Modeling of Frontotemporal Dementia Using iPSC Technology
Abstract
1. Introduction
2. Results
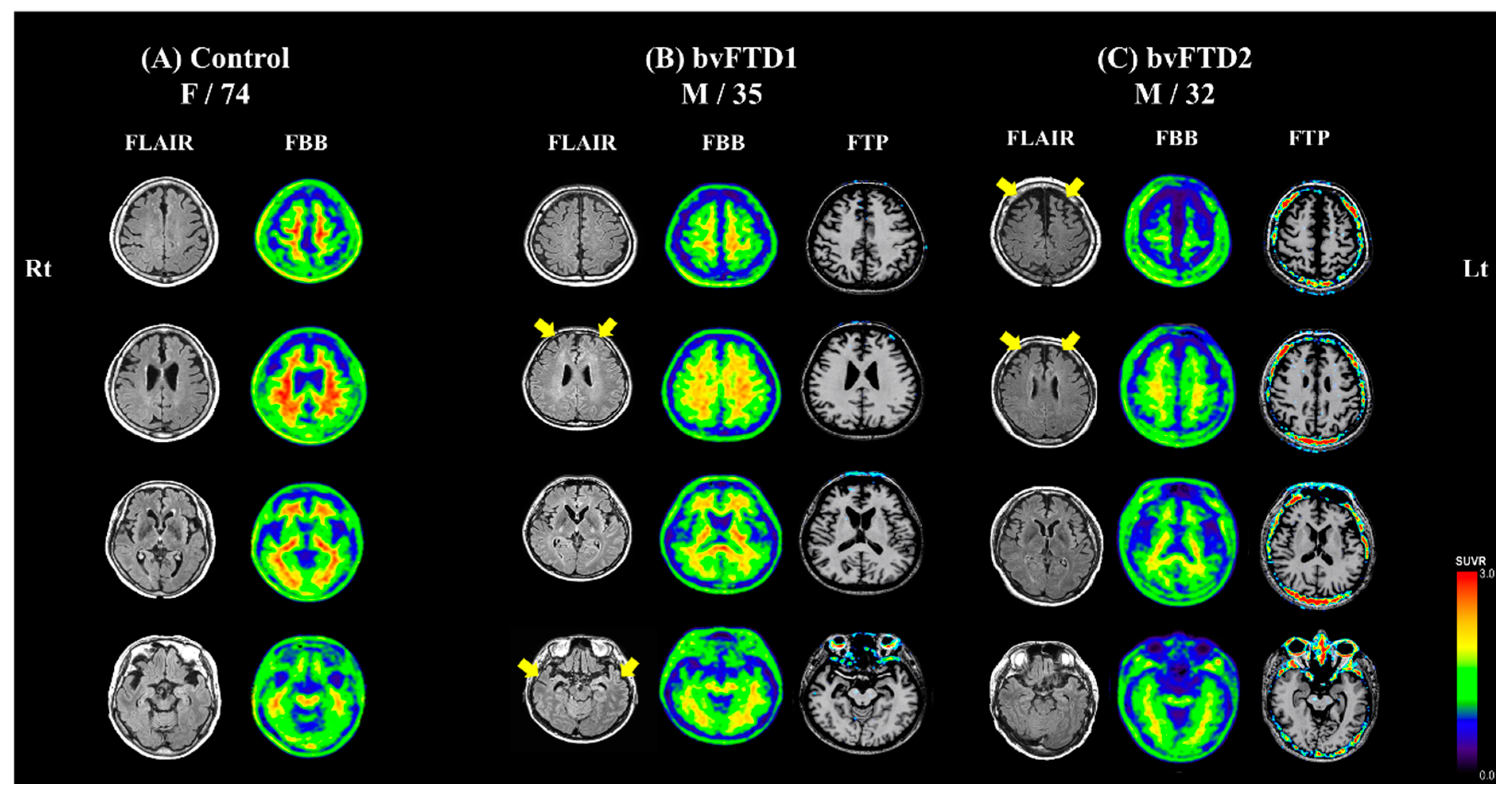
2.1. Case Description
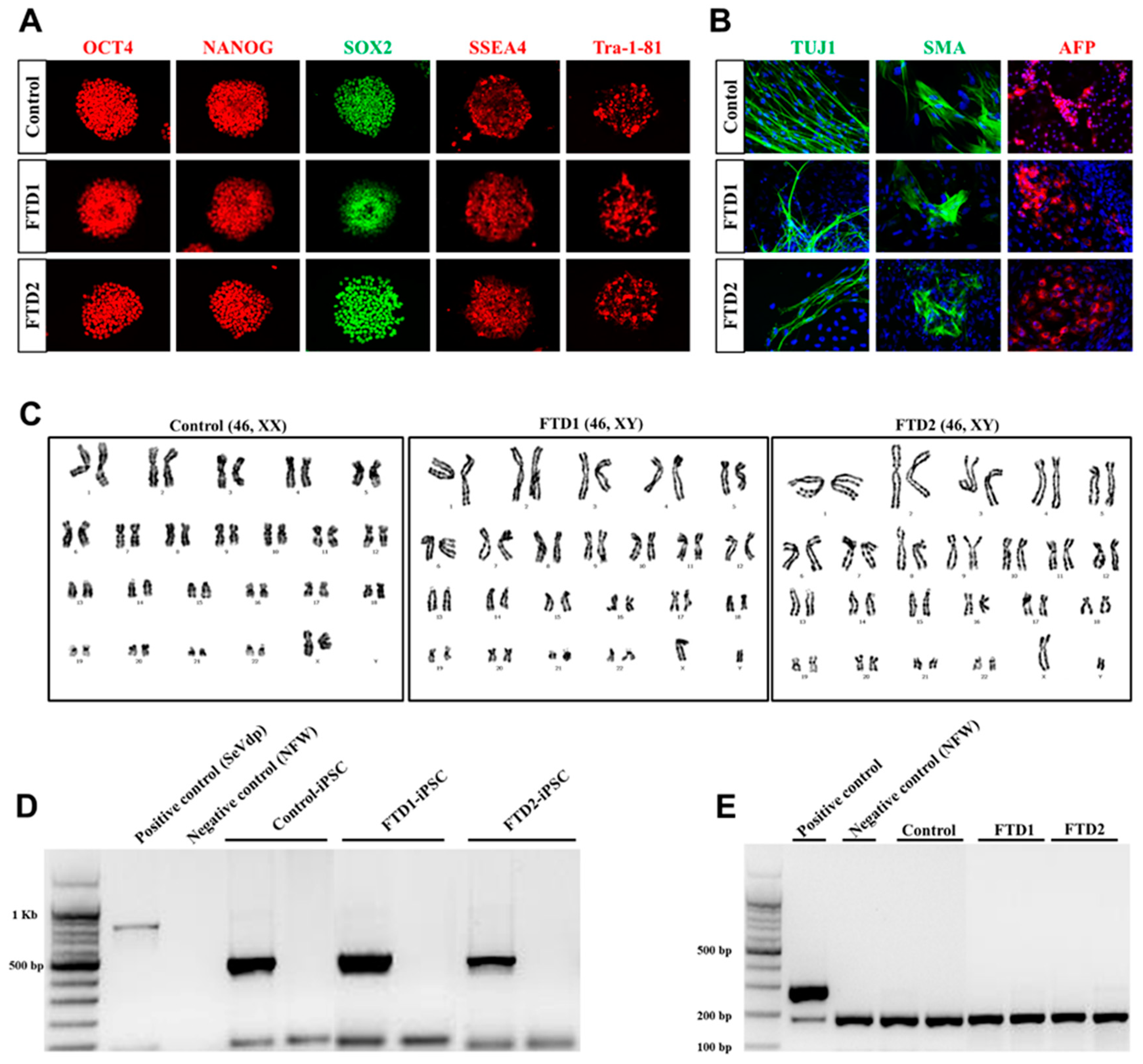
2.2. Generation and Characterization of the bvFTD Patient-Derived iPSCs
2.3. Neuronal Differentiation of bvFTD-Patients-Derived iPSCs
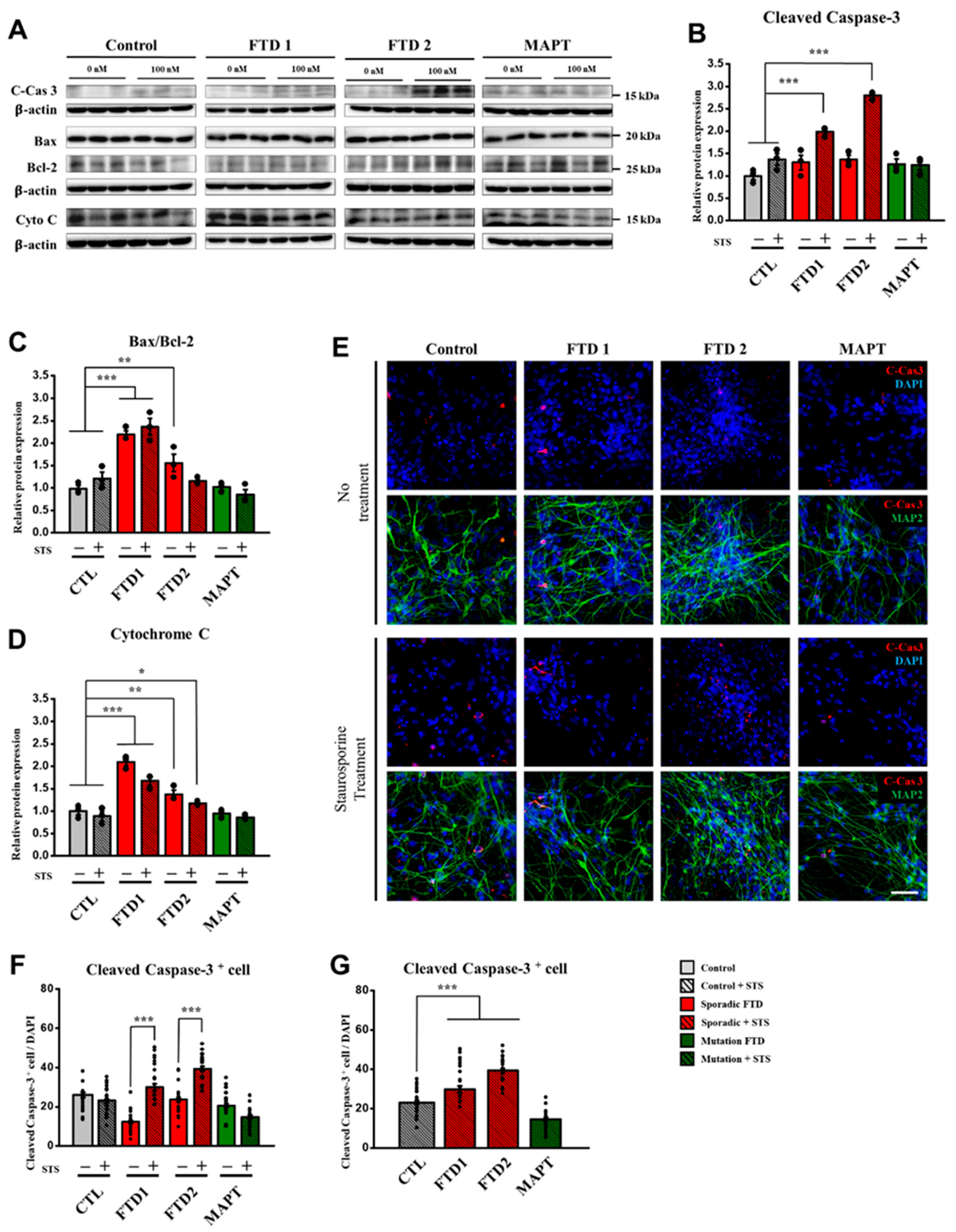
2.4. Cell Death Marker Expression in the bvFTD iPSC-Derived Neurons (iPSNs)
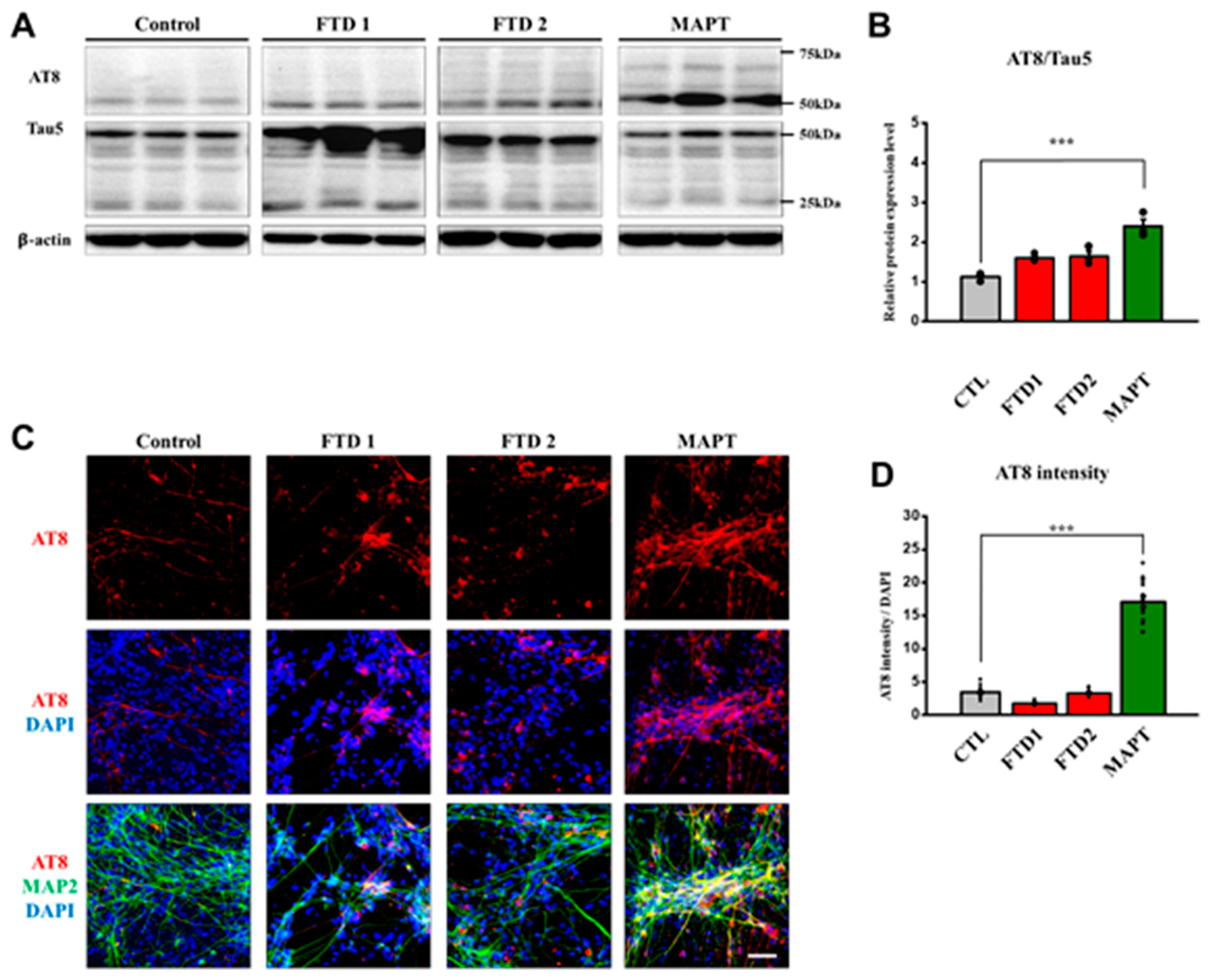
2.5. Amyloid Beta Secretion and p-Tau Expression in the bvFTD iPSC-Derived Neurons
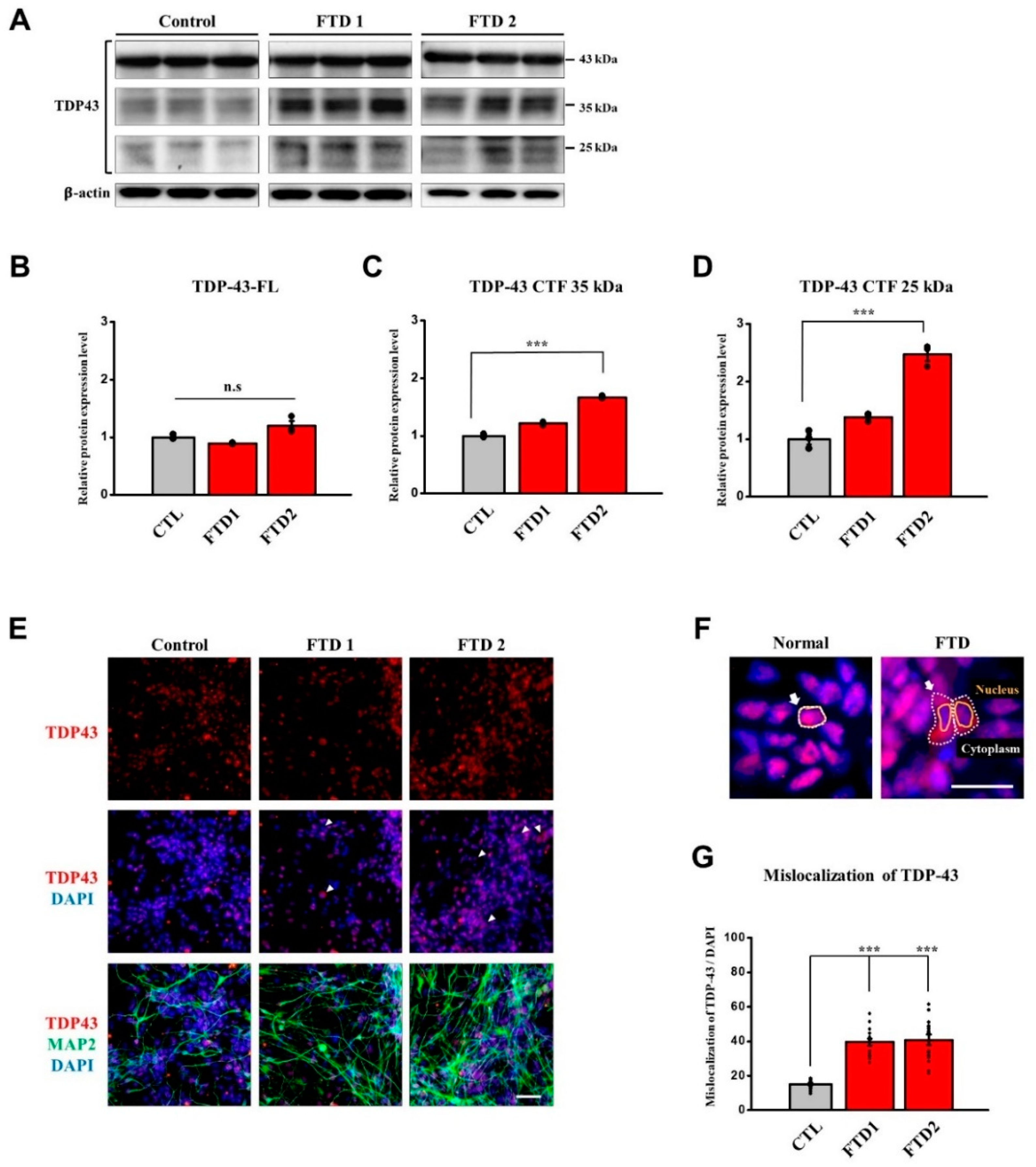
2.6. TDP-43 and FUS Expression in the bvFTD iPSC-Derived Neurons
3. Discussion
4. Materials and Methods
4.1. Participants
4.2. Whole Exome Sequencing and Data Analysis
4.3. Generation of iPSCs Using Sendai Virus Vector and iPSC Culture
4.4. Detection of Residual Sendai Virus Expression and Mycoplasma Detection PCR
4.5. Neural Induction
4.6. Post-Mitotic Neuronal Differentiation
4.7. Cellular Stress-Induced Caspase-3 Activation Assay
4.8. Immunocytochemistry
4.9. Image Acquisition and Quantifications
4.10. Western Blot Analysis
4.11. Extracellular Aβ ELISA
4.12. Statistical Analysis
4.13. Ethics Approval and Consent to Participate
