Development of CRISPR Interference (CRISPRi) Platform for Metabolic Engineering of Leuconostoc citreum and Its Application for Engineering Riboflavin Biosynthesis
Abstract
1. Introduction
2. Results and Discussion
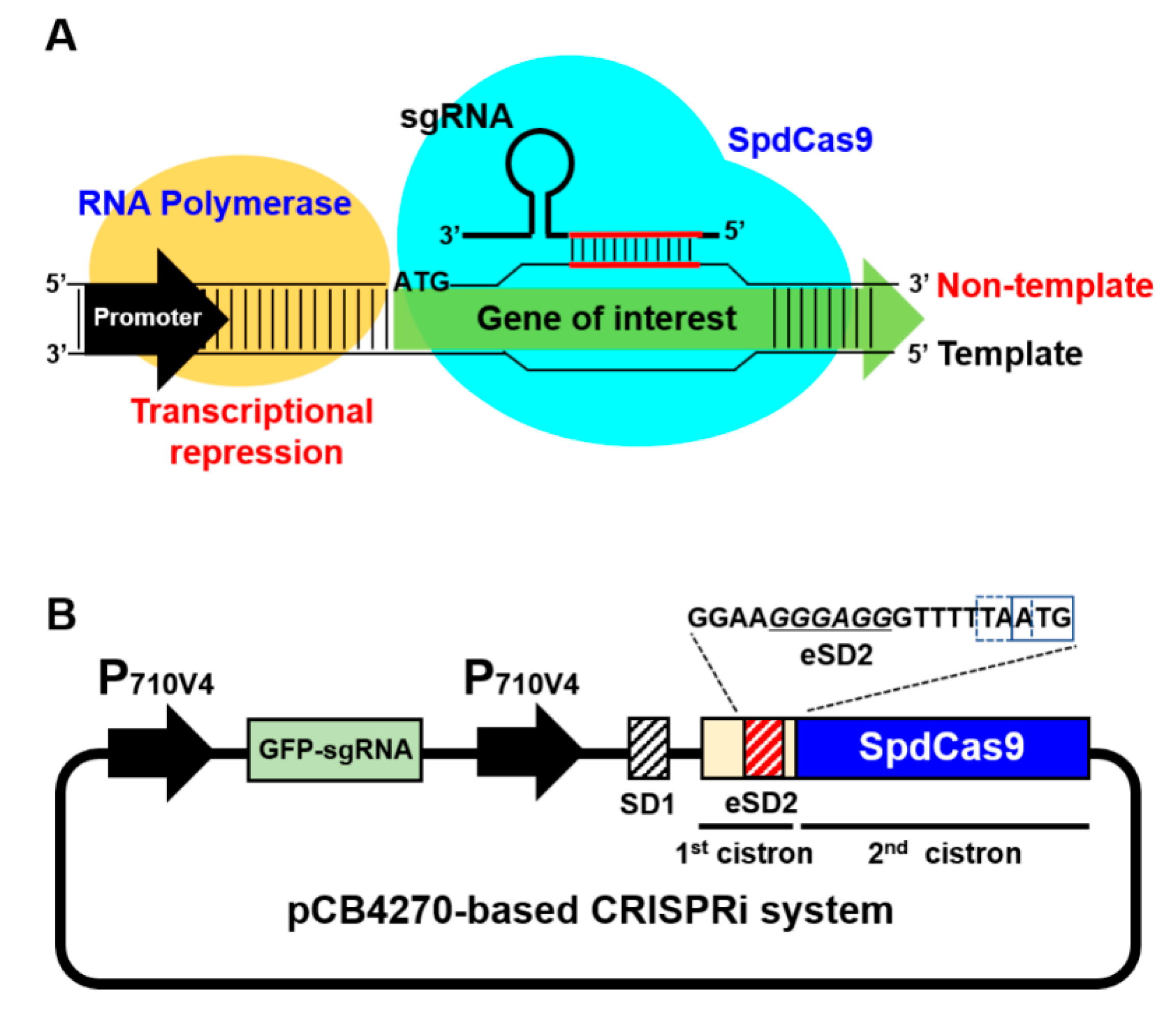
2.1. Construction of CRISPRi in the BCD Platform
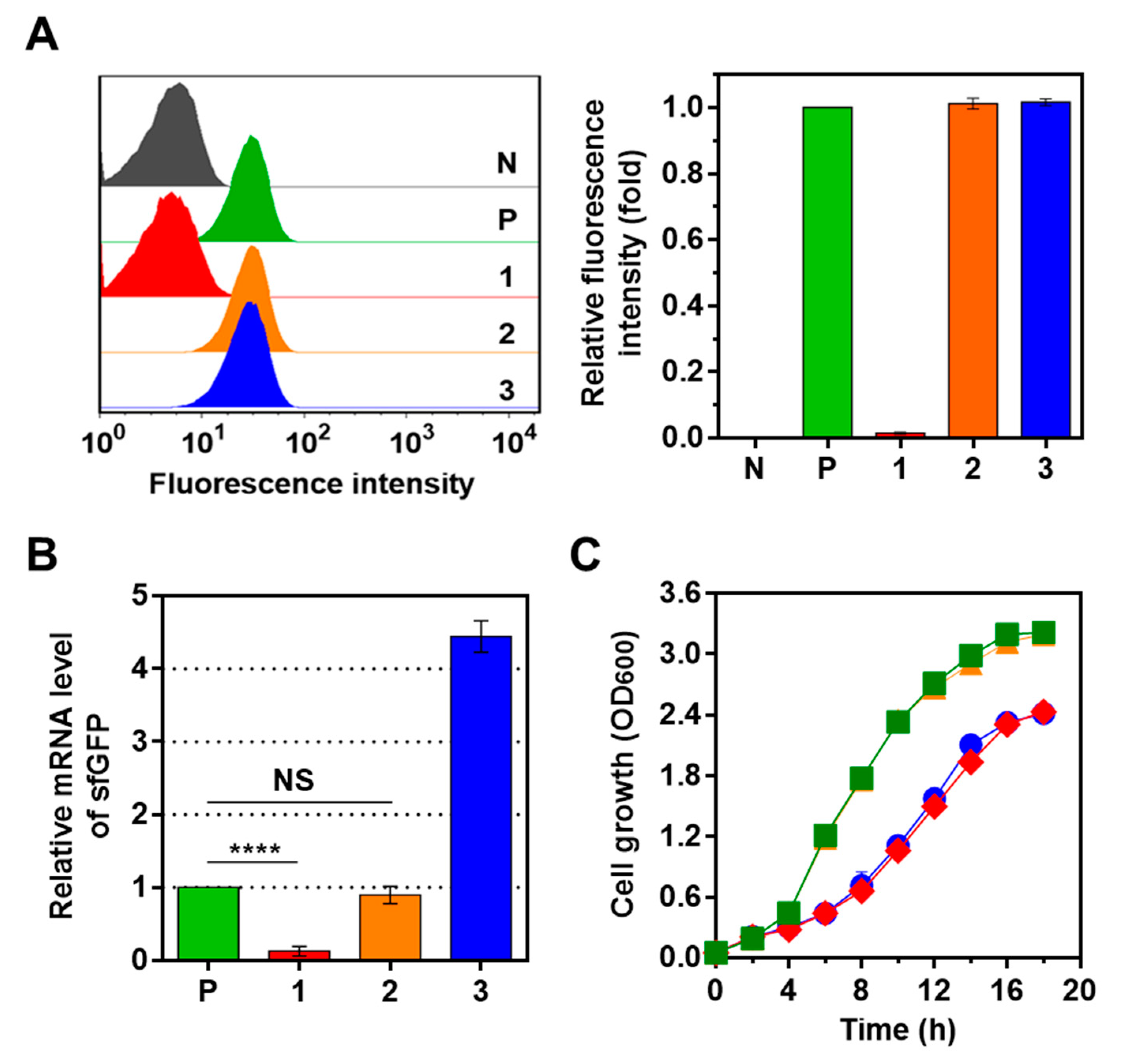
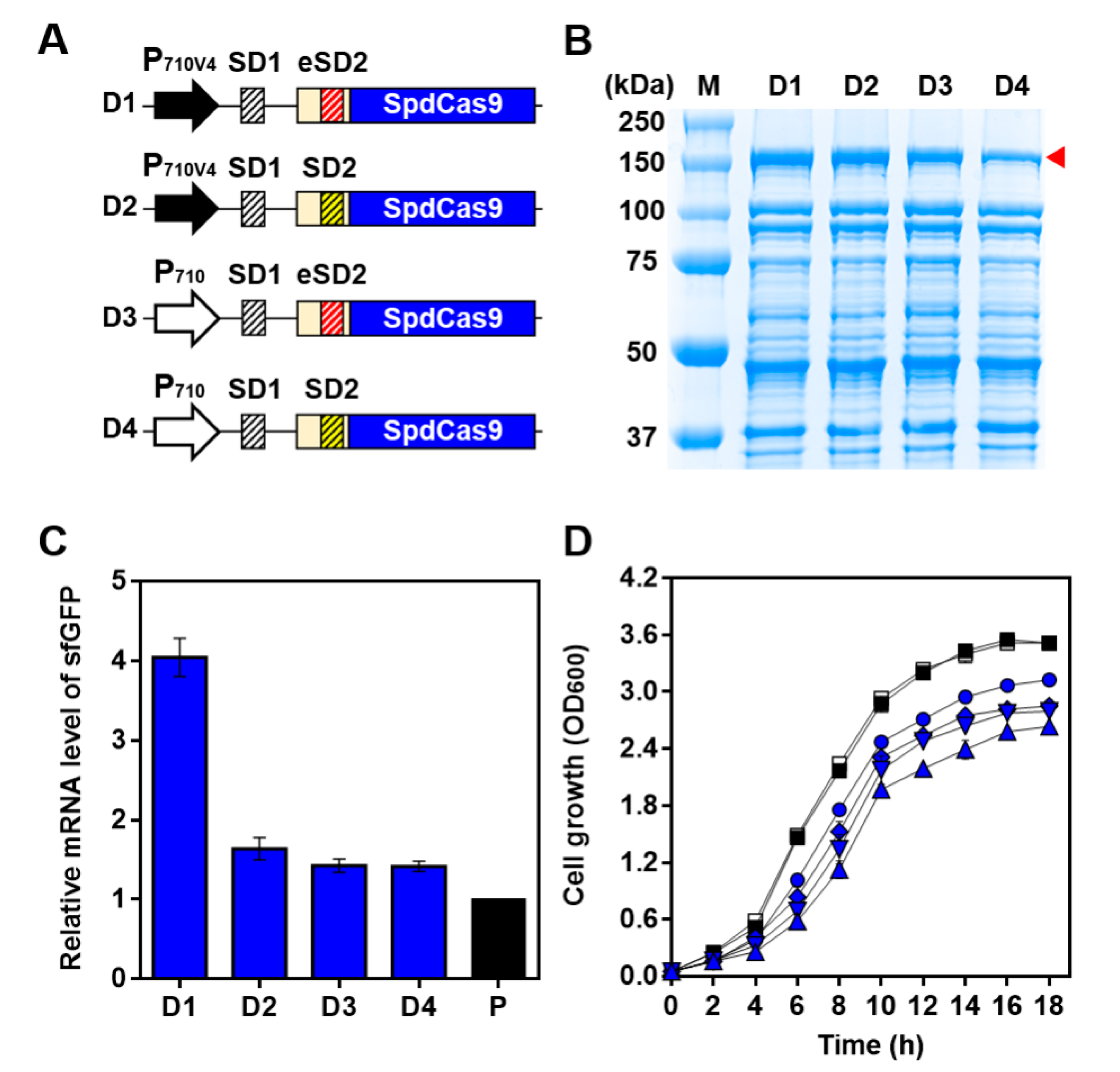
2.2. Optimization of CRISPRi System Using Synthetic Parts
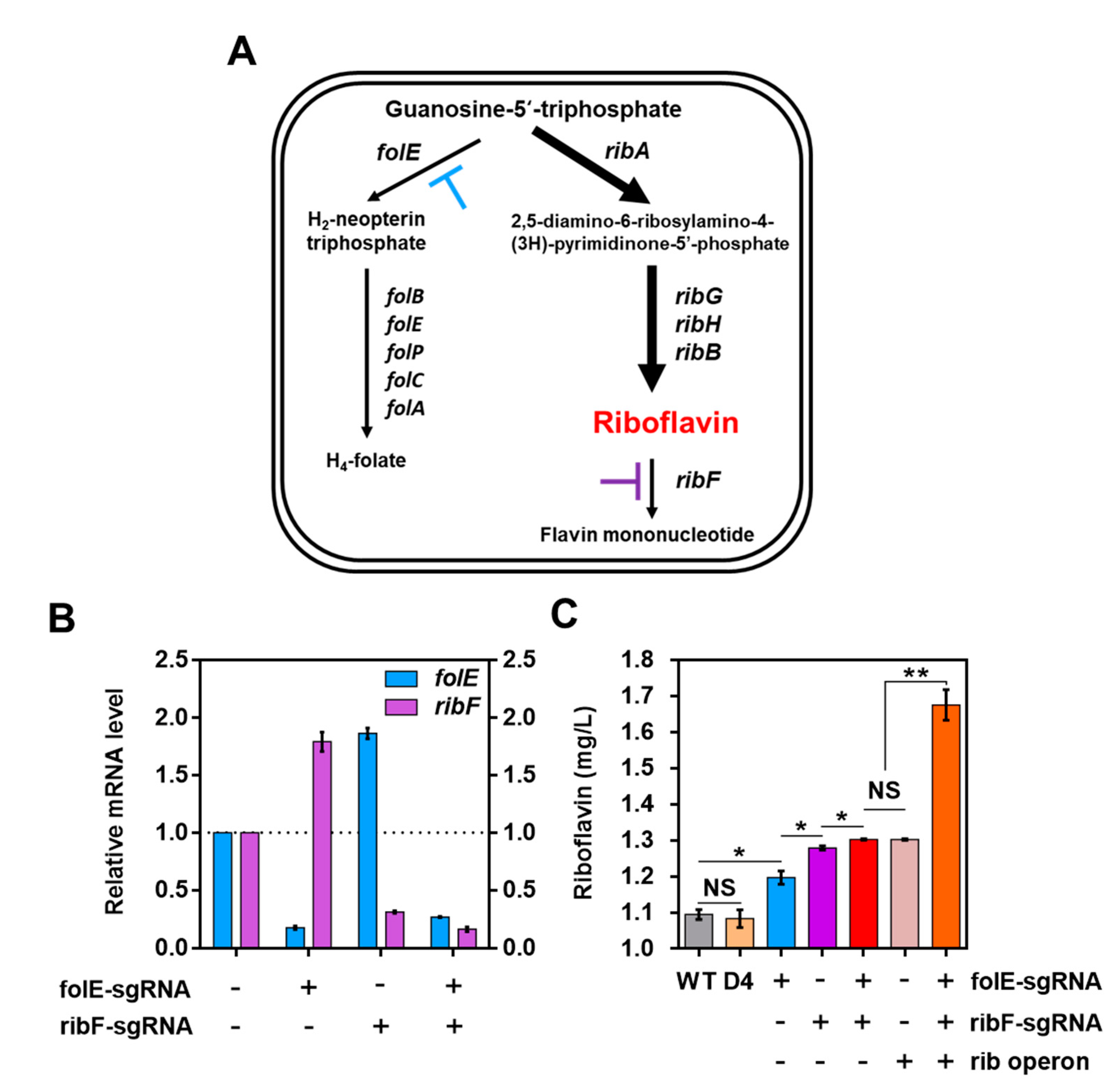
2.3. Increase in Riboflavin Production via Optimized CRISPRi System for L. citreum
2.4. Increase in Riboflavin Production by Co-Expression of the Rib Operon in L. citreum
3. Conclusions
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Plasmid Construction and Chromosome Integration
4.3. Fluorescence-Activated Cell-Sorting (FACS) Analysis
4.4. Protein Preparation and Analysis
4.5. Quantitative Reverse Transcription PCR (qRT-PCR)
4.6. High-Performance Liquid Chromatography (HPLC) Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| LAB | lactic acid bacteria |
| CRISPRi | CRISPR interference |
| sgRNA | single guide RNA |
| SpdCas9 | catalytically deactivated Cas9 of Streptococcus pyogenes |
| BCD | bicistronic design |
| RBS | ribosome binding sites |
| sfGFP | superfolder green fluorescent protein |
| PAM | protospacer adjacent motif |
| qRT-PCR | quantitative reverse transcription PCR |
| HPLC | high performance liquid chromatography |
| PBS | phosphate-buffered saline |
| TBS-T | Tris-buffered saline with Tween-20 solution |
References
