MiR-93/miR-375: Diagnostic Potential, Aggressiveness Correlation and Common Target Genes in Prostate Cancer
Abstract
1. Introduction
2. Results
2.1. Patient Characteristics
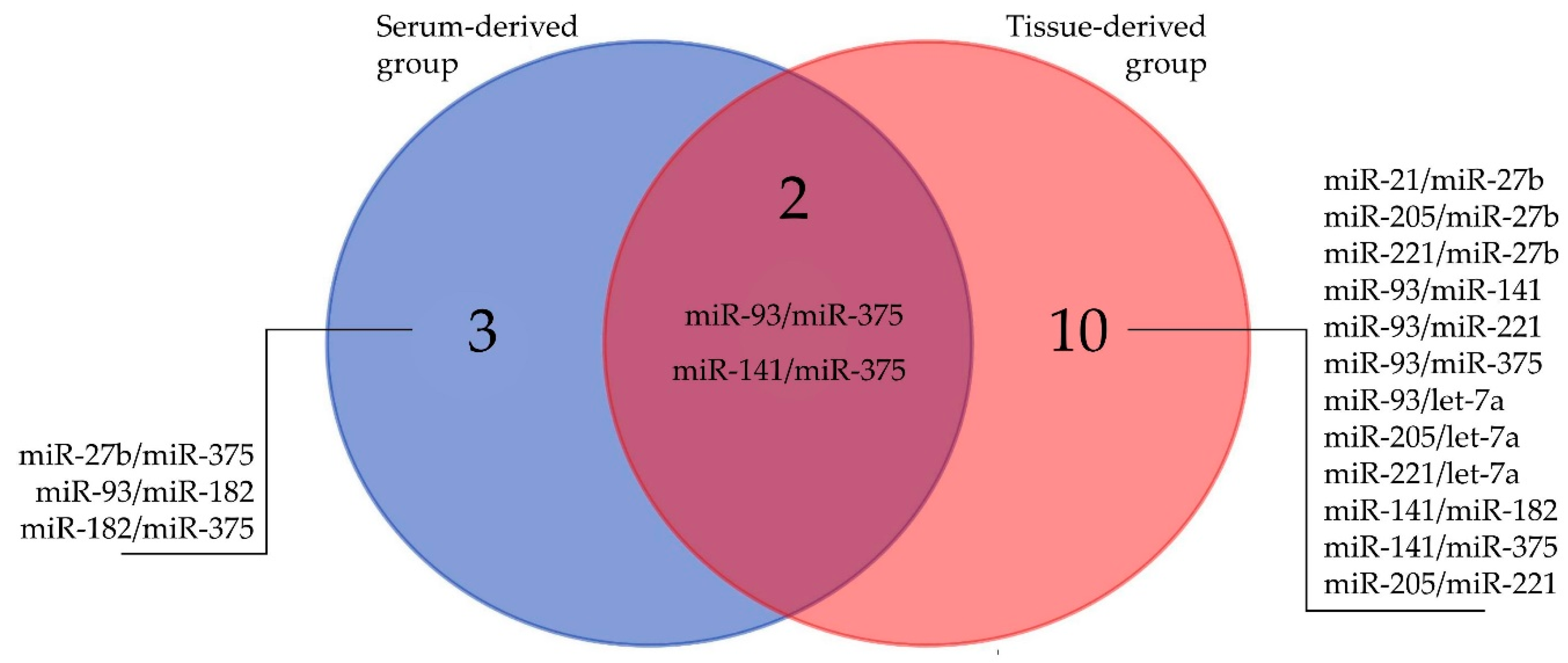
2.2. Literature Search
2.3. MiRNA Expression in Serum and Tissue
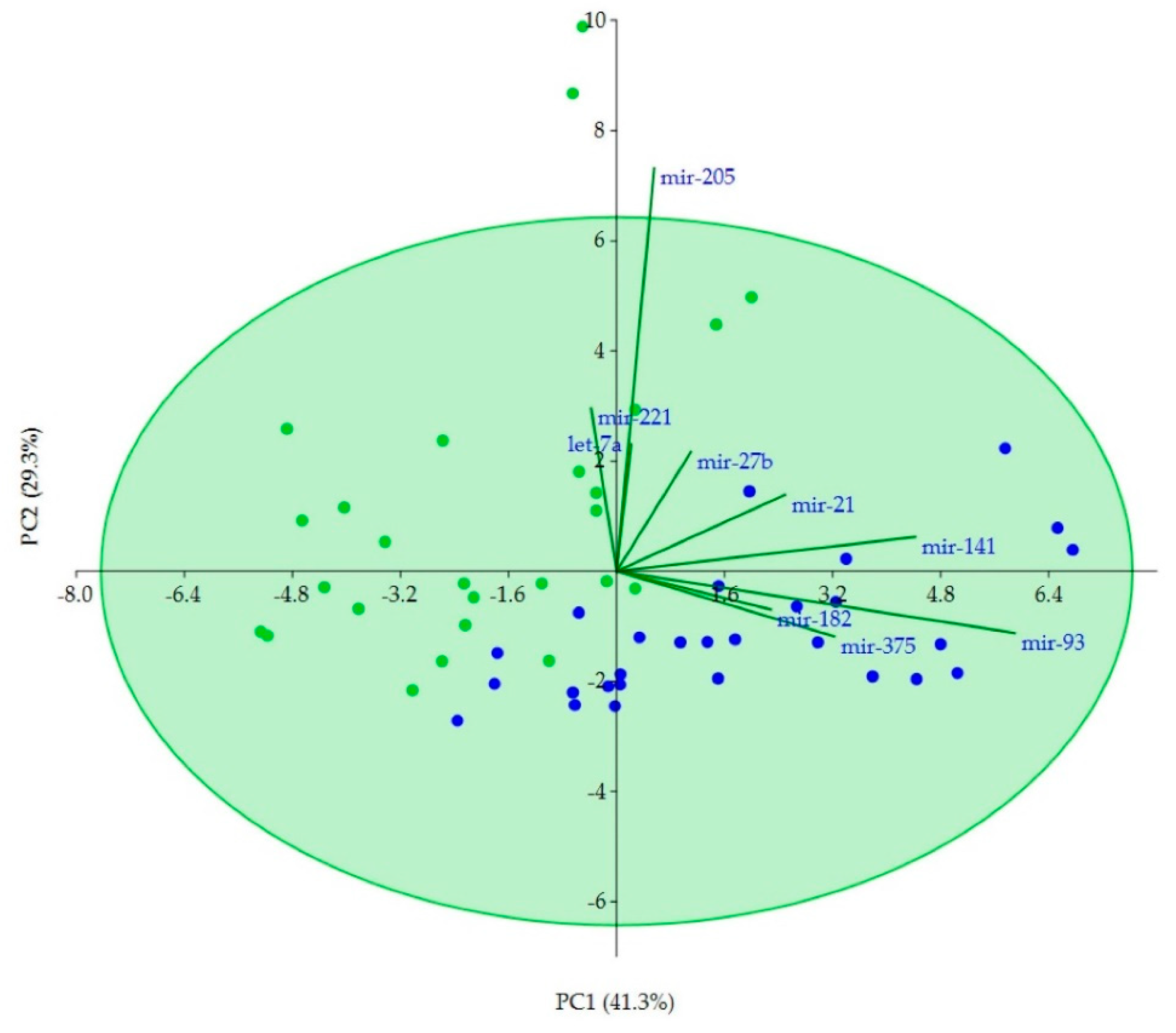
2.4. Correlations of Clinicopathological Variables with miRNAs and Correlations between miRNAs
2.5. Discriminatory Potential of miRNAs
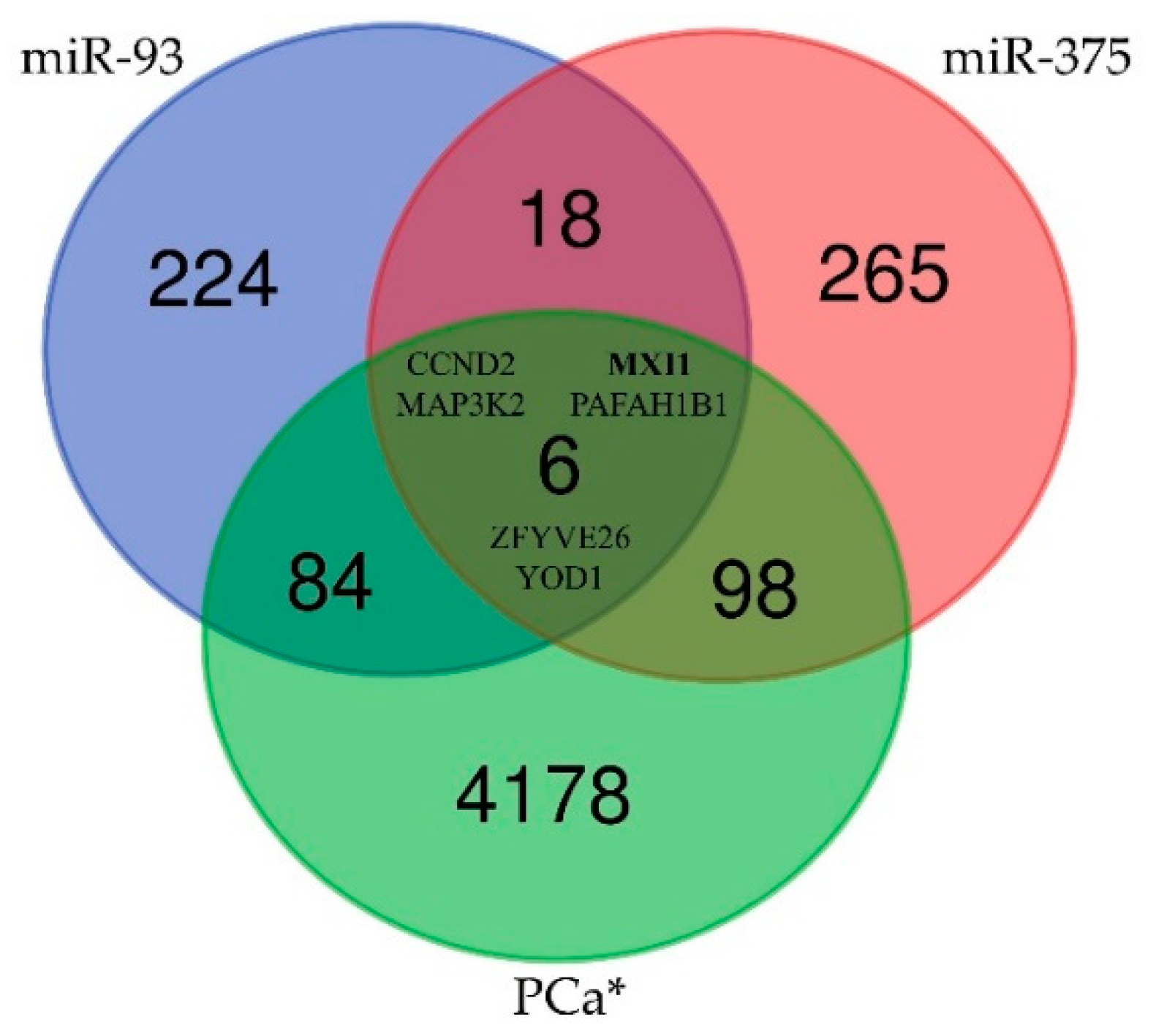
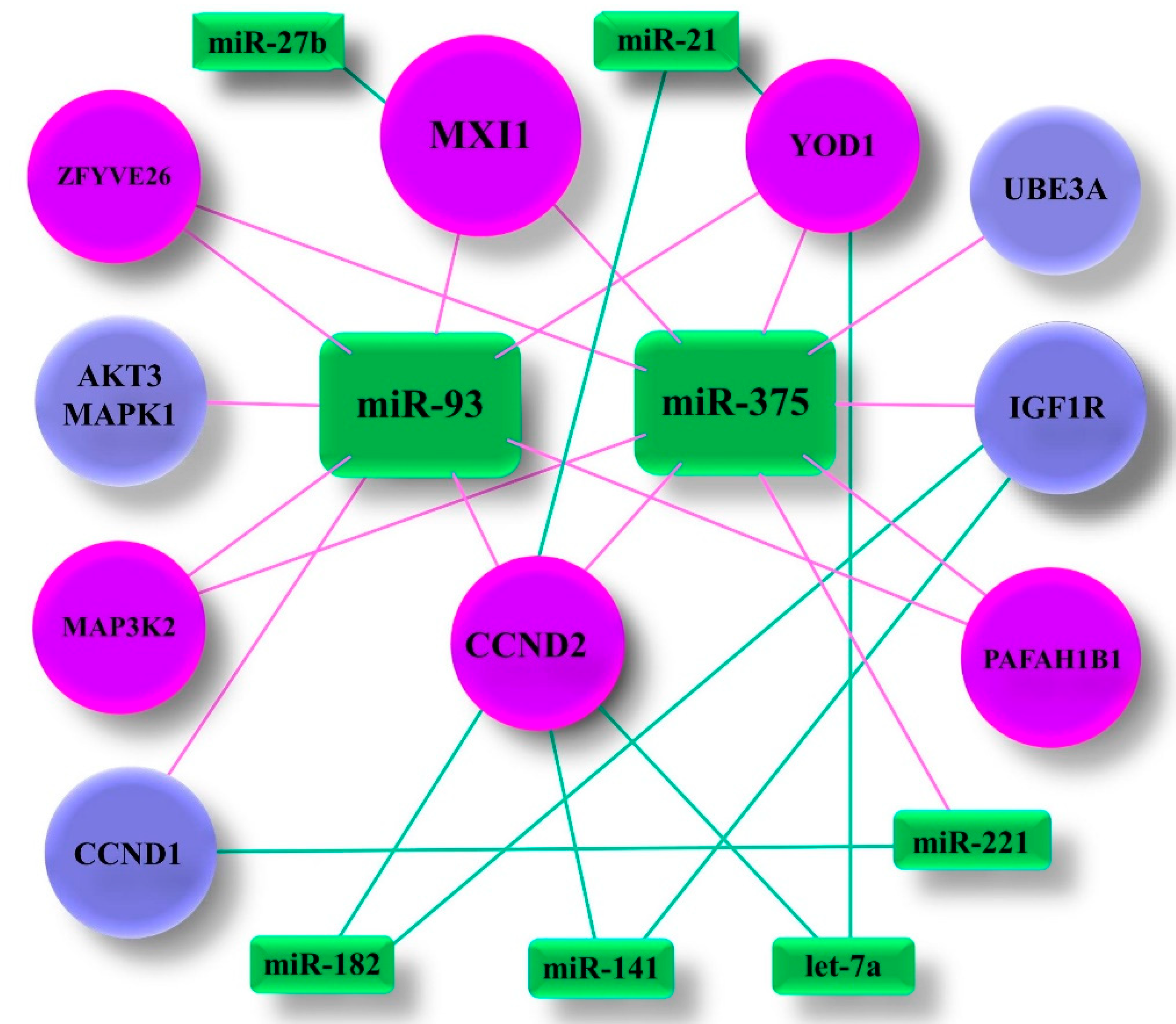
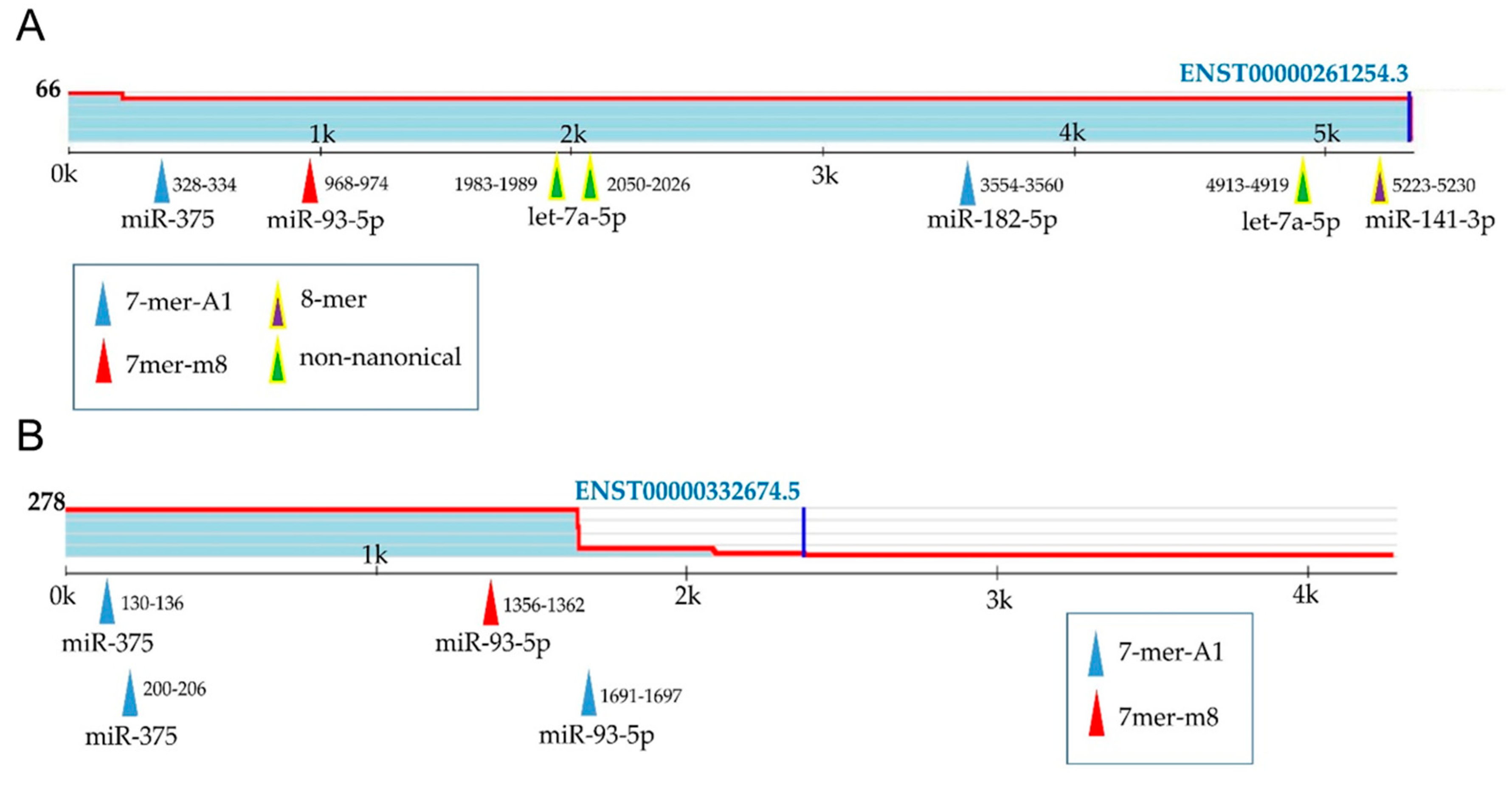
2.6. Prediction of Target Genes and Functional Links between Selected miRNAs and Prostate Cancer
3. Discussion
4. Materials and Methods
4.1. Patients Selection and Data Collection
4.2. Literature Search
4.3. RNA Extraction from Tissue and Serum
4.4. cDNA Synthesis
4.5. miRNA Expression Analysis
4.6. Prediction of the Target Genes of Selected miRNAs
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Richardsen, E.; Andersen, S.; Melbø-Jørgensen, C.; Rakaee, M.; Ness, N.; Al-Saad, S.; Nordby, Y.; Pedersen, M.I.; Dønnem, T.; Bremnes, R.M.; et al. MicroRNA 141 is associated to outcome and aggressive tumor characteristics in prostate cancer. Sci. Rep. 2019, 9, 386. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality world-wide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Mottet, N.; Cornford, P.; Van den Bergh, R.C.N.; Briers, E.; De Santis, M.; Fanti, S.; Gillessen, S.; Grummet, J.; Henry, A.M.; Lam, T.B.; et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on prostate cancer. Eur. Assoc. Urol. 2020, 1–182. [Google Scholar]
- Adhyam, M.; Gupta, A.K. A review on the clinical utility of PSA. Indian J. Surg. Oncol. 2012, 3, 120–129. [Google Scholar] [CrossRef]
- Bokhorst, L.P.; Valdagni, R.; Rannikko, A.; Kakehi, Y.; Pickles, T.; Bangma, C.H.; Roobol, M.J. PRIAS study group. A decade of active surveillance in the PRIAS study: An update and evaluation of the criteria used to recommend a switch to active treatment. Eur. Urol. 2016, 70, 954–960. [Google Scholar] [CrossRef]
- Zedan, A.H.; Hansen, T.F.; Assenholt, J.; Pleckaitis, M.; Madsen, J.S.; Osther, P.J.S. microRNA expression in tumour tissue and plasma in patients with newly diagnosed metastatic prostate cancer. Tumour Biol. 2018, 40, 1010428318775864. [Google Scholar] [CrossRef]
- Takudome, S.; Ando, R.; Koda, Y. Discoveries and application of prostate-specific antigen, and some proposals to optimize prostate cancer screening. Cancer Manag. Res. 2016, 8, 45–47. [Google Scholar] [CrossRef]
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in control of gene expression: An overview of nuclear functions. Int. J. Mol. Sci. 2016, 17, 1712. [Google Scholar] [CrossRef]
- Zedan, A.H.; Hansen, T.F.; Assenholt, J.; Madsen, J.S.; Osther, P.J.S. Circulating miRNAs in localized/locally advanced prostate cancer patients after radical prostatectomy and radiotherapy. Prostate 2019, 79, 425–432. [Google Scholar] [CrossRef]
- Chen, C.Z. MicroRNAs as Oncogenes and Tumor Suppressors. N. Engl. J. Med. 2005, 353, 1768–1771. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Vernooy, S.Y.; Guo, M.; Hay, B.A. The drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 2003, 13, 790–795. [Google Scholar] [CrossRef]
