics. Trends Pharmacol. Sci. 2018, 39, 402–423. [Google Scholar] [CrossRef] [PubMed]
- Chapman, K.L.; Kinsella, G.K.; Cox, A.; Donnelly, D.; Findlay, J.B. Interactions of the melanocortin-4 receptor with the peptide agonist NDP-MSH. J. Mol. Biol 2010, 401, 433–450. [Google Scholar] [CrossRef]
- Schioth, H.B.; Muceniece, R.; Mutulis, F.; Bouifrouri, A.A.; Mutule, I.; Wikberg, J.E. Further pharmacological characterization of the selective melanocortin 4 receptor antagonist HS014: Comparison with SHU9119. Neuropeptides 1999, 33, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chen, M.; Lai, Y.; Gantz, I.; Georgeson, K.E.; Harmon, C.M. Molecular determinants of human melanocortin-4 receptor responsible for antagonist SHU9119 selective activity. J. Biol. Chem. 2002, 277, 20328–20335. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Cai, M.; Aprahamian, C.J.; Georgeson, K.E.; Hruby, V.; Harmon, C.M.; Yang, Y. Contribution of the conserved amino acids of the melanocortin-4 receptor in [corrected] [Nle4,D-Phe7]-α-melanocyte-stimulating [corrected] hormone binding and signaling. J. Biol. Chem. 2007, 282, 21712–21719. [Google Scholar] [CrossRef]
- Nickolls, S.A.; Cismowski, M.I.; Wang, X.; Wolff, M.; Conlon, P.J.; Maki, R.A. Molecular determinants of melanocortin 4 receptor ligand binding and MC4/MC3 receptor selectivity. J. Pharmacol. Exp. Ther. 2003, 304, 1217–1227. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.X.; Segaloff, D.L. Functional characterization of melanocortin-4 receptor mutations associated with childhood obesity. Endocrinology 2003, 144, 4544–4551. [Google Scholar] [CrossRef] [PubMed]
- Kopanchuk, S.; Veiksina, S.; Mutulis, F.; Mutule, I.; Yahorava, S.; Mandrika, I.; Petrovska, R.; Rinken, A.; Wikberg, J.E. Kinetic evidence for tandemly arranged ligand binding sites in melanocortin 4 receptor complexes. Neurochem. Int. 2006, 49, 533–542. [Google Scholar] [CrossRef]
- Srisai, D.; Yin, T.C.; Lee, A.A.; Rouault, A.A.J.; Pearson, N.A.; Grobe, J.L.; Sebag, J.A. MRAP2 regulates ghrelin receptor signaling and hunger sensing. Nat. Commun. 2017, 8, 713. [Google Scholar] [CrossRef]
- Rouault, A.A.J.; Lee, A.A.; Sebag, J.A. Regions of MRAP2 required for the inhibition of orexin and prokineticin receptor signaling. Biochim. Biophys. Acta 2017, 1864, 2322–2329. [Google Scholar] [CrossRef]
- Chaly, A.L.; Srisai, D.; Gardner, E.E.; Sebag, J.A. The melanocortin receptor accessory protein 2 promotes food intake through inhibition of the prokineticin receptor-1. eLife 2016, 5, e12397. [Google Scholar] [CrossRef]
- Milligan, G.; Ward, R.J.; Marsango, S. GPCR homo-oligomerization. Curr. Opin. Cell Biol. 2019, 57, 40–47. [Google Scholar] [CrossRef]
- Ferre, S.; Ciruela, F.; Casado, V.; Pardo, L. Oligomerization of G protein-coupled receptors: Still doubted? Prog. Mol. Biol. Transl. Sci. 2020, 169, 297–321. [Google Scholar] [PubMed]
- Gassmann, M.; Shaban, H.; Vigot, R.; Sansig, G.; Haller, C.; Barbieri, S.; Humeau, Y.; Schuler, V.; Muller, M.; Kinzel, B.; et al. Redistribution of GABAB(1) protein and atypical GABAB responses in GABAB(2)-deficient mice. J. Neurosci. 2004, 24, 6086–6097. [Google Scholar] [CrossRef] [PubMed]
- Rashid, A.J.; So, C.H.; Kong, M.M.; Furtak, T.; El-Ghundi, M.; Cheng, R.; O’Dowd, B.F.; George, S.R. D1-D2 dopamine receptor heterooligomers with unique pharmacology are coupled to rapid activation of Gq/11 in the striatum. Proc. Natl. Acad. Sci. USA 2007, 104, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Waldhoer, M.; Fong, J.; Jones, R.M.; Lunzer, M.M.; Sharma, S.K.; Kostenis, E.; Portoghese, P.S.; Whistler, J.L. A heterodimer-selective agonist shows in vivo relevance of G protein-coupled receptor dimers. Proc. Natl. Acad. Sci. USA 2005, 102, 9050–9055. [Google Scholar] [CrossRef] [PubMed]