- Hemme, D.; Foucaud-Scheunemann, C. Leuconostoc, characteristics, use in dairy technology and prospects in functional foods. Int. Dairy J. 2004, 14, 467–494. [Google Scholar] [CrossRef]
- Sybesma, W.; Starrenburg, M.; Tijsseling, L.; Hoefnagel, M.H.; Hugenholtz, J. Effects of cultivation conditions on folate production by lactic acid bacteria. Appl. Environ. Microbiol. 2003, 69, 4542–4548. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.-K.; Ahn, S.-B.; Lee, S.-M.; Shon, M.-Y.; Kim, S.-Y.; Shin, M.-S. Immune-enhancing effects of Leuconostoc strains isolated from Kimchi. J. Biomed. Res. 2012, 13, 353–356. [Google Scholar]
- Li, L.; Shin, S.Y.; Lee, K.; Han, N. Production of natural antimicrobial compound d-phenyllactic acid using Leuconostoc mesenteroides ATCC 8293 whole cells involving highly active d-lactate dehydrogenase. Lett. Appl. Microbiol. 2014, 59, 404–411. [Google Scholar] [CrossRef] [PubMed]
- de Vos, W.M. Systems solutions by lactic acid bacteria: From paradigms to practice. Microb. Cell. Fact. 2011, 10, S2. [Google Scholar] [CrossRef]
- Eom, H.-J.; Moon, J.-S.; Seo, E.-Y.; Han, N.S. Heterologous expression and secretion of Lactobacillus amylovorus α-amylase in Leuconostoc citreum. Biotechnol. Lett. 2009, 31, 1783. [Google Scholar] [CrossRef] [PubMed]
- García-Fruitós, E. Lactic acid bacteria: A promising alternative for recombinant protein production. Microb. Cell. Fact. 2012, 11, 157. [Google Scholar] [CrossRef]
- Caggianiello, G.; Kleerebezem, M.; Spano, G. Exopolysaccharides produced by lactic acid bacteria: From health-promoting benefits to stress tolerance mechanisms. Appl. Microbiol. Biotechnol. 2016, 100, 3877–3886. [Google Scholar] [CrossRef]
