Abstract
Uveal melanoma (UM), the most common intraocular malignancy in adults, is a rare subset of melanoma. Despite effective primary therapy, around 50% of patients will develop the metastatic disease. Several clinical trials have been evaluated for patients with advanced UM, though outcomes remain dismal due to the lack of efficient therapies. Epigenetic dysregulation consisting of aberrant DNA methylation, histone modifications, and small non-coding RNA expression, silencing tumor suppressor genes, or activating oncogenes, have been shown to play a significant role in UM initiation and progression. Given that there is no evidence any approach improves results so far, adopting combination therapies, incorporating a new generation of epigenetic drugs targeting these alterations, may pave the way for novel promising therapeutic options. Furthermore, the fusion of effector enzymes with nuclease-deficient Cas9 (dCas9) in clustered regularly interspaced short palindromic repeats (CRISPR) associated protein 9 (Cas9) system equips a potent tool for locus-specific erasure or establishment of DNA methylation as well as histone modifications and, therefore, transcriptional regulation of specific genes. Both, CRISPR-dCas9 potential for driver epigenetic alterations discovery, and possibilities for their targeting in UM are highlighted in this review.
1. Biology and Molecular Subtypes of Uveal Melanoma
Uveal melanoma (UM) is the most common primary cancer of the eye, causing fatal liver metastasis in up to half of the patients [1,2]. The average annual incidence varies widely according to ethnicity or latitude between less than 1 to more than 9 per million population per year, with the highest incidence in white Caucasians [3,4]. Primary UM is treated with either surgery or radiation with a low local recurrence rate. However, there are no efficient therapies for metastatic UM, and as a result, most of the patients survive less than 12 months after metastases diagnosis [5]. A recent meta-analysis including 912 metastatic UM patients reported the median OS 10.2 months (95% CI 9.5–11.0) [6]. The UM tumors arise from melanocytes located in the uveal layer of the eye, with the choroid the most frequent site (82%), followed by the ciliary body (15%) and iris (3%) [7]. G protein subunit alpha q (GNAQ) and G protein subunit alpha 11 (GNA11) hotspot mutations, present in 83% of UM, are considered to be initiating events in UM tumorigenesis [8,9]. Recurrent cytogenetic abnormalities, including the most significant loss of one copy of chromosome 3 (M3) as well as 8p loss/8q gain, 6p gains, and 1p deletion, also hold prognostic potential [10]. Loss-of-function mutations in the BRCA1 associated protein 1 (BAP1) gene, located on 3p21, accompanied by decreased BAP1 mRNA and protein expression, have been identified in M3-UM, indicating that BAP1 abnormalities are highly correlated with the development of UM metastases. Patients with BAP1 mutations are generally younger, between 30 and 59 years, compared to the mean age at diagnosis, 62 years [11,12]. Gene expression profiling is considered an important prognostic tool that can predict metastatic risk with higher certainty than clinical stage or chromosome 3 status. A commercially available expression panel of 15 genes, established by Castle Biosciences, categorizes patients as Class 1 (low metastatic risk) or Class 2 (high metastatic risk) [13]. Class 1 patients are subdivided into Class 1a and Class 1b categories based on the key mutations and the number of 8q and 6p copies [2]. The 5-year screening of patients revealed significant differences in metastatic risk of Class 1a, 1b, and Class 2 UMs, which are 2%, 21%, and 72%, respectively [14]. Recently, Robertson and colleagues identified four molecularly distinct UM subtypes, two associated with poor-prognosis M3 and two associated with better-prognosis disomy 3 (D3). Two subgroups of M3 tumors possess diverse clinical outcomes and are related to different biological pathways. In the first one, DNA damage repair, hypoxia-inducible factor 1 alpha (HIF1A), and MYC signaling are more prominent, while high levels of mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase (PI3K)/Akt are more noticeable in the other subgroup [15]. This highlights the necessity for tailored therapeutic strategies that target these subtype-specific molecular changes. Moreover, unsupervised consensus clustering of the most variable 1% of CpG probes brought in four methylation clusters strongly associated with prognostic groups. UM in DNA methylation clusters 2 and 3 were highly enriched (12 of 16 tumors) in SF3B1/SRFR2 mutations, whereas EIF1AX mutant tumors were only present in DNA methylation cluster 1. Thus, D3 UM with EIF1AX versus SF3B1/SRFR2 mutations conveys diverse DNA methylation patterns. M3 BAP1-aberrant UM tumors offered a single global DNA methylation profile [15].
2. Role of Epigenetic Changes in UM Progression
Epigenetic alterations can result in aberrant gene regulation, thereby playing an essential role in tumorigenesis. Therefore, a complete overview of the epigenomic landscape is required to track the molecular events involved in the initiation and progression of a particular disease. Epigenetic changes that, for example, silence tumor suppressor genes or activate oncogenes include DNA methylation, histone modifications, and small non-coding RNAs. Many of them have been associated with UM initiation and progression (Figure 1) [16].
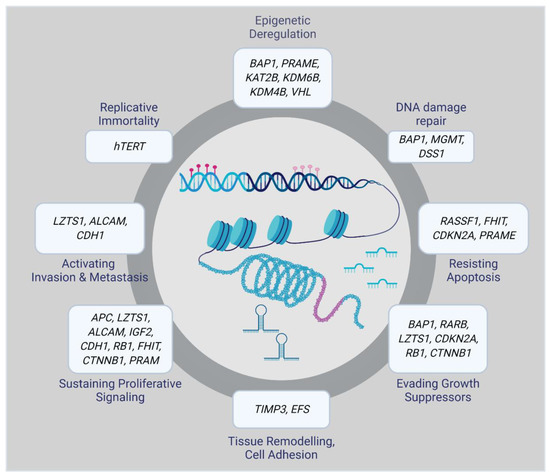
Figure 1.
Epigenetically altered genes in uveal melanoma (UM).
2.1. DNA Methylation
DNA methylation is a covalent modification with the addition of a methyl group [-CH3] to the cytosine residue in the CpG dinucleotide sequence. Methylation/demethylation is an essential mechanism in maintaining cell- or tissue-specific gene expression [17]. Correlation between gene expression profiles with global DNA methylome clusters identified so far in prognostically distinct UM tumors, suggests an epigenetic contribution to the underlying molecular pathology that produces this transcriptome [15,18].
Preferentially Expressed Antigen in Melanoma (PRAME) is an emerging epigenetic biomarker of metastasis in low-risk UM tumors [19]. It was shown that hypomethylation of specific CpG sites nearby the PRAME promoter resulted in its transcriptional activation, correlated with high metastatic risk in both classes 1 UMs [20]. There was a significant association between the high expression level of the Deleted Split hand/Split foot 1 (DSS1) gene and some clinical-pathological features. A recent study discovered that 64% of UMs showed higher expression of DSS1 than healthy tissues. It was demonstrated that there is an inverse correlation between DSS1 expression activity and the methylation status of its promoter [21].
Aberrant promoter hypermethylation of CpG islands plays a critical role in the inactivation of tumor suppressor genes in cancer [22]. Hypermethylation of p16, TIMP3, RASSF1A, RASEF, hTERT, and EFS genes have been reported in UM [23,24,25,26,27,28].
The Ras association domain family 1 isoform A (RASSF1A) gene is located on chromosome 3p21.3, and its absence or inactivation has been proved to be a contributing factor in UM tumor formation and progression [29]. It plays a crucial role in cell-cycle regulation, apoptosis, and microtubule stability [30]. Methylation of promoter sites of this gene control entry at the retinoblastoma checkpoint and inhibits cyclin D1 protein accumulation at the post-transcriptional level, leading to cell-cycle progression block from the G1 to the S phase. Though the methylation of RASSF1A may not be wholly responsible for UM development, it could be a contributing factor in UM tumorigenesis [29]. M3, which is related to the tumor’s metastatic capacity, has been reported in approximately half of all UMs. Considering the position of RASSF1A on the p21.3 region of chromosome 3, it could serve as a tumor suppressor gene whose silencing by methylation acts as a ‘second hit’ after monosomy occurs [26].
The Ras and EF-hand domain-containing (RASEF) gene, located on chromosome 9, region q21 is a candidate tumor suppressor gene. In 2007, UM cell lines and primary UM samples were screened for mutations in the RASEF gene region. The authors discovered that all cell lines and samples that did not express RASEF contained a methylated promoter, although those with RASEF expression lacked this methylation. They also demonstrated that methylation not only co-occur with low expression but also with a homozygous genotype. These findings propose that a combination of methylation and loss of heterozygosity may be the mechanism for loss of RASEF expression [31]. Homozygous tumors with a methylated RASEF promoter region tend to display reduced survival compared with heterozygous tumors without methylation, suggesting loss of heterozygosity might be related to the aggressive behavior of the tumor [31].
A study by Venza and colleagues in 2015 showed that DNA methyltransferase 1 (Dnmt1) and Dnmt3b have a preeminent role in P16INK4A (alias CDKN2A) repression. They demonstrated that epigenetic alterations in the P16INK4A and P14ARF (the alternative reading frame protein product of the CDKN2A locus) genes were frequently associated with cutaneous as well as UMs [32]. Moreover, it was demonstrated that P16INK4A is frequently inactivated by hypermethylation in both primary UM and UM cell lines, accompanied by a down-regulated expression of P16INK4A [23]. Both these studies also reported that loss of P16INK4A expression, attributable to CpG methylation, could be reversed when treated with the demethylating drug 5-aza-2′-deoxycytidine (decitabine). Interestingly, in UM patients who possess a tumor with a methylated P16INK4A promoter, metastasis tends to be more common. Therefore, modulation of abnormal methylation could be considered a valid target for UM treatment [23].
A recent study by Field and colleagues indicates that hypermethylation on chromosome 3 correlated with down-regulated gene expression at several loci, including 3p21 where BAP1 is located. All Class 2 tumors contained a novel hypermethylated site within the BAP1 locus, which reveals that BAP1 itself is epigenetically regulated. In functional validation experiments, Bap1 knockdown in UM cell lines consisted of a similar methylomic repatterning with UM tumors, enhanced for genes involved in axon guidance, melanogenesis, and development [33]. This study provides evidence that BAP1 loss leads to large-scale methylomic repatterning resulting in the Class 2 phenotype. Deciphering the role of epigenetic deregulation could explain the loss of melanocytic differentiation and gain of neural crest-like migratory behavior in Class 2 UMs.