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Karageorgiou, E.; Miller, B.L. Frontotemporal lobar degeneration: A clinical approach. Semin. Neurol. 2014, 34, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Fumagalli, L.; Prior, R.; Van Den Bosch, L. Current Advances and Limitations in Modeling ALS/FTD in a Dish Using Induced Pluripotent Stem Cells. Front. Neurosci. 2017, 11, 671. [Google Scholar] [CrossRef] [PubMed]
- Rascovsky, K.; Hodges, J.R.; Knopman, D.; Mendez, M.F.; Kramer, J.H.; Neuhaus, J.; van Swieten, J.C.; Seelaar, H.; Dopper, E.G.; Onyike, C.U.; et al. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain 2011, 134, 2456–2477. [Google Scholar] [CrossRef] [PubMed]
- Lagier-Tourenne, C.; Polymenidou, M.; Cleveland, D.W. TDP-43 and FUS/TLS: Emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet. 2010, 19, R46–R64. [Google Scholar] [CrossRef]
- Umoh, M.E.; Dammer, E.B.; Dai, J.; Duong, D.M.; Lah, J.J.; Levey, A.I.; Gearing, M.; Glass, J.D.; Seyfried, N.T. A proteomic network approach across the ALS-FTD disease spectrum resolves clinical phenotypes and genetic vulnerability in human brain. EMBO Mol. Med. 2018, 10, 48–62. [Google Scholar] [CrossRef]
- Ferrari, R.; Manzoni, C.; Hardy, J. Genetics and molecular mechanisms of frontotemporal lobar degeneration: An update and future avenues. Neurobiol. Aging 2019, 78, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Bang, J.; Spina, S.; Miller, B.L. Frontotemporal dementia. Lancet 2015, 386, 1672–1682. [Google Scholar] [CrossRef]
