Synergetic Effects of Intronic Mature miR-944 and ΔNp63 Isoforms on Tumorigenesis in a Cervical Cancer Cell Line
Abstract
1. Introduction
2. Results
2.1. Association between Expression Levels of miR-944 and TAp63 mRNA and ΔNp63 mRNA in Cervical Cancer Tissue
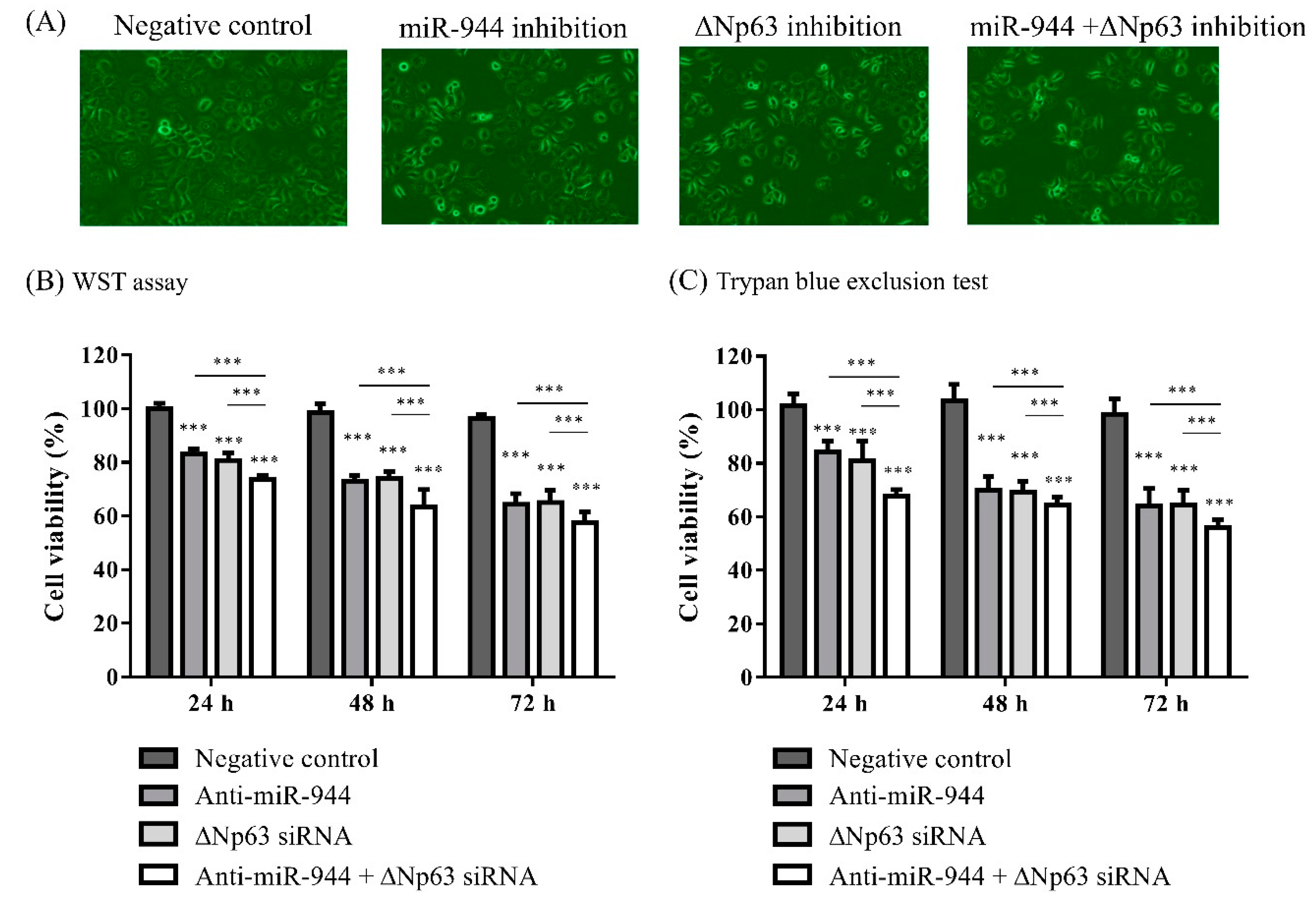
2.2. Cell Viability According to Inhibition of miR-944 and ΔNp63 in ME-180 Cells
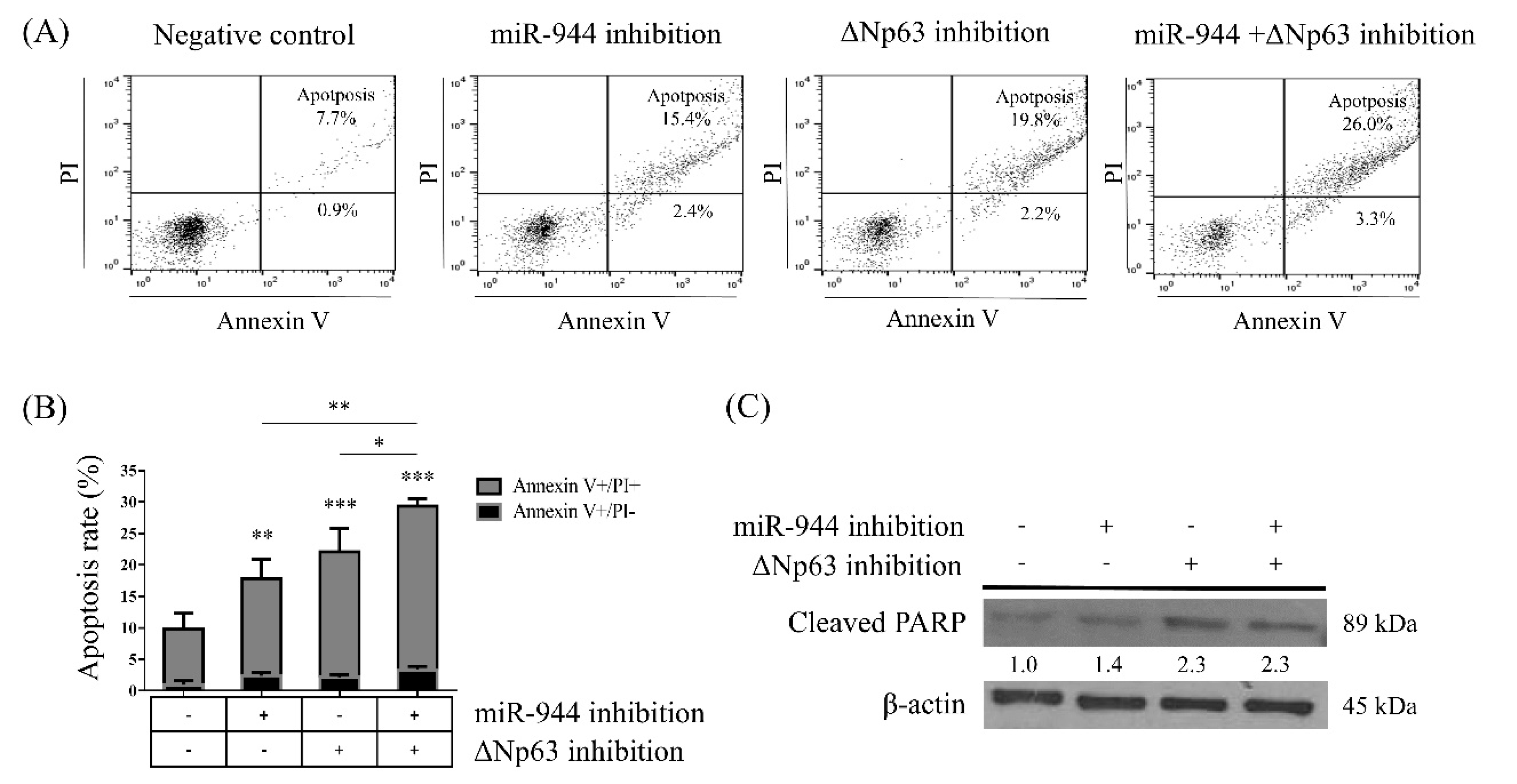
2.3. Combined miR-944 and ΔNp63 Inhibition Increases ME-180 Cell Apoptosis
2.4. Combined miR-944 and ΔNp63 Inhibition Decreases Migration in ME-180 Cells
2.5. Combined miR-944 and ΔNp63 Inhibition Affects EMT in ME-180 Cells
2.6. Identification of Differentially Expressed Genes (DEGs) According to miR-944, ΔNp63, and Combined miR-944 and ΔNp63 Inhibition
3. Discussion
4. Materials and Methods
4.1. Clinical Samples
4.2. Cell Culture
4.3. Transfection of Anti-miRNA and siRNA
4.4. Cell Viability Assay Using EZ-Cytox
4.5. Trypan Blue Exclusion Assay
4.6. Cell Apoptosis Analysis
4.7. Wound Healing and Transwell Invasion Assay
4.8. Protein Isolation and Western Blot Analysis
4.9. Total RNA Extraction
4.10. TP63 mRNA Expression Analysis
4.11. miR-944 Expression Analysis
4.12. RNA Sequence Analysis
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| cDNA | Complementary DNA |
| CT | Cycle threshold |
| DAVID | Database for annotation, visualization and integrated discovery |
| DEG | Differentially expressed gene |
| EMT | Epithelial–mesenchymal transition |
| FDR | False discovery rate |
| GAPDH | Glyceraldehyde-3-phsphate dehydrogenase |
| GO | Gene Ontology |
| miRNA | MicroRNA |
| PI | Propidium iodide |
| RPMI | Roswell Park Memorial Institute |
| RT | Reverse transcriptase |
| TA | Transactivation |
| TJ | Tight junction |
| TP63 | Tumor protein p63 |
| WST | Water-soluble tetrazolium salt |
References
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Bouyssou, J.M.; Manier, S.; Huynh, D.; Issa, S.; Roccaro, A.M.; Ghobrial, I.M. Regulation of microRNAs in cancer metastasis. Biochim. Biophys. Acta 2014, 1845, 255–265. [Google Scholar] [CrossRef]
