Structural Analysis of the SANT/Myb Domain of FLASH and YARP Proteins and Their Complex with the C-Terminal Fragment of NPAT by NMR Spectroscopy and Computer Simulations
Abstract
1. Introduction
2. Results
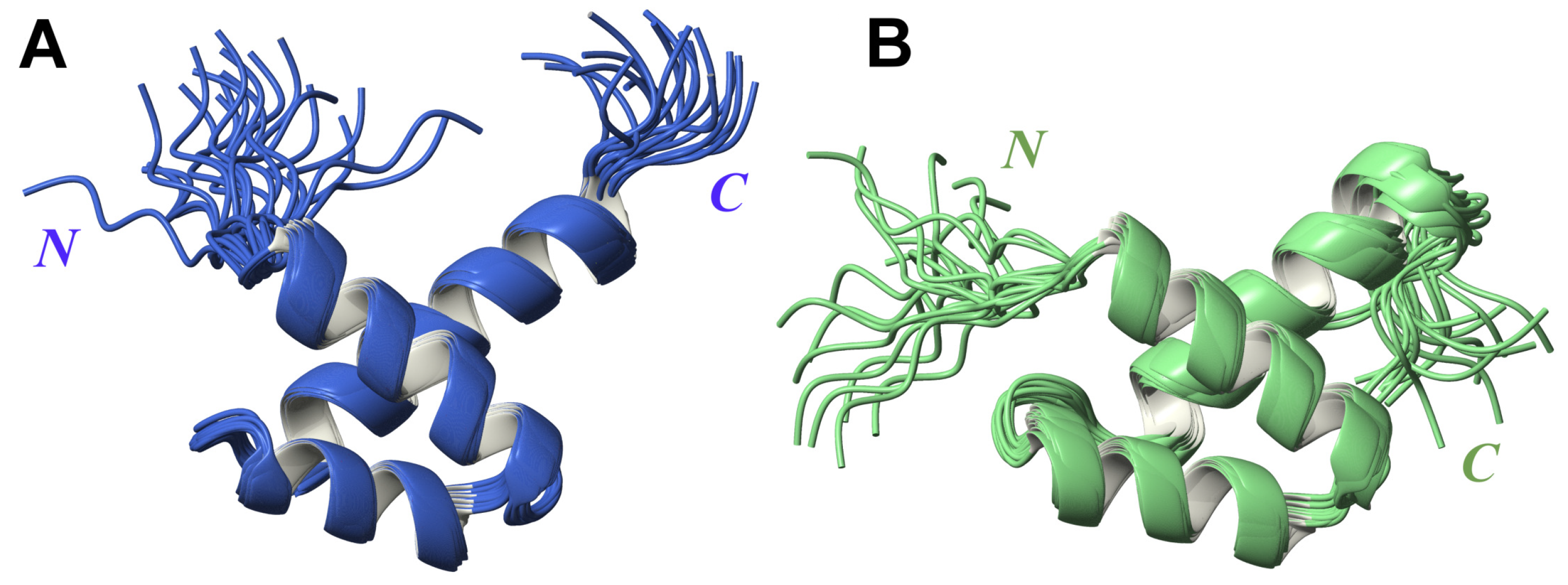
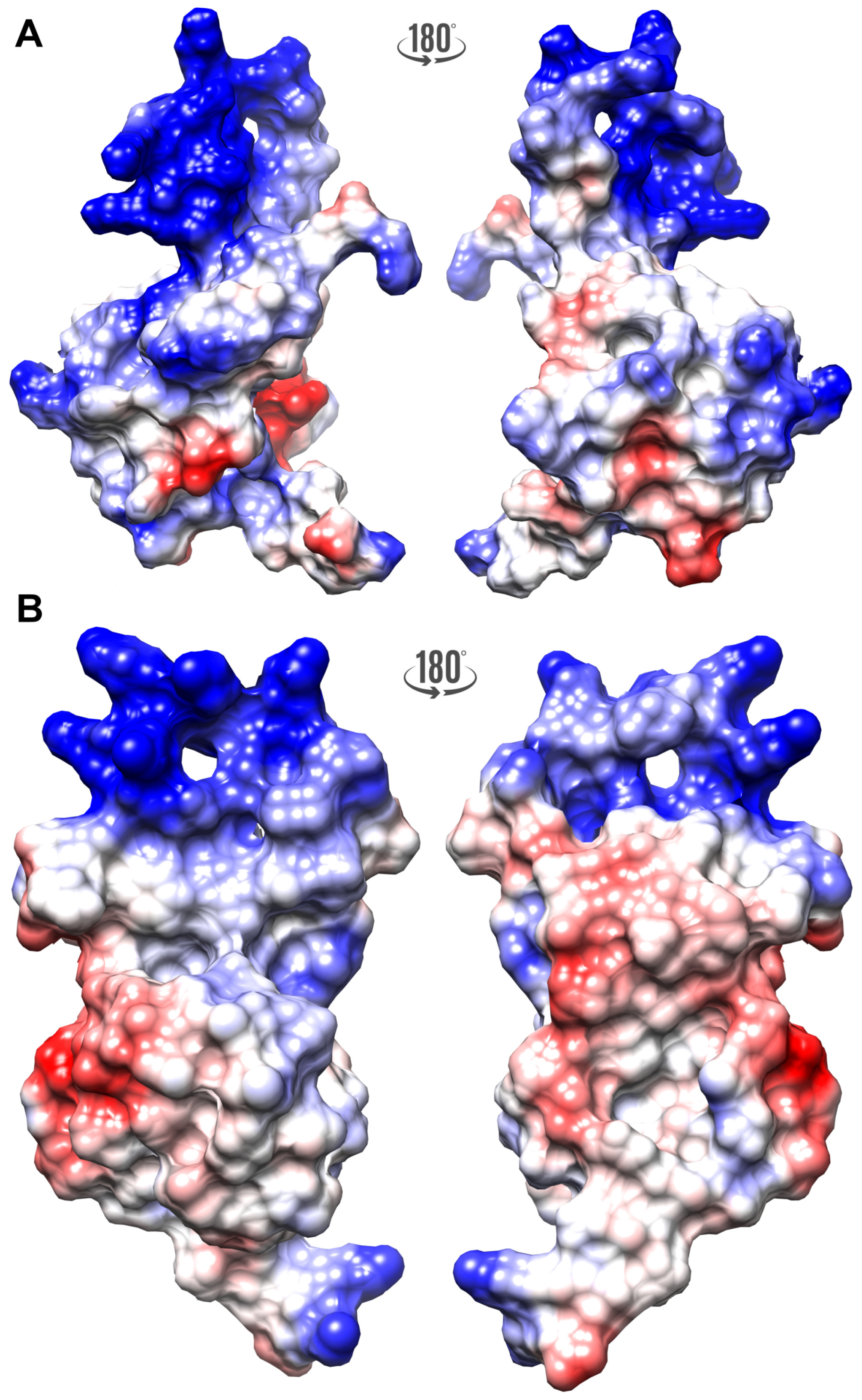
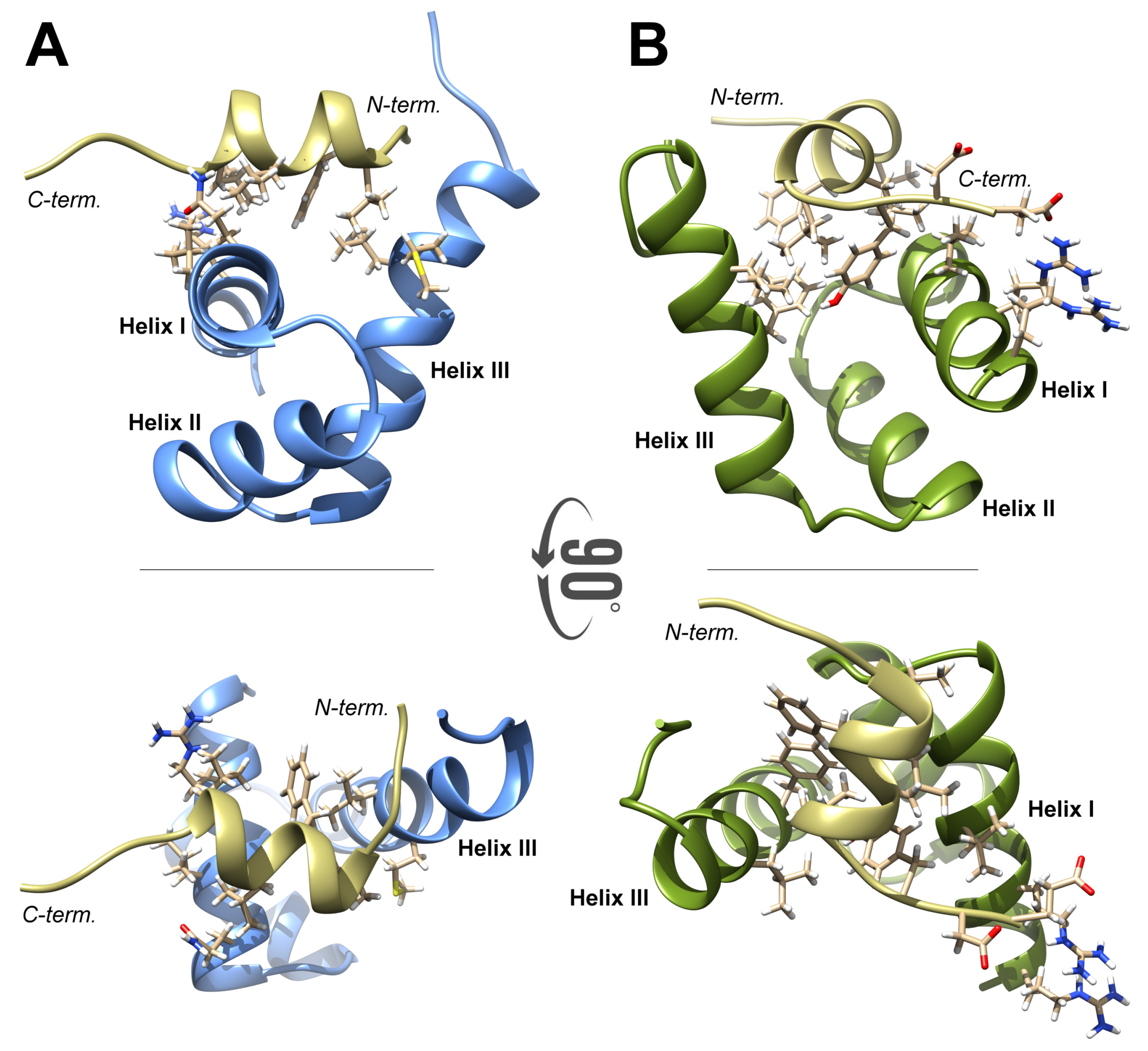
2.1. High-Resolution 3D Structure of FLASH and YARP Fragments
2.2. Backbone Dynamics of the FLASH and YARP Fragments Derived from N Relaxation Data
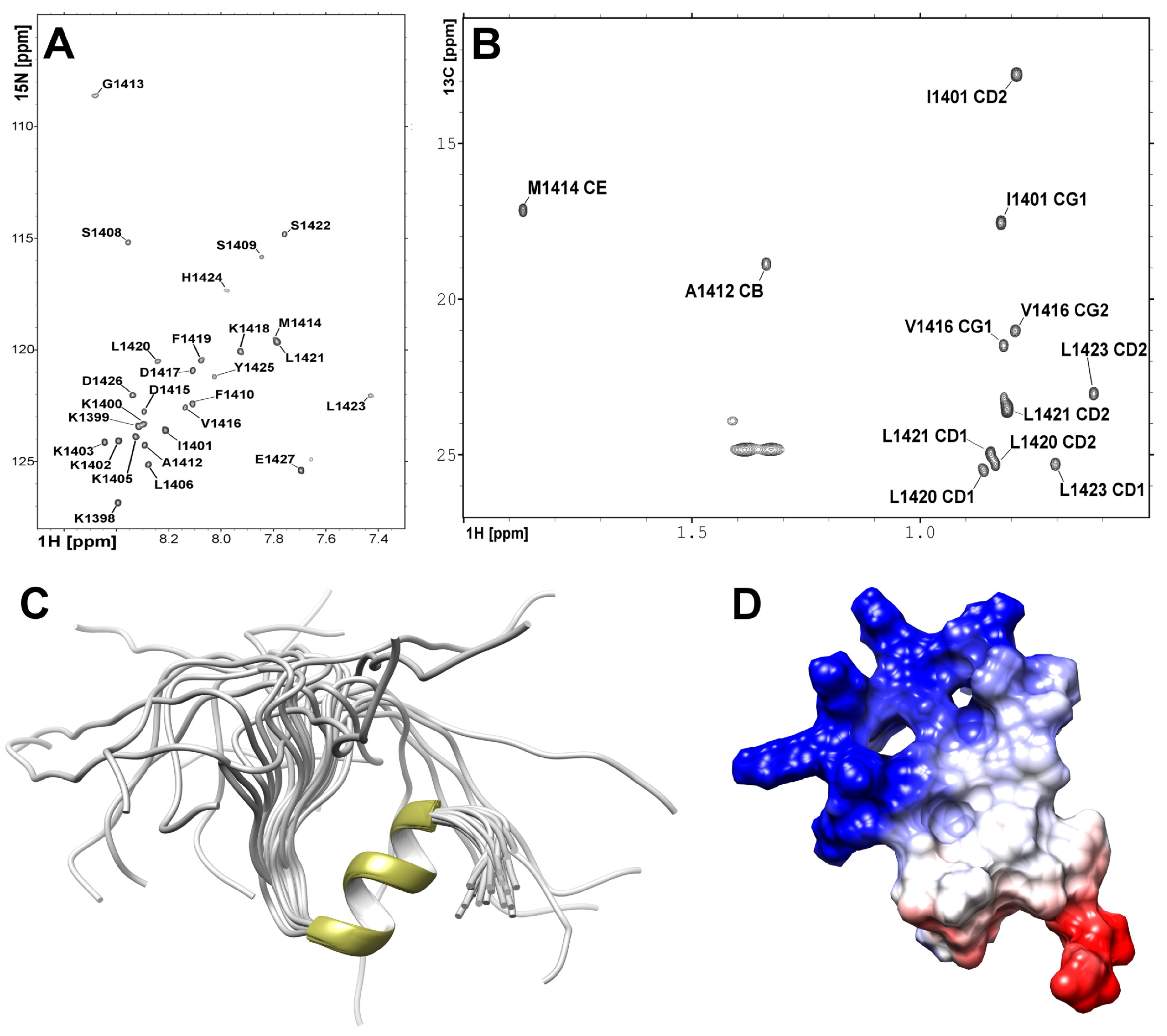
2.3. Structure Analysis of the NPAT Peptide in Solution
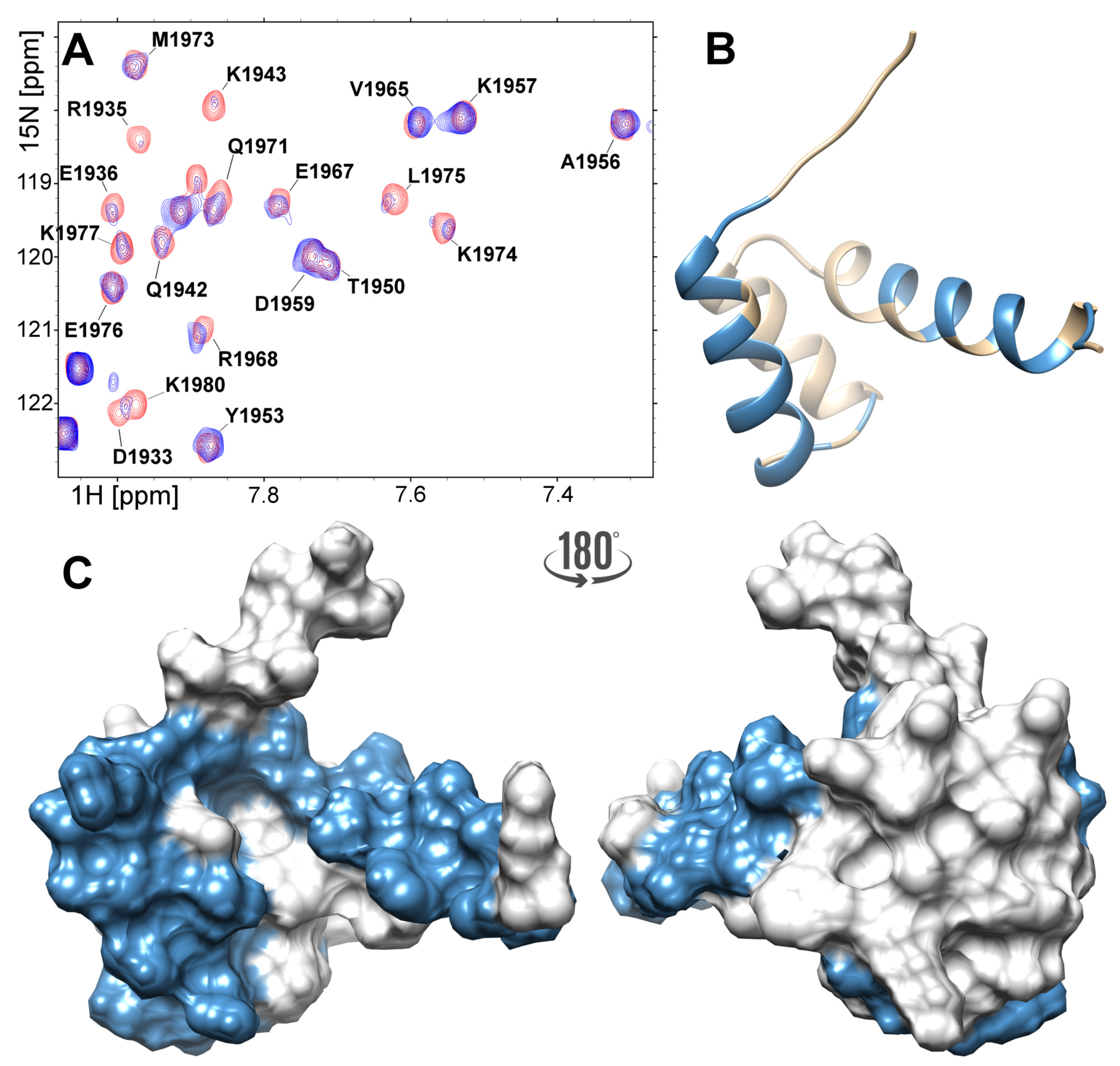
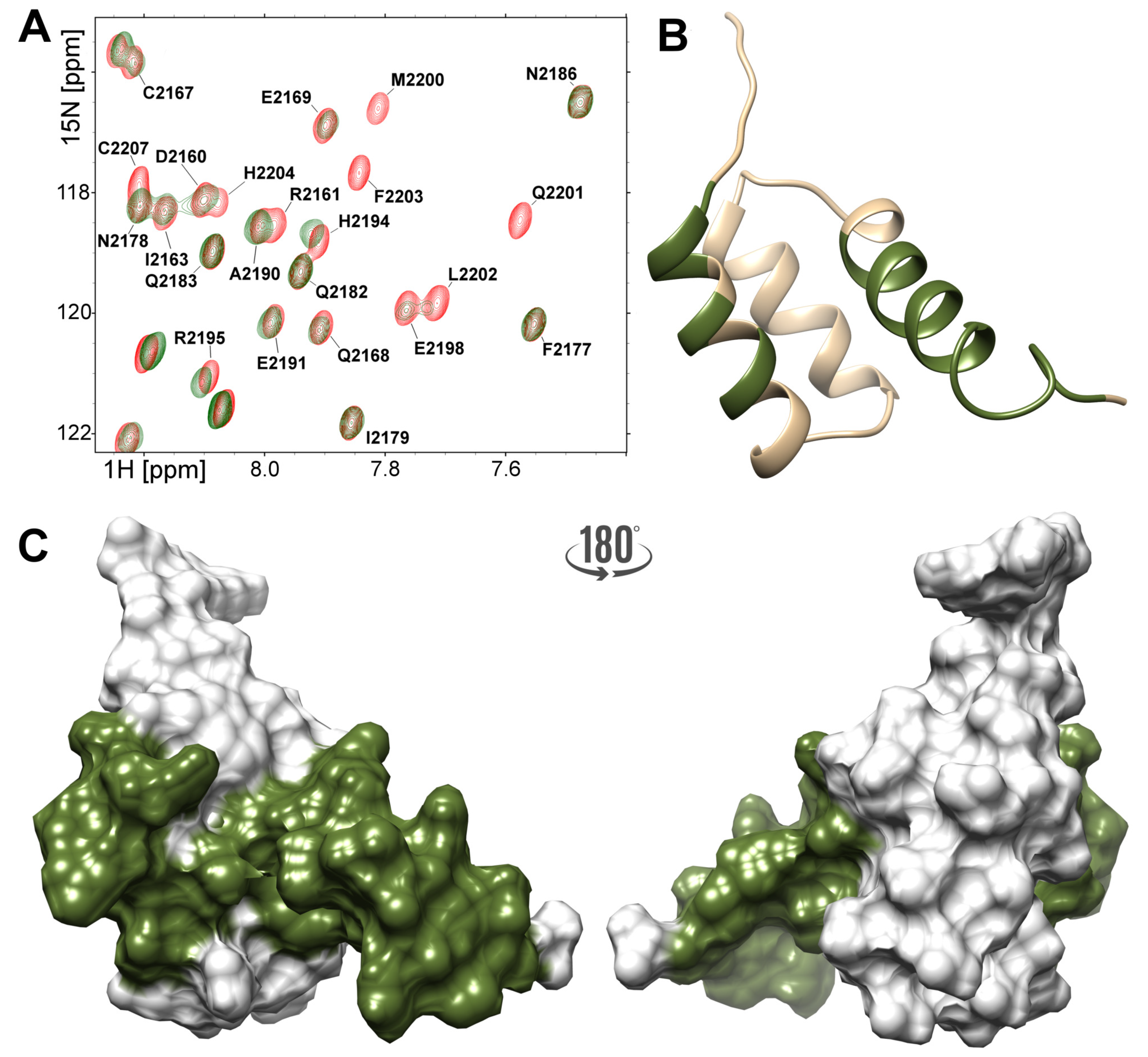
2.4. Interactions of the NPAT Peptide with FLASH and YARP Fragments
2.5. Modeling of FLASH-NPAT and YARP-NPAT Complexes
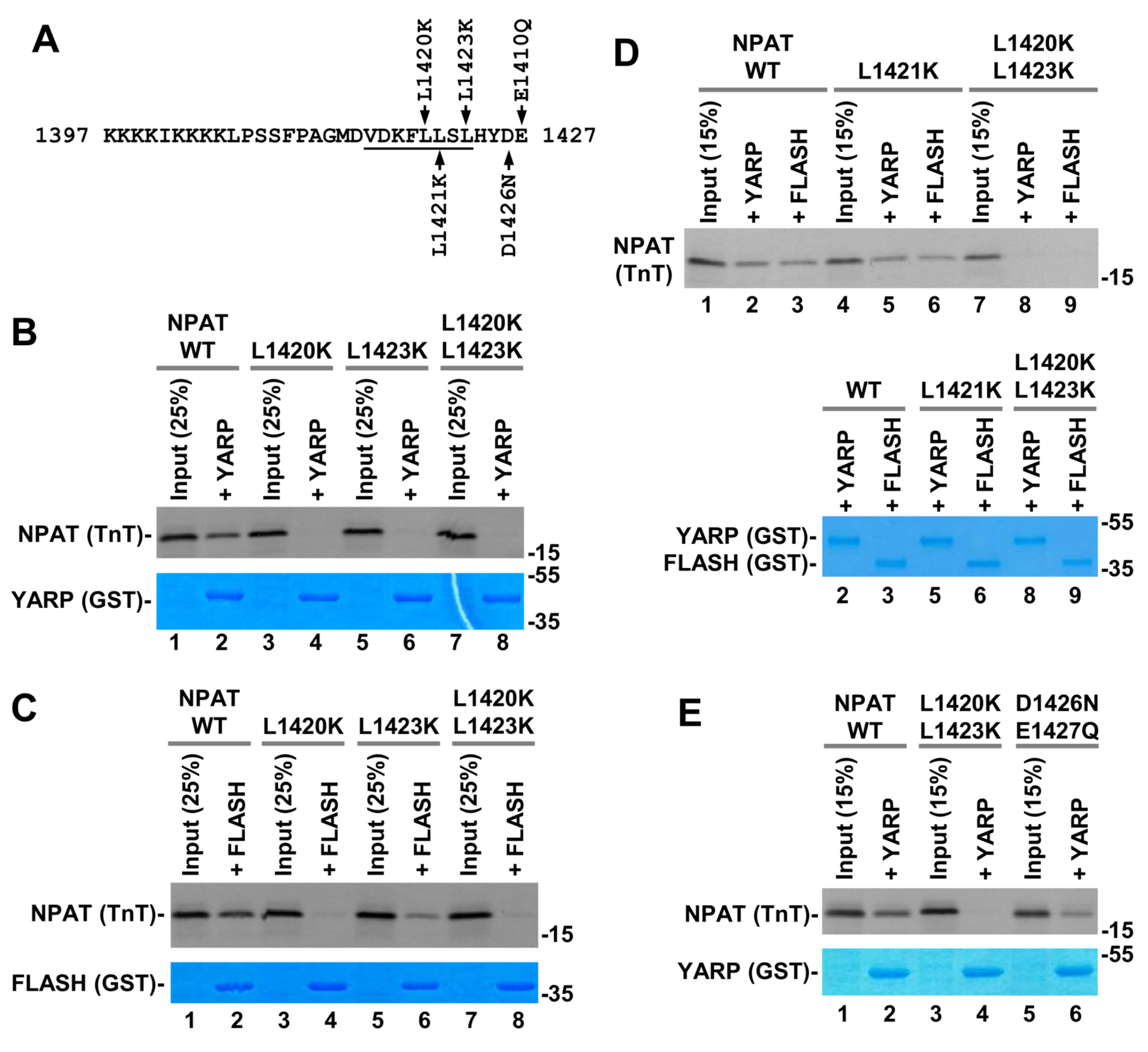
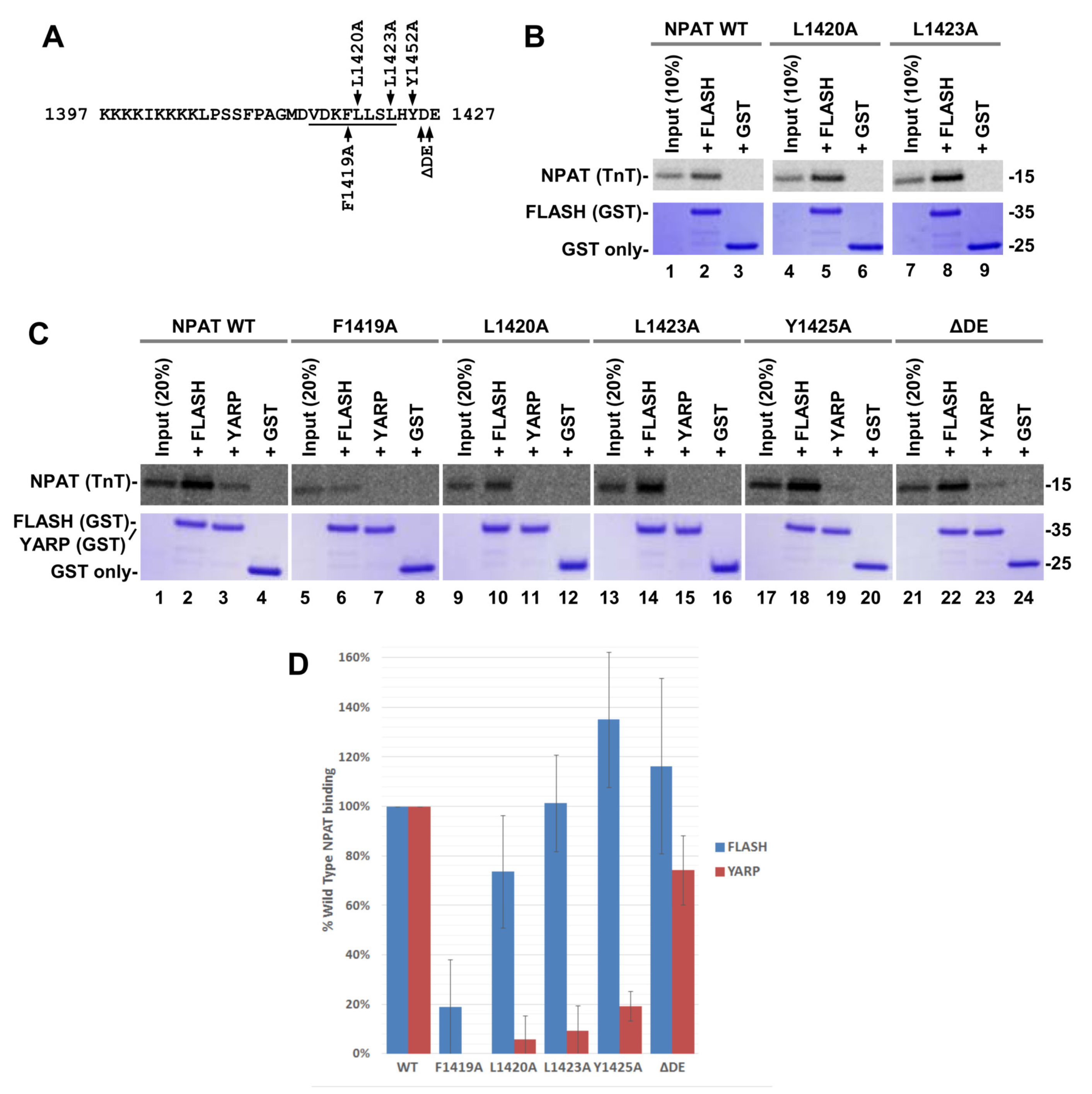
2.6. In Vitro Binding
2.7. Can FLASH and YARP Bind DNA?
3. Discussion
3.1. FLASH, YARP and Their Complexes with NPAT
3.2. Myb and SANT Domains Are Multi-Functional Binding Modules
3.3. FLASH-NPAT Complex as a Platform That Recognizes Histone Gene Clusters
4. Materials and Methods
4.1. Expression and Purification of the C-Terminal Fragments of FLASH and YARP for NMR Studies
4.2. Synthesis and Purification of the C-terminal NPAT Peptide
4.3. Multidimensional NMR Spectroscopy
4.4. Evaluation of High-Resolution 3D Structures of the FLASH, YARP, and NPAT Fragments
4.5. N Relaxation of the Backbone Amide Groups
4.6. Solution 3D Structures of the FLASH-NPAT and YARP-NPAT Complexes
4.7. Modeling the Interactions of FLASH–NPAT and YARP–NPAT Complexes with DNA
4.8. GST Pull-Down Assay
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| FLASH | FLICE-associated huge protein |
| YARP | Yin Yang 1-Associated Protein-Related Protein |
| NPAT | Nuclear Protein, Ataxia-Telangiectasia Locus |
| HLB | histone locus bodies |
| DBD | DNA binding domain |
| SANT | (SWI3, ADA2, N-CoR and TFIIIB B) domain |
| MYB | MYeloBlastosis DNA-binding domain |
| snRNP | small nuclear RiboNucleoProteins |
| SMRT | Silencing Mediator of Retinoid acid and Thyroid hormone receptor |
| HDAC | Histone Deacetylases |
| NOE | Nuclear Overhauser Effect |
| SDM | Spectral Density Mapping |
| HSQC | Heteronuclear Single-Quantum Correlation |
| CSP | Chemical Shifts Perturbations |
| DSS | sodium 2,2-dimethyl-2-silapentane-5-sulfonate |
| NPT | Isothermal–isobaric ensemble |
| SEC | Size Exclusion Chromatography |
| HPLC | High-Performance Liquid Chromatography |
| RP-HPLC | Reverse Phase High-Performance Liquid Chromatography |
| MALDI-TOF MS | Matrix-Assisted Laser Desorption/Ionization Time-Of-Flight Mass Spectrometry |
References
