A Foundational Study for Normal F8-Containing Mouse Models for the miRNA Regulation of Hemophilia A: Identification and Analysis of Mouse miRNAs that Downregulate the Murine F8 Gene
Abstract
1. Introduction
2. Results
2.1. Prediction Analysis and MS2-Tagged Affinity Purification Assays Identify miRNAs Associated with Murine F8 3′UTR
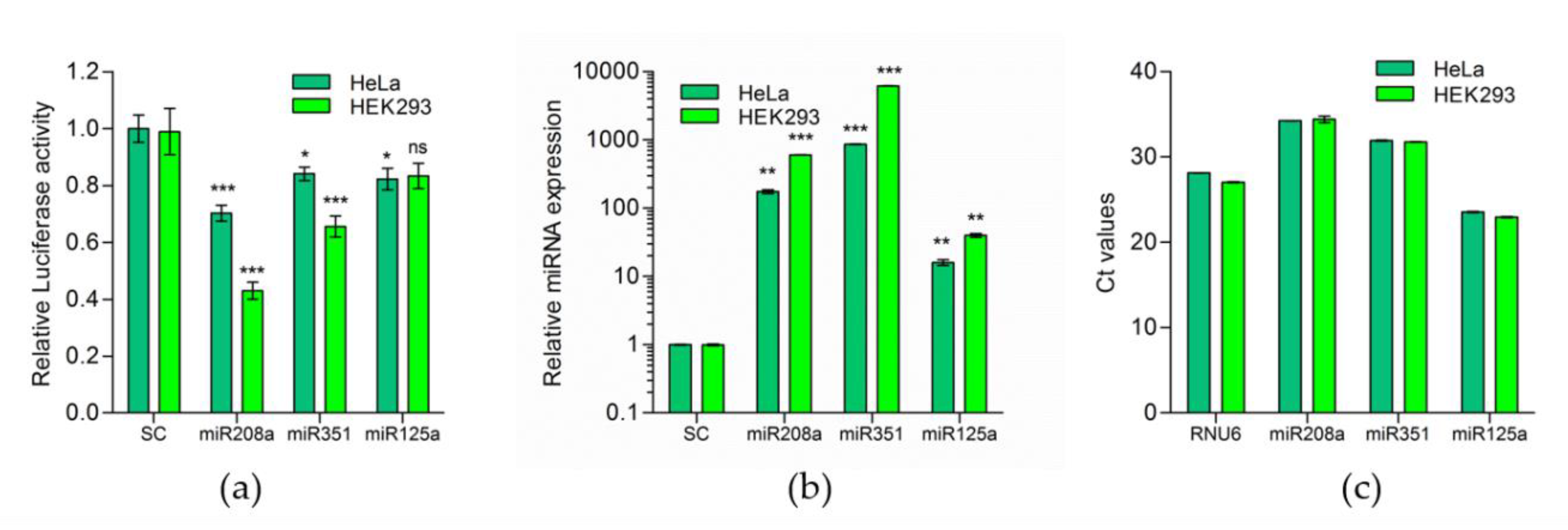
2.2. Predicted miRNAs Indeed Target Murine 3′UTR of F8-mRNA
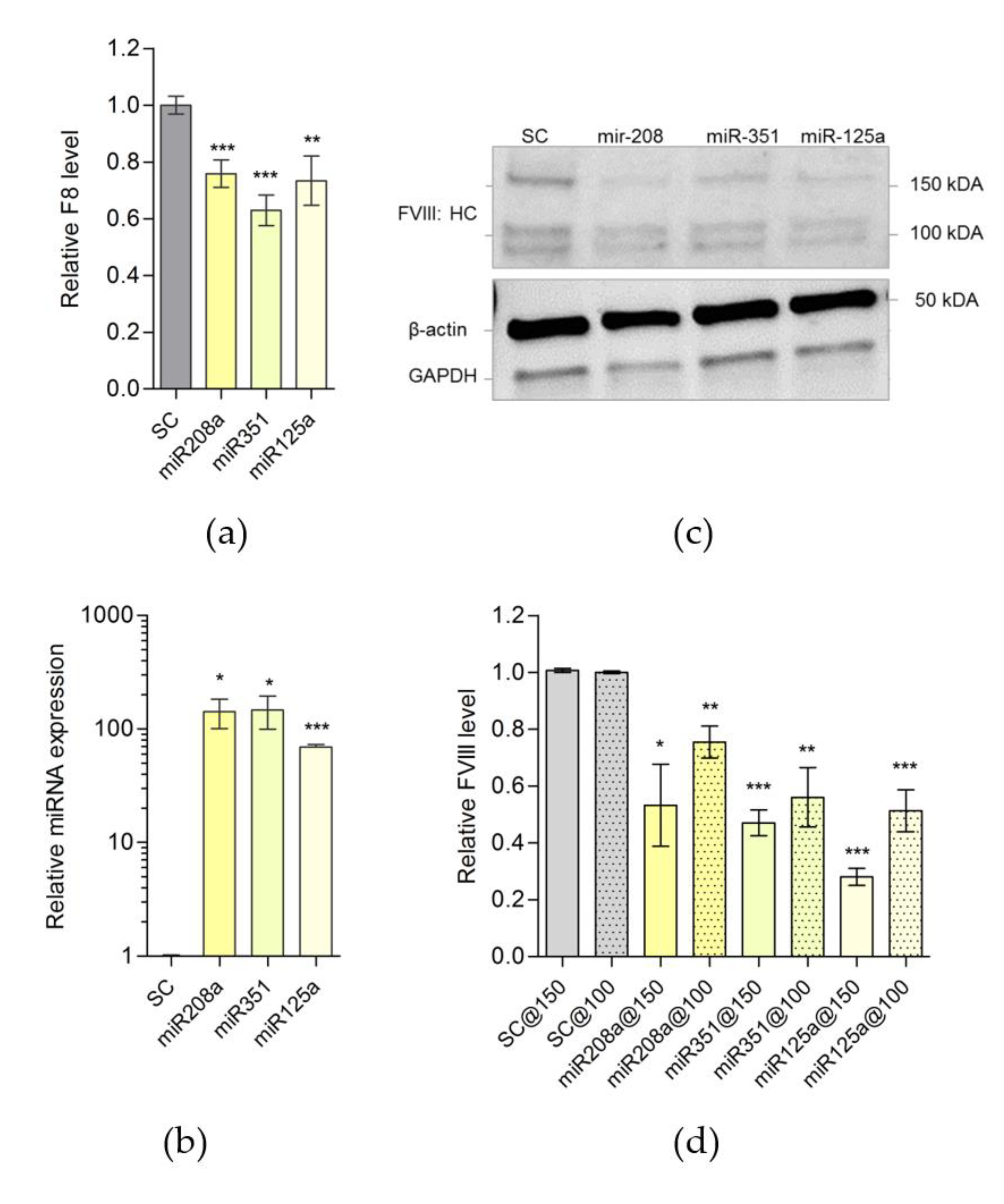
2.3. MiR-208, miR-125a and miR-351 Target F8 3′UTR and Downregulate FVIII Expression in Mouse Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Plasmid Constructs
4.3. Luciferase Assay
4.4. MS2-Tagged RNA Affinity Purification (MS2-TRAP) and Identification of F8 3′UTR-Interacting miRNAs
4.5. Cell Transfection
4.6. RT-qPCR
4.7. Western Blot
4.8. Data Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| DF | Degrees of freedom DF |
| F8 | gene encodes coagulation factor VIII |
| FVIII | coagulation factor VIII |
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase |
| HEK293T | Human embryonic kidney 293 cells |
| HeLa | Human immortal cell line derived from cervical cancer cells |
| HRP | horseradish peroxidase |
| miRNA | micro RNA |
| mmu | Mus musculus (mouse) |
| muF8 | Murine F8 |
| MS1 | MILE SVEN 1 cells |
| MS2 | MS2 plasmid |
| MS2-E | MS2 vector-empty, negative control |
| MS2-muF8 | MS2 vector fused to the 3′UTR of murine F8 |
| MS2-TRAP | MS2-tagged affinity assay |
| NSG | Next generation sequencing |
| qPCR | Quantitative polymerase chain reaction |
| TPBS | Phosphate-buffered saline buffer with Tween 20 |
| UTR | Untranslated region |
References
- Orlova, N.A.; Kovnir, S.V.; Vorobiev, I.I.; Gabibov, A.G.; Vorobiev, A.I. Blood Clotting Factor VIII: From Evolution to Therapy. Acta Nat. 2013, 5, 19–39. [Google Scholar] [CrossRef]