- Kim, E.J.; Kwon, J.C.; Park, K.H.; Park, K.W.; Lee, J.H.; Choi, S.H.; Jeong, J.H.; Kim, B.C.; Yoon, S.J.; Yoon, Y.C.; et al. Clinical and genetic analysis of MAPT, GRN, and C9orf72 genes in Korean patients with frontotemporal dementia. Neurobiol. Aging 2014, 35, e1213–e1217. [Google Scholar] [CrossRef]
- Tang, M.; Gu, X.; Wei, J.; Jiao, B.; Zhou, L.; Zhou, Y.; Weng, L.; Yan, X.; Tang, B.; Xu, J.; et al. Analyses MAPT, GRN, and C9orf72 mutations in Chinese patients with frontotemporal dementia. Neurobiol. Aging 2016, 46, e211–e235. [Google Scholar] [CrossRef]
- Che, X.Q.; Zhao, Q.H.; Huang, Y.; Li, X.; Ren, R.J.; Chen, S.D.; Wang, G.; Guo, Q.H. Genetic Features of MAPT, GRN, C9orf72 and CHCHD10 Gene Mutations in Chinese Patients with Frontotemporal Dementia. Curr. Alzheimer Res. 2017, 14, 1102–1108. [Google Scholar] [CrossRef]
- Kim, E.J.; Kim, Y.E.; Jang, J.H.; Cho, E.H.; Na, D.L.; Seo, S.W.; Jung, N.Y.; Jeong, J.H.; Kwon, J.C.; Park, K.H.; et al. Analysis of frontotemporal dementia, amyotrophic lateral sclerosis, and other dementia-related genes in 107 Korean patients with frontotemporal dementia. Neurobiol. Aging 2018, 72, e181–e186. [Google Scholar] [CrossRef] [PubMed]
- Nierode, G.J.; Perea, B.C.; McFarland, S.K.; Pascoal, J.F.; Clark, D.S.; Schaffer, D.V.; Dordick, J.S. High-Throughput Toxicity and Phenotypic Screening of 3D Human Neural Progenitor Cell Cultures on a Microarray Chip Platform. Stem Cell Rep. 2016, 7, 970–982. [Google Scholar] [CrossRef]
