Phenylpropanoids Are Connected to Cell Wall Fortification and Stress Tolerance in Avocado Somatic Embryogenesis
Abstract
1. Introduction
2. Results
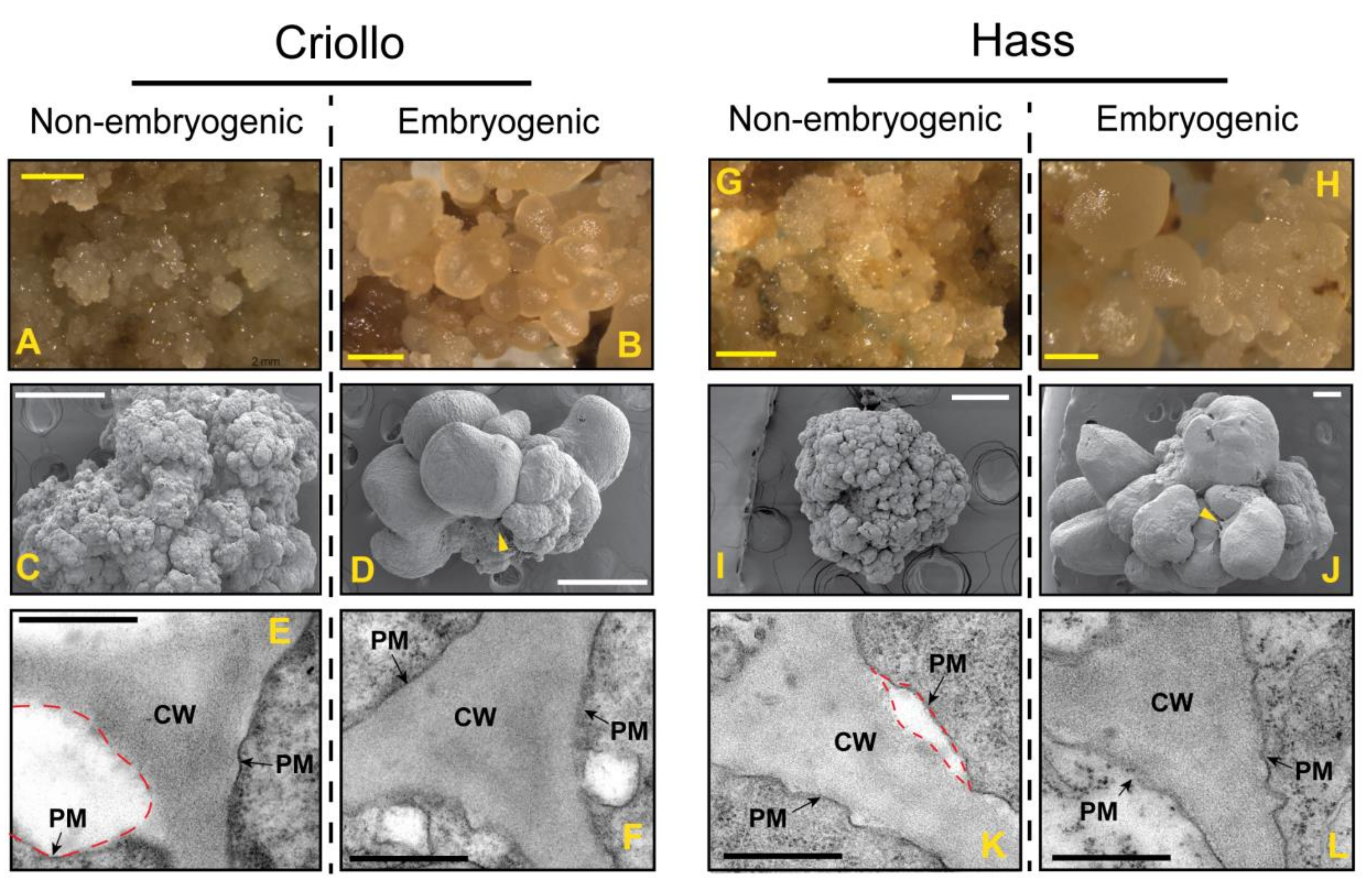
2.1. Establishment of Non-Embryogenic and Embryogenic Cultures of Hass and Criollo Avocado
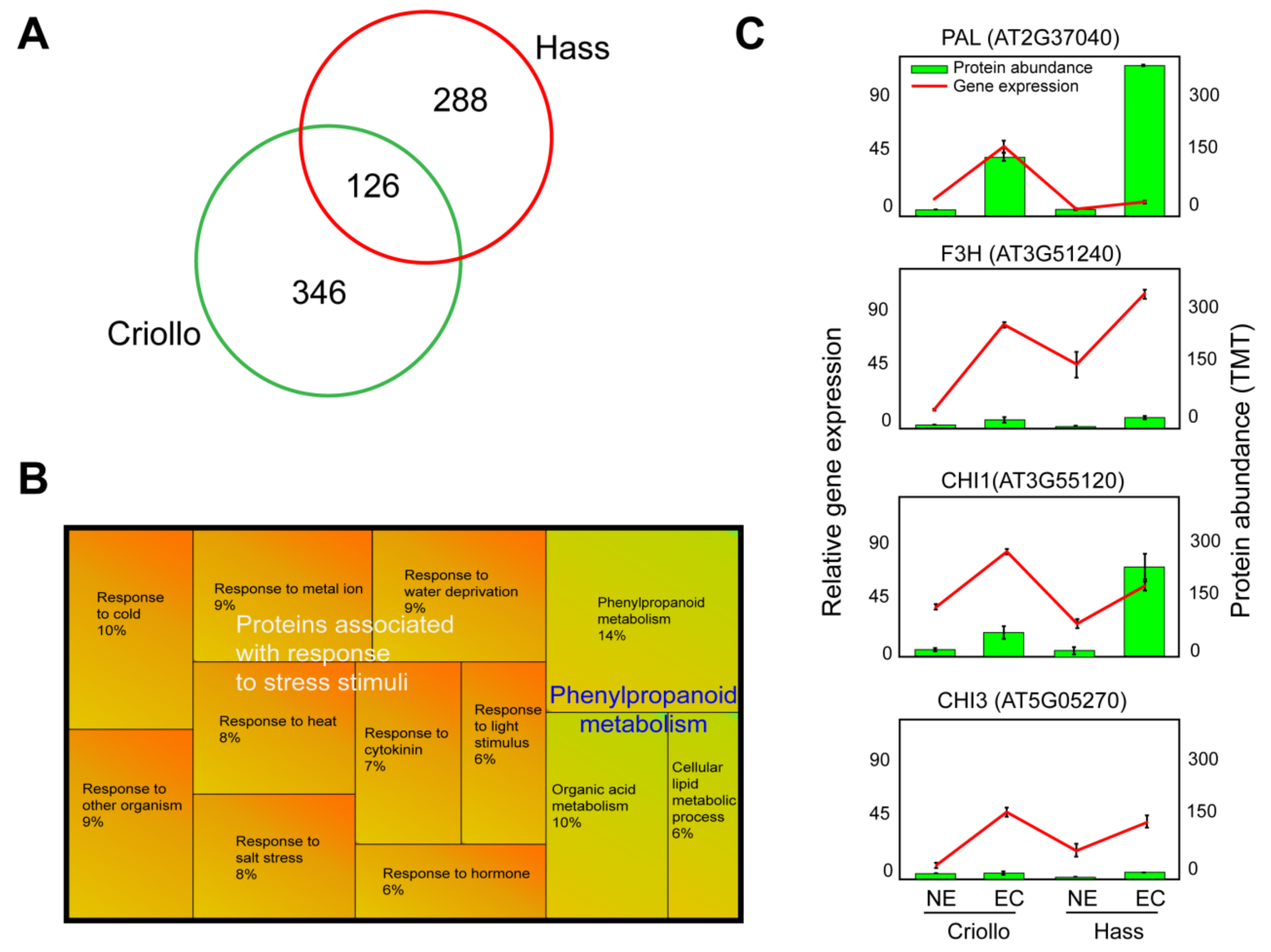
2.2. Comparative Proteomics: Embryogenic vs. Non-Embryogenic Cultures
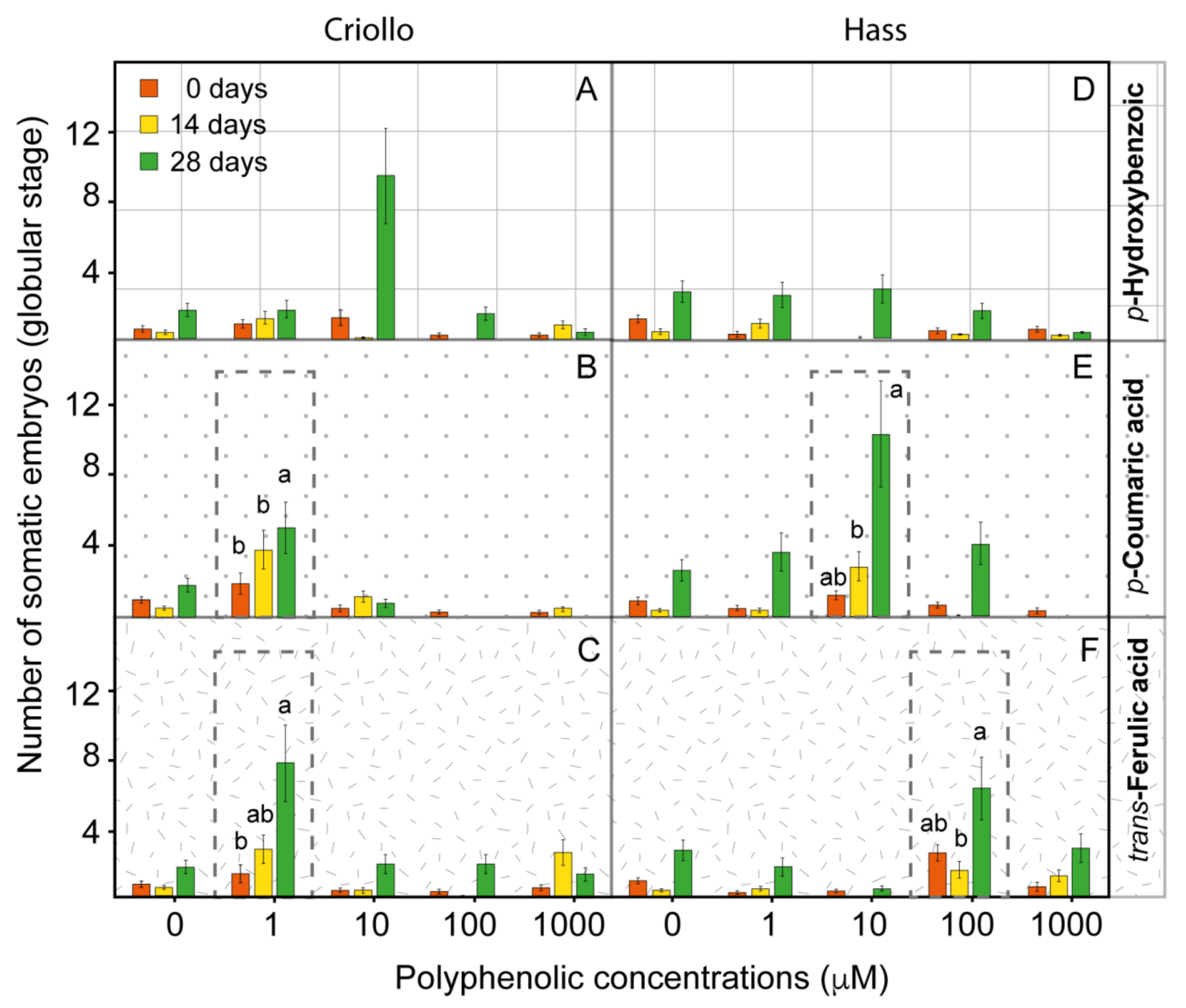
2.3. Phenolic Metabolites Improve the Production of Somatic Embryos in Hass and Criollo Avocado Embryogenetic Cultures
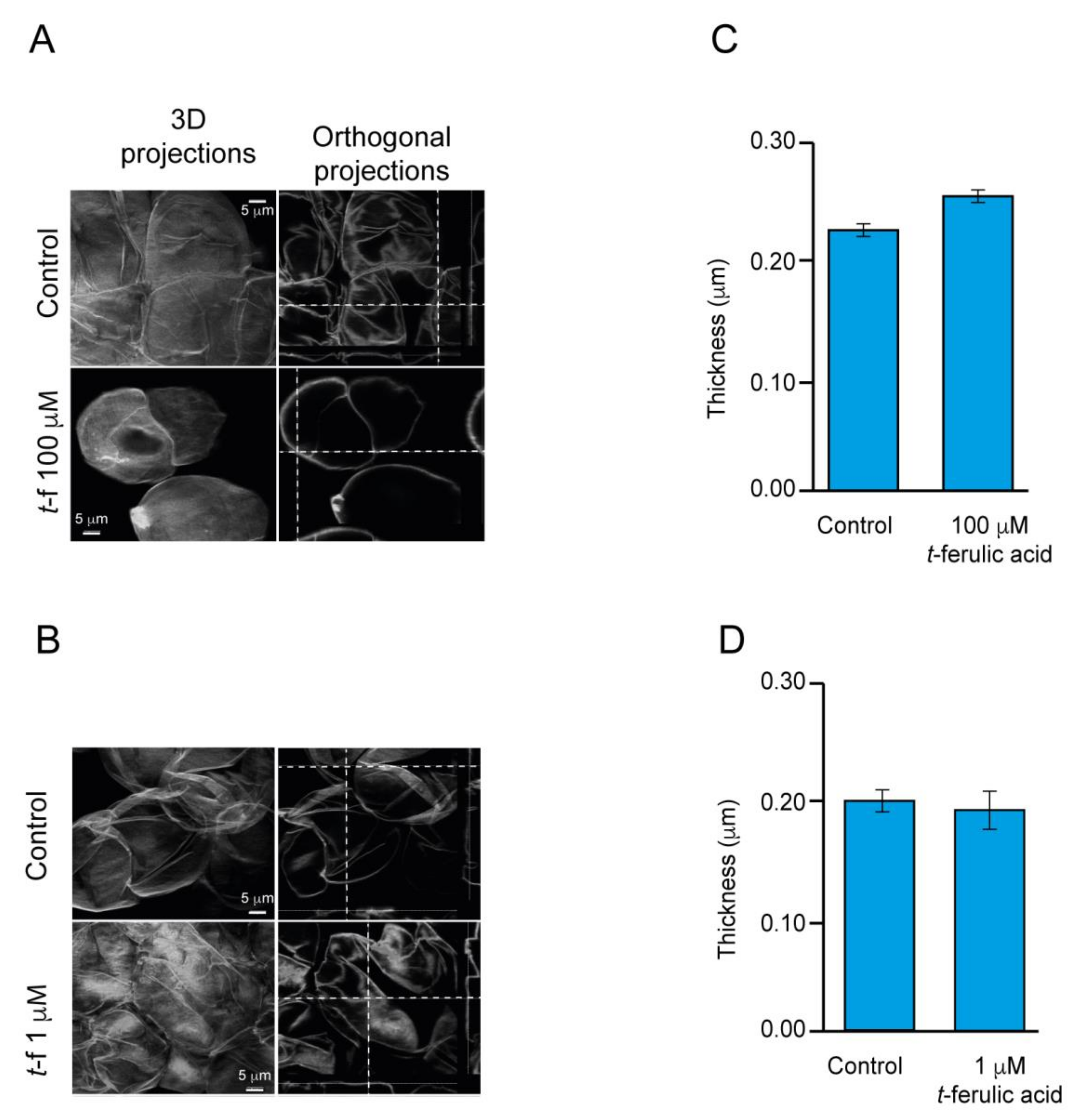
2.4. The Improvement of the Production of Embryos in Avocado EC Is Associated with the Alteration of the Endogenous Content of Polyphenolic Compounds
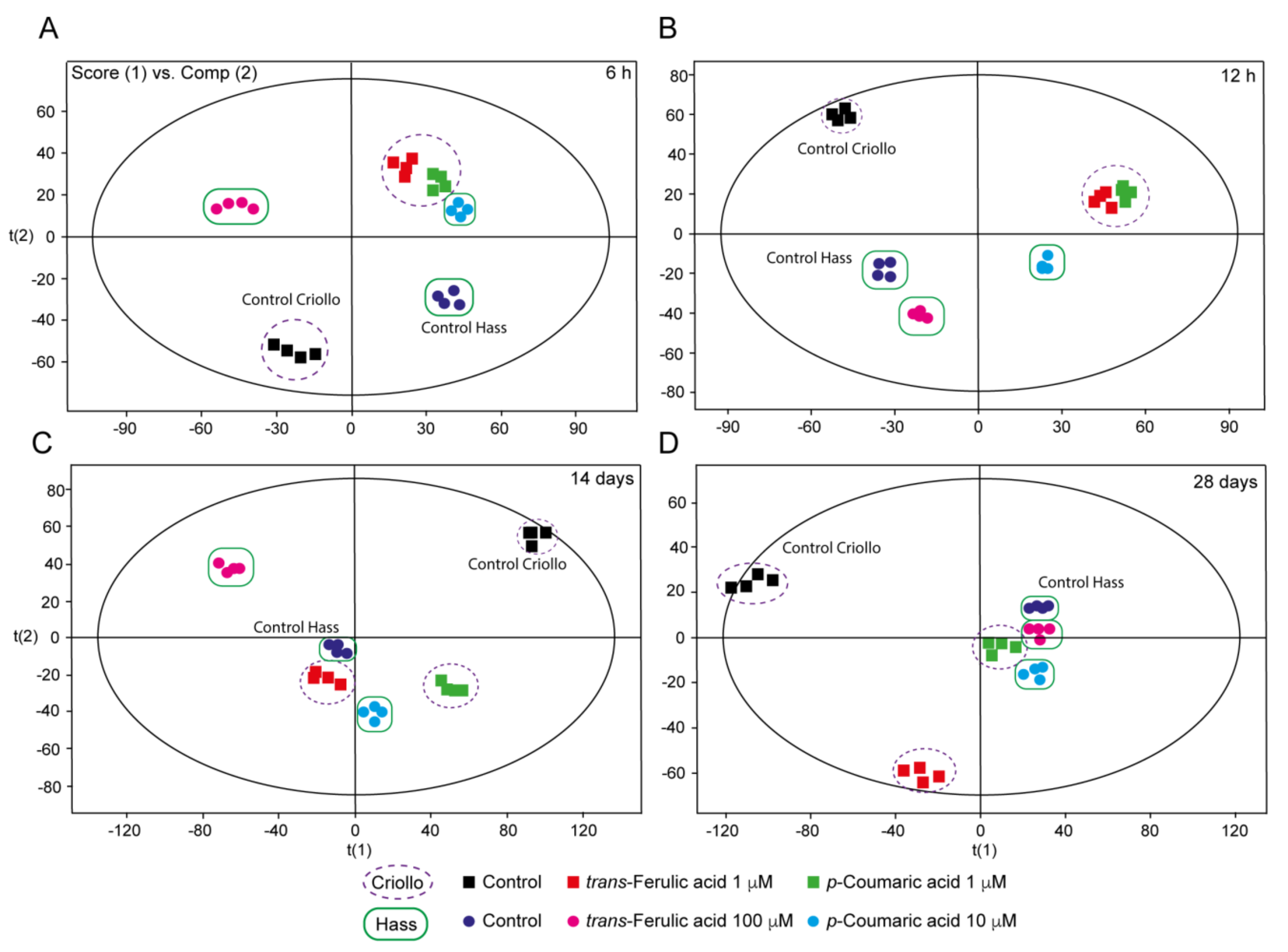
2.5. Untargeted Metabolomics Provides New Clues Related to the Effect of P-Coumaric and Trans-Ferulic Acid on the Improvement of Embryo Production in Avocado
3. Discussion
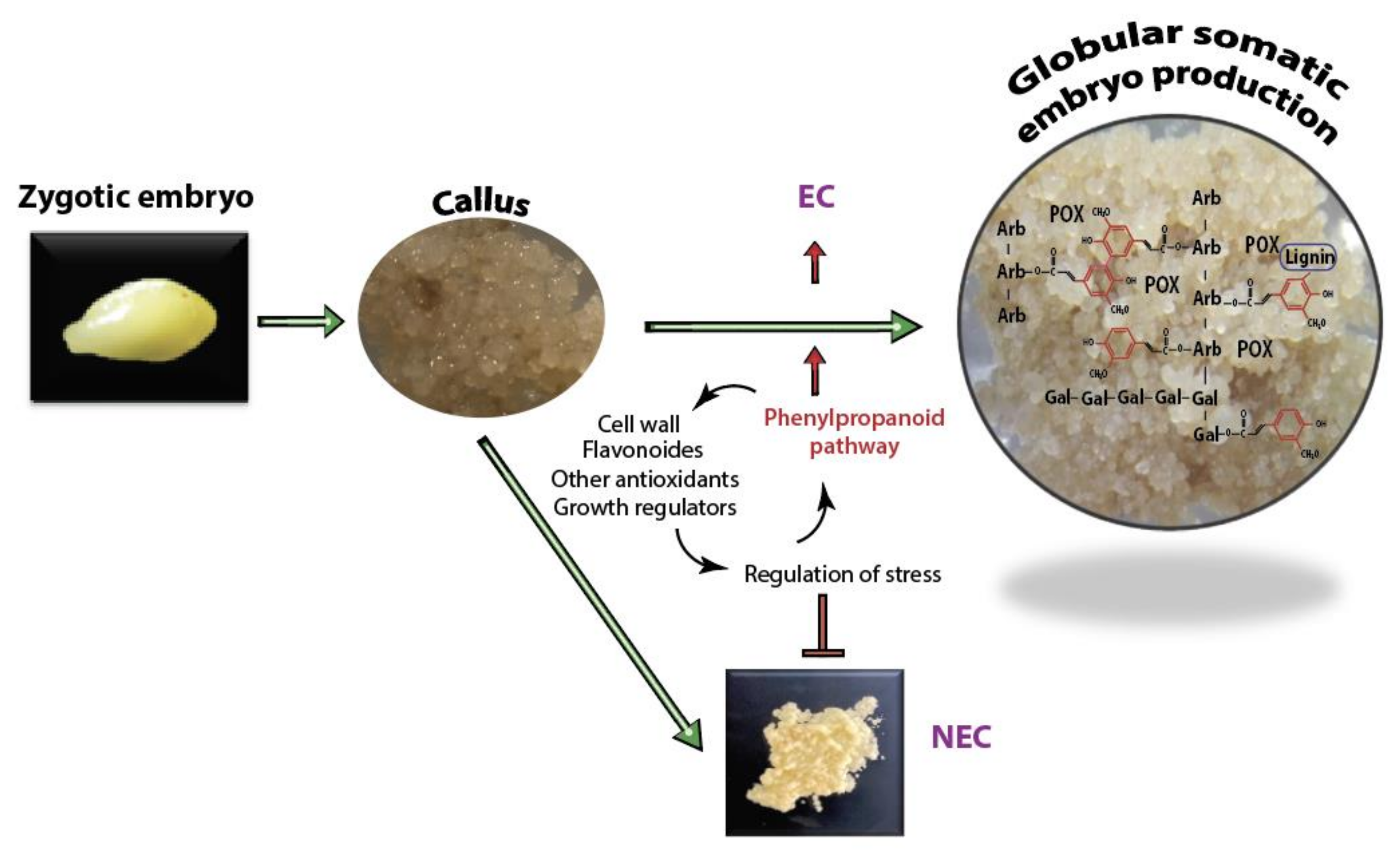
3.1. Stress Regulation and Cell Wall Fortification a Common Feature of Embryogenic Callus and Possible Association with the Improvement of Embryogenesis
3.2. The Improvement of the Production of Somatic Embryos by Polyphenolics Is Linked to the General Reduction in the Level of Endogenous Phenolic Compounds
3.3. Change in Metabolic Flow May Explains the Positive Association between Polyphenolics and the Overproliferation of Avocado Somatic Embryos
4. Materials and Methods
4.1. Chemical Reagents
4.2. Establishment of Embryogenic Cultures
4.3. Protein Extraction
4.4. Proteomics Analysis
4.5. Protein Digestion and Tandem Mass Tags (TMT) Labeling
4.6. NanoLC-MS/MS Analysis and Synchronous Precursor Selection (SPS)-MS3 for TMT Analysis
4.7. Data Analysis and Interpretation
4.8. Phytochemical Extraction
4.9. Determination and Quantification of Phenolic Compounds and Untargeted Metabolomics Analysis
4.10. Stereoscope Analysis
4.11. Scanning Electron. Microscopy Preparation (SEM)
4.12. Transmission Electron. Microscopy (TEM)
4.13. Confocal Microscopy
4.14. Embryogenic Callus Treatments with Phenolic Compounds
4.15. RNA Extraction and Quantitative PCR Assays (qPCR)
4.16. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ALH2C4 | Aldehyde dehydrogenase family 2-member C4 |
| c-CA | cis-cinnamic acid |
| p-CA | p-coumaric acid |
| t-CA | trans-cinnamic acid |
| CAD9 | Cinnamyl alcohol dehydrogenase 9 |
| BSA | Bovine serum albumin |
| CCoAOMT | Caffeoyl-CoA O-methyltransferase |
| CCoAOMT1 | Caffeoyl-CoA O-methyltransferase 1 |
| C4H/ CYP73A5 | Cinnamic acid 4-hydroxylases |
| 4CL1 | 4-coumarate: CoA ligase 1 |
| EC | Embryogenic callus |
| EMSN | Extracellular matrix surface Network |
| EP | Embryogenic potency |
| t-FA | trans-ferulic acid |
| GLM | General linear model |
| LDOX | Leucoanthocyanidin dioxygenase |
| MS | Murashige and Skoog |
| MSP | Murashige and Skoog medium supplemented with 0.1 mg L−1 picloram |
| NEC | Non-embryogenic callus |
| OMT1 | Flavone 3’-O-methyltransferase 1 |
| PAL1 | Phenylalanine ammonia-lyase 1 |
| PCA | Principal component analysis |
| PHBA | p-hydroxybenzoic acid |
| POX52 | Peroxidase 52 |
| PP | Phenylpropanoid pathway |
| ROS | Reactive oxygen species |
| SA | Sinapic acid |
| SE | Somatic embryogenesis |
| TCEP | tris (2-carboxyethyl) phosphine |
| TMT | Tandem mass tags |
| VBE | Vanillyl benzyl ether |
| VA | Vanillin |
Appendix A
References