- Domingos-Lopes, M.; Lamosa, P.; Stanton, C.; Ross, R.; Silva, C. Isolation and characterization of an exopolysaccharide-producing Leuconostoc citreum strain from artisanal cheese. Lett. Appl. Microbiol. 2018, 67, 570–578. [Google Scholar] [CrossRef]
- Yang, Y.; Feng, F.; Zhou, Q.; Zhao, F.; Du, R.; Zhou, Z.; Han, Y. Isolation, purification, and characterization of exopolysaccharide produced by Leuconostoc citreum N21 from dried milk cake. Trans. Tianjin Univ. 2019, 25, 161–168. [Google Scholar] [CrossRef]
- Oh, Y.H.; Choi, J.W.; Kim, E.Y.; Song, B.K.; Jeong, K.J.; Park, K.; Kim, I.-K.; Woo, H.M.; Lee, S.H.; Park, S.J. Construction of synthetic promoter-based expression cassettes for the production of cadaverine in recombinant Corynebacterium glutamicum. Appl. Biochem. Biotechnol. 2015, 176, 2065–2075. [Google Scholar] [CrossRef] [PubMed]
- Ko, Y.J.; You, S.K.; Kim, M.; Lee, E.; Shin, S.K.; Park, H.M.; Oh, Y.; Han, S.O. Enhanced Production of 5-aminolevulinic Acid via Flux Redistribution of TCA Cycle toward l-Glutamate in Corynebacterium glutamicum. Biotechnol. Bioprocess Eng. 2019, 24, 915–923. [Google Scholar] [CrossRef]
- Van Pijkeren, J.-P.; Britton, R.A. High efficiency recombineering in lactic acid bacteria. Nucleic Acids Res. 2012, 40, e76. [Google Scholar] [CrossRef] [PubMed]
- Pines, G.; Freed, E.F.; Winkler, J.D.; Gill, R.T. Bacterial recombineering: Genome engineering via phage-based homologous recombination. ACS Synth. Biol. 2015, 4, 1176–1185. [Google Scholar] [CrossRef]