- Almeida, S.; Zhang, Z.; Coppola, G.; Mao, W.; Futai, K.; Karydas, A.; Geschwind, M.D.; Tartaglia, M.C.; Gao, F.; Gianni, D.; et al. Induced pluripotent stem cell models of progranulin-deficient frontotemporal dementia uncover specific reversible neuronal defects. Cell Rep. 2012, 2, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Fong, H.; Wang, C.; Knoferle, J.; Walker, D.; Balestra, M.E.; Tong, L.M.; Leung, L.; Ring, K.L.; Seeley, W.W.; Karydas, A.; et al. Genetic correction of tauopathy phenotypes in neurons derived from human induced pluripotent stem cells. Stem Cell Rep. 2013, 1, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Freibaum, B.D.; Lu, Y.; Lopez-Gonzalez, R.; Kim, N.C.; Almeida, S.; Lee, K.H.; Badders, N.; Valentine, M.; Miller, B.L.; Wong, P.C.; et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 2015, 525, 129–133. [Google Scholar] [CrossRef]
- Raitano, S.; Ordovas, L.; De Muynck, L.; Guo, W.; Espuny-Camacho, I.; Geraerts, M.; Khurana, S.; Vanuytsel, K.; Toth, B.I.; Voets, T.; et al. Restoration of progranulin expression rescues cortical neuron generation in an induced pluripotent stem cell model of frontotemporal dementia. Stem Cell Rep. 2015, 4, 16–24. [Google Scholar] [CrossRef]