- Chan, S.H.; Wang, L.H. Regulation of cancer metastasis by microRNAs. J. Biomed. Sci. 2015, 22, 9. [Google Scholar] [CrossRef] [PubMed]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, P.; Palanichamy, J.K.; Singh, A.; Das, P.; Bhagat, M.; Kassab, M.A.; Sinha, S.; Chattopadhyay, P. Biogenesis of intronic miRNAs located in clusters by independent transcription and alternative splicing. RNA 2014, 20, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Yao, S. MicroRNA biogenesis and their functions in regulating stem cell potency and differentiation. Biol. Proced. Online 2016, 18, 8. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Li, Z.; Chen, P.; Huang, H.; Hurst, L.D.; Chen, J. Young intragenic miRNAs are less coexpressed with host genes than old ones: Implications of miRNA-host gene coevolution. Nucleic Acids Res. 2012, 40, 4002–4012. [Google Scholar] [CrossRef]
- Hinske, L.C.; Galante, P.A.; Kuo, W.P.; Ohno-Machado, L. A potential role for intragenic miRNAs on their hosts’ interactome. BMC Genom. 2010, 11, 533. [Google Scholar] [CrossRef]
- Lin, S.L.; Miller, J.D.; Ying, S.Y. Intronic microRNA (miRNA). J. Biomed. Biotechnol. 2006, 2006, 26818. [Google Scholar] [CrossRef]
- Gao, X.; Qiao, Y.; Han, D.; Zhang, Y.; Ma, N. Enemy or partner: Relationship between intronic micrornas and their host genes. IUBMB Life 2012, 64, 835–840. [Google Scholar] [CrossRef]
- Liu, M.; Roth, A.; Yu, M.; Morris, R.; Bersani, F.; Rivera, M.N.; Lu, J.; Shioda, T.; Vasudevan, S.; Ramaswamy, S.; et al. The IGF2 intronic miR-483 selectively enhances transcription from IGF2 fetal promoters and enhances tumorigenesis. Genes Dev. 2013, 27, 2543–2548. [Google Scholar] [CrossRef]
- Franca, G.S.; Vibranovski, M.D.; Galante, P.A. Host gene constraints and genomic context impact the expression and evolution of human microRNAs. Nat. Commun. 2016, 7, 11438. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Lu, M.; Miao, J.; Li, T.; Wang, E.; Cui, Q. Cepred: Predicting the co-expression patterns of the human intronic microRNAs with their host genes. PLoS ONE 2009, 4, e4421. [Google Scholar] [CrossRef] [PubMed]
- Sowa, N.; Horie, T.; Kuwabara, Y.; Baba, O.; Watanabe, S.; Nishi, H.; Kinoshita, M.; Takanabe-Mori, R.; Wada, H.; Shimatsu, A.; et al. MicroRNA 26b encoded by the intron of small CTD phosphatase (SCP) 1 has an antagonistic effect on its host gene. J. Cell Biochem. 2012, 113, 3455–3465. [Google Scholar] [CrossRef] [PubMed]