- Johnsen, J.M.; Fletcher, S.N.; Huston, H.; Roberge, S.; Martin, B.K.; Kircher, M.; Josephson, N.C.; Shendure, J.; Ruuska, S.; Koerper, M.A.; et al. Novel approach to genetic analysis and results in 3000 hemophilia patients enrolled in the My Life, Our Future initiative. Blood Adv. 2017, 1, 824–834. [Google Scholar] [CrossRef] [PubMed]
- El-Maarri, O.; Herbiniaux, U.; Graw, J.; Schroder, J.; Terzic, A.; Watzka, M.; Brackmann, H.H.; Schramm, W.; Hanfland, P.; Schwaab, R.; et al. Analysis of mRNA in hemophilia A patients with undetectable mutations reveals normal splicing in the factor VIII gene. J. Thromb. Haemost. 2005, 3, 332–339. [Google Scholar] [CrossRef] [PubMed]
- Sarachana, T.; Dahiya, N.; Simhadri, V.L.; Pandey, G.S.; Saini, S.; Guelcher, C.; Guerrera, M.F.; Kimchi-Sarfaty, C.; Sauna, Z.E.; Atreya, C.D. Small ncRNA Expression-Profiling of Blood from Hemophilia A Patients Identifies miR-1246 as a Potential Regulator of Factor 8 Gene. PLoS ONE 2015, 10, e0132433. [Google Scholar] [CrossRef] [PubMed]
- Jankowska, K.I.; McGill, J.; Pezeshkpoor, B.; Oldenburg, J.; Atreya, C.D.; Sauna, Z.E. Clinical manifestation of hemophilia A in the absence of mutations in the F8 gene that encodes FVIII: Role of microRNAs. Transfusion 2020, 60, 401–413. [Google Scholar] [CrossRef]
- Selvaraj, S.R.; Pipe, S.W. Not in the genotype: Can unexplained hemophilia A result from “micro (RNA) management”? Transfusion 2020, 60, 227–228. [Google Scholar] [CrossRef]
- Sun, T.; Li, M.Y.; Li, P.F.; Cao, J.M. MicroRNAs in Cardiac Autophagy: Small Molecules and Big Role. Cells 2018, 7, 104. [Google Scholar] [CrossRef]
- Yen, C.T.; Fan, M.N.; Yang, Y.L.; Chou, S.C.; Yu, I.S.; Lin, S.W. Current animal models of hemophilia: The state of the art. Thromb. J. 2016, 14, 22. [Google Scholar] [CrossRef] [PubMed]
- Lenting, P.J.; Van Mourik, J.A.; Mertens, K. The life cycle of coagulation factor VIII in view of its structure and function. Blood 1998, 92, 3983–3996. [Google Scholar] [CrossRef]