- Osley, M. The regulation of histone synthesis in the cell cycle. Annu. Rev. Biochem. 1991, 60, 827–861. [Google Scholar] [CrossRef]
- Heintz, N. The regulation of histone gene expression during the cell cycle. Biochim. Biophys. Acta (BBA)-Gene Struct. Expr. 1991, 1088, 327–339. [Google Scholar] [CrossRef]
- Stein, G.S.; Stein, J.L.; Van Wijnen, A.J.; Lian, J.B. Transcriptional control of cell cycle progression: The histone gene is a paradigm for the G1/S phase and proliferation/differentiation transitions. Cell Biol. Int. 1996, 20, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Marzluff, W.F. Metazoan replication-dependent histone mRNAs: A distinct set of RNA polymerase II transcripts. Curr. Opin. Cell Biol. 2005, 17, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Marzluff, W.F.; Duronio, R.J. Histone mRNA expression: Multiple levels of cell cycle regulation and important developmental consequences. Curr. Opin. Cell Biol. 2002, 14, 692–699. [Google Scholar] [CrossRef]
- Zhao, J.; Kennedy, B.K.; Lawrence, B.D.; Barbie, D.A.; Matera, A.G.; Fletcher, J.A.; Harlow, E. NPAT links cyclin E—Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 2000, 14, 2283–2297. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Van Tine, B.A.; Wei, Y.; Garrett, M.D.; Nelson, D.; Adams, P.D.; Wang, J.; Qin, J.; Chow, L.T.; Harper, J.W. Cell cycle—Regulated phosphorylation of p220NPAT by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 2000, 14, 2298–2313. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Dynlacht, B.; Imai, T.; Hori, T.A.; Harlow, E. Expression of NPAT, a novel substrate of cyclin E–CDK2, promotes S-phase entry. Genes Dev. 1998, 12, 456–461. [Google Scholar] [CrossRef]
- DeRan, M.; Pulvino, M.; Greene, E.; Su, C.; Zhao, J. Transcriptional activation of histone genes requires NPAT-dependent recruitment of TRRAP-Tip60 complex to histone promoters during the G1/S phase transition. Mol. Cell. Biol. 2008, 28, 435–447. [Google Scholar] [CrossRef]
- Adams, P.D. Regulation of the retinoblastoma tumor suppressor protein by cyclin/cdks. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2001, 1471, M123–M133. [Google Scholar] [CrossRef]
- Rubin, S.M. Deciphering the retinoblastoma protein phosphorylation code. Trends Biochem. Sci. 2013, 38, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Dyson, N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998, 12, 2245–2262. [Google Scholar] [CrossRef] [PubMed]
- Harbour, J.W.; Dean, D.C. The Rb/E2F pathway: Expanding roles and emerging paradigms. Genes Dev. 2000, 14, 2393–2409. [Google Scholar] [CrossRef] [PubMed]
- Nizami, Z.; Deryusheva, S.; Gall, J.G. The Cajal body and histone locus body. Cold Spring Harb. Perspect. Biol. 2010, 2, a000653. [Google Scholar] [CrossRef]