- Moll, U.M.; Slade, N. p63 and p73: Roles in development and tumor formation. Mol. Cancer Res. 2004, 2, 371–386. [Google Scholar] [PubMed]
- Billant, O.; Leon, A.; Le Guellec, S.; Friocourt, G.; Blondel, M.; Voisset, C. The dominant-negative interplay between p53, p63 and p73: A family affair. Oncotarget 2016, 7, 69549–69564. [Google Scholar] [CrossRef]
- Massion, P.P.; Taflan, P.M.; Jamshedur Rahman, S.M.; Yildiz, P.; Shyr, Y.; Edgerton, M.E.; Westfall, M.D.; Roberts, J.R.; Pietenpol, J.A.; Carbone, D.P.; et al. Significance of p63 amplification and overexpression in lung cancer development and prognosis. Cancer Res. 2003, 63, 7113–7121. [Google Scholar]
- Nekulova, M.; Holcakova, J.; Coates, P.; Vojtesek, B. The role of p63 in cancer, stem cells and cancer stem cells. Cell Mol. Biol. Lett. 2011, 16, 296–327. [Google Scholar] [CrossRef]
- Qian, L.; Xu, F.; Wang, X.; Jiang, M.; Wang, J.; Song, W.; Wu, D.; Shen, Z.; Feng, D.; Ling, B.; et al. LncRNA expression profile of DeltaNp63alpha in cervical squamous cancers and its suppressive effects on LIF expression. Cytokine 2017, 96, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Xu, Q.; Ling, B.; Xiao, W.; Liu, P. Reduced expression of DeltaNup63alpha in cervical squamous cell carcinoma. Clin. Investig. Med. 2011, 34, E184–E191. [Google Scholar] [CrossRef]
- Kim, K.H.; Cho, E.G.; Yu, S.J.; Kang, H.; Kim, Y.J.; Kim, S.H.; Lee, T.R. DeltaNp63 intronic miR-944 is implicated in the DeltaNp63-mediated induction of epidermal differentiation. Nucleic Acids Res. 2015, 43, 7462–7479. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Lee, S.; Kim, J.; Kim, G.; Park, K.H.; Kim, T.U.; Chung, D.; Lee, H. DeltaNp63 to TAp63 expression ratio as a potential molecular marker for cervical cancer prognosis. PLoS ONE 2019, 14, e0214867. [Google Scholar]
- Xie, H.; Lee, L.; Scicluna, P.; Kavak, E.; Larsson, C.; Sandberg, R.; Lui, W.O. Novel functions and targets of miR-944 in human cervical cancer cells. Int. J. Cancer 2015, 136, 230–241. [Google Scholar] [CrossRef]
- He, Z.; Xu, H.; Meng, Y.; Kuang, Y. miR-944 acts as a prognostic marker and promotes the tumor progression in endometrial cancer. Biomed. Pharm. 2017, 88, 902–910. [Google Scholar] [CrossRef] [PubMed]