- Chao, B.N.; Baldwin, W.H.; Healey, J.F.; Parker, E.T.; Shafer-Weaver, K.; Cox, C.; Jiang, P.; Kanellopoulou, C.; Lollar, P.; Meeks, S.L.; et al. Characterization of a genetically engineered mouse model of hemophilia A with complete deletion of the F8 gene. J. Thromb. Haemost. 2016, 14, 346–355. [Google Scholar] [CrossRef]
- Sabatino, D.E.; Nichols, T.C.; Merricks, E.; Bellinger, D.A.; Herzog, R.W.; Monahan, P.E. Animal models of hemophilia. Prog. Mol. Biol. Transl. Sci. 2012, 105, 151–209. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Gorospe, M. Identification of mRNA-Interacting Factors by MS2-TRAP (MS2-Tagged RNA Affinity Purification). Methods Mol. Biol. 2016, 1421, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Srikantan, S.; Gorospe, M. MS2-TRAP (MS2-tagged RNA affinity purification): Tagging RNA to identify associated miRNAs. Methods 2012, 58, 81–87. [Google Scholar] [CrossRef]
- Dahiya, N.; Atreya, C.D. RAP1 Downregulation by miR-320c Reduces Platelet Activation in Ex-vivo Storage. Microrna 2019, 8, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Kumaran, V.; Benten, D.; Follenzi, A.; Joseph, B.; Sarkar, R.; Gupta, S. Transplantation of endothelial cells corrects the phenotype in hemophilia A mice. J. Thromb. Haemost. 2005, 3, 2022–2031. [Google Scholar] [CrossRef] [PubMed]
- Nourse, J.; Braun, J.; Lackner, K.; Huttelmaier, S.; Danckwardt, S. Large-scale identification of functional microRNA targeting reveals cooperative regulation of the hemostatic system. J. Thromb. Haemost. 2018, 16, 2233–2245. [Google Scholar] [CrossRef]
- Wang, X.; Sundquist, K.; Svensson, P.J.; Rastkhani, H.; Palmer, K.; Memon, A.A.; Sundquist, J.; Zöller, B. Association of recurrent venous thromboembolism and circulating microRNAs. Clin. Epigenet. 2019, 11, 28. [Google Scholar] [CrossRef]