- Bongiorno-Borbone, L.; De Cola, A.; Vernole, P.; Finos, L.; Barcaroli, D.; Knight, R.A.; Melino, G.; De Laurenzi, V. FLASH and NPAT positive but not Coilin positive Cajal Bodies correlate with cell ploidy. Cell Cycle 2008, 7, 2357–2367. [Google Scholar] [CrossRef]
- Birnstiel, M.L.; Busslinger, M.; Strub, K. Transcription termination and 3’ processing: The end is in site! Cell 1985, 41, 349–359. [Google Scholar] [CrossRef]
- Dominski, Z.; Carpousis, A.J.; Clouet-d’Orval, B. Emergence of the β-CASP ribonucleases: Highly conserved and ubiquitous metallo-enzymes involved in messenger RNA maturation and degradation. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2013, 1829, 532–551. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, Y.; Aik, W.S.; Yang, X.C.; Marzluff, W.F.; Walz, T.; Dominski, Z.; Tong, L. Structure of an active human histone pre-mRNA 3’-end processing machinery. Science 2020, 367, 700–703. [Google Scholar] [CrossRef]
- Schümperli, D.; Pillai, R. The special Sm core structure of the U7 snRNP: Far-reaching significance of a small nuclear ribonucleoprotein. Cell. Mol. Life Sci. CMLS 2004, 61, 2560–2570. [Google Scholar] [CrossRef]
- Dominski, Z.; Marzluff, W.F. Formation of the 3’ end of histone mRNA: Getting closer to the end. Gene 2007, 396, 373–390. [Google Scholar] [CrossRef]
- Yang, X.C.; Burch, B.D.; Yan, Y.; Marzluff, W.F.; Dominski, Z. FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3’ end processing of histone pre-mRNAs. Mol. Cell 2009, 36, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.C.; Sabath, I.; Dębski, J.; Kaus-Drobek, M.; Dadlez, M.; Marzluff, W.F.; Dominski, Z. A complex containing the CPSF73 endonuclease and other polyadenylation factors associates with U7 snRNP and is recruited to histone pre-mRNA for 3’-end processing. Mol. Cell. Biol. 2013, 33, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Aik, W.S.; Lin, M.H.; Tan, D.; Tripathy, A.; Marzluff, W.F.; Dominski, Z.; Chou, C.Y.; Tong, L. The N-terminal domains of FLASH and Lsm11 form a 2: 1 heterotrimer for histone pre-mRNA 3’-end processing. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Dominski, Z.; Yang, X.C.; Marzluff, W.F. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell 2005, 123, 37–48. [Google Scholar] [CrossRef]
- Marzluff, W.F.; Gongidi, P.; Woods, K.R.; Jin, J.; Maltais, L.J. The human and mouse replication-dependent histone genes. Genomics 2002, 80, 487–498. [Google Scholar] [CrossRef]
- Liu, J.L.; Murphy, C.; Buszczak, M.; Clatterbuck, S.; Goodman, R.; Gall, J.G. The drosophila melanogaster cajal body. J. Cell Biol. 2006, 172, 875–884. [Google Scholar] [CrossRef]