- Zhu, D.; Zhao, K.; Xu, H.; Zhang, X.; Bai, Y.; Saris, P.E.; Qiao, M. Construction of thyA deficient Lactococcus lactis using the Cre-loxP recombination system. Ann. Microbiol. 2015, 65, 1659–1665. [Google Scholar] [CrossRef]
- Oh, J.-H.; van Pijkeren, J.-P. CRISPR–Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res. 2014, 42, e131. [Google Scholar] [CrossRef]
- Berlec, A.; Škrlec, K.; Kocjan, J.; Olenic, M.; Štrukelj, B. Single plasmid systems for inducible dual protein expression and for CRISPR-Cas9/CRISPRi gene regulation in lactic acid bacterium Lactococcus lactis. Sci. Rep. 2018, 8, 1009. [Google Scholar] [CrossRef]
- Zheng, Y.; Su, T.; Qi, Q. Microbial CRISPRi and CRISPRa Systems for Metabolic Engineering. Biotechnol. Bioprocess Eng. 2019, 24, 579–591. [Google Scholar] [CrossRef]
- Son, Y.J.; Ryu, A.J.; Li, L.; Han, N.S.; Jeong, K.J. Development of a high-copy plasmid for enhanced production of recombinant proteins in Leuconostoc citreum. Microb. Cell. Fact. 2016, 15, 12. [Google Scholar] [CrossRef]
- Jang, S.H.; Cha, J.W.; Han, N.S.; Jeong, K.J. Development of bicistronic expression system for the enhanced and reliable production of recombinant proteins in Leuconostoc citreum. Sci. Rep. 2018, 8, 8852. [Google Scholar] [CrossRef]
- Larson, M.H.; Gilbert, L.A.; Wang, X.; Lim, W.A.; Weissman, J.S.; Qi, L.S. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 2013, 8, 2180. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [PubMed]