- Kang, S.H.; Park, Y.H.; Lee, D.; Kim, J.P.; Chin, J.; Ahn, Y.; Park, S.B.; Kim, H.J.; Jang, H.; Jung, Y.H.; et al. The Cortical Neuroanatomy Related to Specific Neuropsychological Deficits in Alzheimer’s Continuum. Dement. Neurocogn. Disord. 2019, 18, 77–95. [Google Scholar] [CrossRef]
- Valdez, C.; Wong, Y.C.; Schwake, M.; Bu, G.; Wszolek, Z.K.; Krainc, D. Progranulin-mediated deficiency of cathepsin D results in FTD and NCL-like phenotypes in neurons derived from FTD patients. Hum. Mol. Genet. 2017, 26, 4861–4872. [Google Scholar] [CrossRef]
- Dafinca, R.; Scaber, J.; Ababneh, N.; Lalic, T.; Weir, G.; Christian, H.; Vowles, J.; Douglas, A.G.; Fletcher-Jones, A.; Browne, C.; et al. C9orf72 Hexanucleotide Expansions Are Associated with Altered Endoplasmic Reticulum Calcium Homeostasis and Stress Granule Formation in Induced Pluripotent Stem Cell-Derived Neurons from Patients with Amyotrophic Lateral Sclerosis and Frontotemporal Dementia. Stem Cells 2016, 34, 2063–2078. [Google Scholar] [CrossRef]
- Ehrlich, M.; Hallmann, A.L.; Reinhardt, P.; Arauzo-Bravo, M.J.; Korr, S.; Ropke, A.; Psathaki, O.E.; Ehling, P.; Meuth, S.G.; Oblak, A.L.; et al. Distinct Neurodegenerative Changes in an Induced Pluripotent Stem Cell Model of Frontotemporal Dementia Linked to Mutant TAU Protein. Stem Cell Rep. 2015, 5, 83–96. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Eiraku, M.; Watanabe, K.; Matsuo-Takasaki, M.; Kawada, M.; Yonemura, S.; Matsumura, M.; Wataya, T.; Nishiyama, A.; Muguruma, K.; Sasai, Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 2008, 3, 519–532. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Roh, J.H.; Chang, E.H.; Lee, Y.; Lee, S.; Kim, M.; Koh, W.; Chang, J.W.; Kim, H.J.; Nakanishi, M.; et al. iPSC Modeling of Presenilin1 Mutation in Alzheimer’s Disease with Cerebellar Ataxia. Exp. Neurobiol. 2018, 27, 350–364. [Google Scholar] [CrossRef] [PubMed]
- Rosen, H.J.; Gorno-Tempini, M.L.; Goldman, W.P.; Perry, R.J.; Schuff, N.; Weiner, M.; Feiwell, R.; Kramer, J.H.; Miller, B.L. Patterns of brain atrophy in frontotemporal dementia and semantic dementia. Neurology 2002, 58, 198–208. [Google Scholar] [CrossRef]
- Strang, K.H.; Golde, T.E.; Giasson, B.I. MAPT mutations, tauopathy, and mechanisms of neurodegeneration. Lab. Investig. 2019, 99, 912–928. [Google Scholar] [CrossRef]
- Mackenzie, I.R.A.; Rademakers, R.; Neumann, M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 2010, 9, 995–1007. [Google Scholar] [CrossRef]
- Iovino, M.; Agathou, S.; Gonzalez-Rueda, A.; Del Castillo Velasco-Herrera, M.; Borroni, B.; Alberici, A.; Lynch, T.; O’Dowd, S.; Geti, I.; Gaffney, D.; et al. Early maturation and distinct tau pathology in induced pluripotent stem cell-derived neurons from patients with MAPT mutations. Brain 2015, 138, 3345–3359. [Google Scholar] [CrossRef]