- Ghule, P.N.; Dominski, Z.; Lian, J.B.; Stein, J.L.; Van Wijnen, A.J.; Stein, G.S. The subnuclear organization of histone gene regulatory proteins and 3’ end processing factors of normal somatic and embryonic stem cells is compromised in selected human cancer cell types. J. Cell. Physiol. 2009, 220, 129–135. [Google Scholar] [CrossRef]
- White, A.E.; Burch, B.D.; Yang, X.c.; Gasdaska, P.Y.; Dominski, Z.; Marzluff, W.F.; Duronio, R.J. Drosophila histone locus bodies form by hierarchical recruitment of components. J. Cell Biol. 2011, 193, 677–694. [Google Scholar] [CrossRef]
- Barcaroli, D.; Dinsdale, D.; Neale, M.; Bongiorno-Borbone, L.; Ranalli, M.; Munarriz, E.; Sayan, A.; McWilliam, J.; Smith, T.; Fava, E.; et al. FLASH is an essential component of Cajal bodies. Proc. Natl. Acad. Sci. USA 2006, 103, 14802–14807. [Google Scholar] [CrossRef]
- Yang, X.C.; Sabath, I.; Kunduru, L.; van Wijnen, A.J.; Marzluff, W.F.; Dominski, Z. A conserved interaction that is essential for the biogenesis of histone locus bodies. J. Biol. Chem. 2014, 289, 33767–33782. [Google Scholar] [CrossRef]
- Lu, P.; Hankel, I.L.; Hostager, B.S.; Swartzendruber, J.A.; Friedman, A.D.; Brenton, J.L.; Rothman, P.B.; Colgan, J.D. The developmental regulator protein Gon4l associates with protein YY1, co-repressor Sin3a, and histone deacetylase 1 and mediates transcriptional repression. J. Biol. Chem. 2011, 286, 18311–18319. [Google Scholar] [CrossRef] [PubMed]
- Barr, J.Y.; Goodfellow, R.X.; Colgan, D.F.; Colgan, J.D. Early B cell progenitors deficient for GON4L fail to differentiate due to a block in mitotic cell division. J. Immunol. 2017, 198, 3978–3988. [Google Scholar] [CrossRef] [PubMed]
- Bulchand, S.; Menon, S.D.; George, S.E.; Chia, W. Muscle wasted: A novel component of the Drosophila histone locus body required for muscle integrity. J. Cell Sci. 2010, 123, 2697–2707. [Google Scholar] [CrossRef]
- Boyer, L.A.; Latek, R.R.; Peterson, C.L. The SANT domain: A unique histone-tail-binding module? Nat. Rev. Mol. Cell Biol. 2004, 5, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Sattler, M.; Schleucher, J.; Griesinger, C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution. Prog. Nucl. Magn. Reson. Spectrosc. 1999, 34, 93–158. [Google Scholar] [CrossRef]
- Sharma, D.; Rajarathnam, K. 13C NMR chemical shifts can predict disulfide bond formation. J. Biomol. NMR 2000, 18, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Bax, A. Prediction of Xaa-Pro peptide bond conformation from sequence and chemical shifts. J. Biomol. NMR 2010, 46, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Bax, A. Protein structural information derived from NMR chemical shift with the neural network program TALOS-N. In Artificial Neural Networks; Springer: Berlin, Germany, 2015; pp. 17–32. [Google Scholar]