- Bikard, D.; Jiang, W.; Samai, P.; Hochschild, A.; Zhang, F.; Marraffini, L.A. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013, 41, 7429–7437. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, E.; Thakur, P.; Pareek, M.; Agarwal, N. Gene silencing by CRISPR interference in mycobacteria. Nat. Commun. 2015, 6, 1–11. [Google Scholar] [CrossRef]
- Kim, S.K.; Han, G.H.; Seong, W.; Kim, H.; Kim, S.-W.; Lee, D.-H.; Lee, S.-G. CRISPR interference-guided balancing of a biosynthetic mevalonate pathway increases terpenoid production. Metab. Eng. 2016, 38, 228–240. [Google Scholar] [CrossRef]
- Su, K.-C.; Tsang, M.-J.; Emans, N.; Cheeseman, I.M. CRISPR/Cas9-based gene targeting using synthetic guide RNAs enables robust cell biological analyses. Mol. Biol. Cell 2018, 29, 2370–2377. [Google Scholar] [CrossRef]
- Nielsen, A.A.; Voigt, C.A. Multi-input CRISPR/Cas genetic circuits that interface host regulatory networks. Mol. Syst. Biol. 2014, 10, 763. [Google Scholar] [CrossRef]
- Cho, S.; Choe, D.; Lee, E.; Kim, S.C.; Palsson, B.; Cho, B.-K. High-level dCas9 expression induces abnormal cell morphology in Escherichia coli. ACS Synth. Biol. 2018, 7, 1085–1094. [Google Scholar] [CrossRef]
- Zhang, S.; Voigt, C.A. Engineered dCas9 with reduced toxicity in bacteria: Implications for genetic circuit design. Nucleic Acids Res. 2018, 46, 11115–11125. [Google Scholar] [CrossRef]
- Wu, X.; Kriz, A.J.; Sharp, P.A. Target specificity of the CRISPR-Cas9 system. Quant Biol. 2014, 2, 59–70. [Google Scholar] [CrossRef]
- LeBlanc, J.; Laiño, J.E.; del Valle, M.J.; Vannini, V.V.; van Sinderen, D.; Taranto, M.P.; de Valdez, G.F.; de Giori, G.S.; Sesma, F. B-Group vitamin production by lactic acid bacteria–current knowledge and potential applications. J. Appl. Microbiol. 2011, 111, 1297–1309. [Google Scholar] [CrossRef] [PubMed]