- Teruel, R.; Corral, J.; Perez-Andreu, V.; Martinez-Martinez, I.; Vicente, V.; Martinez, C. Potential role of miRNAs in developmental haemostasis. PLoS ONE 2011, 6, e17648. [Google Scholar] [CrossRef]
- Jankowska, K.I.; Sauna, Z.E.; Atreya, C.D. Role of microRNAs in Hemophilia and Thrombosis in Humans. Int. J. Mol. Sci. 2020, 21, 3598. [Google Scholar] [CrossRef] [PubMed]
- Lozier, J.N.; Nichols, T.C. Animal models of hemophilia and related bleeding disorders. Semin. Hematol. 2013, 50, 175–184. [Google Scholar] [CrossRef]
- Monahan, P.E. The expanding menagerie: Animal models of hemophilia A. J. Thromb. Haemost. 2010, 8, 2469–2471. [Google Scholar] [CrossRef]
- Bril, W.S.; Van Helden, P.M.W.; Hausl, C.; Zuurveld, M.G.; Ahmad, R.U.; Hollestelle, M.J.; Reitsma, P.H.; Fijnvandraat, K.; Van Lier, R.A.W.; Schwarz, H.P.; et al. Tolerance to factor VIII in a transgenic mouse expressing human factor VIII cDNA carrying an Arg(593) to Cys substitution. Thromb. Haemost. 2006, 95, 341–347. [Google Scholar]
- Jin, D.-Y.; Zhang, T.-P.; Gui, T.; Stafford, D.W.; Monahan, P.E. Creation of a mouse expressing defective human factor IX. Blood 2004, 104, 1733–1739. [Google Scholar] [CrossRef] [PubMed]
- Meeks, S.L.; Healey, J.F.; Parker, E.T.; Barrow, R.T.; Lollar, P. Non-classical anti-factor VIII C2 domain antibodies are pathogenic in a murine in vivo bleeding model. J. Thromb. Haemost. 2009, 7, 658–664. [Google Scholar] [CrossRef] [PubMed]
- Furie, B.; Furie, B.C. In vivo thrombus formation. J. Thromb. Haemost. 2007, 5, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Sachs, U.J.; Nieswandt, B. In vivo thrombus formation in murine models. Circ. Res. 2007, 100, 979–991. [Google Scholar] [CrossRef] [PubMed]
- Broze, G.J.; Yin, Z.F., Jr.; Lasky, N. A tail vein bleeding time model and delayed bleeding in hemophiliac mice. Thromb. Haemost. 2001, 85, 747–748. [Google Scholar]

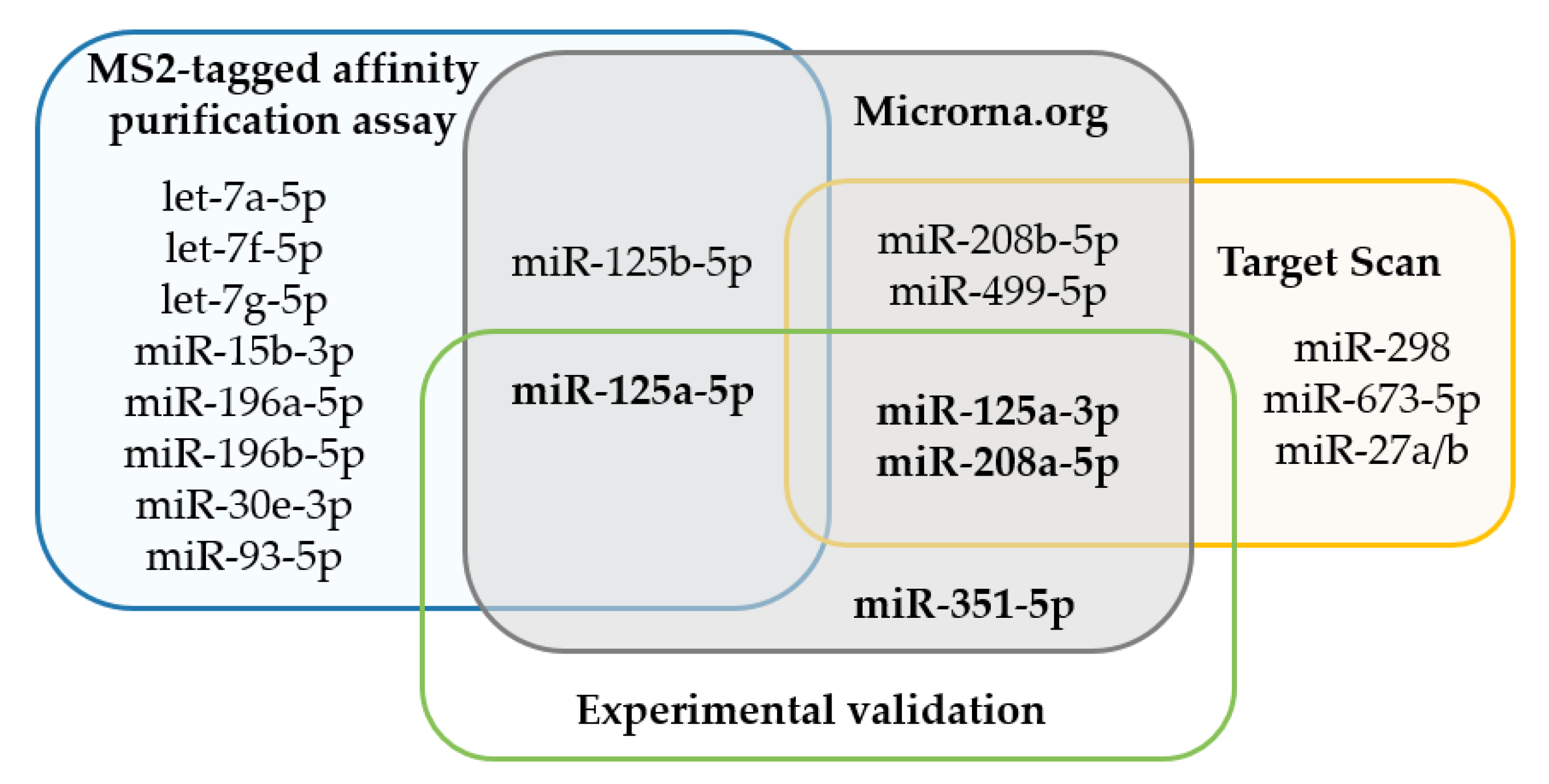
| miRNA | Precursor | MS2-E | MS2-muF8 | Fold Change |