2.2. Histone Modifications
Histone modification refers to the process of acetylation, phosphorylation, histone methylation, polyadenylation, ubiquitination, and ADP ribosylation, achieved by the relevant enzymatic activity [34]. Depletion of Bap1 protein trigger hyperubiquitination of H2A in melanoma cells and melanocytes, bringing about loss of differentiation along with the gain of stem-like properties [35,36]. On the contrary, treatment with histone deacetylase inhibitors (HDACis) in vivo in a xenograft model reversed the H2A hyperubiquitination, which may have therapeutic potential for inducing prolonged dormancy of micrometastatic UM disease [35]. While Hdac4 is localized to the nucleus in BAP1-mutant UM cells and the cells in which a BAP1 mutation was introduced using CRISPR-Cas9, it is restricted mainly to the cytoplasm in BAP1 wild-type UM cells and in normal human uveal melanocytes. Hence, Bap1 can inhibit the epigenetic function of Hdac4, at least in part, by diminishing its localization to the nucleus. Besides, short hairpin RNA (shRNA)-mediated depletion of Hdac4 in BAP1-mutant UM cells significantly impeded cell proliferation [37]. These findings suggest novel insights into the role of BAP1 in development and cancer and propose HDACis as potential therapeutic agents for BAP1-mutant cancer’s treatment.
2.3. miRNA-Based Epigenetic Mechanism
Micro RNAs (miRNA) are among the most studied non-coding RNAs. They are short (17–22 nucleotides in length), phylogenetically preserved single-stranded RNA molecules involved in the gene expression regulation. Their dysregulation has been ascertained to confer resistance to apoptosis, promote cell-cycle progression, and enhance invasiveness and metastasis of many cancers [38]. A significant number of miRNAs have been shown to be differentially expressed in UM cell lines and tumor tissues [39]. The expression level of let-7b, miR-143, miR-193b, miR-199a, and miR-652 were proved to be increased in Class 2 UMs, so they can be used to differentiate between Class 1 and Class 2 UM tumors [40]. Radhakrishnan and colleagues identified distinct miRNAs present in metastatic UM and absent in non-metastasizing tumors [41].
Moreover, 96 miRNAs were reported altered in UM cell lines. Among them, 65 were downregulated, 28 upregulated, and 3 exhibited a different expression pattern [42]. The pleiotropic nature makes miRNAs particularly attractive drug targets. MiR-27a is an oncogenic miRNA overexpressed in various cancer types. Genistein was found to inhibit miR-27a expression in highly aggressive UM cells in vitro and in vivo, thus increasing the expression of its target gene ZBTB10 significantly. Therefore, the authors hypothesized that genistein growth inhibitory activities can be mediated via miR-27a regulatory mechanism [43].
Treatment of UM cells with a DNA hypomethylating agent decitabine and HDACi trichostatin A (TSA), can regulate miR-124a expression level via epigenetic mechanisms [44]. Furthermore, decitabine was capable of enhancing miR-137 expression, which is generally epigenetically silenced during UM initiation [45], proving that individual epigenetic mechanisms can interact with each other. The list of UM-associated miRNAs (reviewed in [46,47]) is continually expanding along with the development of experimental methods and miRNA research tools. However, the identification of clinically relevant target genes and corresponding biological pathways will pave the way for their therapeutic targeting.
3. Current Therapeutic Approaches Available for UM
3.1. Primary UM Therapies
A historical approach to definitive, local treatment in primary UM is enucleation. This method is still appropriate for large tumors with extensive extraocular growth and a low probability of retaining vision. However, since in 2006, when the COMS study failed to prove a survival gain with enucleation compared to brachytherapy, there has generally been a movement toward vision- and eye-preserving modalities [48,49]. Tumor size and location, retinal detachment or invasion, and patient age and health status are the main factors affecting clinical management [2].
Two surgical approaches which offer better eye-preserving outcome are transretinal and transscleral endoresection, even though local recurrence rates are higher with transscleral resection than with brachytherapy or enucleation. Kivela and colleagues reported 6.1% of tumor recurrence after brachytherapy and 32.6% after transscleral resection similar to the other studies [50,51,52].
Radiation therapy, including brachytherapy, charged particle radiotherapy, and stereotactic radiotherapy, is the most commonly globe-preserving technique in the treatment of UM [2,53,54] Iodine-125 and ruthenium-106 are the most frequently used radioisotopes. Beside, Palladium 103 is rarely employed, and cobalt 60 (60Co) was used in the past [55,56]. However, brachytherapy could cause critical side effects, as well. Therefore, regular ophthalmologic follow-up must be carried out to monitor probable detrimental effects, including neovascular glaucoma, radiation-induced retinopathy, cataracts, and macular edema that can appear up to 5 years after therapy [56]. In 2014 Shah and colleagues showed that utilization of intravitreal anti-vascular endothelial growth factor (VEGF) after brachytherapy could weaken or delay the rate of moderate vision loss, macular edema, and poor visual acuity [57]. The use of charged-particle radiotherapy can improve local control, eye preservation, and disease-free survival in medium to large tumors or those in a location that may not be amenable to plaque brachytherapy [58]. Furthermore, a high rate of local tumor control (>95% at 15 years) [59] can be achieved without significantly worse complications than plaque brachytherapy.
Laser photocoagulation, photodynamic therapy, and transpupillary thermotherapy are laser methods that could be employed as primary therapy for small choroidal lesions [60]. However, their clinical utility is limited due to conflicting results and possible side effects [2].
Several clinical trials were performed for adjuvant UM settings, however, with no or marginal survival benefit. These include, for example sequential treatment with low-dose dacarbazine and interferon alfa-2b (NCT01100528) [61], fotemustine (NCT02843386) [62], adjuvant intra-arterial fotemustine [63], adjuvant interferon alfa-2a [64], or Bacillus Calmette–Guérin injections [65]. A recent phase I/II clinical trial of adjuvant ipilimumab in 10 high-risk UM patients showed that at 36 months follow-up, 80% of patients had no evidence of distant disease (NCT01585194) [66]. Adjuvant sunitinib treatment assessed retrospectively on the sample of 64 UM patients was associated with better overall survival (OS), especially in the group of patients under 60 years of age [67]. However, small sample size and retrospective design warrant further investigation of these results. Four adjuvant trials phase II/III are ongoing, focusing on combination immunotherapy (NCT02519322, NCT03528408), dendritic cells plus autologous tumor RNA (NCT01983748) and HDACi valproic acid (VPA) in comparison to sunitinib (NCT02068586).
3.2. Metastatic UM Therapies
Systemic chemotherapy, immunotherapy, liver-directed therapies, and targeted agents against the MAPK pathway are among treatments that have been evaluated in clinical trials for patients with advanced disease [2]. However, response rates are usually less than 10%, and no therapy has been confirmed to improve OS [68,69]. As a result, median progression-free survival (PFS) and OS for the metastatic patients are disappointingly low (3.29 months and 10.2 months, respectively) [53,70]. UM most commonly (93%) metastasizes to the liver [71]. Resection of hepatic lesions may offer long-term survival and cure in selected cases. Various other techniques e.g., radiofrequency ablation, stereotactic radiotherapy, regional chemotherapy such as hepatic intra-arterial infusion, and isolated hepatic perfusion or embolization are other liver-directed approaches that were tested with limited success [70]. Similarly, chemotherapy regimens such as dacarbazine, cisplatin, treosulfan, temozolomide, fotemustine, and various combinations, have been utilized in metastatic UM with poor outcomes. None of these agents has been shown to prolong survival response rates, which varied from 0% to 15% [72,73,74,75,76].
While progress in immunotherapy, especially immune checkpoint inhibitors targeting cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4) and programmed cell death-1 (PD-1), have had a significant therapeutic benefit for metastatic cutaneous melanoma, it has not same clinical efficiency in metastatic UM [69]. The poor mutational burden observed in UM may, in some way, contribute to the limited success of the immune checkpoint blockade. Furthermore, the upregulation of immunosuppressive factors such as IDO1 and TIGIT might be partly responsible for treatment resistance suggesting a role for combination immune therapies [15]. Although retrospective analysis of 89 metastatic UM patients treated with ipilimumab plus nivolumab confirmed that dual checkpoint inhibition yields higher response rates than single-agent immunotherapy, 92% of patients discontinued treatment due to toxicity or progressive disease [77].
Targeted agents against the MAPK pathway is one of the therapeutic approaches in metastatic UM. GNAQ/GNA11 mutations are fundamental for the activation of the RAS-ERK pathway. However, BRAF or NRAS mutations, which mediate sensitivity to the BRAF inhibitors vemurafenib and dabrafenib in cutaneous melanoma [78,79], are not common in UM [80]. The molecular profile of UM suggests treatments that target downstream components of the molecular pathways driving tumor growth, such as MEK and protein kinase C (PKC). Although there are no adequate results so far (response rates generally less than 10%). Two clinical trials are currently underway assessing the safety and anti-tumor efficacy of orally available PKC inhibitor LXS196 in patients with solid tumors harboring GNAQ/11 mutations (NCT03947385) and metastatic UM (NCT02601378). Preliminary results were published for 17 UM patients, of whom a partial response was achieved in two, stable disease (>6 months) in seven, while the progressive disease was detected in seven patients [81].
One of the highly selective potential inhibitors of MEK is selumetinib. Enhanced clinical outcomes with selumetinib compared to chemotherapy (temozolomide or dacarbazine) was demonstrated in a randomized, phase II study in 101 metastatic UM patients [82]. Patients who received selumetinib gained significantly longer PFS than those who received standard chemotherapy (15.9 vs. 7 weeks, p < 0.001). Subsequently, the phase III study enrolled UM patients who were categorized into two groups, selumetinib plus dacarbazine vs. dacarbazine alone. Unfortunately, this study did not reach its primary endpoint; there was no significant improvement in median PFS in the selumetinib plus dacarbazine than the dacarbazine alone (2.8 vs. 1.8 months, p = 0.32). Furthermore, objective response rate was not significantly increased (3.1 vs. 0%, p = 0.36). Efforts to optimize the effectiveness of MEK inhibition are continuing.