- Jaremko, Ł; Jaremko, M; Elfaki, I; Mueller, J.W.; Ejchart, A; Bayer, P; Zhukov, I. Structure and dynamics of the first archaeal parvulin reveal a new functionally important loop in parvulin-type prolyl isomerases. J. Biol. Chem. 2011, 286, 6554–6565. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Rullmann, J.A.C.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef]
- Vriend, G. WHAT IF: A molecular modeling and drug design program. J. Mol. Graph. 1990, 8, 52–56. [Google Scholar] [CrossRef]
- Krieger, E.; Koraimann, G.; Vriend, G. Increasing the precision of comparative models with YASARA NOVA—A self-parameterizing force field. Proteins Struct. Funct. Bioinform. 2002, 47, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Dolinsky, T.J.; Czodrowski, P.; Li, H.; Nielsen, J.E.; Jensen, J.H.; Klebe, G.; Baker, N.A. PDB2PQR: Expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007, 35, W522–W525. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Robertson, A.D.; Jensen, J.H. Very fast empirical prediction and rationalization of protein pKa values. Proteins Struct. Funct. Bioinform. 2005, 61, 704–721. [Google Scholar] [CrossRef] [PubMed]
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; McCammon, J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 2001, 98, 10037–10041. [Google Scholar] [CrossRef] [PubMed]
- Skrajna, A.; Yang, X.c.; Tarnowski, K.; Fituch, K.; Marzluff, W.F.; Dominski, Z.; Dadlez, M. Mapping the interaction network of key proteins involved in histone mRNA generation: A hydrogen/deuterium exchange study. J. Mol. Biol. 2016, 428, 1180–1196. [Google Scholar] [CrossRef]
- Biedenkapp, H.; Borgmeyer, U.; Sippel, A.E.; Klempnauer, K.H. Viral myb oncogene encodes a sequence-specific DNA-binding activity. Nature 1988, 335, 835–837. [Google Scholar] [CrossRef]
- Ogata, K.; Kanai, H.; Inoue, T.; Sekikawa, A.; Sasaki, M.; Nagadoi, A.; Sarai, A.; Ishii, S.; Nishimura, Y. Solution structures of Myb DNA-binding domain and its complex with DNA. Nucleic Acids Symp. Ser. 1993, 29, 201–202. [Google Scholar]
- Aasland, R.; Stewart, A.F.; Gibson, T. The SANT domain: A putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional corepressor N-CoR and TFIIIB. Trends Biochem. Sci. 1996, 3, 87–88. [Google Scholar] [CrossRef]
- Boyer, L.A.; Langer, M.R.; Crowley, K.A.; Tan, S.; Denu, J.M.; Peterson, C.L. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol. Cell 2002, 10, 935–942. [Google Scholar] [CrossRef]
- Sterner, D.E.; Wang, X.; Bloom, M.H.; Simon, G.M.; Berger, S.L. The SANT domain of Ada2 is required for normal acetylation of histones by the yeast SAGA complex. J. Biol. Chem. 2002, 277, 8178–8186. [Google Scholar] [CrossRef]