- Flores-Perez, A.; Marchat, L.A.; Rodriguez-Cuevas, S.; Bautista, V.P.; Fuentes-Mera, L.; Romero-Zamora, D.; Maciel-Dominguez, A.; de la Cruz, O.H.; Fonseca-Sanchez, M.; Ruiz-Garcia, E.; et al. Suppression of cell migration is promoted by miR-944 through targeting of SIAH1 and PTP4A1 in breast cancer cells. BMC Cancer 2016, 16, 379. [Google Scholar] [CrossRef]
- Park, S.; Kim, J.; Eom, K.; Oh, S.; Kim, S.; Kim, G.; Ahn, S.; Park, K.H.; Chung, D.; Lee, H. microRNA-944 overexpression is a biomarker for poor prognosis of advanced cervical cancer. BMC Cancer 2019, 19, 419. [Google Scholar] [CrossRef]
- Bergholz, J.; Xiao, Z.X. Role of p63 in development, tumorigenesis and cancer progression. Cancer Microenviron. 2012, 5, 311–322. [Google Scholar] [CrossRef]
- Rocco, J.W.; Leong, C.O.; Kuperwasser, N.; DeYoung, M.P.; Ellisen, L.W. p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell 2006, 9, 45–56. [Google Scholar] [CrossRef]
- DeYoung, M.P.; Johannessen, C.M.; Leong, C.O.; Faquin, W.; Rocco, J.W.; Ellisen, L.W. Tumor-specific p73 up-regulation mediates p63 dependence in squamous cell carcinoma. Cancer Res. 2006, 66, 9362–9368. [Google Scholar] [CrossRef]
- Giacobbe, A.; Compagnone, M.; Bongiorno-Borbone, L.; Antonov, A.; Markert, E.K.; Zhou, J.H.; Annicchiarico-Petruzzelli, M.; Melino, G.; Peschiaroli, A. p63 controls cell migration and invasion by transcriptional regulation of MTSS1. Oncogene 2016, 35, 1602–1608. [Google Scholar] [CrossRef]
- Lee, K.B.; Ye, S.; Park, M.H.; Park, B.H.; Lee, J.S.; Kim, S.M. p63-Mediated activation of the beta-catenin/c-Myc signaling pathway stimulates esophageal squamous carcinoma cell invasion and metastasis. Cancer Lett. 2014, 353, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Kumakura, Y.; Rokudai, S.; Iijima, M.; Altan, B.; Yoshida, T.; Bao, H.; Yokobori, T.; Sakai, M.; Sohda, M.; Miyazaki, T.; et al. Elevated expression of ΔNp63 in advanced esophageal squamous cell carcinoma. Cancer Sci. 2017, 108, 2149–2155. [Google Scholar] [CrossRef]
- Hoellen, F.; Waldmann, A.; Banz-Jansen, C.; Holtrich, U.; Karn, T.; Oberlander, M.; Habermann, J.K.; Hormann, M.; Koster, F.; Ribbat-Idel, J. Claudin-1 expression in cervical cancer. Mol. Clin. Oncol. 2017, 7, 880–884. [Google Scholar] [CrossRef]