- Smith, S.M.; Gomez, D.F.; Beaver, R.A.; Hulcr, J.; Cognato, A.I. Reassessment of the species in the Euwallacea fornicatus (Coleoptera: Curculionidae: Scolytinae) complex after the rediscovery of the “lost” type specimen. Insects 2019, 10, 261. [Google Scholar] [CrossRef]
- Encina, C.L.; Parisi, A.; O’Brien, C.; Mitter, N. Enhancing somatic embryogenesis in avocado (Persea americana Mill.) using a two-step culture system and including glutamine in the culture medium. Sci. Hortic. 2014, 165, 44–50. [Google Scholar] [CrossRef]
- Lavi, U.; Lahav, E.; Genizi, A.; Degani, C.; Gazit, S.; Hillel, J. Quantitative genetic analysis of traits in Avocado cultivars. Plant Breed. 1991, 106, 149–160. [Google Scholar] [CrossRef]
- Van Nocker, S.; Gardiner, S.E. Breeding better cultivars, faster: Applications of new technologies for the rapid deployment of superior horticultural tree crops. Hortic. Res. 2014, 1, 14022. [Google Scholar] [CrossRef] [PubMed]
- Pliego-Alfaro, F.; Barceló-Muñoz, A.; Simón-Pérez, E.; de la Viña-Nieto, G.; Sánchez-Romero, C.; Perán-Quesada, R. La micropropagación en la mejora de patrones de aguacate (Persea americana Mill.): Problemas y limitaciones. Rev. Chapingo Ser. Hortic. 1999, 5, 239–244. [Google Scholar]
- Palomo-Ríos, E.; Perez, C.; Mercado, J.A.; Pliego-Alfaro, F. Enhancing frequency of regeneration of somatic embryos of avocado (Persea americana Mill.) using semi-permeable cellulose acetate membranes. Plant Cell Tissue Organ Cult. 2013, 115, 199–207. [Google Scholar] [CrossRef]
- Pliego-Alfaro, F.; Barceló-Muñoz, A.; Lopez-Gomez, R.; Ibarra-Laclette, E.; Herrera-Estrella, L.; Palomo-Ríos, E.; Mercado, J.A.; Litz, R.E. Biotechnology. In The Avocado: Botany, Production and Uses; CABI Publishing: Wallingford, Oxfordshire, UK, 2013; pp. 268–300. [Google Scholar]
- O’Brien, C.; Hiti-Bandaralage, J.C.A.; Hayward, A.; Mitter, N. Avocado (Persea americana Mill.). In Biological Nitrogen Fixation in Forest Ecosystems: Foundations and Applications; Springer Science and Business Media LLC: Berlin, Germany, 2018; Volume II, pp. 305–328. [Google Scholar]
- Pliego-Alfaro, F.; Murashige, T. Somatic embryogenesis in avocado (Persea americana Mill.) in vitro. Plant Cell Tissue Organ Cult. 1988, 12, 61–66. [Google Scholar] [CrossRef]
- Litz, R.E.; Litz, W. Maturation of avocado somatic embryos and plant recovery. Plant Cell Tissue Organ Cult. 1999, 58, 141–148. [Google Scholar] [CrossRef]
- Sánchez-Romero, C.; Márquez-Martín, B.; Pliego-Alfaro, F. Somatic and zygotic embryogenesis in avocado. In The Plant Plasma Membrane; Springer Science and Business Media LLC: Berlin, Germany, 2005; Volume 2, pp. 271–284. [Google Scholar]
- Perán-Quesada, R.; Sánchez-Romero, C.; Barceló-Muñoz, A.; Pliego-Alfaro, F. Factors affecting maturation of avocado somatic embryos. Sci. Hortic. 2004, 102, 61–73. [Google Scholar] [CrossRef]
- Márquez-Martín, B.; Sesmero, R.; Quesada, M.A.; Pliego-Alfaro, F.; Sánchez-Romero, C. Water relations in culture media influence maturation of avocado somatic embryos. J. Plant Physiol. 2011, 168, 2028–2034. [Google Scholar] [CrossRef] [PubMed]
- Guzmán-García, E.; Sánchez-Romero, C.; Panis, B.; Carpentier, S. The use of 2D-DIGE to understand the regeneration of somatic embryos in avocado. Proteomics 2013, 13, 3498–3507. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Yang, X.; Guo, K.; Deng, J.; Xu, J.; Gao, W.; Lindsey, K.; Zhang, X. ROS homeostasis regulates somatic embryogenesis via the regulation of auxin signaling in cotton. Mol. Cell. Proteom. 2016, 15, 2108–2124. [Google Scholar] [CrossRef] [PubMed]