- Mo, X.; Kowenz-Leutz, E.; Laumonnier, Y.; Xu, H.; Leutz, A. Histone H3 tail positioning and acetylation by the c-Myb but not the v-Myb DNA-binding SANT domain. Genes Dev. 2005, 19, 2447–2457. [Google Scholar] [CrossRef] [PubMed]
- Dini, P.W.; Lipsick, J.S. Oncogenic truncation of the first repeat of c-Myb decreases DNA binding in vitro and in vivo. Mol. Cell. Biol. 1993, 13, 7334–7348. [Google Scholar] [CrossRef] [PubMed]
- Hanaoka, S.; Nagadoi, A.; Yoshimura, S.; Aimoto, S.; Li, B.; de Lange, T.; Nishimura, Y. NMR structure of the hRap1 Myb motif reveals a canonical three-helix bundle lacking the positive surface charge typical of Myb DNA-binding domains. J. Mol. Biol. 2001, 312, 167–175. [Google Scholar] [CrossRef]
- Guenther, M.G.; Barak, O.; Lazar, M.A. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol. 2001, 21, 6091–6101. [Google Scholar] [CrossRef] [PubMed]
- Codina, A.; Love, J.D.; Li, Y.; Lazar, M.A.; Neuhaus, D.; Schwabe, J.W. Structural insights into the interaction and activation of histone deacetylase 3 by nuclear receptor corepressors. Proc. Natl. Acad. Sci. USA 2005, 102, 6009–6014. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Li, Y.; Ishizuka, T.; Guenther, M.G.; Lazar, M.A. A SANT motif in the SMRT corepressor interprets the histone code and promotes histone deacetylation. EMBO J. 2003, 22, 3403–3410. [Google Scholar] [CrossRef]
- Andrés, M.E.; Burger, C.; Peral-Rubio, M.J.; Battaglioli, E.; Anderson, M.E.; Grimes, J.; Dallman, J.; Ballas, N.; Mandel, G. CoREST: A functional corepressor required for regulation of neural-specific gene expression. Proc. Natl. Acad. Sci. USA 1999, 96, 9873–9878. [Google Scholar] [CrossRef]
- Liu, J.L.; Buszczak, M.; Gall, J.G. Nuclear bodies in the Drosophila germinal vesicle. Chromosome Res. 2006, 14, 465–475. [Google Scholar] [CrossRef]
- Kiriyama, M.; Kobayashi, Y.; Saito, M.; Ishikawa, F.; Yonehara, S. Interaction of FLASH with arsenite resistance protein 2 is involved in cell cycle progression at S phase. Mol. Cell. Biol. 2009, 29, 4729–4741. [Google Scholar] [CrossRef]
- Jiang, I.; Tsai, C.K.; Chen, S.C.; Wang, S.h.; Amiraslanov, I.; Chang, C.F.; Wu, W.J.; Tai, J.H.; Liaw, Y.C.; Huang, T.H. Molecular basis of the recognition of the ap65-1 gene transcription promoter elements by a Myb protein from the protozoan parasite Trichomonas vaginalis. Nucleic Acids Res. 2011, 39, 8992–9008. [Google Scholar] [CrossRef]
- Nishikawa, T.; Okamura, H.; Nagadoi, A.; König, P.; Rhodes, D.; Nishimura, Y. Solution structure of a telomeric DNA complex of human TRF1. Structure 2001, 9, 1237–1251. [Google Scholar] [CrossRef]
- Cutler, G.; Perry, K.M.; Tjian, R. Adf-1 is a nonmodular transcription factor that contains a TAF-binding Myb-like motif. Mol. Cell. Biol. 1998, 18, 2252–2261. [Google Scholar] [CrossRef] [PubMed]