- Kaneko, Y.; Kohno, T.; Kakuki, T.; Takano, K.I.; Ogasawara, N.; Miyata, R.; Kikuchi, S.; Konno, T.; Ohkuni, T.; Yajima, R.; et al. The role of transcriptional factor p63 in regulation of epithelial barrier and ciliogenesis of human nasal epithelial cells. Sci. Rep. 2017, 7, 10935. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, R.; Arora, H.; Rizvi, M.A. EMT in cervical cancer: Its role in tumour progression and response to therapy. Cancer Lett. 2015, 356, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Hollestelle, A.; Peeters, J.K.; Smid, M.; Timmermans, M.; Verhoog, L.C.; Westenend, P.J.; Heine, A.A.; Chan, A.; Sieuwerts, A.M.; Wiemer, E.A.; et al. Loss of E-cadherin is not a necessity for epithelial to mesenchymal transition in human breast cancer. Breast Cancer Res. Treat. 2013, 138, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Ren, D.; Guo, W.; Huang, S.; Wang, Z.; Li, Q.; Du, H.; Song, L.; Peng, X. N-cadherin promotes epithelial-mesenchymal transition and cancer stem cell-like traits via ErbB signaling in prostate cancer cells. Int. J. Oncol. 2016, 48, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, S.; Doi, R.; Toyoda, E.; Tsuji, S.; Wada, M.; Koizumi, M.; Tulachan, S.S.; Ito, D.; Kami, K.; Mori, T.; et al. N-cadherin expression and epithelial-mesenchymal transition in pancreatic carcinoma. Clin. Cancer Res. 2004, 10, 4125–4133. [Google Scholar] [CrossRef] [PubMed]
- Elloul, S.; Elstrand, M.B.; Nesland, J.M.; Trope, C.G.; Kvalheim, G.; Goldberg, I.; Reich, R.; Davidson, B. Snail, Slug, and Smad-interacting protein 1 as novel parameters of disease aggressiveness in metastatic ovarian and breast carcinoma. Cancer 2005, 103, 1631–1643. [Google Scholar] [CrossRef]
- Grzegrzolka, J.; Biala, M.; Wojtyra, P.; Kobierzycki, C.; Olbromski, M.; Gomulkiewicz, A.; Piotrowska, A.; Rys, J.; Podhorska-Okolow, M.; Dziegiel, P. Expression of EMT markers SLUG and TWIST in breast cancer. Anticancer Res. 2015, 35, 3961–3968. [Google Scholar]