- Causevic, A.; Delaunay, A.; Ounnar, S.; Righezza, M.; Delmotte, F.; Brignolas, F.; Hagège, D.; Maury, S. DNA methylating and demethylating treatments modify phenotype and cell wall differentiation state in sugarbeet cell lines. Plant Physiol. Biochem. 2005, 43, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, N.; Kobayashi, H.; Togashi, T.; Mori, Y.; Kikuchi, K.; Kuriyama, K.; Tokuji, Y. Formation of embryogenic cell clumps from carrot epidermal cells is suppressed by 5-azacytidine, a DNA methylation inhibitor. J. Plant Physiol. 2005, 162, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Custard, K.D.; Brown, R.C.; Lemmon, B.E.; Harada, J.J.; Goldberg, R.B.; Fischer, R.L. DNA Methylation is critical for Arabidopsis embryogenesis and seed viability. Plant Cell 2006, 18, 805–814. [Google Scholar] [CrossRef]
- Valledor, L.; Meijón, M.; Hasbun, R.; Cañal, M.J.; Rodríguez, R. Variations in DNA methylation, acetylated histone H4, and methylated histone H3 during Pinus radiata needle maturation in relation to the loss of in vitro organogenic capability. J. Plant Physiol. 2010, 167, 351–357. [Google Scholar] [CrossRef]
- Viejo, M.; Rodríguez, R.; Valledor, L.; Pérez, M.; Cañal, M.-J.; Hasbún, R. DNA methylation during sexual embryogenesis and implications on the induction of somatic embryogenesis in Castanea sativa Miller. Sex. Plant Reprod. 2010, 23, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Fehér, A. Somatic embryogenesis — Stress-induced remodeling of plant cell fate. Biochim. Biophys. Acta (BBA) Bioenerg. 2015, 1849, 385–402. [Google Scholar] [CrossRef]
- Nic-Can, G.; Galaz-Ávalos, R.M.; De-La-Peña, C.; Magaña, A.A.; Wróbel, K.; Loyola-Vargas, V.M. Somatic embryogenesis: Identified factors that lead to embryogenic repression. A case of species of the same genus. PLoS ONE 2015, 10, e0126414. [Google Scholar] [CrossRef]
- Us-Camas, R.; Rivera-Solis, G.; Duarte-Aké, F.; De-La-Peña, C. In vitro culture: An epigenetic challenge for plants. Plant Cell Tissue Organ Cult. 2014, 118, 187–201. [Google Scholar] [CrossRef]
- De-La-Peña, C.; Nic-Can, G.; Galaz-Ávalos, R.M.; Avilez-Montalvo, R.; Loyola-Vargas, V.M. The role of chromatin modifications in somatic embryogenesis in plants. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Duarte-Aké, F.; De-La-Peña, C. Epigenetic advances in somatic embryogenesis in sequenced genome crops. In Somatic Embryogenesis: Fundamental Aspects and Applications; Springer Science and Business Media LLC: Berlin, Germany, 2016; pp. 81–102. [Google Scholar]
- Cvikrová, M.; Malá, J.; Hrubcová, M.; Eder, J.; Zoń, J.; Macháčková, I. Effect of inhibition of biosynthesis of phenylpropanoids on sessile oak somatic embryogenesis. Plant Physiol. Biochem. 2003, 41, 251–259. [Google Scholar] [CrossRef]
- Misra, B.B.; Dey, S. Accumulation patterns of phenylpropanoids and enzymes in East Indian sandalwood tree undergoing developmental progression ‘in vitro’. Aust. J. Crop. Sci. 2013, 7, 681–690. [Google Scholar]
- Kobayashi, T.; Higashi, K.; Sasaki, K.; Asami, T.; Yoshida, S.; Kamada, H. Purification from conditioned medium and chemical identification of a factor that inhibits somatic embryogenesis in carrot. Plant Cell Physiol. 2000, 41, 268–273. [Google Scholar] [CrossRef]
- Alemanno, L.; Ramos, T.; Gargadenec, A.; Andary, C.; Ferriere, N. Localization and identification of phenolic compounds in Theobroma cacao L. somatic embryogenesis. Ann. Bot. 2003, 92, 613–623. [Google Scholar] [CrossRef]