- Croce, C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009, 10, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Mens, M.M.J.; Ghanbari, M. Cell cycle regulation of stem cells by microRNAs. Stem Cell Rev. 2018, 14, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Ji, W.; Sun, B.; Su, C. Targeting MicroRNAs in Cancer Gene Therapy. Genes 2017, 8, 21. [Google Scholar] [CrossRef] [PubMed]
- Sita-Lumsden, A.; Dart, D.A.; Waxman, J.; Bevan, C.L. Circulating microRNAs as potential new biomarkers for prostate cancer. Br. J. Cancer 2013, 108, 1925–1930. [Google Scholar] [CrossRef]
- Xu, W.; Liu, M.; Peng, X.; Zhou, P.; Zhou, J.; Xu, K.; Xu, H.; Jiang, S. miR-24-3p and miR-27a-3p promote cell proliferation in glioma cells via cooperative regulation of MXI1. Int. J. Oncol 2013, 42, 757–766. [Google Scholar] [CrossRef]
- Wei, J.; Yin, Y.; Deng, Q.; Zhou, J.; Wang, Y.; Yin, G.; Yang, J.; Tang, Y. Integrative Analysis of MicroRNA and Gene Interactions for Revealing Candidate Signatures in Prostate Cancer. Front. Genet. 2020, 11, 176. [Google Scholar] [CrossRef]
- Filella, X.; Foj, L. MiRNAs as a novel biomarkers in the management of prostate cancer. Clin. Chem. Lab. Med. 2017, 55, 715–736. [Google Scholar] [CrossRef]
- Liu, R.S.C.; Olkhov-Mitsel, E.; Jeyapala, R.; Zhao, F.; Commisso, K.; Klotz, L.; Loblaw, A.; Liu, S.K.; Vesprini, D.; Fleshner, N.E.; et al. Assessment of serum microRNA biomarkers to predict reclassification of prostate cancer in patients on active surveillance. J. Urol. 2018, 199, 1475–1481. [Google Scholar] [CrossRef]
- Wang, D.; Lu, G.; Shao, Y.; Xu, D. MiR-182 promotes prostate cancer progression through activating Wnt/β-catenin signal pathway. Biomed. Pharmacother. 2018, 99, 334–339. [Google Scholar] [CrossRef]
- Baumann, B.; Acosta, A.M.; Richards, Z.; Deaton, R.; Sapatynska, A.; Murphy, A.; Kajdacsy-Balla, A.; Gann, P.H.; Nonn, L. Association of high miR-182 levels with low-risk prostate cancer. Am. J. Pathol. 2019, 189, 911–923. [Google Scholar] [CrossRef] [PubMed]
- Gandellini, P.; Giannoni, E.; Casamichele, A.; Taddei, M.L.; Callari, M.; Piovan, C.; Valdagni, R.; Pierotti, M.A.; Zaffaroni, N.; Chiarugi, P. miR-205 hinders the malignant interplay between prostate cancer cells and associated fibroblasts. Antioxid. Redox Signal. 2014, 20, 1045–1059. [Google Scholar] [CrossRef] [PubMed]
- Ippolito, L.; Marini, A.; Cavallini, L.; Morandi, A.; Pietrovito, L.; Pintus, G.; Giannoni, E.; Schrader, T.; Puhr, M.; Chiarugi, P.; et al. Metabolic shift toward oxidative phosphorylation in docetaxel resistant prostate cancer cells. Oncotarget 2016, 7, 61890–61904. [Google Scholar] [CrossRef] [PubMed]
- Kotb, S.; Mosharafa, A.; Essawi, M.; Hassan, H.; Meshref, A.; Morsy, A. Circulating miRNAs 21 and 221 as biomarkers for early diagnosis of prostate cancer. Tumour Biol. 2014, 35, 12613–12617. [Google Scholar] [CrossRef]
- Guo, X.; Han, T.; Hu, P.; Guo, X.; Zhu, C.; Wang, Y.; Chang, S. Five microRNAs in serum potential biomarkers for prostate risk assessment and therapeutic intervention. Int. Urol. Nephrol. 2018, 50, 2193–2200. [Google Scholar] [CrossRef]
- Zhang, H.L.; Yang, L.F.; Zhu, Y.; Yao, X.D.; Zhang, S.L.; Dai, B.; Zhu, Y.P.; Shen, Y.J.; Shi, G.H.; Ye, D.W. Serum miRNA-21: Elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate 2011, 71, 326–331. [Google Scholar] [CrossRef]
