Silencing of TaCKX1 Mediates Expression of Other TaCKX Genes to Increase Yield Parameters in Wheat
Abstract
1. Introduction
2. Results
2.1. Expression Levels of Silenced TaCKX1 in Segregating T1 and T2 Plants
2.2. Co-Expression of Silenced TaCKX1 with Other TaCKX Genes in T1 and T2 and CKX Enzyme Activity
2.3. Influence of TaCKX1 Silencing on Phenotypic Traits and Chlorophyll Content in Flag Leaves of T1 and T2 Plants
2.4. Phytohormone Content in 7 DAP Spikes of T2
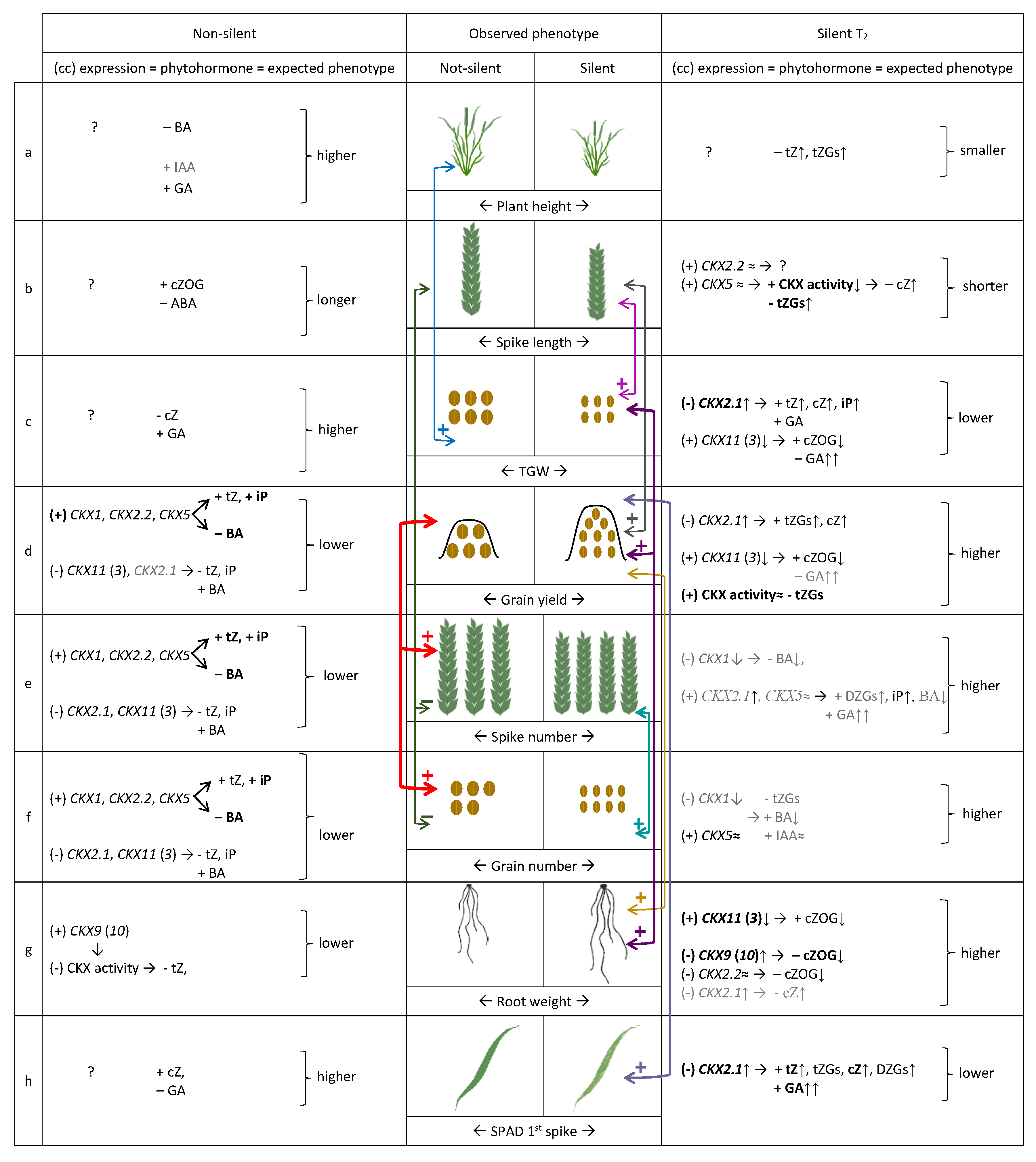
2.5. Coordinated Effect of TaCKX1 Silencing on Expression of Other TaCKX Genes and Phytohormone Level in 7 DAP Spikes as Well as Phenotype in T2
2.6. Models of Co-Regulation of Phytohormone Levels and Phenotype Traits by Coordinated Expression of TaCKX Genes in Non-Silenced and Silenced T2 Plants
3. Discussion
3.1. Various Levels of TaCKX1 Silencing Influence Different Models of Co-Expression with Other TaCKX Genes and Parameters of Yield-Related Traits
3.2. Co-Operating Effect of TaCKX on the Level of Active CKs in Silenced Plants
3.3. Cross Talk of CKs with Other Phytohormones
3.4. Coordinated Effect of TaCKX Gene Expression on the Content of CKs, Other Phytohormones and Yield-Related Traits
4. Materials and Methods
4.1. Vector Construction
4.2. Plant Material, Agrobacterium-Mediated Transformation and In-Vitro Culture
4.3. PCR Analysis
4.4. RNA Extraction and cDNA Synthesis
4.5. Quantitative RT-qPCR
4.6. Quantification of ABA, Auxins, Cytokinins and GA3
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Reynolds, M.; Braun, H. Wheat breeding benefits to low-input agriculture. Nat. Plants 2019, 5, 652–653. [Google Scholar] [CrossRef] [PubMed]
- Borrill, P.; Harrington, S.A.; Uauy, C. Applying the latest advances in genomics and phenomics for trait discovery in polyploid wheat. Plant J. 2019, 97, 56–72. [Google Scholar] [CrossRef] [PubMed]
- Nadolska-Orczyk, A.; Rajchel, I.K.; Orczyk, W.; Gasparis, S. Major genes determining yield-related traits in wheat and barley. Theor. Appl. Genet. 2017, 130, 1081–1098. [Google Scholar] [CrossRef] [PubMed]