Some of the other agents targeting pathways, apart from MAPK, are gefitinib (an epidermal growth factor inhibitor) [83], lenalidomide [84], thalidomide (immunomodulator) [85], bevacizumab (VEGF-blocking antibody) plus interferon-α [86], bevacizumab plus temozolomide [87], aflibercept (a “decoy” receptor binding circulating VEGF) [88], imatinib (a KIT inhibitor) [89,90] carboplatin/paclitaxel/sorafenib (a multikinase inhibitor) [91] and sunitinib (a multiple receptor tyrosine kinase inhibitor) [92]. Nevertheless, none of these agents or combinations provided worthwhile outcome in metastatic UM.
4. Potential of Epigenetic Therapies in UM
Epigenetic processes, specifically DNA promoter methylation and histone modifications, are vital cellular events during tumorigenesis [93]. Though significantly explored in hematologic malignancies, in solid tumors, epigenetic therapeutic approaches remain in the rear [94]. Epigenetic dysregulation plays a crucial role in UM pathogenesis, by its genetic simplicity [18]. Recently it has been uncovered that a distinct global DNA methylation state is associated with the poor-prognosis subtype characterized by M3 and BAP1 mutations [15]. HDACis induce morphologic and transcriptomic changes along with cell-cycle arrest, associated with lower metastatic risk in preclinical models [35]. Some epigenetic drugs have been used in preclinical studies (Table 1) and clinical trials (Table 2) for UM. Given the critical impact of epigenetic regulatory mechanisms in triggering metastasis, there is a rationale for adopting the epigenetic approach as a novel therapy for UM treatment that could potentially add to the recent progress in immune and targeted therapies.

Table 1.
Preclinical studies focused on epigenetic drug anti-cancer effects in uveal melanoma.

Table 2.
Clinical trials of epigenetic drugs in solid tumors, including uveal melanoma.
4.1. Inhibitors of DNA Methylation
During DNA methylation, gene silencing occurs due to the adding methyl group to cytosine residue in CpG islands located principally in promoter regions. This phenomenon is regulated by the affected promoter. Aberrant gene silencing by DNA methylation has shown the ability to modulate cancer biology and cause drug resistance [104]. Decitabine is a powerful DNMT inhibitor (DNMTi) that appears to diminish the methylation process in numerous cancer cell lines (including melanoma), allowing the re-expression of genes that malignant cells are trying to turn off [105].
A recent examination investigated the safety of utilizing the demethylating agent decitabine by hepatic arterial infusion in patients with unresectable liver metastases. A study including 9 eligible patients, initiated treatment in a dose-escalation phase I clinical trial. The primary tumor types consist of four UMs, four colorectal carcinomas, one skin melanoma, and one epithelial ovarian cancer. Decitabine was applied at three different doses (two patients at dose 10, four at 15 and six at a 20 mg decitabine/m2/day). It was administered as a 1-h hepatic arterial infusion every 4 weeks for five successive days. Expression levels of 30 cancer test antigens (CTA) in pre-treatment and post-treatment biopsies from all cohorts were analyzed. The expression of 21 out of 30 CTAs after treatment escalated. Predominant treatment-related adverse events were grades 1 and 2 hematological toxicity. No patients experienced treatment-limiting detriment [98]. However, during study treatment or post-study exposure to immune checkpoint therapy, no objective tumor responses were observed in four patients with UM liver metastases.
4.2. Histone Deacetylase Inhibitors
Given that DNA is wrapped around histones, the acetylation of which mediates chromatin relaxation associated with a higher level of gene transcription, histone acetylation and deacetylation represent an essential part of the epigenetic regulatory mechanism. Hence, increased activity of HDAC, one of the main characteristics of oncogenic changes, can be potentially restored by HDACis. HDACis are compounds that were shown to induce growth arrest of transformed cells, cell death, angiogenesis inhibition, and terminal differentiation [106,107]. Human HDACs, grouped into four classes based on the similarity of DNA sequences and function include 18 different substances [108]. It has been proven that HDACis induce dormancy of micrometastatic disease through differentiation of UM cells and shift UM cells from Class 2 to the Class 1 signature [35]. Some HDACis such as VPA, TSA, tenovin-6, panobinostat, entinostat, depsipeptide, vorinostat, quisinostat, NaB, JSL-1, MC1568, and MC1575 have shown promising results in preclinical UM models [109].
VPA, which is now in the clinical trial [NCT02068586], is a well-characterized HDACi used for almost 40 years in epilepsy treatment and as an anti-cancer agent recently [110]. Gene expression profiling, allowing enrollment of high-risk Class 2 patients, about half of whom will develop overt metastatic disease within three years of eye tumor diagnosis, can be highly relevant for clinical testing [111]. The ability of VPA to reverse the H2A hyperubiquitination caused by Bap1 loss, and to shift the gene expression profile of Class 2 cells toward a Class 1 profile, inducing changes compatible with melanocytic differentiation suggests that Bap1 normally maintains melanocyte differentiation in UM cells and its function can be at least partially reversed in lack of Bap1 by increasing histone H3 acetylation [35].
Similarly, panobinostat has been demonstrated to induce morphological differentiation, G1 cell-cycle arrest, and a shift to a differentiated, melanocytic gene-expression profile in cultured UM cells. Not only does it inhibits the proliferation of UM cells, but also it significantly reduces the fraction of viable cells [35].
The effect of vorinostat or suberanilohydroxamic acid (SAHA) on acetylation of histone H3 and H4 to P14ARF and P16INK4A promoters in UM cells have been evaluated by Venza et al. SAHA treatment-induced H3 and H4 hyperacetylation at the P14ARF promoter, followed by increased P14ARF expression, caused significant reduction in UM cell growth, migration, and invasion [101]. Vorinostat was also tested in a phase II clinical trial for treating patients with metastatic or unresectable melanoma [NCT00121225]. Despite stable disease in a high proportion of patients and some early responses, the primary endpoint of response has not met yet [112].
TSA upregulates the expression level of miR-137, a tumor suppressor gene acting through the down-regulation of it targets MITF and CDK6. A similar effect was caused by the transient transfection of miR-124a, down-regulated in UM cells. The transfection inhibited cell growth, migration, and invasion, via repression of the potential targets of miR-124a CDK4, CDK6, CCND2, and EZH2 in UM cells [44].
Depsipeptide is a very potent HDACi that inhibits cell growth, induces apoptosis, and declines the migration of viable UM cells in both primary and metastatic UM cell lines [95,96].
Tenovin-6 suppressed UM cells’ growth in vitro through the deacetylation of SIRT1 and SIRT2 in UM cells, thus activating p53 expression and inducing apoptosis. A combination of tenovin-6 with the conventional vinblastine, a chemotherapeutic agent used for systemic therapy of UM patients, inhibited the viability of UM cells. Furthermore, tenovin-6, decreased the level of intracellularly active β-catenin, blocks WNT/β-catenin signaling, and diminished the population of cancer stem cells [97].
In a recent research JSL-1, a novel HDACi was studied in vitro and in vivo. It effectively inhibited cell proliferation. JSL-1 increases the expression of pro-apoptotic BH3-only protein (BIM) in UM cells, bringing about the induction of apoptosis. JSL-1 suppressed migration and invasion of UM cells with MMP-2 reduction. JSL-1 also inhibited the growth of UM xenografts in NOD-SCID mice. Furthermore, it eliminated UM cancer stem-like cells, which are considered seeds of metastasis by blocking the canonical Wnt/β-catenin pathway, impairing self-renewal capacity, and declining percentage of ALDH+ cells [100].
4.3. Combination Therapy Using Epigenetic Drugs
Epigenetic drugs such as DNMTis, HDACis, histone methyltransferase inhibitors (HMTis) e.g., enhancer of Zeste homolog 2 inhibitors (EZH2is), and modifiers of miRNA expression such as antagomirs were shown to reduce the resistance of tumor cells to the natural killer and cytotoxic T cells, and enhancing the functions of antigen-presenting cells [113]. At present, increasing attention is paid to the newly combined therapeutic approaches employing epigenetic drugs or new molecular inhibitors and other therapies to promote the efficacy of cancer treatment [114].
The safety and tolerability combination of dual epigenetic therapy (DNMT and HDAC inhibition) with chemotherapy was explored in metastatic melanoma. In this trial, decitabine and panobinostat were combined with a known standard agent for metastatic melanoma temozolomide. 17 melanoma patients, including 11 cutaneous, 4 ocular, and 2 mucosal melanomas, underwent therapy with this combination. Despite limited efficacy, this trial was generally well-tolerated by the treated groups and appeared safe to be continued to Phase II. None of the patients experienced dose-limiting toxicity. A maximum tolerated dose was not reached, as well [115].
MEK inhibitors (MEKis) appear to have anti-proliferative activity in metastatic UM patients, though responses are short-lived. Gonçalves and colleagues recently evaluated a group of epigenetic inhibitors, including Disruptor of telomeric silencing 1-like inhibitors (DOT1Lis), EZH2is, lysine-specific demethylase 1 inhibitors (LSD1is), DNMTis, and histone acetyltransferase inhibitors (HATis) as a strategy to diminish escape from MEKi therapy in vitro [99]. The authors proved that decitabine dramatically enhanced the anti-proliferative activity of trametinib in cell viability, 3D organoid, and colony formation assays. A combination of two drugs MEKi-DNMTi principally affected the expression of genes involved in G1 and G2/2M checkpoints, cell survival, chromosome segregation, and the mitotic spindle formation. Induction of DNA repair or senescence does not differ from either drug alone. Instead, the expression level of the CDK inhibitor p21, and the BIM were increased. Likewise, the DNMTi-MEKi combination more efficiently suppressed the growth of MP41 cells in UM xenografts than monotherapy. Thus, DNMTi may improve the activity of MEKi in UM [99].