- Melbø-Jørgensen, C.; Ness, N.; Andersen, S.; Valkov, A.; Dønnem, T.; Al-Saad, S.; Kiselev, Y.; Berg, T.; Nordby, Y.; Bremnes, R.M.; et al. Stromal expression of miR-21 predicts biochemical failure in prostate cancer patients with Gleason score 6. PLoS ONE 2014, 9, e113039. [Google Scholar] [CrossRef]
- Leite, K.R.M.; Reis, S.T.; Viana, N.; Morais, D.R.; Moura, C.M.; Silva, I.A.; Pontes, J.; Katz, B.; Srougi, M. Controlling RECK miR-21 promotes tumor cell invasion and is related to biochemical recurrence in prostate cancer. J. Cancer 2015, 6, 292–301. [Google Scholar] [CrossRef]
- Porzycki, P.; Ciszkowicz, E.; Semik, M.; Tyrka, M. Combination of three miRNA (miR-141, miR-21, and miR-375) as potential diagnostic tool for prostate cancer recognition. Int. Urol. Nephrol. 2018, 50, 1619–1626. [Google Scholar] [CrossRef]
- Kurul, N.O.; Ates, F.; Yilmaz, I.; Narli, G.; Yesildal, C.; Senkul, T. The association of let-7c, miR-21, miR-145, miR-182, and miR-221 with clinicopathologic parameters of prostate cancer in patients diagnosed with low-risk disease. Prostate 2019, 79, 1125–1132. [Google Scholar] [CrossRef] [PubMed]
- Goto, Y.; Kojima, S.; Nishikawa, R.; Enokida, H.; Chiyomaru, T.; Kinoshita, T.; Nakagawa, M.; Naya, Y.; Ichikawa, T.; Seki, N. The microRNA-23b/27b/24-1 cluster is a disease progression marker and tumor suppressor in prostate cancer. Oncotarget 2014, 5, 7748–7759. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Sun, X.; Liu, Y. miR-27b expression in diagnosis and evaluation prognosis of prostate cancer. Int. J. Clin. Exp. Pathol. 2017, 10, 11415–11424. [Google Scholar]
- Pimenta, R.C.A.; Viana, N.I.; Amaral, G.Q.; Park, R.; Morais, D.R.; Pontes, J., Jr.; Guimaraes, V.R.; Camargo, J.A.; Leite, K.R.M.; Nahas, W.C.; et al. MicroRNA-23b and microRNA-27b plus flutamide treatment enhances apoptosis rate and decreases CCNG1 expression in a castration -resistant prostate cancer cell line. Tumor Biol. 2018, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Moltzahn, F.; Olshen, A.B.; Baehner, L.; Peek, A.; Fong, L.; Stöppler, H.; Simko, J.; Hilton, J.F.; Carroll, P.; Blelloch, R. Microfluidic-based multiplex qRT-PCR identifies diagnostic and prognostic microRNA signatures in the sera of prostate cancer patients. Cancer Res. 2011, 71, 550–560. [Google Scholar] [CrossRef]
- Liu, M.X.; Liao, J.; Xie, M.; Gao, Z.K.; Wang, X.H.; Zhang, Y.; Shang, M.H.; Yin, L.H.; Pu, Y.P.; Liu, R. Mir-93-5p transferred by exosomes promotes the proliferation of esophageal cancer cells via intercellular communication by targeting PTEN. Biomed. Environ. Sci. 2018, 31, 171–185. [Google Scholar]
- Mihelich, B.L.; Maranville, J.C.; Nolley, R.; Peehl, D.M.; Nonn, L. Elevated serum microRNA levels associate with absence of high-grade prostate cancer in a retrospective cohort. PLoS ONE 2015, 10, e0124245. [Google Scholar] [CrossRef]
- Yang, K.; Li, Y.W.; Gao, Z.Y.; Xiao, W.; Li, T.Q.; Song, W.; Zheng, J.; Chen, H.; Chen, G.H.; Zou, H.Y. MiR-93 functions as a tumor promoter in prostate cancer by targeting disabled homolog 2 (DAB2) and an antitumor polysaccharide from green tea (Camellia sinensis) on their expression. Int. J. Biol. Makromol. 2019, 125, 557–565. [Google Scholar] [CrossRef]