- Lee, M.H.; Cho, Y.; Kim, D.H.; Woo, H.J.; Yang, J.Y.; Kwon, H.J.; Yeon, M.J.; Park, M.; Kim, S.H.; Moon, C.; et al. Menadione induces G2/M arrest in gastric cancer cells by down-regulation of CDC25C and proteasome mediated degradation of CDK1 and cyclin B1. Am. J. Transl. Res. 2016, 8, 5246–5255. [Google Scholar] [PubMed]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Turner, A.; Aggarwal, P.; Matter, A.; Storvick, E.; Arnett, D.K.; Broeckel, U. Comprehensive evaluation of AmpliSeq transcriptome, a novel targeted whole transcriptome RNA sequencing methodology for global gene expression analysis. BMC Genom. 2015, 16, 1069. [Google Scholar] [CrossRef]
- Papp, A.C.; Azad, A.K.; Pietrzak, M.; Williams, A.; Handelman, S.K.; Igo, R.P., Jr.; Stein, C.M.; Hartmann, K.; Schlesinger, L.S.; Sadee, W. AmpliSeq transcriptome analysis of human alveolar and monocyte-derived macrophages over time in response to Mycobacterium tuberculosis infection. PLoS ONE 2018, 13, e0198221. [Google Scholar] [CrossRef] [PubMed]
- Termo Fisher Scientific. Available online: https://www.thermofisher.com/kr/ko/home/life-science/microarray-analysis/microarray-analysis-instruments-software-services/microarray-analysis-software/affymetrix-transcriptome-analysis-console-software.html (accessed on 5 August 2020).




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Park, S.; Chang, Y.; Park, K.H.; Lee, H. Synergetic Effects of Intronic Mature miR-944 and ΔNp63 Isoforms on Tumorigenesis in a Cervical Cancer Cell Line. Int. J. Mol. Sci. 2020, 21, 5612. https://doi.org/10.3390/ijms21165612
Kim J, Park S, Chang Y, Park KH, Lee H. Synergetic Effects of Intronic Mature miR-944 and ΔNp63 Isoforms on Tumorigenesis in a Cervical Cancer Cell Line. International Journal of Molecular Sciences. 2020; 21(16):5612. https://doi.org/10.3390/ijms21165612
Chicago/Turabian StyleKim, Jungho, Sunyoung Park, Yunhee Chang, Kwang Hwa Park, and Hyeyoung Lee. 2020. "Synergetic Effects of Intronic Mature miR-944 and ΔNp63 Isoforms on Tumorigenesis in a Cervical Cancer Cell Line" International Journal of Molecular Sciences 21, no. 16: 5612. https://doi.org/10.3390/ijms21165612
APA StyleKim, J., Park, S., Chang, Y., Park, K. H., & Lee, H. (2020). Synergetic Effects of Intronic Mature miR-944 and ΔNp63 Isoforms on Tumorigenesis in a Cervical Cancer Cell Line. International Journal of Molecular Sciences, 21(16), 5612. https://doi.org/10.3390/ijms21165612