|---|---|---|---|---|
| let-7a-5p | let-7a-1 | 222.0 | 343.0 | 1.55 |
| miR-93-5p | mir-93 | 189.0 | 305.0 | 1.61 |
| miR-196b-5p | mir-196b | 46.0 | 202.0 | 4.39 |
| miR-196a-5p | mir-196a-1 | 92.0 | 188.0 | 2.04 |
| miR-125a-5p | mir-125a | 97.0 | 150.0 | 1.55 |
| let-7f-5p | let-7f-1 | 55.0 | 147.0 | 2.67 |
| miR-30e-3p | mir-30e | 26.0 | 119.0 | 4.58 |
| miR-15b-3p | mir-15b | 31.0 | 83.0 | 2.68 |
| let-7g-5p | let-7g | 16.0 | 71.0 | 4.44 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jankowska, K.I.; Chattopadhyay, M.; Sauna, Z.E.; Atreya, C.D. A Foundational Study for Normal F8-Containing Mouse Models for the miRNA Regulation of Hemophilia A: Identification and Analysis of Mouse miRNAs that Downregulate the Murine F8 Gene. Int. J. Mol. Sci. 2020, 21, 5621. https://doi.org/10.3390/ijms21165621
Jankowska KI, Chattopadhyay M, Sauna ZE, Atreya CD. A Foundational Study for Normal F8-Containing Mouse Models for the miRNA Regulation of Hemophilia A: Identification and Analysis of Mouse miRNAs that Downregulate the Murine F8 Gene. International Journal of Molecular Sciences. 2020; 21(16):5621. https://doi.org/10.3390/ijms21165621
Chicago/Turabian StyleJankowska, Katarzyna I., Maitreyi Chattopadhyay, Zuben E. Sauna, and Chintamani D. Atreya. 2020. "A Foundational Study for Normal F8-Containing Mouse Models for the miRNA Regulation of Hemophilia A: Identification and Analysis of Mouse miRNAs that Downregulate the Murine F8 Gene" International Journal of Molecular Sciences 21, no. 16: 5621. https://doi.org/10.3390/ijms21165621
APA StyleJankowska, K. I., Chattopadhyay, M., Sauna, Z. E., & Atreya, C. D. (2020). A Foundational Study for Normal F8-Containing Mouse Models for the miRNA Regulation of Hemophilia A: Identification and Analysis of Mouse miRNAs that Downregulate the Murine F8 Gene. International Journal of Molecular Sciences, 21(16), 5621. https://doi.org/10.3390/ijms21165621




