NMR Characterization of Conformational Interconversions of Lys48-Linked Ubiquitin Chains
Abstract
1. Introduction
2. Results and Discussion
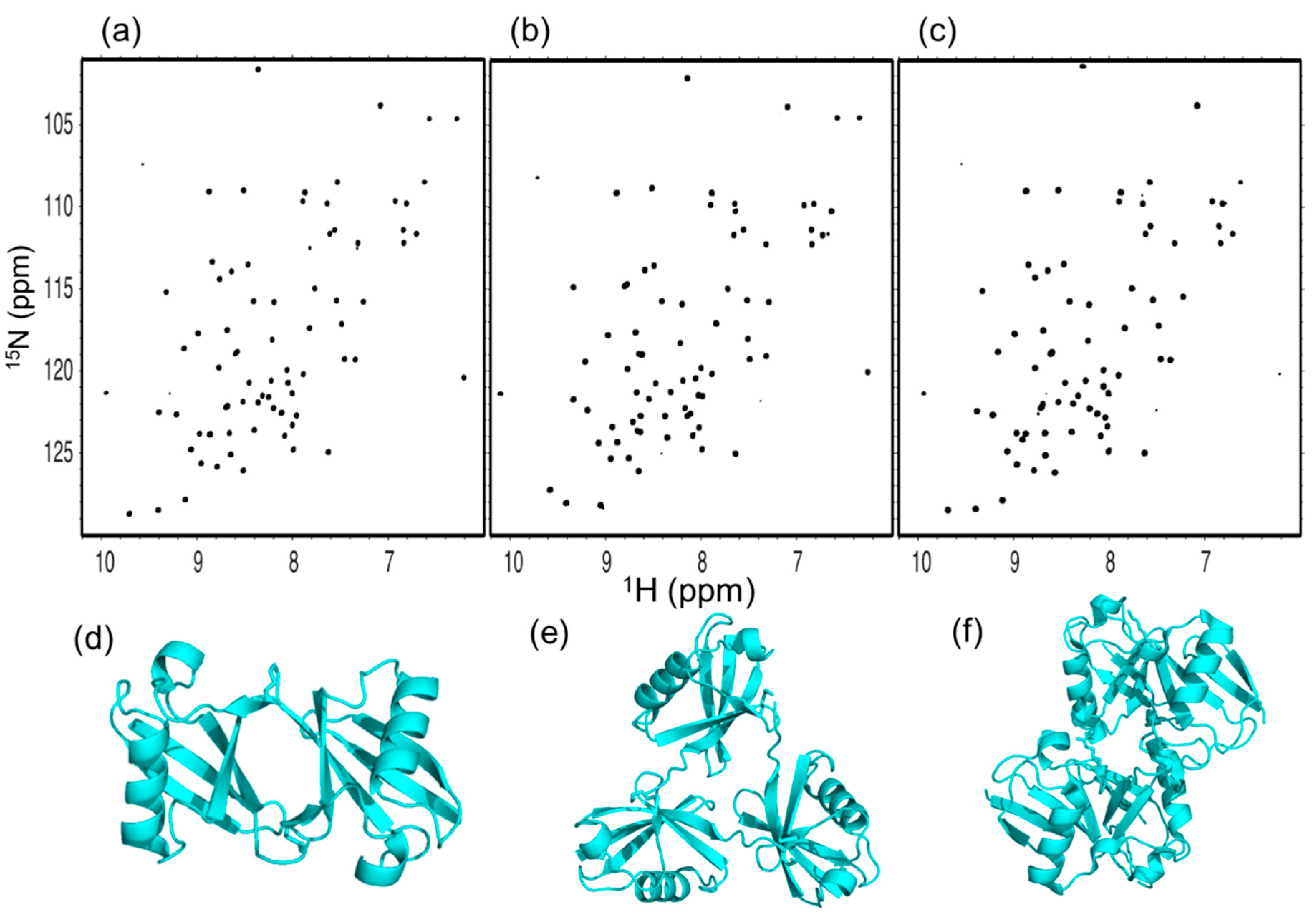
2.1. Spectral Comparison Among Cyclic Forms of Lys48-Linked diUb, triUb, and tetraUb
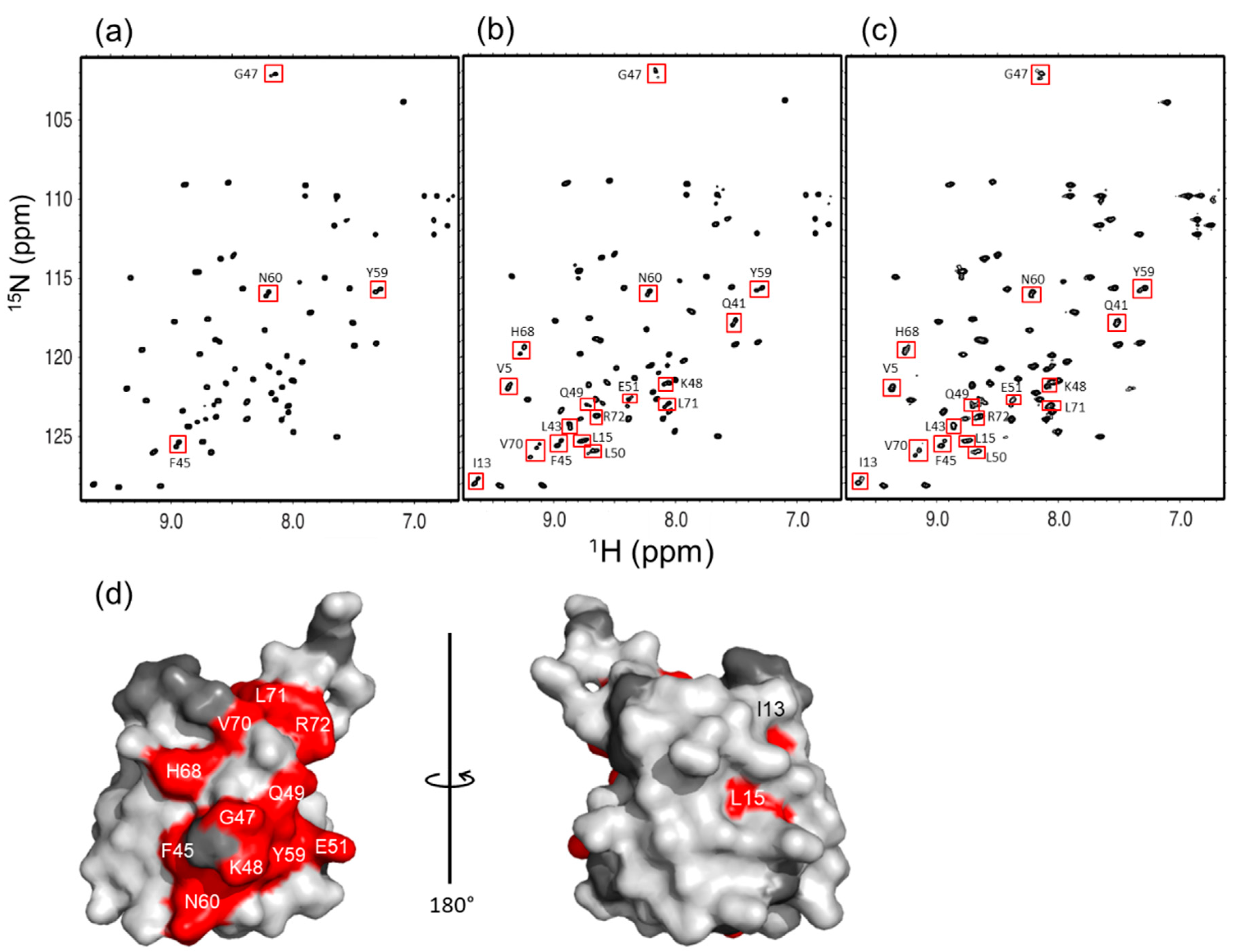
2.2. Spectral Comparison among Native Forms of Lys48-Linked diUb, triUb, and tetraUb
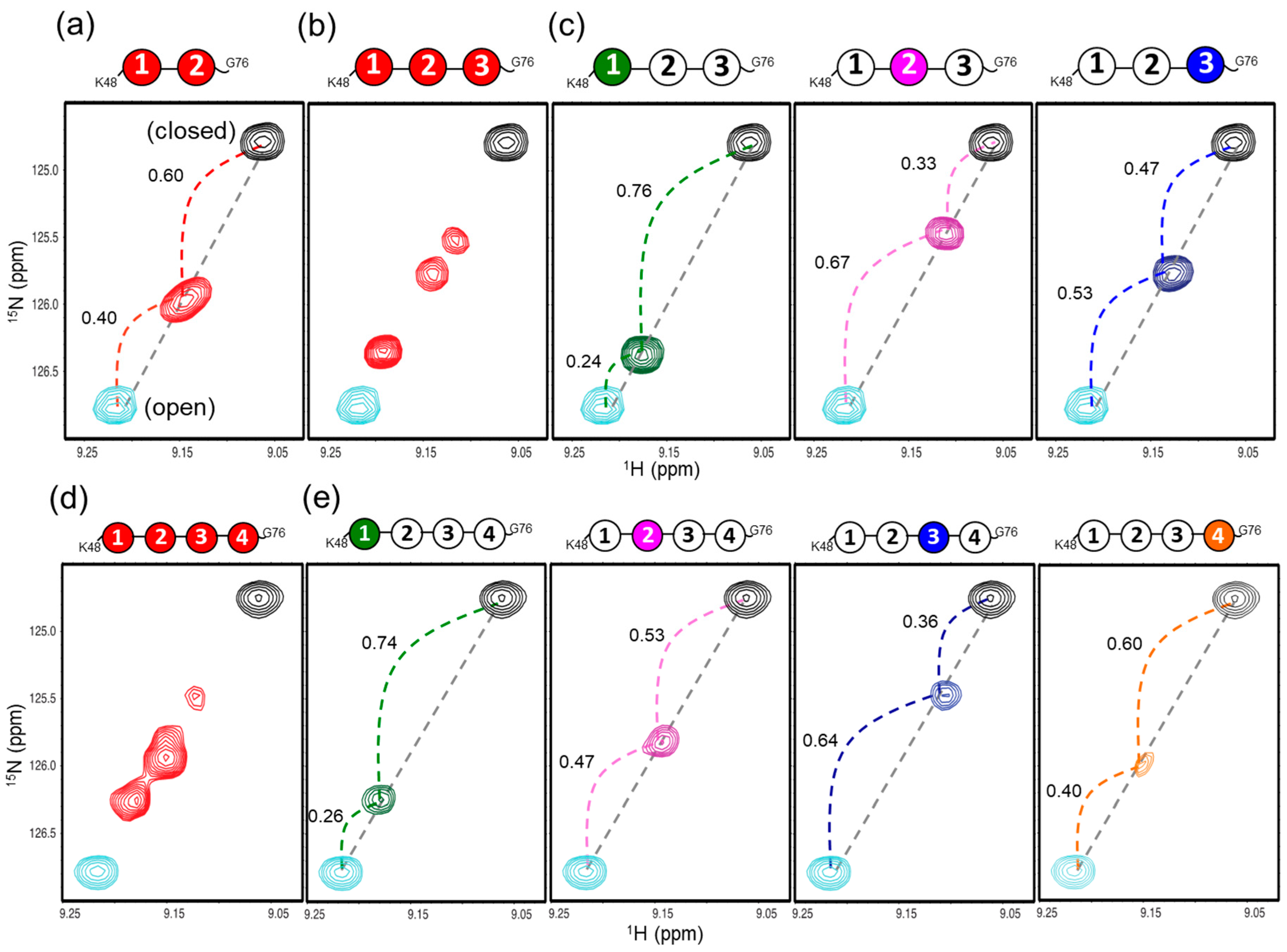
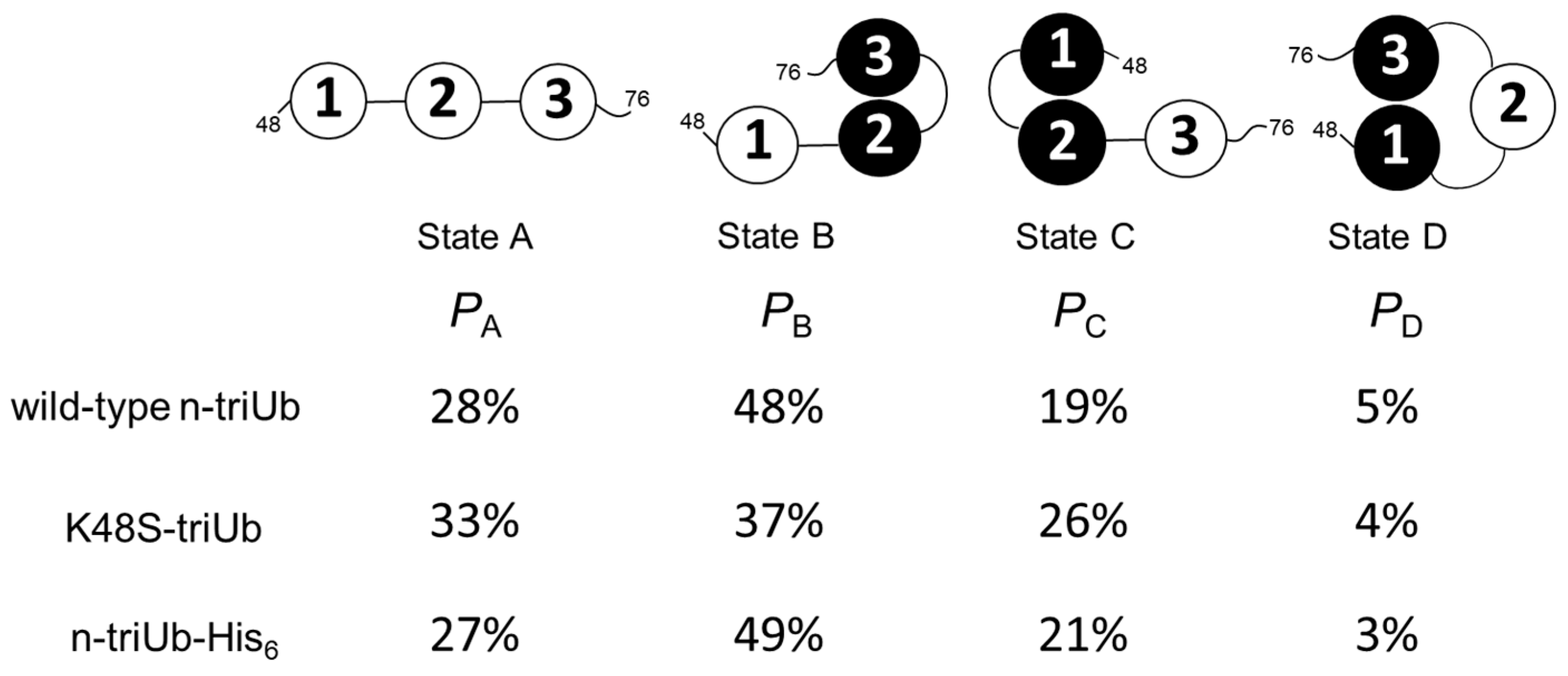
2.3. Conformational Equilibrium of the Native Forms of Lys48-Linked triUb and tetraUb
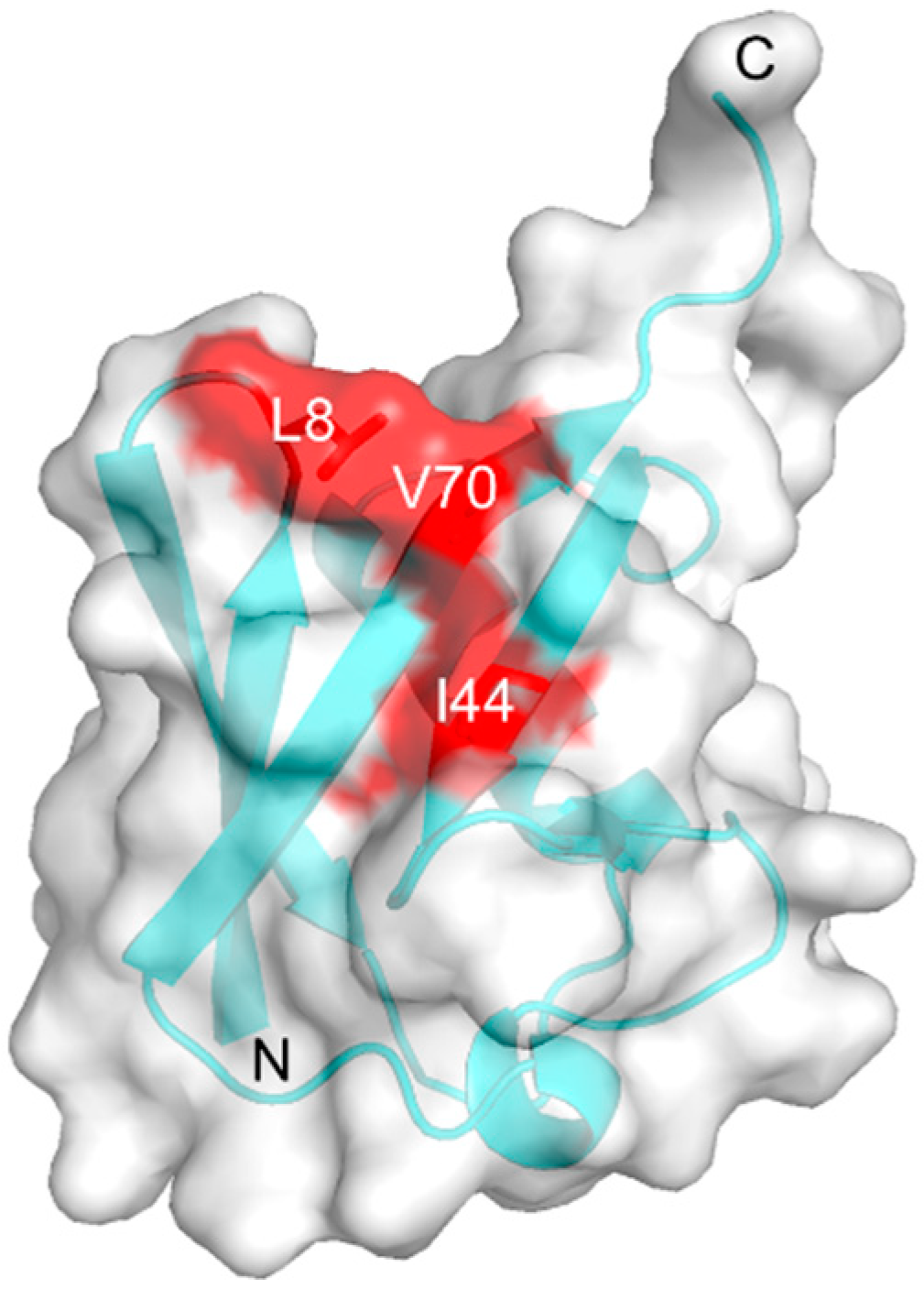
2.4. Inter-Subunit Interactions of Lys48-Linked triUb
3. Materials and Methods
3.1. Expression and Purification of Human Ub and Derivatives
3.2. Expression and Purification of Ub-Related Enzymes
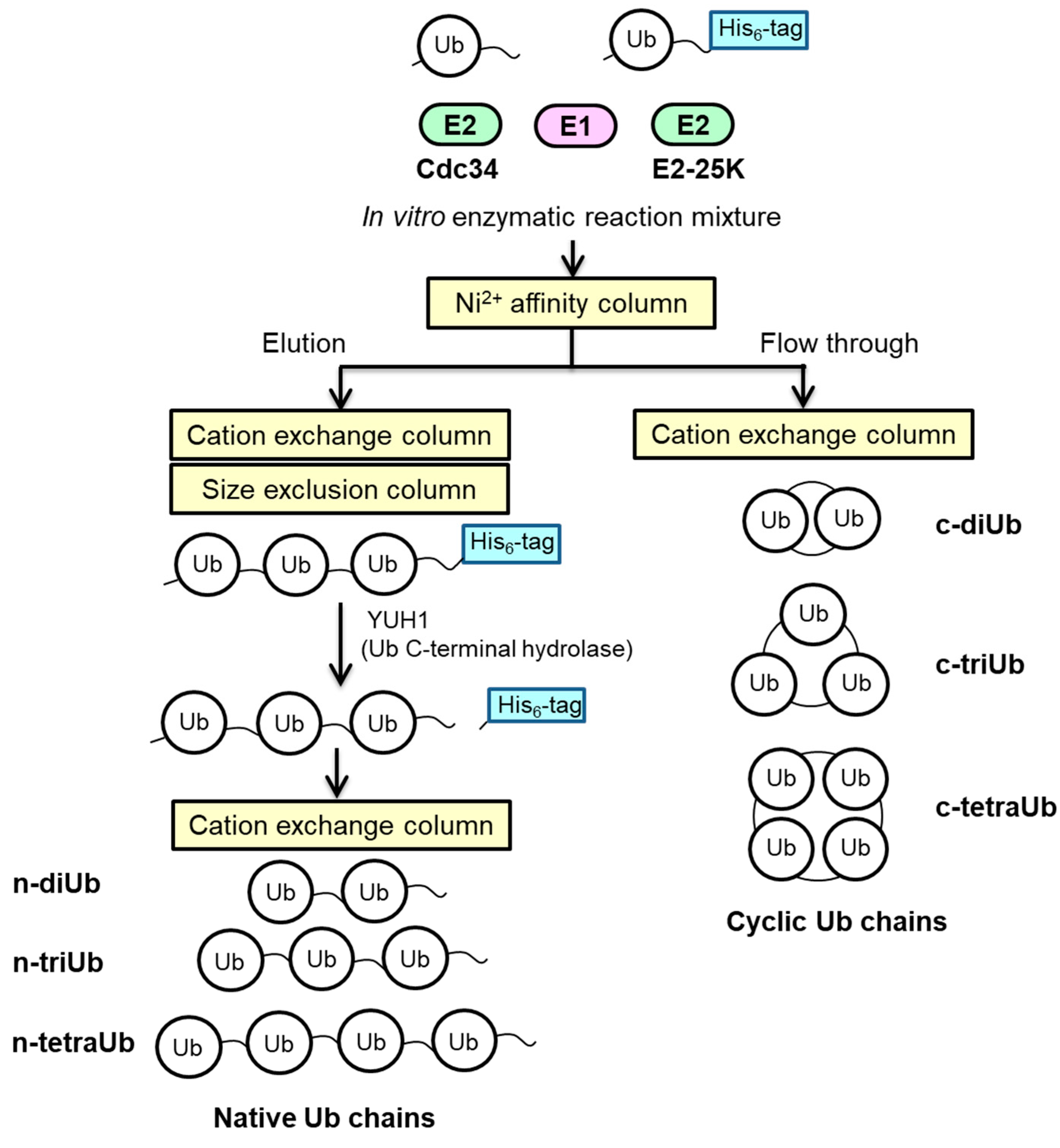
3.3. Enzymatic Synthesis of Lys48-Linked tri-Ub
3.4. NMR Measurements
3.5. Crystallization, X-Ray Data Collection, and Structure Determination
3.6. Accession Number
4. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| DTT | Dithiothreitol |
| GST | Glutathione S-transferase |
| HSQC | Heteronuclear Single Quantum Correlation |