- Umehara, M.; Ogita, S.; Sasamoto, H.; Koshino, H.; Asami, T.; Fujioka, S.; Yoshida, S.; Kamada, H. Identification of a Novel factor, Vanillyl Benzyl Ether, Which Inhibits Somatic Embryogenesis of Japanese Larch (Larix leptolepis Gordon). Plant Cell Physiol. 2005, 46, 445–453. [Google Scholar] [CrossRef]
- Umehara, M.; Ogita, S.; Sasamoto, H.; Koshino, H.; Nakamura, T.; Asami, T.; Yoshida, S.; Kamada, H. Identification of a factor that complementarily inhibits somatic embryogenesis with vanillyl benzyl ether in Japanese larch. Vitr. Cell. Dev. Boil. Anim. 2007, 43, 203–208. [Google Scholar] [CrossRef]
- Rendón-Anaya, M.; Ibarra-Laclette, E.; Méndez-Bravo, A.; Lan, T.; Zheng, C.; Carretero-Paulet, L.; Perez-Torres, C.A.; Chacón-López, A.; Hernandez-Guzmán, G.; Chang, T.-H.; et al. The avocado genome informs deep angiosperm phylogeny, highlights introgressive hybridization, and reveals pathogen-influenced gene space adaptation. Proc. Natl. Acad. Sci. USA 2019, 116, 17081–17089. [Google Scholar] [CrossRef]
- Jiang, H.; Wood, K.V.; A Morgan, J. Metabolic engineering of the phenylpropanoid pathway in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2005, 71, 2962–2969. [Google Scholar] [CrossRef]
- Qualley, A.V.; Widhalm, J.R.; Adebesin, F.; Kish, C.M.; Dudareva, N. Completion of the core -oxidative pathway of benzoic acid biosynthesis in plants. Proc. Natl. Acad. Sci. USA 2012, 109, 16383–16388. [Google Scholar] [CrossRef]
- Smertenko, A.; Bozhkov, P.V. Somatic embryogenesis: Life and death processes during apical–basal patterning. J. Exp. Bot. 2014, 65, 1343–1360. [Google Scholar] [CrossRef] [PubMed]
- Nic-Can, G.; Avilez-Montalvo, J.R.; Aviles-Montalvo, R.N.; Márquez-López, R.E.; Mellado-Mojica, E.; Galaz-Ávalos, R.M.; Loyola-Vargas, V.M. The Relationship between stress and somatic embryogenesis. In Somatic Embryogenesis: Fundamental Aspects and Applications; Springer Science and Business Media LLC: Berlin, Germany, 2016; pp. 151–170. [Google Scholar]
- Agati, G.; Azzarello, E.; Pollastri, S.; Tattini, M. Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci. 2012, 196, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Mouradov, A.; Spangenberg, G. Flavonoids: A metabolic network mediating plants adaptation to their real estate. Front. Plant Sci. 2014, 5, 620. [Google Scholar] [CrossRef] [PubMed]
- Takeda, H.; Kotake, T.; Nakagawa, N.; Sakurai, N.; Nevins, D.J. Expression and function of cell wall-bound cationic peroxidase in asparagus somatic embryogenesis. Plant Physiol. 2003, 131, 1765–1774. [Google Scholar] [CrossRef] [PubMed]
- Gallego, P.; Martin, L.; Blázquez, A.; Guerra, H.; Villalobos, N. Involvement of peroxidase activity in developing somatic embryos of Medicago arborea L. Identification of an isozyme peroxidase as biochemical marker of somatic embryogenesis. J. Plant Physiol. 2014, 171, 78–84. [Google Scholar] [CrossRef]
- Novaković, L.; Guo, T.; Bacic, A.; Sampathkumar, A.; Johnson, K.L. Hitting the Wall—sensing and signaling pathways involved in plant cell wall remodeling in response to abiotic stress. Plants 2018, 7, 89. [Google Scholar] [CrossRef]
- Pilarska, M.; Knox, J.; Konieczny, R. Arabinogalactan-protein and pectin epitopes in relation to an extracellular matrix surface network and somatic embryogenesis and callogenesis in Trifolium nigrescens Viv. Plant Cell Tissue Organ Cult. 2013, 115, 35–44. [Google Scholar] [CrossRef]