- Capozzi, V.; Menga, V.; Digesu, A.M.; Vita, P.D.; van Sinderen, D.; Cattivelli, L.; Fares, C.; Spano, G. Biotechnological production of vitamin B2-enriched bread and pasta. J. Agric. Food. Chem. 2011, 59, 8013–8020. [Google Scholar] [CrossRef] [PubMed]
- Samaniego-Vaesken, M.d.; Alonso-Aperte, E.; Varela-Moreiras, G. Vitamin food fortification today. Food Nutr. Res. 2012, 56, 5459. [Google Scholar] [CrossRef] [PubMed]
- Laiño, J.E.; del Valle, M.J.; de Giori, G.S.; LeBlanc, J.G.J. Development of a high folate concentration yogurt naturally bio-enriched using selected lactic acid bacteria. LWT Food. Sci. Technol. 2013, 54, 1–5. [Google Scholar] [CrossRef]
- LeBlanc, J.; Burgess, C.; Sesma, F.; de Giori, G.S.; van Sinderen, D. Ingestion of milk fermented by genetically modified Lactococcus lactis improves the riboflavin status of deficient rats. J. Dairy Sci. 2005, 88, 3435–3442. [Google Scholar] [CrossRef]
- Thakur, K.; Tomar, S.K.; De, S. Lactic acid bacteria as a cell factory for riboflavin production. Microb. Biotechnol. 2016, 9, 441–451. [Google Scholar] [CrossRef]
- Perkins, J.; Sloma, A.; Hermann, T.; Theriault, K.; Zachgo, E.; Erdenberger, T.; Hannett, N.; Chatterjee, N.; Williams, V., II; Rufo, G.J. Genetic engineering of Bacillus subtilis for the commercial production of riboflavin. J. Ind. Microbiol. Biotechnol. 1999, 22, 8–18. [Google Scholar] [CrossRef]
- Otgonbayar, G.-E.; Eom, H.-J.; Kim, B.S.; Ko, J.-H.; Han, N.S. Mannitol production by Leuconostoc citreum KACC 91348P isolated from kimchi. J. Microbiol. Biotechnol. 2011, 21, 968–971. [Google Scholar] [CrossRef]