4.4. Third Generation of Epigenetic Drugs
So far, epigenetic drugs consisting of DNMTis, HDACis, HMTis, and modifiers of miRNA expression appear to be more efficient for hematological cancers rather than solid tumors, possibly since solid tumors originate from more-differentiated or even terminally differentiated cells with a reduced capability for epigenetic reprogramming [94]. Development of first-generation and second-generation epigenetic drugs, which are almost exclusively DNMTis or HDACis, following a ‘one size fits all’ approach together with the lack of predictive biomarkers for patient selection thereby, resulted in disappointingly low efficacy in patients with solid tumors. However, success with certain third-generation epigenetic drugs, used according to precision medicine paradigms, has provided new hope in epigenetic therapy. This group includes, among others, bromodomain and extra-terminal domain inhibitors (BETis), histone demethylase inhibitors (HDMis), HMTis, or protein arginine methyltransferase inhibitors (PRMTis) [114]. As they might have therapeutic potential in the treatment of UM, there is an increasing need to evaluate their efficiency. The first such attempt was made with BETi, which provides a novel therapeutic approach in UM treatment. The BET family of proteins, including BRD2, BRD3, BRD4, and BRDT bind to acetylated lysine residues on histone tails to direct the assembly of nuclear complexes regulate chromatin remodeling, DNA replication, and transcription [116,117]. More precisely, BRD4 can regulate oncogenic drivers such as MYC by binding to super-enhancers and non-coding DNA regions that are densely occupied by master transcription factors responsible for cell identity [102,118]. Since MYC amplification plays a crucial role in UM, it was hypothesized that BET inhibition therapeutic activity might be regulated via the down-regulation of MYC and MYC-dependent genes. Phase I and phase II clinical trial of BRD4 inhibitor (PLX2853) in various advanced malignancies, including UM is recruiting now [NCT03297424].
5. Epigenetic Editing
Due to the reversible nature of epigenetic changes, converting them to a “normal-like” chromatin landscape is feasible using “designed modular tools” [119]. The fundamental structure of editing tools includes a programmable DNA-binding domain and an epigenetic effector domain of interest. The zinc finger nucleases (ZFN), transcription activator-like effector (TALE), and nuclease-deficient Cas9 (dCas9) are three different programmable DNA-recognition domains which have been broadly used in epigenome editing, the most promising derivative technology of genome editing [120]. The epigenetic effectors are diverse, ranging from the writers, erasers, and readers, depositing, removing, or detecting DNA and histone modification marks as well as artificial transcription factors [121].
Epigenetic Editing Using CRISPR/dCas9
The clustered regularly interspaced short palindromic repeats (CRISPR) bacteria primitively used associated protein 9 (Cas9) system for their defense against bacteriophages. Recently this system is receiving remarkable attention by its rising role in the treatment of genetic disorders and cancer [122]. CRISPR system consists of an RNA-guided Cas9 endonuclease protein, which has the potential to be repurposed for editing both the genome and epigenome with significant efficiency [123]. This system can dramatically accelerate cancer research progress, either by screening for novel therapeutic targets, developing cancer therapies or by functional genome/epigenome editing. Therefore, it can be considered a robust weapon in the arsenal of future cancer treatment [122,123].
Two de novo DNA methyltransferases (Dnmt3a/b) are responsible for establishing DNA methylation in mammalian cells and Dnmt1 for its maintaining. Furthermore, demethylation is feasible through oxidation of the methyl group by TET (ten–eleven translocation) dioxygenases to build 5-hydroxymethylcytosine, and then its recovery into unmodified cytosine by whether DNA glycosylase-initiated base excision repair or DNA replication-dependent dilution [124].
It was demonstrated that fusion of mutant form of Cas9 without endonuclease activity known as dCas9, with the Dnmt3a or Tet1 enzyme could provide a potent tool for targeted erasure or establishment of DNA methylation, respectively. In 2016 Rudolf Jaenisch group successfully repurposed the CRISPR/Cas9 system to rearrange the targeted genomic sequences’ methylation status. dCas9 protein was fused either to the Dnmt3a (dCas9-Dnmt3a) or the catalytic domain of Tet1 (dCas9-Tet1) to predictably edit the epigenetic state of specific sequences [125].
To methylate genomic sites of interest by raising the local Dnmt3a concentration dCas9 protein was fused to repetitive peptide epitopes (SunTag), recruiting multiple copies of antibody-fused Dnmt3a (dCas9-SunTag-Dnmt3a). The authors reported that dCas9-SunTag-Dnmt3a can significantly boost CpG methylation at the HOXA5 locus in human embryonic kidney (HEK293T) cells and thus inhibit HOXA5 gene expression, not to mention its minimal impact on the global DNA methylome and transcriptome [126].
By gaining insight into the crucial role of molecular abnormalities, including epigenetic changes in cancer initiation, progression, and metastasis, there is a great demand for modern technologies to restore these aberrations. Therefore, in the coming years, the CRISPR-Cas system will play a notable role in cancer research. Repurposing this system to modulate the epigenome of cancer cells grant novel cancer therapies, albeit the employment of this platform, poses enormous challenges [123]. Taking together, dCas9-Dnmt3a/Tet1 could be an appropriate tool for implementing the new therapeutic strategies for UM (Figure 2) and other cancer types.
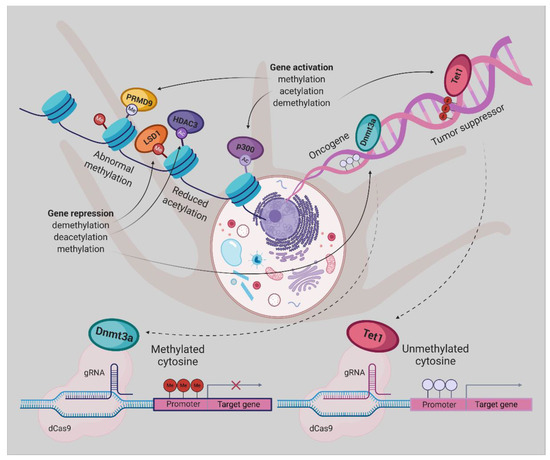
Figure 2.
Epigenetic editing by CRISPR/dCas9, allowing for locus-specific control of epigenetically regulated gene expression, provides a more specific alternative to epigenetic drugs. DNA methylation or histone modifications can be restored using dCas9 protein fused or non-covalently bound to epigenetic effectors, derived from writers or erasers. Gene expression has been activated by DNA demethylation using Tet1, H3K27 acetylation by p300, or H3K4 trimethylation by PRDM9. The antagonistic effect can be achieved either by promoter methylation employing Dnmt3a, removal of a methyl group from H3K4me1/2 and H3K9me2 by LSD1 or deacetylation of H3K27ac by HDAC3 [127].
6. Conclusions
The overall mortality rate of UM remains high, regardless of progress in diagnosis and the therapy of the primary tumor. Thus, there is an extreme necessity for further research into alternative and modern forms of prevention and treatment. Epigenetics and epigenomics are new emerging research fields that start to unfold an era of contemporary approaches to improve clinical treatment and decline metastasis risk in UM patients. Although epigenetic therapies have been evolved to treat many human diseases, including cancer for more than 30 years, they more notably have relied on drugs that ubiquitously altered epigenetic marks. Notwithstanding, these epigenetic drugs could have severe side effects, since off-target genes may be affected. Moreover, clinical studies reported their disappointing efficacy in UM patients, so far.
Therefore, the establishment of new methods for restoring epigenetic modifications associated with high-risk UM is to be noted. Their success will be determined by the biomarker-directed approach, combining a new generation of epigenetic drugs with traditional and new therapies and the implementation of personalized strategies in clinical trials. Moreover, modern technologies such as dCas9-Dnmt3a/Tet1 system have the potential to start a new era in the treatment of UM and other cancers.
Author Contributions
Conceptualization, B.S.; software, B.S.; writing—original draft preparation, P.C.B., writing—review and editing, Z.K., A.F.; visualization, B.S.; project administration, B.S., A.F., funding acquisition, B.S., A.F. All authors have read and agreed to the published version of the manuscript.
Funding
Projects APVV-17-0369 and KEGA 016UK-4/2018 funded this research.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the writing of the manuscript.