- Mathew, S.; Abraham, T.E. Ferulic acid: An antioxidant found naturally in plant cell walls and feruloyl esterases involved in its release and their applications. Crit. Rev. Biotechnol. 2004, 24, 59–83. [Google Scholar] [CrossRef]
- Lenucci, M.S.; Piro, G.; Dalessandro, G. In muro feruloylation and oxidative coupling in monocots. Plant Signal. Behav. 2009, 4, 228–230. [Google Scholar] [CrossRef][Green Version]
- Schoch, G.; Goepfert, S.; Morant, M.; Hehn, A.; Meyer, D.; Ullmann, P.; Werck-Reichhart, D. CYP98A3 from Arabidopsis thaliana is a 3′-Hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J. Biol. Chem. 2001, 276, 36566–36574. [Google Scholar] [CrossRef]
- Franke, R.; Humphreys, J.M.; Hemm, M.R.; Denault, J.W.; Ruegger, M.O.; Cusumano, J.C.; Chapple, C.C.S. The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J. 2002, 30, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Tahir, N.; Pollet, B.; Ehlting, J.; Larsen, K.; Asnaghi, C.; Ronseau, S.; Proux, C.; Erhardt, M.; Seltzer, V.; Renou, J.-P.; et al. A coumaroyl-ester-3-hydroxylase insertion mutant reveals the existence of nonredundant meta-hydroxylation pathways and essential roles forphenolic precursors in cell expansion and plant growth. Plant Physiol. 2005, 140, 30–48. [Google Scholar] [CrossRef]
- Zhang, Y.; Butelli, E.; De Stefano, R.; Schoonbeek, H.-J.; Magusin, A.; Pagliarani, C.; Wellner, N.; Hill, L.; Orzaez, D.; Granell, A.; et al. Anthocyanins double the shelf life of tomatoes by delaying overripening and reducing susceptibility to gray mold. Curr. Boil. 2013, 23, 1094–1100. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, C.-J. Multifaceted Regulations of gateway enzyme phenylalanine ammonia-lyase in the biosynthesis of phenylpropanoids. Mol. Plant 2015, 8, 17–27. [Google Scholar] [CrossRef]
- Palacio, L.; Cantero, J.J.; Cusido, R.M.; Goleniowski, M.E. Phenolic compound production in relation to differentiation in cell and tissue cultures of Larrea divaricata (Cav.). Plant Sci. 2012, 193, 1–7. [Google Scholar] [CrossRef]
- Szopa, A.; Ekiert, H. Production of biologically active phenolic acids in Aronia melanocarpa (Michx.) Elliott in vitro cultures cultivated on different variants of the Murashige and Skoog medium. Plant Growth Regul. 2013, 72, 51–58. [Google Scholar] [CrossRef]
- Moyo, M.; Amoo, S.O.; Aremu, A.O.; Gruz, J.; Šubrtová, M.; Dolezal, K.; Van Staden, J. Plant regeneration and biochemical accumulation of hydroxybenzoic and hydroxycinnamic acid derivatives in Hypoxis hemerocallidea organ and callus cultures. Plant Sci. 2014, 227, 157–164. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, T.; Wu, Y.; Zhou, Y.; Jiang, Y.; Zhang, L. Effect of elicitors on the metabolites in the suspension cell culture of Salvia miltiorrhiza Bunge. Physiol. Mol. Boil. Plants 2018, 25, 229–242. [Google Scholar] [CrossRef]
- Newton, R.P.; Smith, C.J. Cyclic nucleotides. Phytochemistry 2004, 65, 2423–2437. [Google Scholar] [CrossRef]
- Steenackers, W.J.; Klíma, P.; Quareshy, M.; Cesarino, I.; Kumpf, R.P.; Corneillie, S.; Araújo, P.; Viaene, T.; Goeminne, G.; Nowack, M.K.; et al. cis-Cinnamic acid is a novel, natural auxin efflux inhibitor that promotes lateral root formation. Plant Physiol. 2016, 173, 552–565. [Google Scholar] [CrossRef]
- Kurepa, J.; Shull, T.E.; Karunadasa, S.S.; Smalle, J.A. Modulation of auxin and cytokinin responses by early steps of the phenylpropanoid pathway. BMC Plant Biol. 2018, 18, 278. [Google Scholar] [CrossRef] [PubMed]
- Trewavas, A. Plant cyclic AMP comes in from the cold. Nature 1997, 390, 657–658. [Google Scholar] [CrossRef] [PubMed]