| NMR | Nuclear magnetic resonance |
| Ub | Ubiquitin |
| YUH | Yeast ubiquitin hydrolase |
References
- Vogel, C.; Bashton, M.; Kerrison, N.D.; Chothia, C.; Teichmann, S.A. Structure, function and evolution of multidomain proteins. Curr. Opin. Struct. Biol. 2004, 14, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Göbl, C.; Madl, T.; Simon, B.; Sattler, M. NMR approaches for structural analysis of multidomain proteins and complexes in solution. Prog. Nucl. Magn. Reson. Spectrosc. 2014, 80, 26–63. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef] [PubMed]
- Pickart, C.M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001, 70, 503–533. [Google Scholar] [CrossRef]
- Alfano, C.; Faggiano, S.; Pastore, A. The Ball and Chain of Polyubiquitin Structures. Trends Biochem. Sci. 2016, 41, 371–385. [Google Scholar] [CrossRef]
- Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009, 78, 477–513. [Google Scholar] [CrossRef]
- Hoege, C.; Pfander, B.; Moldovan, G.L.; Pyrowolakis, G.; Jentsch, S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 2002, 419, 135–141. [Google Scholar] [CrossRef]
- Pickart, C.M.; Eddins, M.J. Ubiquitin: Structures, functions, mechanisms. Biochim. Biophys. Acta 2004, 1695, 55–72. [Google Scholar] [CrossRef]
- Varadan, R.; Assfalg, M.; Raasi, S.; Pickart, C.; Fushman, D. Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Mol. Cell 2005, 18, 687–698. [Google Scholar] [CrossRef]
- Varadan, R.; Assfalg, M.; Haririnia, A.; Raasi, S.; Pickart, C.; Fushman, D. Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J. Biol. Chem. 2004, 279, 7055–7063. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Reyes-Turcu, F.; Licchesi, J.D.; Odenwaelder, P.; Wilkinson, K.D.; Barford, D. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009, 10, 466–473. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Gong, Z.; Jiang, W.X.; Yang, J.; Zhu, W.K.; Guo, D.C.; Zhang, W.P.; Liu, M.L.; Tang, C. Lys63-linked ubiquitin chain adopts multiple conformational states for specific target recognition. Elife 2015, 4, e05767. [Google Scholar] [CrossRef] [PubMed]
- Cook, W.J.; Jeffrey, L.C.; Carson, M.; Chen, Z.; Pickart, C.M. Structure of a diubiquitin conjugate and a model for interaction with ubiquitin conjugating enzyme (E2). J. Biol. Chem. 1992, 267, 16467–16471. [Google Scholar]