Abbreviations
| ADP | Adenosine diphosphate |
| BAP1 | BRCA1 associated protein 1 |
| BET | Bromodomain and Extra-Terminal motif |
| BETi | Bromodomain and Extra-Terminal motif inhibitor |
| BRAF | B-Raf Proto-Oncogene, Serine/Threonine Kinase |
| BRD2,3,4 | Bromodomain Containing 2,3,4 |
| BRDT | Bromodomain Testis Associated |
| Cas9 | (CRISPR) associated protein 9 |
| CCND2 | Cyclin D2 |
| CDK4 | Cyclin Dependent Kinase 4 |
| CDK6 | Cyclin Dependent Kinase 6 |
| -CH3 | Methyl group |
| CpG | Cytosines followed by guanine residues |
| COMS | Collaborative Ocular Melanoma Study |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| CTA | Cancer test antigens |
| CTLA-4 | Cytotoxic T-lymphocyte-associated antigen 4 |
| D3 | Disomy 3 |
| dCas9 | Nuclease-deficient Cas9 |
| DNA | Deoxyribonucleic acid |
| DNMT | DNA methyltransferase |
| DNMTi | DNMT inhibitor |
| DOT1Li | Histone H3K79 methyltransferase inhibitor |
| DSS1 | Deleted Split hand/Split foot 1 |
| EFS | Embryonal Fyn-Associated Substrate |
| EIF1AX | Eukaryotic Translation Initiation Factor 1A X-Linked |
| EHMT2 | Histone-lysine N-methyltransferase known as G9A |
| EZH2 | Enhancer of zeste homolog |
| GNAQ | G Protein Subunit Alpha Q |
| GNA11 | G Protein Subunit Alpha 11 |
| HAT | Histone acetyltransferase |
| HDAC | Histone deacetylase |
| HDACi | Histone deacetylase inhibitor |
| HDM | Histone demethylase |
| HDMi | Histone demethylase inhibitor |
| HIF1A | Hypoxia-inducible factor 1 alpha |
| HMT | Histone methyltransferase |
| HOXA5 | Homeobox A5 |
| H2A | Histone H2A |
| H3 | Histone 3 |
| H4 | Histone 4 |
| IDO1 | Indoleamine 2,3-Dioxygenase 1 |
| KDM1Ai | Lysine-specific histone demethylase 1A inhibitor |
| LSDi | Lysine-specific demethylase 1 inhibitor |
| MAPK | Mitogen-activated protein kinase |
| M3 | Monosomy 3 |
| Me | Methylation |
| MEK | Mitogen-activated protein kinase kinase |
| MITF | Melanocyte Inducing Transcription Factor |
| miRNA | MicroRNA |
| MYC | MYC Proto-Oncogene, BHLH Transcription Factor |
| NRAS | NRAS Proto-Oncogene, GTPase |
| OS | Overall survival |
| PD-1 | Programmed cell death-1 |
| PFS | Progression-free survival |
| PI3K | Phosphatidylinositol 3-kinase |
| PKC | Protein kinase C |
| PRMT | Protein arginine methyltransferase |
| PRMTi | Protein arginine methyltransferase inhibitor |
| PRAME | Preferentially expressed antigen in melanoma |
| p16, P16INK4A | Cyclin Dependent Kinase Inhibitor 2A |
| P14ARF | The alternate reading frame protein product of the CDKN2A locus |
| RASEF | RAS and EF-hand domain containing |
| RASSF1A | Ras association domain family member 1 |
| RNA | Ribonucleic acid |
| SAHA | Suberanilohydroxamic acid |
| SF3B1 | Splicing Factor 3b Subunit 1 |
| shRNA | Short hairpin RNA |
| TALE | Transcription activator-like effector |
| TET | Ten-eleven translocation protein |
| TIGIT | T-cell immunoreceptor with Ig and ITIM domains |
| TSA | Trichostatin A |
| TIMP3 | TIMP metallopeptidase inhibitor 3 |
| UM | Uveal melanoma |
| VEGF | Vascular endothelial growth factor |
| VPA | Valproic acid |
| ZBTB10 | Zinc finger and BTB domain containing 10 |
| ZFN | Zinc finger nuclease |
References
- Ramaiya, K.J.; Harbour, J.W. Current management of uveal melanoma. Expert Rev. Ophthalmol. 2007, 2, 939–946. [Google Scholar] [CrossRef]
- Jager, M.J.; Shields, C.L.; Cebulla, C.M.; Abdel-Rahman, M.H.; Grossniklaus, H.E.; Stern, M.-H.; Carvajal, R.D.; Belfort, R.N.; Jia, R.; Shields, J.A. Uveal melanoma. Nat. Rev. Dis. Primers 2020, 6, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Murray, T.G.; Boldt, H.C. Ocular Melanoma: Advances in Diagnostic and Therapeutic Strategies; Future Medicine: London, UK, 2014. [Google Scholar]
- Singh, A.D.; Turell, M.E.; Topham, A.K. Uveal melanoma: Trends in incidence, treatment, and survival. Ophthalmology 2011, 118, 1881–1885. [Google Scholar] [CrossRef] [PubMed]
- Blum, E.S.; Yang, J.; Komatsubara, K.M.; Carvajal, R.D. Clinical management of uveal and conjunctival melanoma. Oncology (Williston Park) 2016, 30, 29–43. [Google Scholar]
- Khoja, L.; Atenafu, E.; Suciu, S.; Leyvraz, S.; Sato, T.; Marshall, E.; Keilholz, U.; Zimmer, L.; Patel, S.; Piperno-Neumann, S. Meta-Analysis in metastatic uveal melanoma to determine progression free and overall survival benchmarks: An international rare cancers initiative (IRCI) ocular melanoma study. Ann. Oncol. 2019, 30, 1370–1380. [Google Scholar] [CrossRef]
- Shields, C.L.; Ganguly, A.; Bianciotto, C.G.; Turaka, K.; Tavallali, A.; Shields, J.A. Prognosis of uveal melanoma in 500 cases using genetic testing of fine-needle aspiration biopsy specimens. Ophthalmology 2011, 118, 396–401. [Google Scholar] [CrossRef]
- Van Raamsdonk, C.D.; Bezrookove, V.; Green, G.; Bauer, J.; Gaugler, L.; O’Brien, J.M.; Simpson, E.M.; Barsh, G.S.; Bastian, B.C. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 2009, 457, 599–602. [Google Scholar] [CrossRef]
- Van Raamsdonk, C.D.; Griewank, K.G.; Crosby, M.B.; Garrido, M.C.; Vemula, S.; Wiesner, T.; Obenauf, A.C.; Wackernagel, W.; Green, G.; Bouvier, N. Mutations in GNA11 in uveal melanoma. N. Engl. J. Med. 2010, 363, 2191–2199. [Google Scholar] [CrossRef]
- Versluis, M.; de Lange, M.J.; van Pelt, S.I.; Ruivenkamp, C.A.; Kroes, W.G.; Cao, J.; Jager, M.J.; Luyten, G.P.; van der Velden, P.A. Digital PCR validates 8q dosage as prognostic tool in uveal melanoma. PLoS ONE 2015, 10, e0116371. [Google Scholar] [CrossRef]
- Harbour, J.W.; Onken, M.D.; Roberson, E.D.; Duan, S.; Cao, L.; Worley, L.A.; Council, M.L.; Matatall, K.A.; Helms, C.; Bowcock, A.M. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 2010, 330, 1410–1413. [Google Scholar] [CrossRef]
- Kalirai, H.; Dodson, A.; Faqir, S.; Damato, B.; Coupland, S. Lack of BAP1 protein expression in uveal melanoma is associated with increased metastatic risk and has utility in routine prognostic testing. Br. J. Cancer 2014, 111, 1373–1380. [Google Scholar] [CrossRef] [PubMed]
- Onken, M.D.; Worley, L.A.; Char, D.H.; Augsburger, J.J.; Correa, Z.M.; Nudleman, E.; Aaberg, T.M., Jr.; Altaweel, M.M.; Bardenstein, D.S.; Finger, P.T. Collaborative Ocular Oncology Group report number 1: Prospective validation of a multi-gene prognostic assay in uveal melanoma. Ophthalmology 2012, 119, 1596–1603. [Google Scholar] [CrossRef] [PubMed]
- Field, M.G.; Harbour, J.W. Recent developments in prognostic and predictive testing in uveal melanoma. Curr. Opin. Ophthalmol. 2014, 25, 234. [Google Scholar] [CrossRef] [PubMed]
- Robertson, A.G.; Shih, J.; Yau, C.; Gibb, E.A.; Oba, J.; Mungall, K.L.; Hess, J.M.; Uzunangelov, V.; Walter, V.; Danilova, L. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell 2017, 32, 204–220. e215. [Google Scholar] [CrossRef]
- Sharma, A.; Stei, M.; Fröhlich, H.; Holz, F.; Loeffler, K.; Herwig-Carl, M. Genetic and epigenetic insights into uveal melanoma. Clin. Genet. 2018, 93, 952–961. [Google Scholar] [CrossRef]
- Jones, P.A.; Baylin, S.B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002, 3, 415–428. [Google Scholar] [CrossRef]
- Herlihy, N.; Dogrusöz, M.; Van Essen, T.H.; Harbour, J.W.; Van Der Velden, P.A.; Van Eggermond, M.C.; Haasnoot, G.W.; Van Den Elsen, P.J.; Jager, M.J. Skewed expression of the genes encoding epigenetic modifiers in high-risk uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2015, 56, 1447–1458. [Google Scholar] [CrossRef]
- Field, M.G.; Decatur, C.L.; Kurtenbach, S.; Gezgin, G.; Van Der Velden, P.A.; Jager, M.J.; Kozak, K.N.; Harbour, J.W. PRAME as an independent biomarker for metastasis in uveal melanoma. Clin. Cancer Res. 2016, 22, 1234–1242. [Google Scholar] [CrossRef]
- Field, M.G.; Durante, M.A.; Decatur, C.L.; Tarlan, B.; Oelschlager, K.M.; Stone, J.F.; Kuznetsov, J.; Bowcock, A.M.; Kurtenbach, S.; Harbour, J.W. Epigenetic reprogramming and aberrant expression of PRAME are associated with increased metastatic risk in Class 1 and Class 2 uveal melanomas. Oncotarget 2016, 7, 59209. [Google Scholar] [CrossRef]
- Venza, M.; Visalli, M.; Catalano, T.; Beninati, C.; Teti, D.; Venza, I. DSS1 promoter hypomethylation and overexpression predict poor prognosis in melanoma and squamous cell carcinoma patients. Hum. Pathol. 2017, 60, 137–146. [Google Scholar] [CrossRef]
- Portela, A.; Esteller, M. Epigenetic modifications and human disease. Nat. Biotechnol. 2010, 28, 1057–1068. [Google Scholar] [CrossRef] [PubMed]
- van der Velden, P.A.; Metzelaar-Blok, J.A.; Bergman, W.; Monique, H.; Hurks, H.; Frants, R.R.; Gruis, N.A.; Jager, M.J. Promoter hypermethylation: A common cause of reduced p16INK4a expression in uveal melanoma. Cancer Res. 2001, 61, 5303–5306. [Google Scholar] [PubMed]