- Nic-Can, G.; Loyola-Vargas, V.M. The role of the auxins during somatic embryogenesis. In Somatic Embryogenesis: Fundamental Aspects and Applications; Springer Science and Business Media LLC: Berlin, Germany, 2016; pp. 171–182. [Google Scholar]
- Ashihara, H.; Stasolla, C.; Loukanina, N.; Thorpe, T.A. Purine metabolism during white spruce somatic embryo development: Salvage of adenine, adenosine, and inosine. Plant Sci. 2001, 160, 647–657. [Google Scholar] [CrossRef]
- Bolwell, G.P.; Cramer, C.L.; Lamb, C.J.; Schuch, W.; Dixon, R.A. L-Phenylalanine ammonia-lyase from Phaseolus vulgaris: Modulation of the levels of active enzyme by trans-cinnamic acid. Planta 1986, 169, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Mavandad, M.; Edwards, R.; Liang, X.; Lamb, C.J.; Dixon, R.A. Effects of trans-cinnamic acid on expression of the bean phenylalanine ammonia-lyase gene family. Plant Physiol. 1990, 94, 671–680. [Google Scholar] [CrossRef]
- O’Neal, D.; Keller, C. Partial purification and some properties of phenylalanine ammonia-lyase of tobacco (Nicotiana tabacum). Phytochemistry 1970, 9, 1373–1383. [Google Scholar] [CrossRef]
- Sato, T.; Kiuchi, F.; Sankawa, U. Inhibition of phenylalanine ammonia-lyase by cinnamic acid derivatives and related compounds. Phytochemistry 1982, 21, 845–850. [Google Scholar] [CrossRef]
- Appert, C.; Logemann, E.; Hahlbrock, K.; Schmid, J.; Amrhein, N. Structural and catalytic properties of the four phenylalanine ammonia-lyase isoenzymes from parsley (Petroselinum Crispum Nym.). JBIC J. Boil. Inorg. Chem. 1994, 225, 491–499. [Google Scholar] [CrossRef]
- Schoch, G.A.; Nikov, G.N.; Alworth, W.L.; Werck-Reichhart, D. Chemical inactivation of the cinnamate 4-hydroxylase allows for the accumulation of salicylic acid in elicited cells. Plant Physiol. 2002, 130, 1022–1031. [Google Scholar] [CrossRef]
- Schilmiller, A.L.; Stout, J.; Weng, J.-K.; Humphreys, J.; Ruegger, M.O.; Chapple, C.C.S. Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J. 2009, 60, 771–782. [Google Scholar] [CrossRef] [PubMed]
- Cvikrová, M.; Malá, J.; Eder, J.; Hrubcová, M.; Vágner, M. Abscisic acid, polyamines and phenolic acids in sessile oak somatic embryos in relation to their conversion potential. Plant Physiol. Biochem. 1998, 36, 247–255. [Google Scholar] [CrossRef]
- Widhalm, J.R.; Dudareva, N. A familiar ring to it: Biosynthesis of plant benzoic acids. Mol. Plant 2015, 8, 83–97. [Google Scholar] [CrossRef]
- Lu, F.; Karlen, S.D.; Regner, M.; Kim, H.; Ralph, S.A.; Sun, R.-C.; Kuroda, K.-I.; Augustin, M.A.; Mawson, R.; Sabarez, H.; et al. Naturally p-hydroxybenzoylated lignins in palms. BioEnergy Res. 2015, 8, 934–952. [Google Scholar] [CrossRef]
- Xu, C.; Zhao, L.; Pan, X.; Šamaj, J. Developmental localization and methylesterification of pectin epitopes during somatic embryogenesis of banana (Musa spp. AAA). PLoS ONE 2011, 6, e22992. [Google Scholar] [CrossRef] [PubMed]
- Dobrowolska, I.; Majchrzak, O.; Baldwin, T.C.; Kurczyńska, E. Differences in protodermal cell wall structure in zygotic and somatic embryos of Daucus carota (L.) cultured on solid and in liquid media. Protoplasma 2011, 249, 117–129. [Google Scholar] [CrossRef]
- Burr, S.J.; Fry, S.C. Feruloylated arabinoxylans are oxidatively cross-Linked by extracellular maize peroxidase but not by horseradish peroxidase. Mol. Plant 2009, 2, 883–892. [Google Scholar] [CrossRef]