- Cook, W.J.; Jeffrey, L.C.; Kasperek, E.; Pickart, C.M. Structure of tetraubiquitin shows how multiubiquitin chains can be formed. J. Mol. Biol. 1994, 236, 601–609. [Google Scholar] [CrossRef]
- Varadan, R.; Walker, O.; Pickart, C.; Fushman, D. Structural properties of polyubiquitin chains in solution. J. Mol. Biol. 2002, 324, 637–647. [Google Scholar] [CrossRef]
- Tenno, T.; Fujiwara, K.; Tochio, H.; Iwai, K.; Morita, E.H.; Hayashi, H.; Murata, S.; Hiroaki, H.; Sato, M.; Tanaka, K.; et al. Structural basis for distinct roles of Lys63- and Lys48-linked polyubiquitin chains. Genes Cells 2004, 9, 865–875. [Google Scholar] [CrossRef]
- Ryabov, Y.; Fushman, D. Interdomain mobility in di-ubiquitin revealed by NMR. Proteins 2006, 63, 787–796. [Google Scholar] [CrossRef]
- Eddins, M.J.; Varadan, R.; Fushman, D.; Pickart, C.M.; Wolberger, C. Crystal structure and solution NMR studies of Lys48-linked tetraubiquitin at neutral pH. J. Mol. Biol. 2007, 367, 204–211. [Google Scholar] [CrossRef]
- Hirano, T.; Serve, O.; Yagi-Utsumi, M.; Takemoto, E.; Hiromoto, T.; Satoh, T.; Mizushima, T.; Kato, K. Conformational dynamics of wild-type Lys-48-linked diubiquitin in solution. J. Biol. Chem. 2011, 286, 37496–37502. [Google Scholar] [CrossRef]