- Gao, Y.; Deng, K.; Liu, X.; Dai, M.; Chen, X.; Chen, J.; Chen, J.; Huang, Y.; Dai, S.; Chen, J. Molecular mechanism and role of microRNA-93 inhuman cancers: A study based on bioinformatics analysis, meta-analysis, and quantitative polymerase chain reaction validation. J. Cell Biochem. 2019, 120, 6370–6383. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosowa-Agadjanyan, R.L.; Peterson, E.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as Stable Blood-Based Markers for Cancer Detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Wach, S.; Nolte, E.; Szczyrba, J.; Stöhr, R.; Hartmann, A.; Ømtoft, T.; Dyrskjøt, L.; Eltze, E.; Wieland, W.; Keck, B.; et al. MicroRNA profiles of prostate carcinoma detected by multiplatform microRNA screening. Int. J. Cancer 2012, 130, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Brase, J.C.; Johannes, M.; Schlomm, T.; Falth, M.; Haese, A.; Steuber, T.; Beissbarth, T.; Kuner, R.; Sültmann, R. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int. J. Cancer 2011, 128, 608–616. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, J.C.; Fink, L.M.; Goodman, O.B.; Symanowski, J.T.; Vogelzang, N.J.; Ward, D.C. Comparison of circulating microRNA 141 to circulating tumor cells, lactate dehydrogenase, and prostate-specific antigen for determining treatment response in patients with metastatic prostate cancer. Clin. Genitourin. Cancer 2011, 9, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Paziewska, A.; Mikula, M.; Dabrowska, M.; Kulecka, M.; Goryca, K.; Antoniewicz, A.; Dobruch, J.; Borowka, A.; Rutkowski, P.; Ostrowski, J. Candidate diagnostic miRNAs that can detect cancer in prostate biopsy. Prostate 2018, 78, 178–185. [Google Scholar] [CrossRef]
- Xu, S.; Ge, J.; Zhang, Z.; Zhou, W. miR-141 inhibits prostatic cancer cell proliferation and migration, and induces cell apoptosis via targeting of RUNX1. Oncol. Rep. 2018, 39, 1454–1460. [Google Scholar] [CrossRef] [PubMed]
- Selth, L.A.; Townley, S.; Gillis, J.L.; Ochnik, A.M.; Murti, K.; Macfarlane, R.J.; Chi, K.N.; Marshall, V.R.; Tilley, W.D.; Butler, L.M. Discovery of circulating microRNAs associated with human prostate cancer using a mouse model of disease. Int. J. Cancer 2012, 131, 652–661. [Google Scholar] [CrossRef]
- Bryant, R.J.; Pawlowski, T.; Catto, J.W.F.; Marsden, G.; Vessella, R.L.; Rhees, B.; Kuslich, C.; Visakorpi, T.; Hamdy, F.C. Changes in circulating microRNA levels associated with prostate cancer. Br. J. Cancer 2012, 106, 768–774. [Google Scholar] [CrossRef]
- Zhao, Z.; Weickmann, S.; Jung, M.; Lein, M.; Kilic, E.; Stephan, C.; Erbersdobler, A.; Fendler, A.; Jung, K. A novel predictor tool of biochemical recurrence after radical prostatectomy based on a five-microRNA tissue signature. Cancers 2019, 11, 1603. [Google Scholar] [CrossRef]
- Cheng, H.H.; Mitchell, P.S.; Kroh, E.M.; Dowell, A.E.; Chéry, L.; Siddiqui, J.; Nelson, P.S.; Vessella, R.L.; Knudsen, B.S.; Chinnaiyan, A.M.; et al. Circulating microRNA profiling identifies a subset metastatic prostate cancer patients with evidence cancer-associated hypoxia. PLoS ONE 2013, 8, e69239. [Google Scholar] [CrossRef]
- Nguyen, P.V.; Srihari, S.; Leong, H.W. Identifying conserved protein complexes between species by constructing interolog networks. BMC Bioinform. 2013, 14. [Google Scholar] [CrossRef]