- Van der Velden, P.A.; Zuidervaart, W.; Hurks, M.H.; Pavey, S.; Ksander, B.R.; Krijgsman, E.; Frants, R.R.; Tensen, C.P.; Willemze, R.; Jager, M.J. Expression profiling reveals that methylation of TIMP3 is involved in uveal melanoma development. Int. J. Cancer 2003, 106, 472–479. [Google Scholar] [CrossRef]
- Zeschnigk, M.; Tschentscher, F.; Lich, C.; Brandt, B.; Horsthemke, B.; Lohmann, D.R. Methylation analysis of several tumour suppressor genes shows a low frequency of methylation of CDKN2A and RARB in uveal melanomas. Int. J. Genom. 2003, 4, 329–336. [Google Scholar]
- Maat, W.; van der Velden, P.A.; Out-Luiting, C.; Plug, M.; Dirks-Mulder, A.; Jager, M.J.; Gruis, N.A. Epigenetic inactivation of RASSF1a in uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2007, 48, 486–490. [Google Scholar] [CrossRef][Green Version]
- Merhavi, E.; Cohen, Y.; Avraham, B.C.R.; Frenkel, S.; Chowers, I.; Pe’er, J.; Goldenberg-Cohen, N. Promoter methylation status of multiple genes in uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2007, 48, 4403–4406. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Moulin, A.P.; Clément, G.; Bosman, F.T.; Zografos, L.; Benhattar, J. Methylation of CpG island promoters in uveal melanoma. Br. J. Ophthalmol. 2008, 92, 281–285. [Google Scholar] [CrossRef]
- Calipel, A.; Abonnet, V.; Nicole, O.; Mascarelli, F.; Coupland, S.E.; Damato, B.; Mouriaux, F. Status of RASSF1A in uveal melanocytes and melanoma cells. Mol. Cancer Res. 2011, 9, 1187–1198. [Google Scholar] [CrossRef][Green Version]
- Pfeifer, G.P.; Yoon, J.-H.; Liu, L.; Tommasi, S.; Wilczynski, S.P.; Dammann, R. Methylation of the RASSF1A gene in human cancers. Biol. Chem. 2002, 383, 907–914. [Google Scholar] [CrossRef]
- Maat, W.; Beiboer, S.H.; Jager, M.J.; Luyten, G.P.; Gruis, N.A.; van der Velden, P.A. Epigenetic regulation identifies RASEF as a tumor-suppressor gene in uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2008, 49, 1291–1298. [Google Scholar] [CrossRef]
- Venza, M.; Visalli, M.; Beninati, C.; Biondo, C.; Teti, D.; Venza, I. Role of genetics and epigenetics in mucosal, uveal, and cutaneous melanomagenesis. Anti-Cancer Agents Med. Chem. (Former. Curr. Med. Chem. -Anti-Cancer Agents) 2016, 16, 528–538. [Google Scholar] [CrossRef] [PubMed]
- Field, M.G.; Kuznetsov, J.N.; Bussies, P.L.; Cai, L.Z.; Alawa, K.A.; Decatur, C.L.; Kurtenbach, S.; Harbour, J.W. BAP1 loss is associated with DNA methylomic repatterning in highly aggressive Class 2 uveal melanomas. Clin. Cancer Res. 2019, 25, 5663–5673. [Google Scholar] [CrossRef]
- Li, B.; Carey, M.; Workman, J.L. The role of chromatin during transcription. Cell 2007, 128, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Landreville, S.; Agapova, O.A.; Matatall, K.A.; Kneass, Z.T.; Onken, M.D.; Lee, R.S.; Bowcock, A.M.; Harbour, J.W. Histone deacetylase inhibitors induce growth arrest and differentiation in uveal melanoma. Clin. Cancer Res. 2012, 18, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Matatall, K.A.; Agapova, O.A.; Onken, M.D.; Worley, L.A.; Bowcock, A.M.; Harbour, J.W. BAP1 deficiency causes loss of melanocytic cell identity in uveal melanoma. BMC Cancer 2013, 13, 371. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, J.N.; Aguero, T.H.; Owens, D.A.; Kurtenbach, S.; Field, M.G.; Durante, M.A.; Rodriguez, D.A.; King, M.L.; Harbour, J.W. BAP1 regulates epigenetic switch from pluripotency to differentiation in developmental lineages giving rise to BAP1-mutant cancers. Sci. Adv. 2019, 5, eaax1738. [Google Scholar] [CrossRef]
- Deng, S.; Calin, G.A.; Croce, C.M.; Coukos, G.; Zhang, L. Mechanisms of microRNA deregulation in human cancer. Cell Cycle 2008, 7, 2643–2646. [Google Scholar] [CrossRef]
- Li, Y.; Jia, R.; Ge, S. Role of epigenetics in uveal melanoma. Int. J. Biol. Sci. 2017, 13, 426. [Google Scholar] [CrossRef]
- Worley, L.A.; Long, M.D.; Onken, M.D.; Harbour, J.W. Micro-RNAs associated with metastasis in uveal melanoma identified by multiplexed microarray profiling. Melanoma Res. 2008, 18, 184–190. [Google Scholar] [CrossRef]
- Radhakrishnan, A.; Badhrinarayanan, N.; Biswas, J.; Krishnakumar, S. Analysis of chromosomal aberration (1, 3, and 8) and association of microRNAs in uveal melanoma. Mol. Vis. 2009, 15, 2146. [Google Scholar]
- Venza, M.; Dell’Aversana, C.; Visalli, M.; Altucci, L.; Teti, D.; Venza, I. Identification of microRNA expression patterns in cutaneous and uveal melanoma cell lines. Tumori J. 2014, 100, 4–7. [Google Scholar] [CrossRef]
- Sun, Q.; Cong, R.; Yan, H.; Gu, H.; Zeng, Y.; Liu, N.; Chen, J.; Wang, B. Genistein inhibits growth of human uveal melanoma cells and affects microRNA-27a and target gene expression. Oncol. Rep. 2009, 22, 563–567. [Google Scholar] [PubMed]
- Chen, X.; He, D.; Da Dong, X.; Dong, F.; Wang, J.; Wang, L.; Tang, J.; Hu, D.-N.; Yan, D.; Tu, L. MicroRNA-124a is epigenetically regulated and acts as a tumor suppressor by controlling multiple targets in uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2013, 54, 2248–2256. [Google Scholar] [CrossRef]
- Chen, X.; Wang, J.; Shen, H.; Lu, J.; Li, C.; Hu, D.-N.; Da Dong, X.; Yan, D.; Tu, L. Epigenetics, microRNAs, and carcinogenesis: Functional role of microRNA-137 in uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2011, 52, 1193–1199. [Google Scholar] [CrossRef] [PubMed]
- Smolkova, B.; Horvathova, V.K.; Zmetakova, I.; Kalinkova, L.; Czanner, G.; Markova, A.; Furdova, A. Role of epigenetic deregulation in hematogenous dissemination of malignant uveal melanoma. Neoplasma 2018, 65, 840–854. [Google Scholar] [CrossRef]
- Li, Z.; Yu, X.; Shen, J.; Jiang, Y. MicroRNA dysregulation in uveal melanoma: A new player enters the game. Oncotarget 2015, 6, 4562. [Google Scholar] [CrossRef]
- Melia, M.; Moy, C.S.; Reynolds, S.M.; Hayman, J.A.; Murray, T.G.; Hovland, K.R.; Earle, J.D.; Kurinij, N.; Dong, L.M.; Miskala, P.H. Quality of life after iodine 125 brachytherapy vs enucleation for choroidal melanoma: 5-year results from the Collaborative Ocular Melanoma Study: COMS QOLS Report No. 3. Arch. Ophthalmol. (Chicago Ill.: 1960) 2006, 124, 226–238. [Google Scholar]
- Furdova, A.; Sramka, M.; Chorvath, M.; Kralik, G.; Furda, R.; Gregus, M. Clinical experience of stereotactic radiosurgery at a linear accelerator for intraocular melanoma. Melanoma Res. 2017, 27, 463–468. [Google Scholar] [CrossRef]
- Puusaari, I.; Damato, B.; Kivelä, T. Transscleral local resection versus iodine brachytherapy for uveal melanomas that are large because of tumour height. Graefes Arch. Clin. Exp. Ophthalmol. 2007, 245, 522–533. [Google Scholar] [CrossRef]
- Bechrakis, N.E.; Petousis, V.; Willerding, G.; Krause, L.; Wachtlin, J.; Stroux, A.; Foerster, M.H. Ten-Year results of transscleral resection of large uveal melanomas: Local tumour control and metastatic rate. Br. J. Ophthalmol. 2010, 94, 460–466. [Google Scholar] [CrossRef]
- Kivelä, T.; Puusaari, I.; Damato, B. Transscleral resection versus iodine brachytherapy for choroidal malignant melanomas 6 mm or more in thickness: A matched case–Control study. Ophthalmology 2003, 110, 2235–2244. [Google Scholar] [CrossRef] [PubMed]
- Furdova, A.; Sramka, M. Uveal Malignant Melanoma and Stereotactic Radiosurgery: Intraocular Uveal Melanoma and One-Day Session Stereotactic Radiosurgery at Linear Accelerator, 1st ed.; LAP LAMBERT Academic Publishing: Saarbrücken, Germany, 2014; p. 183. [Google Scholar]
- Furdova, A.; Strmen, P.; Waczulikova, I.; Chorvath, M.; Sramka, M.; Slezak, P. One-Day session LINAC–based stereotactic radiosurgery of posterior uveal melanoma. Eur. J. Ophthalmol. 2012, 22, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Damato, B. Progress in the management of patients with uveal melanoma. The 2012 Ashton Lecture. Eye 2012, 26, 1157–1172. [Google Scholar] [CrossRef] [PubMed]
- Sagoo, M.S.; Shields, C.L.; Mashayekhi, A.; Freire, J.; Emrich, J.; Reiff, J.; Komarnicky, L.; Shields, J.A. Plaque radiotherapy for juxtapapillary choroidal melanoma: Tumor control in 650 consecutive cases. Ophthalmology 2011, 118, 402–407. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.U.; Shields, C.L.; Bianciotto, C.G.; Iturralde, J.; Al-Dahmash, S.A.; Say, E.A.T.; Badal, J.; Mashayekhi, A.; Shields, J.A. Intravitreal bevacizumab at 4-month intervals for prevention of macular edema after plaque radiotherapy of uveal melanoma. Ophthalmology 2014, 121, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Mishra, K.K.; Quivey, J.M.; Daftari, I.K.; Weinberg, V.; Cole, T.B.; Patel, K.; Castro, J.R.; Phillips, T.L.; Char, D.H. Long-term Results of the UCSF-LBNL Randomized Trial: Charged Particle With Helium Ion Versus Iodine-125 Plaque Therapy for Choroidal and Ciliary Body Melanoma. Int. J. Radiat. Oncol. Biol. Phys. 2015, 92, 376–383. [Google Scholar] [CrossRef]