- Borroto-Escuela, D.O.; Fuxe, K. Oligomeric receptor complexes and their allosteric receptor-receptor interactions in the plasma membrane represent a new biological principle for integration of signals in the CNS. Front. Mol. Neurosci. 2019, 12, 230. [Google Scholar] [CrossRef]
- McGraw, D.W.; Mihlbachler, K.A.; Schwarb, M.R.; Rahman, F.F.; Small, K.M.; Almoosa, K.F.; Liggett, S.B. Airway smooth muscle prostaglandin-EP1 receptors directly modulate β2-adrenergic receptors within a unique heterodimeric complex. J. Clin. Invest. 2006, 116, 1400–1409. [Google Scholar] [CrossRef]
- Tadagaki, K.; Jockers, R.; Kamal, M. History and biological significance of GPCR heteromerization in the neuroendocrine system. Neuroendocrinology 2012, 95, 223–231. [Google Scholar] [CrossRef]
- Tadagaki, K.; Tudor, D.; Gbahou, F.; Tschische, P.; Waldhoer, M.; Bomsel, M.; Jockers, R.; Kamal, M. Human cytomegalovirus-encoded UL33 and UL78 heteromerize with host CCR5 and CXCR4 impairing their HIV coreceptor activity. Blood 2012, 119, 4908–4918. [Google Scholar] [CrossRef]
- Tschische, P.; Tadagaki, K.; Kamal, M.; Jockers, R.; Waldhoer, M. Heteromerization of human cytomegalovirus encoded chemokine receptors. Biochem. Pharmacol. 2011, 82, 610–619. [Google Scholar] [CrossRef]
- Quitterer, U.; Abdalla, S. Discovery of pathologic GPCR aggregation. Front. Med. 2019, 6, 9. [Google Scholar] [CrossRef] [PubMed]
- Levoye, A.; Dam, J.; Ayoub, M.A.; Guillaume, J.L.; Couturier, C.; Delagrange, P.; Jockers, R. The orphan GPR50 receptor specifically inhibits MT1 melatonin receptor function through heterodimerization. EMBO J. 2006, 25, 3012–3023. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.J. Dimerization in GPCR mobility and signaling. Curr. Opin. Pharmacol. 2010, 10, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Wouters, E.; Marin, A.R.; Dalton, J.A.R.; Giraldo, J.; Stove, C. Distinct dopamine D2 receptor antagonists differentially impact D2 receptor oligomerization. Int. J. Mol. Sci. 2019, 20, 1686. [Google Scholar] [CrossRef] [PubMed]
- Breit, A.; Lagace, M.; Bouvier, M. Hetero-oligomerization between β2- and β3-adrenergic receptors generates a β-adrenergic signaling unit with distinct functional properties. J. Biol. Chem. 2004, 279, 28756–28765. [Google Scholar] [CrossRef]
- Bouvier, M. Oligomerization of G-protein-coupled transmitter receptors. Nat. Rev. Neurosci. 2001, 2, 274–286. [Google Scholar] [CrossRef]
- George, S.R.; O’Dowd, B.F.; Lee, S.P. G-protein-coupled receptor oligomerization and its potential for drug discovery. Nat. Rev. Drug Discov. 2002, 1, 808–820. [Google Scholar] [CrossRef]
- Prasad, S.; Ponimaskin, E.; Zeug, A. Serotonin receptor oligomerization regulates cAMP-based signaling. J. Cell Sci. 2019, 132, jcs.230334. [Google Scholar] [CrossRef]
- Uberti, M.A.; Hague, C.; Oller, H.; Minneman, K.P.; Hall, R.A. Heterodimerization with β2-adrenergic receptors promotes surface expression and functional activity of α1D-adrenergic receptors. J. Pharmacol. Exp. Ther. 2005, 313, 16–23. [Google Scholar] [CrossRef]
- Ward, R.J.; Xu, T.R.; Milligan, G. GPCR oligomerization and receptor trafficking. Methods Enzymol. 2013, 521, 69–90. [Google Scholar]
- Ge, B.; Lao, J.; Li, J.; Chen, Y.; Song, Y.; Huang, F. Single-molecule imaging reveals dimerization/oligomerization of CXCR4 on plasma membrane closely related to its function. Sci. Rep. 2017, 7, 16873. [Google Scholar] [CrossRef] [PubMed]