- Terzo, E.A.; Lyons, S.M.; Poulton, J.S.; Temple, B.R.; Marzluff, W.F.; Duronio, R.J. Distinct self-interaction domains promote Multi Sex Combs accumulation in and formation of the Drosophila histone locus body. Mol. Biol. Cell 2015, 26, 1559–1574. [Google Scholar] [CrossRef] [PubMed]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef]
- Lee, W.; Tonelli, M.; Markley, J.L. NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 2015, 31, 1325–1327. [Google Scholar] [CrossRef]
- Wishart, D.S.; Bigam, C.G.; Yao, J.; Abildgaard, F.; Dyson, H.J.; Oldfield, E.; Markley, J.L.; Sykes, B.D. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR 1995, 6, 135–140. [Google Scholar] [CrossRef]
- Güntert, P.; Qian, Y.Q.; Otting, G.; Müller, M.; Gehring, W.; Wüthrich, K. Structure determination of the Antp (C39->S) homeodomain from nuclear magnetic resonance data in solution using a novel strategy for the structure calculation with the programs DIANA, CALIBA, HABAS and GLOMSA. J. Mol. Biol. 1991, 217, 531–540. [Google Scholar] [CrossRef]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D: Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef]
- Koradi, R.; Billeter, M.; Wüthrich, K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996, 14, 51–55. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Farrow, N.A.; Muhandiram, R.; Singer, A.U.; Pascal, S.M.; Kay, C.M.; Gish, G.; Shoelson, S.E.; Pawson, T.; Forman-Kay, J.D.; Kay, L.E. Backbone dynamics of a free and a phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 1994, 33, 5984–6003. [Google Scholar] [CrossRef] [PubMed]
- Kay, L.E.; Nicholson, L.K.; Delaglio, F.; Bax, A.; Torchia, D. Pulse sequences for removal of the effects of cross correlation between dipolar and chemical-shift anisotropy relaxation mechanisms on the measurement of heteronuclear T1 and T2 values in proteins. J. Magn. Reson. 1992, 97, 359–375. [Google Scholar] [CrossRef]
- D’Auvergne, E.J.; Gooley, P.R. Optimisation of NMR dynamic models I. Minimisation algorithms and their performance within the model-free and Brownian rotational diffusion spaces. J. Biomol. NMR 2008, 40, 107. [Google Scholar] [CrossRef] [PubMed]
- Van Zundert, G.; Rodrigues, J.; Trellet, M.; Schmitz, C.; Kastritis, P.; Karaca, E.; Melquiond, A.; van Dijk, M.; De Vries, S.; Bonvin, A. The HADDOCK2. 2 web server: User-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX web server: An online force field. Nucleic Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef]

| FLASH | YARP | NPAT | |
|---|---|---|---|
| NOESY distance constraints | |||
| Intraresidue | 219 | 224 | 129 |