- Gragoudas, E.; Li, W.; Goitein, M.; Lane, A.M.; Munzenrider, J.E.; Egan, K.M. Evidence-based estimates of outcome in patients irradiated for intraocular melanoma. Arch. Ophthalmol. 2002, 120, 1665–1671. [Google Scholar] [CrossRef]
- Turcotte, S.; Bergeron, D.; Rousseau, A.P.; Mouriaux, F. Primary transpupillary thermotherapy for choroidal indeterminate melanocytic lesions. Can. J. Ophthalmol. 2014, 49, 464–467. [Google Scholar] [CrossRef]
- Binkley, E.; Triozzi, P.L.; Rybicki, L.; Achberger, S.; Aldrich, W.; Singh, A. A prospective trial of adjuvant therapy for high-Risk uveal melanoma: Assessing 5-Year survival outcomes. Br. J. Ophthalmol. 2020, 104, 524–528. [Google Scholar] [CrossRef]
- Piperno-Neumann, S.; Rodrigues, M.J.; Servois, V.; Pierron, G.; Gastaud, L.; Negrier, S.; Levy-Gabriel, C.; Lumbroso, L.; Cassoux, N.; Bidard, F.-C.; et al. A randomized multicenter phase 3 trial of adjuvant fotemustine versus surveillance in high risk uveal melanoma (UM) patients (FOTEADJ). J. Clin. Oncol. 2017, 35, 9502. [Google Scholar] [CrossRef]
- Voelter, V.; Schalenbourg, A.; Pampallona, S.; Peters, S.; Halkic, N.; Denys, A.; Goitein, G.; Zografos, L.; Leyvraz, S. Adjuvant intra-Arterial hepatic fotemustine for high-Risk uveal melanoma patients. Melanoma Res. 2008, 18, 220–224. [Google Scholar] [CrossRef] [PubMed]
- Lane, A.M.; Egan, K.M.; Harmon, D.; Holbrook, A.; Munzenrider, J.E.; Gragoudas, E.S. Adjuvant interferon therapy for patients with uveal melanoma at high risk of metastasis. Ophthalmology 2009, 116, 2206–2212. [Google Scholar] [CrossRef] [PubMed]
- McLean, I.W.; Berd, D.; Mastrangelo, M.J.; Shields, J.A.; Davidorf, F.H.; Grever, M.; Makley, T.A.; Gamel, J.W. A randomized study of methanol-Extraction residue of bacille Calmette-Guerin as postsurgical adjuvant therapy of uveal melanoma. Am. J. Ophthalmol. 1990, 110, 522–526. [Google Scholar] [CrossRef]
- Fountain, E.; Bassett, R.L.; Cain, S.; Posada, L.; Gombos, D.S.; Hwu, P.; Bedikian, A.; Patel, S.P. Adjuvant Ipilimumab in High-Risk Uveal Melanoma. Cancers 2019, 11, 152. [Google Scholar] [CrossRef] [PubMed]
- Valsecchi, M.E.; Orloff, M.; Sato, R.; Chervoneva, I.; Shields, C.L.; Shields, J.A.; Mastrangelo, M.J.; Sato, T. Adjuvant Sunitinib in High-Risk Patients with Uveal Melanoma: Comparison with Institutional Controls. Ophthalmology 2018, 125, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Carvajal, R.D.; Schwartz, G.K.; Tezel, T.; Marr, B.; Francis, J.H.; Nathan, P.D. Metastatic disease from uveal melanoma: Treatment options and future prospects. Br. J. Ophthalmol. 2017, 101, 38–44. [Google Scholar] [CrossRef]
- Algazi, A.P.; Tsai, K.K.; Shoushtari, A.N.; Munhoz, R.R.; Eroglu, Z.; Piulats, J.M.; Ott, P.A.; Johnson, D.B.; Hwang, J.; Daud, A.I. Clinical outcomes in metastatic uveal melanoma treated with PD-1 and PD-L1 antibodies. Cancer 2016, 122, 3344–3353. [Google Scholar] [CrossRef]
- Khoja, L.; Atenafu, E.G.; Joshua, A.M.; Group, I.R.C.s.I.-O.M. Meta-Analysis of phase II trials in metastatic uveal melanoma (MUM) to determine progression-free (PFS) and overall survival (OS) benchmarks for future phase II trials: An irci-ocular melanoma initiative. Am. Soc. Clin. Oncol. 2016, 34 (Suppl. 15), 9567. [Google Scholar] [CrossRef]
- Willson, J.K.; Albert, D.M.; Diener-West, M.; McCaffrey, L.; Moy, C.S.; Scully, R.E. Assessment of metastatic disease status at death in 435 patients with large choroidal melanoma in the collaborative ocular melanoma study (coms) coms report no. 15. Arch Ophthalmol. 2001, 119, 670–676. [Google Scholar]
- Spagnolo, F.; Grosso, M.; Picasso, V.; Tornari, E.; Pesce, M.; Queirolo, P. Treatment of metastatic uveal melanoma with intravenous fotemustine. Melanoma Res. 2013, 23, 196–198. [Google Scholar] [CrossRef]
- Augsburger, J.J.; Corrêa, Z.M.; Shaikh, A.H. Effectiveness of treatments for metastatic uveal melanoma. Am. J. Ophthalmol. 2009, 148, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Schmittel, A.; Schmidt-Hieber, M.; Martus, P.; Bechrakis, N.; Schuster, R.; Siehl, J.; Foerster, M.; Thiel, E.; Keilholz, U. A randomized phase II trial of gemcitabine plus treosulfan versus treosulfan alone in patients with metastatic uveal melanoma. Ann. Oncol. 2006, 17, 1826–1829. [Google Scholar] [CrossRef] [PubMed]
- Spagnolo, F.; Caltabiano, G.; Queirolo, P. Uveal melanoma. Cancer Treat. Rev. 2012, 38, 549–553. [Google Scholar] [CrossRef] [PubMed]
- Homsi, J.; Bedikian, A.Y.; Papadopoulos, N.E.; Kim, K.B.; Hwu, W.-J.; Mahoney, S.L.; Hwu, P. Phase 2 open-label study of weekly docosahexaenoic acid–paclitaxel in patients with metastatic uveal melanoma. Melanoma Res. 2010, 20, 507–510. [Google Scholar] [CrossRef] [PubMed]
- Najjar, Y.G.; Navrazhina, K.; Ding, F.; Bhatia, R.; Tsai, K.; Abbate, K.; Durden, B.; Eroglu, Z.; Bhatia, S.; Park, S.; et al. Ipilimumab plus nivolumab for patients with metastatic uveal melanoma: A multicenter, retrospective study. J. Immunother. Cancer 2020, 8, e000331. [Google Scholar] [CrossRef] [PubMed]
- Swaika, A.; Crozier, J.A.; Joseph, R.W. Vemurafenib: An evidence-based review of its clinical utility in the treatment of metastatic melanoma. Drug Des. Dev. Ther. 2014, 8, 775. [Google Scholar]
- Trinh, V.A.; Davis, J.E.; Anderson, J.E.; Kim, K.B. Dabrafenib therapy for advanced melanoma. Ann. Pharmacother. 2014, 48, 519–529. [Google Scholar] [CrossRef]
- Cruz, F.; Rubin, B.P.; Wilson, D.; Town, A.; Schroeder, A.; Haley, A.; Bainbridge, T.; Heinrich, M.C.; Corless, C.L. Absence of BRAF and NRAS mutations in uveal melanoma. Cancer Res. 2003, 63, 5761–5766. [Google Scholar]
- Park, J.J.; Diefenbach, R.J.; Byrne, N.; Kefford, R.; Long, G.V.; Scolyer, R.A.; Gray, E.; Carlino, M.S.; Rizos, H. Circulating tumor DNA (ctDNA) in patients (pts) with metastatic uveal melanoma (UM) treated with protein kinase C inhibitor (PKCi). J. Clin. Oncol. 2020, 38, e22054. [Google Scholar] [CrossRef]
- Carvajal, R.D.; Sosman, J.A.; Quevedo, J.F.; Milhem, M.M.; Joshua, A.M.; Kudchadkar, R.R.; Linette, G.P.; Gajewski, T.F.; Lutzky, J.; Lawson, D.H. Effect of selumetinib vs chemotherapy on progression-Free survival in uveal melanoma: A randomized clinical trial. Jama 2014, 311, 2397–2405. [Google Scholar] [CrossRef]
- Patel, S.P.; Kim, K.B.; Papadopoulos, N.E.; Hwu, W.-J.; Hwu, P.; Prieto, V.G.; Bar-Eli, M.; Zigler, M.; Dobroff, A.; Bronstein, Y. A phase II study of gefitinib in patients with metastatic melanoma. Melanoma Res. 2011, 21, 357. [Google Scholar] [CrossRef] [PubMed]
- Zeldis, J.; Heller, C.; Seidel, G.; Yuldasheva, N.; Stirling, D.; Shutack, Y.; Libutti, S. A randomized phase II trial comparing two doses of lenalidomide for the treatment of stage IV ocular melanoma. J. Clin. Oncol. 2009, 27, e20012. [Google Scholar]
- Reiriz, A.B.; Richter, M.F.; Fernandes, S.; Cancela, A.I.; Costa, T.D.; Di Leone, L.P.; Schwartsmann, G. Phase II study of thalidomide in patients with metastatic malignant melanoma. Melanoma Res. 2004, 14, 527–531. [Google Scholar] [CrossRef] [PubMed]
- Guenterberg, K.D.; Grignol, V.P.; Relekar, K.V.; Varker, K.A.; Chen, H.X.; Kendra, K.L.; Olencki, T.E.; Carson III, W.E. A pilot study of bevacizumab and interferon-α2b in ocular melanoma. Am. J. Clin. Oncol. 2011, 34, 87. [Google Scholar] [CrossRef]
- Piperno-Neumann, S.; Servois, V.; Bidard, F.-C.; Mariani, P.; Plancher, C.; Diallo, A.; Vago-Ady, N.; Desjardins, L. BEVATEM: Phase II study of bevacizumab (B) in combination with temozolomide (T) in patients (pts) with first-line metastatic uveal melanoma (MUM): Final results. Am. Soc. Clin. Oncol. 2013, 31 (Suppl. 15), 9057. [Google Scholar] [CrossRef]
- Tarhini, A.A.; Frankel, P.; Margolin, K.A.; Christensen, S.; Ruel, C.; Shipe-Spotloe, J.; Gandara, D.R.; Chen, A.; Kirkwood, J.M. Aflibercept (VEGF Trap) in inoperable stage III or stage iv melanoma of cutaneous or uveal origin. Clin. Cancer Res. 2011, 17, 6574–6581. [Google Scholar] [CrossRef]
- Penel, N.; Delcambre, C.; Durando, X.; Clisant, S.; Hebbar, M.; Negrier, S.; Fournier, C.; Isambert, N.; Mascarelli, F.; Mouriaux, F. O-Mel-Inib: A Cancero-Pole Nord-Ouest multicenter phase II trial of high-Dose imatinib mesylate in metastatic uveal melanoma. Investig. New Drugs 2008, 26, 561–565. [Google Scholar] [CrossRef]
- Hofmann, U.B.; Kauczok-Vetter, C.S.; Houben, R.; Becker, J.C. Overexpression of the KIT/SCF in uveal melanoma does not translate into clinical efficacy of imatinib mesylate. Clin. Cancer Res. 2009, 15, 324–329. [Google Scholar] [CrossRef]