- Schelshorn, D.; Joly, F.; Mutel, S.; Hampe, C.; Breton, B.; Mutel, V.; Lutjens, R. Lateral allosterism in the glucagon receptor family: Glucagon-like peptide 1 induces G-protein-coupled receptor heteromer formation. Mol. Pharmacol. 2012, 81, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Zoenen, M.; Urizar, E.; Swillens, S.; Vassart, G.; Costagliola, S. Evidence for activity-regulated hormone-binding cooperativity across glycoprotein hormone receptor homomers. Nat. Commun. 2012, 3, 1007. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lambert, N.A. GPCR dimers fall apart. Sci. Signal. 2010, 3, pe12. [Google Scholar] [CrossRef]
- Calebiro, D.; Rieken, F.; Wagner, J.; Sungkaworn, T.; Zabel, U.; Borzi, A.; Cocucci, E.; Zurn, A.; Lohse, M.J. Single-molecule analysis of fluorescently labeled G-protein-coupled receptors reveals complexes with distinct dynamics and organization. Proc. Natl. Acad. Sci. USA 2013, 110, 743–748. [Google Scholar] [CrossRef]
- Hern, J.A.; Baig, A.H.; Mashanov, G.I.; Birdsall, B.; Corrie, J.E.; Lazareno, S.; Molloy, J.E.; Birdsall, N.J. Formation and dissociation of M1 muscarinic receptor dimers seen by total internal reflection fluorescence imaging of single molecules. Proc. Natl. Acad. Sci. USA 2010, 107, 2693–2698. [Google Scholar] [CrossRef]
- Kasai, R.S.; Suzuki, K.G.; Prossnitz, E.R.; Koyama-Honda, I.; Nakada, C.; Fujiwara, T.K.; Kusumi, A. Full characterization of GPCR monomer-dimer dynamic equilibrium by single molecule imaging. J. Cell Biol. 2011, 192, 463–480. [Google Scholar] [CrossRef]
- Rediger, A.; Tarnow, P.; Bickenbach, A.; Schaefer, M.; Krude, H.; Gruters, A.; Biebermann, H. Heterodimerization of hypothalamic G-protein-coupled receptors involved in weight regulation. Obes. Facts 2009, 2, 80–86. [Google Scholar] [CrossRef]
- Lensing, C.J.; Freeman, K.T.; Schnell, S.M.; Speth, R.C.; Zarth, A.T.; Haskell-Luevano, C. Developing a biased unmatched bivalent ligand (BUmBL) design strategy to target the GPCR homodimer allosteric signaling (cAMP over β-arrestin 2 recruitment) within the melanocortin receptors. J. Med. Chem. 2018, 62, 144–158. [Google Scholar]
- Busnelli, M.; Kleinau, G.; Muttenthaler, M.; Stoev, S.; Manning, M.; Bibic, L.; Howell, L.A.; McCormick, P.J.; Di Lascio, S.; Braida, D.; et al. Design and characterization of superpotent bivalent ligands targeting oxytocin receptor dimers via a channel-like structure. J. Med. Chem. 2016, 59, 7152–7166. [Google Scholar]
- Schiedel, A.C.; Kose, M.; Barreto, C.; Bueschbell, B.; Morra, G.; Sensoy, O.; Moreira, I.S. Prediction and targeting of interaction interfaces in G-protein coupled receptor oligomers. Curr. Top. Med. Chem. 2018, 18, 714–746. [Google Scholar] [CrossRef] [PubMed]
- Barreto, C.A.V.; Baptista, S.J.; Preto, A.J.; Matos-Filipe, P.; Mourao, J.; Melo, R.; Moreira, I. Prediction and targeting of GPCR oligomer interfaces. Prog. Mol. Biol. Transl. Sci. 2020, 169, 105–149. [Google Scholar] [PubMed]
- Townsend-Nicholson, A.; Altwaijry, N.; Potterton, A.; Morao, I.; Heifetz, A. Computational prediction of GPCR oligomerization. Curr. Opin. Struct. Biol. 2019, 55, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Choe, H.W.; Kim, Y.J.; Park, J.H.; Morizumi, T.; Pai, E.F.; Krauss, N.; Hofmann, K.P.; Scheerer, P.; Ernst, O.P. Crystal structure of metarhodopsin II. Nature 2011, 471, 651–655. [Google Scholar] [CrossRef] [PubMed]
- Scheerer, P.; Park, J.H.; Hildebrand, P.W.; Kim, Y.J.; Krauss, N.; Choe, H.W.; Hofmann, K.P.; Ernst, O.P. Crystal structure of opsin in its G-protein-interacting conformation. Nature 2008, 455, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Scheerer, P.; Hofmann, K.P.; Choe, H.W.; Ernst, O.P. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature 2008, 454, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Szczepek, M.; Beyriere, F.; Hofmann, K.P.; Elgeti, M.; Kazmin, R.; Rose, A.; Bartl, F.J.; Von Stetten, D.; Heck, M.; Sommer, M.E.; et al. Crystal structure of a common GPCR-binding interface for G protein and arrestin. Nat. Commun. 2014, 5, 4801. [Google Scholar] [CrossRef]