- Niedowicz, D.M.; Nelson, P.T.; Murphy, M.P. Alzheimer’s disease: Pathological mechanisms and recent insights. Curr. Neuropharmacol. 2011, 9, 674–684. [Google Scholar] [CrossRef]
- Mackenzie, I.R.; Rademakers, R. The molecular genetics and neuropathology of frontotemporal lobar degeneration: Recent developments. Neurogenetics 2007, 8, 237–248. [Google Scholar] [CrossRef]
- Dormann, D.; Haass, C. TDP-43 and FUS: A nuclear affair. Trends Neurosci. 2011, 34, 339–348. [Google Scholar] [CrossRef]
- Buratti, E.; Baralle, F.E. TDP-43: Gumming up neurons through protein-protein and protein-RNA interactions. Trends Biochem. Sci. 2012, 37, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Xu, Y.F.; Dickey, C.A.; Buratti, E.; Baralle, F.; Bailey, R.; Pickering-Brown, S.; Dickson, D.; Petrucelli, L. Progranulin mediates caspase-dependent cleavage of TAR DNA binding protein-43. J. Neurosci. 2007, 27, 10530–10534. [Google Scholar] [CrossRef]
- Kang, Y.; Na, D.L. Seoul Neuropsychological Screening Battery (SNSB); Human Brain Research & Consulting Co.: Incheon, Korea, 2003. [Google Scholar]
- Ahn, H.J.; Chin, J.; Park, A.; Lee, B.H.; Suh, M.K.; Seo, S.W.; Na, D.L. Seoul Neuropsychological Screening Battery-dementia version (SNSB-D): A useful tool for assessing and monitoring cognitive impairments in dementia patients. J. Korean Med. Sci 2010, 25, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Beel, S.; Herdewyn, S.; Fazal, R.; De Decker, M.; Moisse, M.; Robberecht, W.; Van Den Bosch, L.; Van Damme, P. Progranulin reduces insoluble TDP-43 levels, slows down axonal degeneration and prolongs survival in mutant TDP-43 mice. Mol. Neurodegener. 