- Bhatia, S.; Moon, J.; Margolin, K.A.; Weber, J.S.; Lao, C.D.; Othus, M.; Aparicio, A.M.; Ribas, A.; Sondak, V.K. Phase II trial of sorafenib in combination with carboplatin and paclitaxel in patients with metastatic uveal melanoma: SWOG S0512. PLoS ONE 2012, 7, e48787. [Google Scholar] [CrossRef]
- Mahipal, A.; Tijani, L.; Chan, K.; Laudadio, M.; Mastrangelo, M.J.; Sato, T. A pilot study of sunitinib malate in patients with metastatic uveal melanoma. Melanoma Res. 2012, 22, 440–446. [Google Scholar] [CrossRef]
- Baylin, S.B.; Herman, J.G. DNA hypermethylation in tumorigenesis: Epigenetics joins genetics. Trends Genet. 2000, 16, 168–174. [Google Scholar] [CrossRef]
- Morel, D.; Almouzni, G.; Soria, J.-C.; Postel-Vinay, S. Targeting chromatin defects in selected solid tumors based on oncogene addiction, synthetic lethality and epigenetic antagonism. Ann. Oncol. 2017, 28, 254–269. [Google Scholar] [CrossRef] [PubMed]
- Klisovic, D.D.; Katz, S.E.; Effron, D.; Klisovic, M.I.; Wickham, J.; Parthun, M.R.; Guimond, M.; Marcucci, G. Depsipeptide (FR901228) inhibits proliferation and induces apoptosis in primary and metastatic human uveal melanoma cell lines. Investig. Ophthalmol. Vis. Sci. 2003, 44, 2390–2398. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Klisovic, D.D.; Klisovic, M.I.; Effron, D.; Liu, S.; Marcucci, G.; Katz, S.E. Depsipeptide inhibits migration of primary and metastatic uveal melanoma cell lines in vitro: A potential strategy for uveal melanoma. Melanoma Res. 2005, 15, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Zhou, J.; Jin, B.; Pan, J. Class III-specific HDAC inhibitor Tenovin-6 induces apoptosis, suppresses migration and eliminates cancer stem cells in uveal melanoma. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef]
- Jansen, Y.J.; Verset, G.; Schats, K.; Van Dam, P.-J.; Seremet, T.; Kockx, M.; Van Laethem, J.-L.B.; Neyns, B. Phase I clinical trial of decitabine (5-aza-2′-deoxycytidine) administered by hepatic arterial infusion in patients with unresectable liver-predominant metastases. ESMO Open 2019, 4, e000464. [Google Scholar] [CrossRef]
- Gonçalves, J.; Emmons, M.F.; Faião-Flores, F.; Aplin, A.E.; Harbour, J.W.; Licht, J.D.; Wink, M.R.; Smalley, K.S. Decitabine limits escape from MEK inhibition in uveal melanoma. Pigment Cell Melanoma Res. 2020, 33, 507–514. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, M.; Jin, Y.; Jiang, S.; Pan, J. In vitro and in vivo anti-uveal melanoma activity of JSL-1, a novel HDAC inhibitor. Cancer Lett. 2017, 400, 47–60. [Google Scholar] [CrossRef]
- Venza, M.; Visalli, M.; Biondo, C.; Lentini, M.; Catalano, T.; Teti, D.; Venza, I. Epigenetic regulation of p14ARF and p16INK4A expression in cutaneous and uveal melanoma. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2015, 1849, 247–256. [Google Scholar] [CrossRef]
- Delmore, J.E.; Issa, G.C.; Lemieux, M.E.; Rahl, P.B.; Shi, J.; Jacobs, H.M.; Kastritis, E.; Gilpatrick, T.; Paranal, R.M.; Qi, J. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011, 146, 904–917. [Google Scholar] [CrossRef]
- Ambrosini, G.; Sawle, A.D.; Musi, E.; Schwartz, G.K. BRD4-targeted therapy induces Myc-independent cytotoxicity in Gnaq/11-mutatant uveal melanoma cells. Oncotarget 2015, 6, 33397. [Google Scholar] [CrossRef] [PubMed]
- Plimack, E.R.; Stewart, D.J.; Issa, J.P. Combining epigenetic and cytotoxic therapy in the treatment of solid tumors. J. Clin. Oncol. 2007, 25, 4519–4521. [Google Scholar] [CrossRef] [PubMed]
- Coral, S.; Sigalotti, L.; Covre, A.; Nicolay, H.J.; Natali, P.G.; Maio, M. 5-AZA-2′-deoxycytidine in cancer immunotherapy: A mouse to man story. Cancer Res. 2007, 67, 2900–2901; author reply 2901–2902. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bi, G.; Jiang, G. The molecular mechanism of HDAC inhibitors in anticancer effects. Cell. Mol. Immunol. 2006, 3, 285–290. [Google Scholar]
- Xu, W.; Parmigiani, R.; Marks, P. Histone deacetylase inhibitors: Molecular mechanisms of action. Oncogene 2007, 26, 5541–5552. [Google Scholar] [CrossRef]
- Garmpis, N.; Damaskos, C.; Garmpi, A.; Dimitroulis, D.; Spartalis, E.; Margonis, G.-A.; Schizas, D.; Deskou, I.; Doula, C.; Magkouti, E. Targeting histone deacetylases in malignant melanoma: A future therapeutic agent or just great expectations? Anticancer Res. 2017, 37, 5355–5362. [Google Scholar] [PubMed]
- Moschos, M.M.; Dettoraki, M.; Androudi, S.; Kalogeropoulos, D.; Lavaris, A.; Garmpis, N.; Damaskos, C.; Garmpi, A.; Tsatsos, M. The role of histone deacetylase inhibitors in uveal melanoma: Current evidence. Anticancer Res. 2018, 38, 3817–3824. [Google Scholar] [CrossRef]
- Tan, J.; Cang, S.; Ma, Y.; Petrillo, R.L.; Liu, D. Novel histone deacetylase inhibitors in clinical trials as anti-cancer agents. J. Hematol. Oncol. 2010, 3, 5. [Google Scholar] [CrossRef]
- Onken, M.D.; Worley, L.A.; Tuscan, M.D.; Harbour, J.W. An accurate, clinically feasible multi-gene expression assay for predicting metastasis in uveal melanoma. J. Mol. Diagn. 2010, 12, 461–468. [Google Scholar] [CrossRef]
- Haas, N.; Quirt, I.; Hotte, S.; McWhirter, E.; Polintan, R.; Litwin, S.; Adams, P.; McBryan, T.; Wang, L.; Martin, L. Phase II trial of vorinostat in advanced melanoma. Investig. New Drugs 2014, 32, 526–534. [Google Scholar] [CrossRef]
- Venza, M.; Visalli, M.; Catalano, T.; Beninati, C.; Teti, D.; Venza, I. Epidrugs in the immunotherapy of cutaneous and uveal melanoma. Anti-Cancer Agents Med. Chem. (Former. Curr. Med. Chem. -Anti-Cancer Agents) 2017, 17, 190–205. [Google Scholar] [CrossRef] [PubMed]
- Morel, D.; Jeffery, D.; Aspeslagh, S.; Almouzni, G.; Postel-Vinay, S. Combining epigenetic drugs with other therapies for solid tumours—Past lessons and future promise. Nat. Rev. Clin. Oncol. 2019, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Leon-Ferre, R.; Laux, D.; Deutsch, J.; Smith, B.J.; Frees, M.; Milhem, M. Treatment of resistant metastatic melanoma using sequential epigenetic therapy (decitabine and panobinostat) combined with chemotherapy (temozolomide). Cancer Chemother. Pharmacol. 2014, 74, 691–697. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.-L.; Tian, M.; Li, X.; Li, J.-J.; Huang, J.; Ouyang, L.; Zhang, Y.; Liu, B. Inhibition of BET bromodomains as a therapeutic strategy for cancer drug discovery. Oncotarget 2015, 6, 5501. [Google Scholar] [CrossRef] [PubMed]
- Filippakopoulos, P.; Qi, J.; Picaud, S.; Shen, Y.; Smith, W.B.; Fedorov, O.; Morse, E.M.; Keates, T.; Hickman, T.T.; Felletar, I. Selective inhibition of BET bromodomains. Nature 2010, 468, 1067–1073. [Google Scholar] [CrossRef]
- Lovén, J.; Hoke, H.A.; Lin, C.Y.; Lau, A.; Orlando, D.A.; Vakoc, C.R.; Bradner, J.E.; Lee, T.I.; Young, R.A. Selective inhibition of tumor oncogenes by disruption of super-Enhancers. Cell 2013, 153, 320–334. [Google Scholar] [CrossRef]
- Strub, T.; Ballotti, R.; Bertolotto, C. The “ART” of Epigenetics in Melanoma: From histone “Alterations, to Resistance and Therapies”. Theranostics 2020, 10, 1777. [Google Scholar] [CrossRef]
- Nakade, S.; Yamamoto, T.; Sakuma, T. Cancer induction and suppression with transcriptional control and epigenome editing technologies. J. Hum. Genet. 2018, 63, 187–194. [Google Scholar] [CrossRef]
- Allis, C.D.; Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016, 17, 487. [Google Scholar] [CrossRef]
- Akram, F.; Haq, I.U.; Ahmed, Z.; Khan, H.; Ali, M. CRISPR-Cas9, A Promising Therapeutic Tool for Cancer Therapy: A Review. Protein Pept. Lett. 2020. [Google Scholar] [CrossRef]
- Moses, C.; Garcia-Bloj, B.; Harvey, A.R.; Blancafort, P. Hallmarks of cancer: The CRISPR generation. Eur. J. Cancer 2018, 93, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhang, Y. Reversing DNA methylation: Mechanisms, genomics, and biological functions. Cell 2014, 156, 45–68. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.S.; Wu, H.; Ji, X.; Stelzer, Y.; Wu, X.; Czauderna, S.; Shu, J.; Dadon, D.; Young, R.A.; Jaenisch, R. Editing DNA methylation in the mammalian genome. Cell 2016, 167, 233–247. e217. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-H.; Su, J.; Lei, Y.; Brunetti, L.; Gundry, M.C.; Zhang, X.; Jeong, M.; Li, W.; Goodell, M.A. DNA epigenome editing using CRISPR-Cas SunTag-directed DNMT3A. Genome Biol. 2017, 18, 176. [Google Scholar] [CrossRef] [PubMed]
- Syding, L.A.; Nickl, P.; Kasparek, P.; Sedlacek, R. CRISPR/Cas9 Epigenome Editing Potential for Rare Imprinting Diseases: A Review. Cells 2020, 9, 993. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).