| Sequential (|i − j| = 1) | 147 | 201 | 106 |
| Medium range (|i − j| ≤ 5) | 94 | 141 | 38 |
| Long range (|i − j| > 5) | 81 | 40 | 9 |
| Hydrogen bonds | 188 | 160 | 10 |
| Torsion angles restraints | |||
| Backbone & side chain ( / / ) | 129 | 126 | 50 |
| RMSD to main structure | |||
| Ordered backbone atoms () | 0.53 ± 0.14 | 0.66 ± 0.19 | 0.17 ± 0.05 |
| Ordered heavy atoms () | 1.67 ± 0.20 | 1.63 ± 0.23 | 0.95 ± 0.26 |
| Ramachandran plot | |||
| Residues in most favored regions (%) | 97.1 | 91.1 | 88.3 |
| Residues in additionally allowed regions (%) | 2.9 | 7.7 | 11.5 |
| Residues in generously allowed regions (%) | 0.0 | 0.1 | 0.2 |
| Residues in disallowed regions (%) | 0.01 | 0.1 | 0.0 |
| RMS Z-score | |||
| Bond lengths () | 0.253 ± 0.038 | −0.502 ± 0.100 | 0.056 ± 0.082 |
| Bond angles | 0.985 ± 0.115 | −0.534 ± 0.111 | 0.062 ± 0.105 |
| Dihedral angles | 0.589 ± 0.054 | 0.594 ± 0.111 | −0.137 ± 0.178 |
| Side chains planarity | 1.242 ± 0.013 | 0.835 ± 0.075 | 0.267 ± 0.474 |
| Non-bonded interactions | −0.013 ± 0.102 | −0.205 ± 0.101 | −0.538 ± 0.392 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bucholc, K.; Skrajna, A.; Adamska, K.; Yang, X.-C.; Krajewski, K.; Poznański, J.; Dadlez, M.; Domiński, Z.; Zhukov, I. Structural Analysis of the SANT/Myb Domain of FLASH and YARP Proteins and Their Complex with the C-Terminal Fragment of NPAT by NMR Spectroscopy and Computer Simulations. Int. J. Mol. Sci. 2020, 21, 5268. https://doi.org/10.3390/ijms21155268
Bucholc K, Skrajna A, Adamska K, Yang X-C, Krajewski K, Poznański J, Dadlez M, Domiński Z, Zhukov I. Structural Analysis of the SANT/Myb Domain of FLASH and YARP Proteins and Their Complex with the C-Terminal Fragment of NPAT by NMR Spectroscopy and Computer Simulations. International Journal of Molecular Sciences. 2020; 21(15):5268. https://doi.org/10.3390/ijms21155268
Chicago/Turabian StyleBucholc, Katarzyna, Aleksandra Skrajna, Kinga Adamska, Xiao-Cui Yang, Krzysztof Krajewski, Jarosław Poznański, Michał Dadlez, Zbigniew Domiński, and Igor Zhukov. 2020. "Structural Analysis of the SANT/Myb Domain of FLASH and YARP Proteins and Their Complex with the C-Terminal Fragment of NPAT by NMR Spectroscopy and Computer Simulations" International Journal of Molecular Sciences 21, no. 15: 5268. https://doi.org/10.3390/ijms21155268
APA StyleBucholc, K., Skrajna, A., Adamska, K., Yang, X.-C., Krajewski, K., Poznański, J., Dadlez, M., Domiński, Z., & Zhukov, I. (2020). Structural Analysis of the SANT/Myb Domain of FLASH and YARP Proteins and Their Complex with the C-Terminal Fragment of NPAT by NMR Spectroscopy and Computer Simulations. International Journal of Molecular Sciences, 21(15), 5268. https://doi.org/10.3390/ijms21155268