- Selth, L.A.; Townley, S.L.; Bert, A.G.; Stricker, P.D.; Sutherland, P.D.; Horwath, L.G.; Goodall, G.J.; Butler, L.M.; Tilley, W.D. Circulating microRNAs predict biochemical recurrence in prostate cancer patients. Br. J. Cancer 2013, 109, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Haldrup, C.; Kosaka, N.; Ochiya, T.; Borre, M.; Høyer, S.; Orntoft, T.F.; Sorensen, K.D. Profiling of circulating microRNAs for prostate cancer biomarker discovery. Drug Deliv. Transl. Res. 2014, 4, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.; Pollex, T.; Hanna, K.; Tuorto, F.; Meusburger, M.; Helm, M.; Lyko, F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010, 24, 1590–1595. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, H.; Thomsen, A.R.; Haldrup, C.; Dyrskjøt, L.; Høyer, S.; Borre, M.; Mouritzen, P.; Ørntoft, T.F.; Sørensen, K.D. Novel diagnostic and prognostic classifiers for prostate cancer identified by genome-wide microRNA profiling. Oncotarget 2016, 7, 30760–30771. [Google Scholar] [CrossRef] [PubMed]
- Gandellini, P.; Folini, M.; Longoni, N.; Pennati, M.; Binda, M.; Colecchia, M.; Salvioni, R.; Supino, R.; Moretti, R.; Limonta, P.; et al. miR-205 exerts tumor-suppressive functions in human prostate through down-regulation of protein kinase Cε. Cancer Res. 2009, 69, 2287–2295. [Google Scholar] [CrossRef]
- Srivastava, A.; Goldberger, H.; Dimtchev, A.; Ramalinga, M.; Chijioke, J.; Marian, C.; Oermann, E.K.; Uhm, S.; Kim, J.S.; Chen, L.N.; et al. MicroRNA profiling in prostate cancer-the diagnostic potential of urinary miR-205 and miR-214. PLoS ONE 2013, 8, e76994. [Google Scholar] [CrossRef]
- Verdoodt, B.; Neid, M.; Vogt, M.; Kuhn, V.; Liffers, S.T.; Palisaar, R.J.; Noldus, J.; Tannapfel, A.; Mirmohammadsadegh, A. MicroRNA-205, a novel regulator of the anti-apoptotic protein Bcl2, is downregulated in prostate cancer. Int. J. Oncol 2013, 43, 307–314. [Google Scholar] [CrossRef]
- Hagman, Z.; Haflidadóttir, B.; Ceder, J.; Larne, O.; Bjartell, A.; Lilja, H.; Edsjö, A.; Ceder, Y. miR-205 negatively regulates the androgen receptor and is associated with adverse outcome of prostate cancer patients. Br. J. Cancer 2013, 108, 1668–1676. [Google Scholar] [CrossRef]
- Porkka, K.P.; Pfeiffer, M.J.; Waltering, K.K.; Vessella, R.L.; Tammela, T.L.; Visakorpi, T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007, 67, 6130–6135. [Google Scholar] [CrossRef]
- Dülgeroğlu, Y.; Eroğlu, O. Diagnostic performance of microRNAs in the circulation in differential diagnosis of BPH, chronic prostatitis and prostate cancer. Turk. J. Biochem. 2018, 44, 1–9. [Google Scholar] [CrossRef]
- Kelly, B.D.; Miller, N.; Sweeney, K.J.; Durkan, G.C.; Rogers, E.; Walsh, K.; Kerin, M.J. A circulating microRNA signature as a biomarker for prostate cancer in a high risk group. J. Clin. Med. 2015, 4, 1369–1379. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Huo, N.; Li, Y.; He, Z. let-7a and its target, insulin-like growth factor 1 receptor, are differentially expressed in recurrent prostate cancer. Int. J. Mol. Med. 2015, 36, 1409–1416. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Meng, P.; Wang, T.; Qin, W.; Qin, W.; Wang, F.; Yuan, J.; Chen, Z.; Yang, A.; Wang, H. MicroRNA Let-7a Inhibits Proliferation of Human Prostate Cancer Cells In Vitro and In Vivo by Targeting E2F2 and CCND2. PLoS ONE 2010, 5, e10147. [Google Scholar] [CrossRef] [PubMed]
- The Human Protein Atlas. Available online: https://www.proteinatlas.org/ (accessed on 15 June 2020).