- Shigeto, J.; Nagano, M.; Fujita, K.; Tsutsumi, Y. Catalytic profile of Arabidopsis peroxidases, AtPrx-2, 25 and 71, contributing to stem lignification. PLoS ONE 2014, 9, e105332. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Monribot-Villanueva, J.L.; Elizalde-Contreras, J.M.; Aluja, M.; Segura-Cabrera, A.; Birke, A.; Guerrero-Analco, J.A.; Ruiz-May, E. Endorsing and extending the repertory of nutraceutical and antioxidant sources in mangoes during postharvest shelf life. Food Chem. 2019, 285, 119–129. [Google Scholar] [CrossRef]
- Juárez-Trujillo, N.; Monribot-Villanueva, J.L.; Jiménez-Fernández, V.M.; Suárez-Montaño, R.; Aguilar-Colorado, Á.S.; Guerrero-Analco, J.A.; Jiménez, M. Phytochemical characterization of Izote (Yucca elephantipes) flowers. J. Appl. Bot. Food Qual. 2018, 91, 202–210. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Smyth, G.K. limma: Linear Models for Microarray Data. In Nonclinical Statistics for Pharmaceutical and Biotechnology Industries; Springer Science and Business Media LLC: Berlin, Germany, 2005; pp. 397–420. [Google Scholar]
| Cultivar Time | Treatment | PHBA | VA | p-CA | Quercetin 3,4’-di-O-glucoside | t-FA | SA | Naringin | t-CA |
|---|---|---|---|---|---|---|---|---|---|
| Criollo 0 h | 0 µM Control | 0.11 ± 0.01 | 0.11 ± 0.01 | 0.42 ± 0.01 | 0.02 ± 0.01 | 0.23 ± 0.01 | 0 | 0.07 ± 0.01 | 0.02 ± 0.01 |
| 1 µM p-CA | 0.11 ± 0.01 | 0.11 ± 0.01 | 0.42 ± 0.01 | 0.02 ± 0.01 | 0.23 ± 0.01 | 0 | 0.07 ± 0.01 | 0.02 ± 0.01 | |
| 1 µM t-FA | 0.11 ± 0.01 | 0.11 ± 0.01 | 0.42 ± 0.01 | 0.02 ± 0.01 | 0.23 ± 0.01 | 0 | 0.07 ± 0.01 | 0.02 ± 0.01 | |
| Hass 0 h | 0 µM Control | 0.21 ± 0.01 | 0.03 ± 0.03 | 0.66 ± 0.02 | 0.01 ± 0.01 | 0.09 ± 0.01 | 0 | 0 | 0.04 ± 0.01 |
| 10 µM p-CA | 0.21 ± 0.01 | 0.03 ± 0.03 | 0.66 ± 0.02 | 0.01 ± 0.01 | 0.09 ± 0.01 | 0 | 0 | 0.04 ± 0.01 | |
| 100 µM t-FA | 0.21 ± 0.01 | 0.03 ± 0.03 | 0.66 ± 0.02 | 0.01 ± 0.01 | 0.09 ± 0.01 | 0 | 0 | 0.04 ± 0.01 | |
| Criollo 6 h | 0 µM Control | 0.58 ± 0.01 *** | 0.22 ± 0.01 *** | 0.85 ± 0.01 *** | 1.66 ± 0.07 *** | 1.12 ± 0.01 *** | 0.7 ± 0.01 *** | 0.34 ± 0.01 *** | 0.06 ± 0.01 ** |
| 1 µM p-CA | 0.64 ± 0.05 *** | 0.06 ± 0.01 ** | 0.29 ± 0.01 ** | 0 * | 0.12 ± 0.01 ** | 0 ** | 0 ** | 0.4 ± 0.01 *** | |
| 1 µM t-FA | 0.38 ± 0.01 ** | 0.05 ± 0.01 ** | 0.18 ± 0.01 ** | 0.03 ± 0.04 ** | 0.28 ± 0.01 ** | 0 ** | 0 ** | 0.11 ± 0.01 *** | |
| Hass 6 h | 0 µM Control | 0.29 ± 0.01 * | 0.06 ± 0.01 * | 0.62 ± 0.01 * | 0.33 ± 0.05 * | 1 ± 0.01 ** | 0.86 ± 0.02 *** | 0.06 ± 0.01 * | 0.03 ± 0 * |
| 10 µM p-CA | 0.25 ± 0.01 * | 0.05 ± 0.001 * | 0.7 ± 0.01 ** | 0.02 ± 0.03 ** | 0.13 ± 0.01 * | 0 * | 0 * | 0.29 ± 0.01 * | |
| 100 µM t-FA | 0.32 ± 0.01 * | 0.24 ± 0.01 *** | 1.39 ± 0.01 *** | 0.62 ± 0.02 *** | 10.6 ± 0.22 *** | 0.64 ± 0.02 ** | 0.18 ± 0.02 *** | 0.98 ± 0.02 *** | |
| Criollo 12 h | 0 µM Control | 0.21 ± 0.01 ** | 0.15 ± 0.01 *** | 0.61 ± 0.01 *** | 0.62 ± 0.04 *** | 0.52 ± 0.01 *** | 0.72 ± 0.02 *** | 0.05 ± 0.01 *** | 0.02 ± 0.01 ** |
| 1 µM p-CA | 0.25 ± 0.01 *** | 0.06 ± 0.01 ** | 0.46 ± 0.01 ** | 0 ** | 0.26 ± 0.01 ** | 0 ** | 0 *** | 0.15 ± 0.01 *** | |
| 1 µM t-FA | 0.18 ± 0 * | 0.04 ± 0.01 * | 0.18 ± 0.01 * | 0 ** | 0.12 ± 0.01 * | 0 ** | 0 *** | 0.03 ± 0.01 ** | |
| Hass 12 h | 0 µM Control | 0.6 ± 0.01 *** | 0.11 ± 0.01 * | 0.94 ± 0.02 *** | 0.18 ± 0.02 *** | 0.86 ± 0.07 *** | 0.42 ± 0.01 *** | 0.21 ± 0.01 *** | 0.12 ± 0.01 ** |
| 10 µM p-CA | 0.2 ± 0 * | 0.03 ± 0.001 * | 0.24 ± 0.01 * | 0.04 ± 0.01 ** | 0.13 ± 0.01 * | 0 * | 0 * | 0.08 ± 0.01 * | |