- Kniss, A.; Schuetz, D.; Kazemi, S.; Pluska, L.; Spindler, P.E.; Rogov, V.V.; Husnjak, K.; Dikic, I.; Guntert, P.; Sommer, T.; et al. Chain Assembly and Disassembly Processes Differently Affect the Conformational Space of Ubiquitin Chains. Structure 2018, 26, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Dong, X.; Yi, H.W.; Yang, J.; Gong, Z.; Wang, Y.; Liu, K.; Zhang, W.P.; Tang, C. Structural basis for the recognition of K48-linked Ub chain by proteasomal receptor Rpn13. Cell Discov. 2019, 5, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, C.; Wang, E.; Wang, J. PolyUbiquitin chain linkage topology selects the functions from the underlying binding landscape. PLoS Comput. Biol. 2014, 10, e1003691. [Google Scholar] [CrossRef] [PubMed]
- Bowerman, S.; Rana, A.; Rice, A.; Pham, G.H.; Strieter, E.R.; Wereszczynski, J. Determining Atomistic SAXS Models of Tri-Ubiquitin Chains from Bayesian Analysis of Accelerated Molecular Dynamics Simulations. J. Chem. Theory Comput. 2017, 13, 2418–2429. [Google Scholar] [CrossRef] [PubMed]