- Dennis, G., Jr.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003, 4, P3. [Google Scholar] [CrossRef] [PubMed]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Decipherng microRNA function with experimental support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef] [PubMed]
- TargetScanHuman. Available online: http://www.targetscan.org/vert_72/ (accessed on 25 June 2020).
- Jiang, J.; Lee, E.J.; Gusev, Y.; Schmittgen, T.D. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 2005, 33, 5394–5403. [Google Scholar] [CrossRef] [PubMed]
- Bidarra, D.; Constâncio, V.; Barros-Silva, D.; Ramalho-Carvalho, J.; Moreira-Barbosa, C.; Antunes, L.; Maurício, J.; Oliveira, J.; Henrique, R.; Jerónimo, C. Circulating microRNAs as biomarkers for prostate cancer detection and metastasis development prediction. Front. Oncol. 2019, 9, 900. [Google Scholar] [CrossRef]
- Lewis, B.P.; Shih, I.H.; Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of mammalian microRNA targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef]
- Bryzgunova, O.E.; Lekchnov, E.A.; Zaripov, M.M.; Yurchenko, Y.B.; Yarmoschuk, S.V.; Pashkovskaya, O.A.; Rykova, E.Y.; Zheravin, A.A.; Laktionov, P.P. Bioinformatics analysis for evaluation of the diagnostic potentialities of miR-19b, -125b and-205 as liquid biopsy markers of prostate cancer. AIP Conf. Proc. 2017, 1882, 020009. [Google Scholar]
- Mattie, M.D.; Benz, C.C.; Bowers, J.; Sensinger, K.; Wong, L.; Scott, G.K.; Fedele, V.; Ginzinger, D.; Getts, R.; Haqq, C. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol. Cancer 2006, 5, 24. [Google Scholar] [CrossRef]
- Davoren, P.A.; McNeill, R.E.; Lowery, A.J.; Kerin, M.J.; Miller, N. Identification of suitable endogenous control genes for microRNA gene expression analysis in human breast cancer. BMC Mol. Biol. 2008, 9, 76. [Google Scholar] [CrossRef] [PubMed]
- Aghdam, S.G.; Ebrazeh, M.; Hemmatzadeh, M.; Seyfizadeh, N.; Shabgah, A.G.; Azizi, G.; Ebrahimi, N.; Babaie, F.; Mohammadi, H. The role of microRNAs in prostate cancer migration, invasion, and metastasis. J. Cell Physiol. 2019, 234, 9927–9942. [Google Scholar] [CrossRef] [PubMed]
- Daniyal, M.; Siddiqui, Z.A.; Akram, M.; Asif, H.M.; Sultana, S.; Khan, A. Epidemiology, etiology, diagnosis and treatment of prostate cancer. Asian Pac. J. Cancer Prev. 2014, 15, 9575–9578. [Google Scholar] [CrossRef] [PubMed]
- Kachakova, D.; Mitkova, A.; Popov, E.; Popov, I.; Vlahova, A.; Dikov, T.; Christova, S.; Mitev, V.; Slavov, C.; Kaneva, R. Combinations of serum prostate-specific antigen and plasma expression levels of let-7c, miR-30c, miR-141, and miR-375 as potential better diagnostic biomarkers for prostate cancer. DNA Cell Biol. 2015, 34, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Trevethan, R. Sensitivity, specificity, and predictive values: Foundations, pliabilities, and pitfalls in research and practice. Front. Public Health 2017, 5, 307. [Google Scholar] [CrossRef]
- Wolf, A.M.D.; Wender, R.C.; Etzioni, R.B.; Thompson, I.M.; D’Amico, A.V.; Volk, R.J.; Brooks, D.D.; Dash, C.; Guessous, I.; Andrews, K.; et al. American Cancer Society guideline for the early detection of prostate cancer. CA Cancer J. Clin. 2010, 60, 70–98. [Google Scholar] [CrossRef]
- American Cancer Society. American Cancer Society Recommendations for Prostate Cancer Early Detection. Available online: https://www.cancer.org/cancer/prostate-cancer/detection-diagnosis-staging/acs-recommendations.html (accessed on 15 May 2020).
- Sun, Y.; Jia, X.; Hou, L.; Liu, X. Screening of differently expressed miRNA and mRNA in prostate cancer by integrated analysis of transcription data. Urology 2016, 94, 313.e1–313.e3136. [Google Scholar] [CrossRef]
- Zhu, C.; Shao, P.; Bao, M.; Li, P.; Zhou, H.; Cai, H.; Cao, Q.; Tao, L.; Meng, X.; Ju, X.; et al. miR-154 inhibits prostate cancer cell proliferation by targeting CCND2. Urol. Oncol. 2014, 32, 31.e9–31.e16. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, Q.; Wang, Q.; Li, J.; Sipeky, C.; Xia, J.; Gao, P.; Hu, Y.; Zhang, H.; Yang, X.; et al. Genetic association analysis of the RTK/ERK pathway with aggressive prostate cancer highlights the potential role of CCND2 in disease progression. Sci. Rep. 2017, 7, 4538. [Google Scholar] [CrossRef]
- UniProt. UniProtKB-P50539 (MXI1_HUMAN). Available online: https://www.uniprot.org/uniprot/P50539 (accessed on 15 June 2020).
- Zhou, J.; Wang, W.; Gao, Z.; Peng, X.; Chen, X.; Chen, W.; Xu, W.; Xu, H.; Lin, M.C.; Jiang, S. MicroRNA-155 Promotes Glioma Cell Proliferation via the Regulation of MXI1. PLoS ONE 2013, 8, e83055. [Google Scholar] [CrossRef]
- Taj, M.M.; Tawil, R.J.; Engstrom, L.D.; Zeng, Z.; Hwang, C.; Sanda, M.G.; Wechsler, D.S. Mxi1, a Myc Antagonist, Suppresses Proliferation of DU145 Human Prostate Cells. Prostate 2001, 47, 194–204. [Google Scholar] [CrossRef]
- Wahli, W.; Martinez, E. Superfamily of steroid nuclear receptors: Positive and negative regulators of gene expression. FASEB J. 1991, 5, 2243–2249. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Lee, D.H.; Han, K.D.; Yoon, H.; Shin, C.M.; Park, Y.S.; Kim, N. Is nonalcoholic fatty liver disease associated with the development of prostate cancer? A nationwide study with 10,516,985 Korean men. PLoS ONE 2018, 13, e0201308. [Google Scholar] [CrossRef] [PubMed]
- Baker, B.G.; Ball, G.R.; Rakha, E.A.; Nolan, C.C.; Caldas, C.; Ellis, I.O.; Green, A.R. Lack of expression of the proteins GMPR2 and PPARα are associated with the basal phenotype and patient outcome in breast cancer. Breast Cancer Res. Treat. 2013, 137, 127–137. [Google Scholar] [CrossRef] [PubMed]
- US National Library of Medicine. Studies for: Prostate Cancer. Available online: https://clinicaltrials.gov/ct2/results?cond=Prostate+Cancer&term=miRNA&cntry=&state=&city=&dist= (accessed on 17 June 2020).