- Raynaud, C.; Sarçabal, P.; Meynial-Salles, I.; Croux, C.; Soucaille, P. Molecular characterization of the 1,3-propanediol (1,3-PD) operon of Clostridium butyricum. Proc. Natl. Acad. Sci. USA 2003, 100, 5010–5015. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Son, J.; Jang, S.H.; Cha, J.W.; Jeong, K.J. Development of CRISPR Interference (CRISPRi) Platform for Metabolic Engineering of Leuconostoc citreum and Its Application for Engineering Riboflavin Biosynthesis. Int. J. Mol. Sci. 2020, 21, 5614. https://doi.org/10.3390/ijms21165614
Son J, Jang SH, Cha JW, Jeong KJ. Development of CRISPR Interference (CRISPRi) Platform for Metabolic Engineering of Leuconostoc citreum and Its Application for Engineering Riboflavin Biosynthesis. International Journal of Molecular Sciences. 2020; 21(16):5614. https://doi.org/10.3390/ijms21165614
Chicago/Turabian StyleSon, Jaewoo, Seung Hoon Jang, Ji Won Cha, and Ki Jun Jeong. 2020. "Development of CRISPR Interference (CRISPRi) Platform for Metabolic Engineering of Leuconostoc citreum and Its Application for Engineering Riboflavin Biosynthesis" International Journal of Molecular Sciences 21, no. 16: 5614. https://doi.org/10.3390/ijms21165614
APA StyleSon, J., Jang, S. H., Cha, J. W., & Jeong, K. J. (2020). Development of CRISPR Interference (CRISPRi) Platform for Metabolic Engineering of Leuconostoc citreum and Its Application for Engineering Riboflavin Biosynthesis. International Journal of Molecular Sciences, 21(16), 5614. https://doi.org/10.3390/ijms21165614