- Berg, A.; Kukharenko, O.; Scheffner, M.; Peter, C. Towards a molecular basis of ubiquitin signaling: A dual-scale simulation study of ubiquitin dimers. PLoS Comput. Biol. 2018, 14, e1006589. [Google Scholar] [CrossRef] [PubMed]
- Vijay-Kumar, S.; Bugg, C.E.; Cook, W.J. Structure of ubiquitin refined at 1.8 A resolution. J. Mol. Biol. 1987, 194, 531–544. [Google Scholar] [CrossRef]
- Satoh, T.; Sakata, E.; Yamamoto, S.; Yamaguchi, Y.; Sumiyoshi, A.; Wakatsuki, S.; Kato, K. Crystal structure of cyclic Lys48-linked tetraubiquitin. BioChem. Biophys. Res. Commun. 2010, 400, 329–333. [Google Scholar] [CrossRef]
- Von Delbruck, M.; Kniss, A.; Rogov, V.V.; Pluska, L.; Bagola, K.; Lohr, F.; Guntert, P.; Sommer, T.; Dotsch, V. The CUE Domain of Cue1 Aligns Growing Ubiquitin Chains with Ubc7 for Rapid Elongation. Mol. Cell 2016, 62, 918–928. [Google Scholar] [CrossRef]
- Goddard, T.D.; Koeller, D.G. Sparky; Version 3.0; University of California: San Francisco, CA, USA, 1993. [Google Scholar]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzym. 1997, 276, 307–326. [Google Scholar]