- Hanna, J.; Hossain, G.S.; Kocerha, J. The Potential for microRNA Therapeutics and Clinical Research. Front. Genet. 2019, 10, 478. [Google Scholar] [CrossRef] [PubMed]
- Van der Kwast, T.; Bubendorf, L.; Mazeroles, C.; Raspollini, M.R.; Van Leenders, G.J.; Pihl, C.-G.; Kujala, P. Guidelines on processing and reporting of prostate biopsies: The 2013 update of the Pathology Committee of the European Randomized Study of Screening for Prostate Cancer (ERSPC). Virchows Arch. 2013, 463, 367–377. [Google Scholar] [CrossRef]
- Alvarez, M.L.; Doné, S.C. SYBR® Green and TaqMan® Quantitative PCR Arrays: Expression Profile Genes Relevant to a Pathway or a Disease State. In RNA Mapping; Alvarez, M., Nourbakhsh, M., Eds.; Humana Press: New York, NY, USA.
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. Past: Paleontological Statistics Software Package for education and data analysis. Palaeo Electron. 2001, 4. [Google Scholar]
- Lu, J.; Clark, A.G. Impact of microRNA regulation on variation in human gene expression. Genome Res. 2012, 22, 1243–1254. [Google Scholar] [CrossRef]
- Wang, X. miRDB: A microRNA target prediction and functional annotation database with a wiki interface. RNA 2008, 14, 1012–1017. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, CD127–CD131. [Google Scholar] [CrossRef]
- Dweep, H.; Gretz, N. miRWalk2.0: A comprehensive atlas of microRNA-target interactions. Nat. Methods 2015, 12, 697. [Google Scholar] [CrossRef] [PubMed]
- Baek, D.; Villén, J.; Shin, C.; Camargo, F.D.; Gygi, S.P.; Bartel, D.P. The impact of microRNAs on protein output. Nature 2008, 455, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 12, e05005. [Google Scholar] [CrossRef]

| Characteristics | Serum Samples | p-Value a | Tissue Samples | p-Value a | ||||
|---|---|---|---|---|---|---|---|---|
| All | BPH | PCa | All | NT | PCa | |||
| n = 102 | n = 62 | n = 40 | n = 54 | n = 28 | n = 26 | |||
| AGE | ||||||||
| average | 67.98 | 67.93 | 68.02 | 0.482 | 66.90 | 67.88 | 65.92 | 0.716 |
| range | 48–85 | 58–85 | 48–78 | 48–79 | 48–79 | 48–79 | ||
| PSA (ng/mL) | ||||||||
| average | 10.99 | 7.10 | 14.88 | 0.0001 | 12.29 | 14.47 | 20.11 | 0.111 |
| range | 2.7–108 | 2.7–20 | 3.8–108 | 3.43–108 | 3.43–52.6 | 4.1–108 | ||
| 2.5–4.0 | 6 | 5 | 1 | 2 | 2 | 0 | ||
| 4.01–10.0 | 73 | 50 | 23 | 29 | 15 | 14 | ||
| 10.01–20.0 | 16 | 7 | 9 | 6 | 3 | 3 | ||
| >20.0 | 7 | 0 | 7 | 17 | 8 | 9 | ||
| GS 6 | 20 (50%) | 11 (42.3%) | ||||||
| GS 7 | 9 (22.5%) | 7 (26.9%) | ||||||
| GS 8 | 5 (12.5%) | 3 (11.5%) | ||||||
| GS 9 | 6 (15%) | 5 (19.2%) | ||||||
| EAU Group Risk | ||||||||
| Low-risk | 15 (37.5%) | 13 (50%) | ||||||
| Intermediate-risk | 11 (27.5%) | 4 (15.4%) | ||||||
| High-risk | 14 (35%) | 9 (34.6%) | ||||||
| miRNA | Differential Expression in Serum | Differential Expression in Tissue | ||||
|---|---|---|---|---|---|---|
| miR-21 | ↑ | 46 PCa/10 BPH | [27] | ↑ | 10 MT/10 AN | [28] |
| ↑ | 10 PCa/10 BPH | [25] | ↑ | 53 PCac | [29] | |
| ↑ | 20 PCa/8 HC | [30] | ↑ | 45 MT/45 AN | [31] | |
| miR-27b | N/R | ↓ | 49 MT/41 AN | [32] | ||
| ↑ | 63 PCa/28 BPH | [33] | ||||
| ↓ | 44 PCa/10 BPH | [34] | ||||
| miR-93 | ↑ | 36 PCa/12 HC | [35] | ↑ | 16 MT/16 AN | [36] |
| ↓ | 100 PCa/50 BPH | [37] | ↑ | 30 MT/30 BPH | [38] | |
| ↑ | 112 PCa/48 BPH | [39] | ||||
| miR-141 | ↑ | 25 MetPCa/25 HC | [40] | ↑ | 76 MT/76 AN | [41] |
| ↑ | 14 PCa/7 MetPCa 1a: 45 PCa, 2a: 72 PCa | [42] | ↑ | 36 MT/36 AN | [42] | |
| ↑ | 21 PCab | [43] | ↑ | 34 PCa/14 BPH | [44] | |
| ↑ | 36 PCa/12 HC | [35] | ↓ | 55 MT/55 AN | [45] | |
| ↑ | 25 mCRPC/25 HC | [46] | ↑ | 535 PCad | [1] | |
| ↑ | 78 PCa/28 HC | [47] | ↑ | 206 MT/29 AN | [48] | |
| ↑ | 25 mCRPC/25 HC | [49] | ||||
| ↑ | 30 mCRPC/60 locPCa | [50] | ||||
| ↑ | 8 PCa BCR/8 PCa no BCR | [51] | ||||
| ↑ | 31 PCa/13 BPH | [52] | ||||
| ↑ | 72 PCa/34 HC | [26] | ||||
| miR-182 | N/R | ↑ | 76 MT/76 AN | [53] | ||
| ↑ | 1e: 127 PCa/13 BPH 2e: 138 MT/19 AN | [54] | ||||
| ↑ | 56 PCa/56 BPH | [22] | ||||
| ↑ | 45 MT/45 AN | [31] | ||||
| miR-205 | ↓ | 72 PCa/34 HC | [26] | ↓ | 31 MT/31 AN | [55] |
| ↓ | 76 MT/76 AN | [53] | ||||
| ↓ | 40 MT/40 AN | [56] | ||||
| ↓ | 111 MT/111 AN | [57] | ||||
| ↓ | 49 PCa/25 BPH | [58] | ||||
| miR-221 | ↑ | 10 PCa/10 BPH | [25] | ↓ | 9 PCa/4 BPH | [59] |
| ↓ | 76 MT/76 AN | [53] | ||||
| ↓ | 40 MT/40 AN | [56] | ||||
| ↓ | 45 MT/45 AN | [31] | ||||
| ↓ | 206 MT/29 AN | [48] | ||||
| miR-375 | ↑ | 14 PCa/7 MetPCa 1a: 45 PCa, 2a: 72 PCa | [42] | ↑ | 76 MT/76 AN | [53] |
| ↑ | 25 mCRPC/25 HC | [46] | ↑ | 76 MT/76 AN | [41] | |
| ↑ | 78 PCa/28 HC | [47] | ↑ | 1e: 127 PCa/13 BPH 2e: 138 MT/19 AN | [54] | |