2018, 13, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, M.; Kim, H.J.; Koh, W.; Li, L.; Heo, H.; Cho, H.; Lyoo, C.H.; Seo, S.W.; Kim, E.-J.; Nakanishi, M.; et al. Modeling of Frontotemporal Dementia Using iPSC Technology. Int. J. Mol. Sci. 2020, 21, 5319. https://doi.org/10.3390/ijms21155319
Kim M, Kim HJ, Koh W, Li L, Heo H, Cho H, Lyoo CH, Seo SW, Kim E-J, Nakanishi M, et al. Modeling of Frontotemporal Dementia Using iPSC Technology. International Journal of Molecular Sciences. 2020; 21(15):5319. https://doi.org/10.3390/ijms21155319
Chicago/Turabian StyleKim, Minchul, Hee Jin Kim, Wonyoung Koh, Ling Li, Hyohoon Heo, Hanna Cho, Chul Hyoung Lyoo, Sang Won Seo, Eun-Joo Kim, Mahito Nakanishi, and et al. 2020. "Modeling of Frontotemporal Dementia Using iPSC Technology" International Journal of Molecular Sciences 21, no. 15: 5319. https://doi.org/10.3390/ijms21155319
APA StyleKim, M., Kim, H. J., Koh, W., Li, L., Heo, H., Cho, H., Lyoo, C. H., Seo, S. W., Kim, E.-J., Nakanishi, M., Na, D. L., & Song, J. (2020). Modeling of Frontotemporal Dementia Using iPSC Technology. International Journal of Molecular Sciences, 21(15), 5319. https://doi.org/10.3390/ijms21155319