| 100 µM t-FA | 0.4 ± 0 * | 0.17 ± 0.01 *** | 0.48 ± 0.01 * | 0.05 ± 0.04 ** | 0.39 ± 0.01 * | 0 * | 0.22 ± 0.01 *** | 0.08 ± 0.01 * | |
| Criollo 14 days | 0 µM Control | 1.12 ± 0.02 *** | 0.06 ± 0.001 *** | 1.28 ± 0.02 *** | 0.3 ± 0.01 *** | 0.23 ± 0.01 * | 0.03 ± 0.02 *** | 0 *** | 0.01 ± 0.02 *** |
| 1 µM p-CA | 0.09 ± 0.001 * | 0.05 ± 0.01 ** | 0.39 ± 0.01 * | 0 ** | 0.28 ± 0.02 ** | 0 ** | 0 *** | 0.01 ± 0.02 *** | |
| 1 µM t-FA | 0.12 ± 0.01 ** | 0.02 ± 0.01 * | 1.18 ± 0.08 ** | 0 ** | 0.56 ± 0.03 *** | 0.02 ± 0.01 ***** | 0 *** | 0.02 ± 0.01 *** | |
| Hass 14 days | 0 µM Control | 0.09 ± 0.01 * | 0.03 ± 0.01 *** | 0.69 ± 0.03 *** | 0 * | 0.4 ± 0.01 ** | 0.01 ± 0.01 ** | 0 *** | 0.02 ± 0.01 *** |
| 10 µM p-CA | 0.11 ± 0.01 ** | 0.03 ± 0.01 *** | 0.24 ± 0.02 * | 0.06 ± 0.01 ** | 0.55 ± 0.03 ** | 0.1 ± 0.02 *** | 0 *** | 0.01 ± 0.01 ** | |
| 100 µM t-FA | 0 * | 0 * | 0 * | 0 * | 7.06 ± 3.63 *** | 0 ** | 0 *** | 0 * | |
| Criollo 28 days | 0 µM Control | 0.62 ± 0.01 * | 0.05 ± 0.01 ** | 0.96 ± 0 *** | 0 *** | 0.72 ± 0.01 *** | 0.23 ± 0.01 ** | 0 *** | 0.5 ± 0 *** |
| 1 µM p-CA | 3.56 ± 0.06 *** | 0 * | 0.05 ± 0.04 * | 0 *** | 0 * | 0 * | 0 *** | 0 * | |
| 1 µM t-FA | 1.35 ± 0.02 ** | 0.08 ± 0 *** | 0.74 ± 0.01 ** | 0.02 ± 0.04 *** | 0.12 ± 0.10 ** | 1.55 ± 0.12 *** | 0 *** | 0.03 ± 0 ** | |
| Hass 28 days | 0 µM Control | 0.4 ± 0 * | 0.05 ± 0 ** | 1.14 ± 0.01 *** | 0 *** | 0.78 ± 0.01 ** | 0.04 ± 0.01 *** | 0 *** | 0.01 ± 0 *** |
| 10 µM p-CA | 1.36 ± 0.03 *** | 0 * | 0.02 ± 0.02 * | 0 *** | 7.75 ± 4.43 *** | 0 * | 0 *** | 0 ** | |
| 100 µM t-FA | 1.12 ± 0.01 ** | 0.03 ± 0 * | 0.16 ± 0.01 * | 0 *** | 0.08 ± 0 ** | 0.01 ± 0.01 ** | 0 *** | 0 ** |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olivares-García, C.A.; Mata-Rosas, M.; Peña-Montes, C.; Quiroz-Figueroa, F.; Segura-Cabrera, A.; Shannon, L.M.; Loyola-Vargas, V.M.; Monribot-Villanueva, J.L.; Elizalde-Contreras, J.M.; Ibarra-Laclette, E.; et al. Phenylpropanoids Are Connected to Cell Wall Fortification and Stress Tolerance in Avocado Somatic Embryogenesis. Int. J. Mol. Sci. 2020, 21, 5679. https://doi.org/10.3390/ijms21165679
Olivares-García CA, Mata-Rosas M, Peña-Montes C, Quiroz-Figueroa F, Segura-Cabrera A, Shannon LM, Loyola-Vargas VM, Monribot-Villanueva JL, Elizalde-Contreras JM, Ibarra-Laclette E, et al. Phenylpropanoids Are Connected to Cell Wall Fortification and Stress Tolerance in Avocado Somatic Embryogenesis. International Journal of Molecular Sciences. 2020; 21(16):5679. https://doi.org/10.3390/ijms21165679
Chicago/Turabian StyleOlivares-García, Carol A., Martín Mata-Rosas, Carolina Peña-Montes, Francisco Quiroz-Figueroa, Aldo Segura-Cabrera, Laura M. Shannon, Victor M. Loyola-Vargas, Juan L. Monribot-Villanueva, Jose M. Elizalde-Contreras, Enrique Ibarra-Laclette, and et al. 2020. "Phenylpropanoids Are Connected to Cell Wall Fortification and Stress Tolerance in Avocado Somatic Embryogenesis" International Journal of Molecular Sciences 21, no. 16: 5679. https://doi.org/10.3390/ijms21165679
APA StyleOlivares-García, C. A., Mata-Rosas, M., Peña-Montes, C., Quiroz-Figueroa, F., Segura-Cabrera, A., Shannon, L. M., Loyola-Vargas, V. M., Monribot-Villanueva, J. L., Elizalde-Contreras, J. M., Ibarra-Laclette, E., Ramirez-Vázquez, M., Guerrero-Analco, J. A., & Ruiz-May, E. (2020). Phenylpropanoids Are Connected to Cell Wall Fortification and Stress Tolerance in Avocado Somatic Embryogenesis. International Journal of Molecular Sciences, 21(16), 5679. https://doi.org/10.3390/ijms21165679