- Foley, J.A.; Ramankutty, N.; Brauman, K.A.; Cassidy, E.S.; Gerber, J.S.; Johnston, M.; Mueller, N.D.; O’Connell, C.; Ray, D.K.; West, P.C.; et al. Solutions for a cultivated planet. Nature 2011, 478, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Kieber, J.J.; Schaller, G.E. Cytokinin signaling in plant development. Development 2018, 478, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Ryu, H.; Cho, Y.H.; Scacchi, E.; Sabatini, S.; Hwang, I. Cytokinin-facilitated proteolysis of ARABIDOPSIS RESPONSE REGULATOR 2 attenuates signaling output in two-component circuitry. Plant J. 2012, 69, 934–945. [Google Scholar] [CrossRef]
- Cerny, M.; Dycka, F.; Bobalova, J.; Brzobohaty, B. Early cytokinin response proteins and phosphoproteins of Arabidopsis thaliana identified by proteome and phosphoproteome profiling. J. Exp. Bot. 2011, 62, 921–937. [Google Scholar] [CrossRef]
- Potter, K.C.; Wang, J.; Schaller, G.E.; Kieber, J.J. Cytokinin modulates context-dependent chromatin accessibility through the type-B response regulators. Nat. Plants 2018, 4, 1102–1111. [Google Scholar] [CrossRef]
- Brenner, W.G.; Ramireddy, E.; Heyl, A.; Schmulling, T. Gene regulation by cytokinin in Arabidopsis. Front. Plant Sci. 2012, 3, 8. [Google Scholar] [CrossRef]
- Jameson, P.E.; Song, J.C. Cytokinin: A key driver of seed yield. J. Exp. Bot. 2016, 67, 593–606. [Google Scholar] [CrossRef]
- Liu, Z.; Lv, Y.; Zhang, M.; Liu, Y.; Kong, L.; Zou, M.; Lu, G.; Cao, J.; Yu, X. Identification, expression, and comparative genomic analysis of the IPT and CKX gene families in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genom. 2013, 14, 594. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yu, G.H.; Wen, W.W.; Ma, X.Q.; Xu, B.; Huang, B.R. Functional characterization and hormonal regulation of the PHEOPHYTINASE gene LpPPH controlling leaf senescence in perennial ryegrass. J. Exp. Bot. 2016, 67, 935–945. [Google Scholar] [CrossRef] [PubMed]
- Panda, B.B.; Sekhar, S.; Dash, S.K.; Behera, L.; Shaw, B.P. Biochemical and molecular characterisation of exogenous cytokinin application on grain filling in rice. BMC Plant Biol. 2018, 18, 89. [Google Scholar] [CrossRef] [PubMed]
- Sakakibara, H. Cytokinins: Activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 2006, 57, 431–449. [Google Scholar] [CrossRef] [PubMed]