| ↑ | 25 mCRPC/25 HC | [49] | ||||
| ↑ | 30 mCRPC/60 locPCa | [50] | ||||
| ↑ | 31 PCa/13 BPH | [52] | ||||
| ↓ | 33 PCa/25 BPH | [60] | ||||
| let-7a | ↑ | 31 PCa/13 BPH | [47] | ↓ | 9 PCa/4 BPH | [59] |
| ↓ | 100 PCa/50 BPH | [37] | ↓ | 75 PCa/27 BPH | [61] | |
| ↓ | 32 PCa BCR/36 PCa no BCR | [62] | ||||
| ↓ | 26 MT/26 AN | [63] | ||||
| (A) Serum miRNAs | ||||||||
|---|---|---|---|---|---|---|---|---|
| miRNAs | AUC | 95% CI | SE | p-Value | Sensitivity | Specificity | PPV | NPV |
| PSA | 0.737 | 0.636–0.838 | 0.0517 | < 0.0001 | 0.60 | 0.79 | 0.65 | 0.75 |
| miR-21 | 0.757 | 0.661–0.853 | 0.0490 | 0.50 | 0.92 | 0.80 | 0.74 | |
| miR-93 | 0.919 | 0.868–0.934 | 0.0167 | 0.95 | 0.79 | 0.75 | 0.96 | |
| miR-141 | 0.849 | 0.778–0.921 | 0.0364 | 0.92 | 0.66 | 0.64 | 0.93 | |
| miR-182 | 0.881 | 0.816–0.946 | 0.0331 | 0.85 | 0.79 | 0.72 | 0.89 | |
| miR-205 | 0.701 | 0.600–0.801 | 0.0514 | 0.90 | 0.52 | 0.55 | 0.89 | |
| miR-375 | 0.892 | 0.833–0.952 | 0.0305 | 0.73 | 0.90 | 0.83 | 0.84 | |
| let-7a | 0.673 | 0.564–0.783 | 0.0561 | 0.0020 | 0.40 | 0.92 | 0.76 | 0.70 |
| miR-93/miR-375 | 0.908 | 0.870–0.946 | 0.019 | < 0.0001 | 0.81 | 0.85 | 0.78 | 0.88 |
| (B) Tissue miRNAs | ||||||||
| miRNAs | AUC | 95% CI | SE | p-Value | Sensitivity | Specificity | PPV | NPV |
| PSA | 0.536 | 0.380–0.693 | 0.0800 | 0.080 | 0.54 | 0.54 | 0.52 | 0.56 |
| miR-21 | 0.705 | 0.562–0.847 | 0.0726 | 0.0048 | 0.36 | 1.00 | 1.00 | 0.64 |
| miR-93 | 0.859 | 0.763–0.954 | 0.0487 | <0.0001 | 0.92 | 0.64 | 0.71 | 0.90 |
| miR-141 | 0.973 | 0.935–1.010 | 0.0193 | 0.89 | 1.00 | 1.00 | 0.90 | |
| miR-182 | 0.946 | 0.893–1.000 | 0.0271 | 1.00 | 0.79 | 0.81 | 1.00 | |
| miR-205 | 0.688 | 0.544–0.832 | 0.0734 | 0.0104 | 0.77 | 0.57 | 0.63 | 0.73 |
| miR-221 | 0.880 | 0.787–0.974 | 0.0478 | <0.0001 | 0.77 | 0.93 | 0.91 | 0.81 |
| miR-375 | 0.952 | 0.904–1.000 | 0.0246 | 0.81 | 0.96 | 0.95 | 0.84 | |
| let-7a | 0.655 | 0.504–0.807 | 0.0772 | 0.0443 | 0.39 | 0.96 | 0.91 | 0.63 |
| miR-93/miR-375 | 0.879 | 0.817–0.942 | 0.032 | <0.0001 | 0.69 | 0.95 | 0.92 | 0.77 |
| (A) PCa serum miRNAs | ||||||||
|---|---|---|---|---|---|---|---|---|
| miRNAs | AUC | 95% CI | SE | p-value | Sensitivity | Specificity | PPV | NPV |
| miR-21/miR-93 | 0.640 | 0.516–0.764 | 0.063 | 0.0274 | 0.50 | 0.80 | 0.81 | 0.49 |
| miR-21/miR-182 | 0.633 | 0.508–0.759 | 0.064 | 0.0372 | 0.68 | 0.60 | 0.74 | 0.53 |
| miR-21/miR-375 | 0.631 | 0.506–0.756 | 0.064 | 0.0403 | 0.52 | 0.77 | 0.79 | 0.49 |
| miR-93/miR-182 | 0.643 | 0.521–0.764 | 0.062 | 0.0213 | 0.70 | 0.57 | 0.73 | 0.53 |
| miR-93/miR-375 | 0.636 | 0.514–0.758 | 0.0283 | 0.54 | 0.73 | 0.77 | 0.49 | |
| miR-182/miR-375 | 0.641 | 0.520–0.763 | 0.0225 | 0.34 | 0.93 | 0.89 | 0.46 | |
| (B) PCa tissue miRNAs | ||||||||
| miRNAs | AUC | 95% CI | SE | p-value | Sensitivity | Specificity | PPV | NPV |
| miR-93 | 0.746 | 0.542–0.950 | 0.104 | 0.0182 | 0.69 | 0.69 | 0.69 | 0.69 |
| miR-375 | 0.828 | 0.653–1.004 | 0.089 | 0.0002 | 0.77 | 0.92 | 0.91 | 0.80 |
| miR-93/miR-141 | 0.710 | 0.562–0.858 | 0.076 | 0.0055 | 0.58 | 0.88 | 0.83 | 0.68 |
| miR-93/miR-182 | 0.692 | 0.545–0.840 | 0.075 | 0.0105 | 0.50 | 0.92 | 0.87 | 0.65 |
| miR-93/miR-375 | 0.766 | 0.635–0.898 | 0.067 | < 0.0001 | 0.65 | 0.81 | 0.77 | 0.70 |
| miR-141/miR-375 | 0.743 | 0.600–0.886 | 0.073 | 0.0009 | 0.58 | 0.92 | 0.88 | 0.69 |
| miR-182/miR-375 | 0.691 | 0.542–0.839 | 0.0757 | 0.0117 | 0.46 | 0.93 | 0.86 | 0.63 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciszkowicz, E.; Porzycki, P.; Semik, M.; Kaznowska, E.; Tyrka, M. MiR-93/miR-375: Diagnostic Potential, Aggressiveness Correlation and Common Target Genes in Prostate Cancer. Int. J. Mol. Sci. 2020, 21, 5667. https://doi.org/10.3390/ijms21165667
Ciszkowicz E, Porzycki P, Semik M, Kaznowska E, Tyrka M. MiR-93/miR-375: Diagnostic Potential, Aggressiveness Correlation and Common Target Genes in Prostate Cancer. International Journal of Molecular Sciences. 2020; 21(16):5667. https://doi.org/10.3390/ijms21165667
Chicago/Turabian StyleCiszkowicz, Ewa, Paweł Porzycki, Małgorzata Semik, Ewa Kaznowska, and Mirosław Tyrka. 2020. "MiR-93/miR-375: Diagnostic Potential, Aggressiveness Correlation and Common Target Genes in Prostate Cancer" International Journal of Molecular Sciences 21, no. 16: 5667. https://doi.org/10.3390/ijms21165667
APA StyleCiszkowicz, E., Porzycki, P., Semik, M., Kaznowska, E., & Tyrka, M. (2020). MiR-93/miR-375: Diagnostic Potential, Aggressiveness Correlation and Common Target Genes in Prostate Cancer. International Journal of Molecular Sciences, 21(16), 5667. https://doi.org/10.3390/ijms21165667