- Wu, H.; Wacker, D.; Mileni, M.; Katritch, V.; Han, G.W.; Vardy, E.; Liu, W.; Thompson, A.A.; Huang, X.P.; Carroll, F.I.; et al. Structure of the human κ-opioid receptor in complex with JDTic. Nature 2012, 485, 327–332. [Google Scholar] [CrossRef]
- Huang, J.; Chen, S.; Zhang, J.J.; Huang, X.Y. Crystal structure of oligomeric β1-adrenergic G protein-coupled receptors in ligand-free basal state. Nat. Struct. Mol. Biol. 2013, 20, 419–425. [Google Scholar] [CrossRef]
- Guo, W.; Shi, L.; Filizola, M.; Weinstein, H.; Javitch, J.A. Crosstalk in G protein-coupled receptors: Changes at the transmembrane homodimer interface determine activation. Proc. Natl. Acad. Sci. USA 2005, 102, 17495–17500. [Google Scholar] [CrossRef]
- Bakker, R.A.; Dees, G.; Carrillo, J.J.; Booth, R.G.; Lopez-Gimenez, J.F.; Milligan, G.; Strange, P.G.; Leurs, R. Domain swapping in the human histamine H1 receptor. J. Pharmacol. Exp. Ther. 2004, 311, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Mancia, F.; Assur, Z.; Herman, A.G.; Siegel, R.; Hendrickson, W.A. Ligand sensitivity in dimeric associations of the serotonin 5HT2c receptor. EMBO Rep. 2008, 9, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Gorinski, N.; Kowalsman, N.; Renner, U.; Wirth, A.; Reinartz, M.T.; Seifert, R.; Zeug, A.; Ponimaskin, E.; Niv, M.Y. Computational and experimental analysis of the transmembrane domain 4/5 dimerization interface of the serotonin 5-HT1A receptor. Mol. Pharmacol. 2012, 82, 448–463. [Google Scholar] [CrossRef] [PubMed]
- George, S.R.; Lee, S.P.; Varghese, G.; Zeman, P.R.; Seeman, P.; Ng, G.Y.; O’Dowd, B.F. A transmembrane domain-derived peptide inhibits D1 dopamine receptor function without affecting receptor oligomerization. J. Biol. Chem. 1998, 273, 30244–30248. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Hu, K.; Liu, T.; Stern, M.K.; Mistry, R.; Challiss, R.A.; Costanzi, S.; Wess, J. Novel structural and functional insights into M3 muscarinic receptor dimer/oligomer formation. J. Biol. Chem. 2013, 288, 34777–34790. [Google Scholar] [CrossRef] [PubMed]
- Yanagawa, M.; Yamashita, T.; Shichida, Y. Comparative fluorescence resonance energy transfer analysis of metabotropic glutamate receptors: Implications about the dimeric arrangement and rearrangement upon ligand bindings. J. Biol. Chem. 2011, 286, 22971–22981. [Google Scholar] [CrossRef]
- Gurevich, V.V.; Gurevich, E.V. GPCRs and signal transducers: Interaction stoichiometry. Trends Pharmacol. Sci. 2018, 39, 672–684. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kleinau, G.; Heyder, N.A.; Tao, Y.-X.; Scheerer, P. Structural Complexity and Plasticity of Signaling Regulation at the Melanocortin-4 Receptor. Int. J. Mol. Sci. 2020, 21, 5728. https://doi.org/10.3390/ijms21165728
Kleinau G, Heyder NA, Tao Y-X, Scheerer P. Structural Complexity and Plasticity of Signaling Regulation at the Melanocortin-4 Receptor. International Journal of Molecular Sciences. 2020; 21(16):5728. https://doi.org/10.3390/ijms21165728
Chicago/Turabian StyleKleinau, Gunnar, Nicolas A. Heyder, Ya-Xiong Tao, and Patrick Scheerer. 2020. "Structural Complexity and Plasticity of Signaling Regulation at the Melanocortin-4 Receptor" International Journal of Molecular Sciences 21, no. 16: 5728. https://doi.org/10.3390/ijms21165728
APA StyleKleinau, G., Heyder, N. A., Tao, Y.-X., & Scheerer, P. (2020). Structural Complexity and Plasticity of Signaling Regulation at the Melanocortin-4 Receptor. International Journal of Molecular Sciences, 21(16), 5728. https://doi.org/10.3390/ijms21165728