- Vagin, A.; Teplyakov, A. MOLREP: An Automated Program for Molecular Replacement. J. Appl. Crystallogr. 1997, 30, 1022–1025. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Murshudov, G.N.; Vagin, A.A.; Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997, 53, 240–255. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef]
- Liu, S.; Chen, Y.; Li, J.; Huang, T.; Tarasov, S.; King, A.; Weissman, A.M.; Byrd, R.A.; Das, R. Promiscuous interactions of gp78 E3 ligase CUE domain with polyubiquitin chains. Structure 2012, 20, 2138–2150. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.G.; Youn, H.S.; Kang, J.Y.; Park, S.Y.; Kidera, A.; Yoo, Y.J.; Eom, S.H. Crystal structure of the Ube2K/E2-25K and K48-linked di-ubiquitin complex provides structural insight into the mechanism of K48-specific ubiquitin chain synthesis. Biochem. Biophys. Res. Commun. 2018, 506, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Kwasna, D.; Rehman, S.A.A.; Natarajan, J.; Matthews, S.; Madden, R.; De Cesare, V.; Weidlich, S.; Virdee, S.; Ahel, I.; Gibbs-Seymour, I.; et al. Discovery and Characterization of ZUFSP/ZUP1, a Distinct Deubiquitinase Class Important for Genome Stability. Mol. Cell 2018, 70, 150–164. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hiranyakorn, M.; Yanaka, S.; Satoh, T.; Wilasri, T.; Jityuti, B.; Yagi-Utsumi, M.; Kato, K. NMR Characterization of Conformational Interconversions of Lys48-Linked Ubiquitin Chains. Int. J. Mol. Sci. 2020, 21, 5351. https://doi.org/10.3390/ijms21155351
Hiranyakorn M, Yanaka S, Satoh T, Wilasri T, Jityuti B, Yagi-Utsumi M, Kato K. NMR Characterization of Conformational Interconversions of Lys48-Linked Ubiquitin Chains. International Journal of Molecular Sciences. 2020; 21(15):5351. https://doi.org/10.3390/ijms21155351
Chicago/Turabian StyleHiranyakorn, Methanee, Saeko Yanaka, Tadashi Satoh, Thunchanok Wilasri, Benchawan Jityuti, Maho Yagi-Utsumi, and Koichi Kato. 2020. "NMR Characterization of Conformational Interconversions of Lys48-Linked Ubiquitin Chains" International Journal of Molecular Sciences 21, no. 15: 5351. https://doi.org/10.3390/ijms21155351
APA StyleHiranyakorn, M., Yanaka, S., Satoh, T., Wilasri, T., Jityuti, B., Yagi-Utsumi, M., & Kato, K. (2020). NMR Characterization of Conformational Interconversions of Lys48-Linked Ubiquitin Chains. International Journal of Molecular Sciences, 21(15), 5351. https://doi.org/10.3390/ijms21155351