- Bajguz, A.; Piotrowska, A. Conjugates of auxin and cytokinin. Phytochemistry 2009, 70, 957–969. [Google Scholar] [CrossRef]
- Brandizzi, F. Divide, expand, differentiate—New insights on plant organ growth through cytokinin signaling. Plant J. 2019, 97, 803–804. [Google Scholar] [CrossRef]
- Kudo, T.; Kiba, T.; Sakakibara, H. Metabolism and Long-distance Translocation of Cytokinins. J. Integr. Plant Biol. 2010, 52, 53–60. [Google Scholar] [CrossRef]
- Brugière, N.; Jiao, S.; Hantke, S.; Zinselmeier, C.; Roessler, J.A.; Niu, X.; Jones, R.J.; Habben, J.E. Cytokinin oxidase gene expression in maize is localized to the vasculare, and is induced by cytokinins, abscisic acid, and abiotic stress. Plant Physiol. 2003, 132, 1228–1240. [Google Scholar] [CrossRef]
- Ashikari, M.; Sakakibara, H.; Lin, S.Y.; Yamamoto, T.; Takashi, T.; Nishimura, A.; Angeles, E.R.; Qian, Q.; Kitano, H.; Matsuoka, M. Cytokinin oxidase regulates rice grain production. Science 2005, 309, 741–745. [Google Scholar] [CrossRef]
- Zalewski, W.; Galuszka, P.; Gasparis, S.; Orczyk, W.; Nadolska-Orczyk, A. Silencing of the HvCKX1 gene decreases the cytokinin oxidase/dehydrogenase level in barley and leads to higher plant productivity. J. Exp. Bot. 2010, 61, 1839–1851. [Google Scholar] [CrossRef]
- Zalewski, W.; Gasparis, S.; Boczkowska, M.; Rajchel, I.K.; Kala, M.; Orczyk, W.; Nadolska-Orczyk, A. Expression patterns of HvCKX genes indicate their role in growth and reproductive development of barley. PLoS ONE 2014, 9, e115729. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhao, J.Q.; Song, J.C.; Jameson, P.E. Cytokinin dehydrogenase: A genetic target for yield improvement in wheat. Plant Biotechnol. J. 2020, 18, 614–630. [Google Scholar] [CrossRef]
- Ogonowska, H.; Barchacka, K.; Gasparis, S.; Jablonski, B.; Orczyk, W.; Dmochowska-Boguta, M.; Nadolska-Orczyk, A. Specificity of expression of TaCKX family genes in developing plants of wheat and their co-operation within and among organs. PLoS ONE 2019, 14, e0214239. [Google Scholar] [CrossRef]
- Zalewski, W.; Orczyk, W.; Gasparis, S.; Nadolska-Orczyk, A. HvCKX2 gene silencing by biolistic or Agrobacterium-mediated transformation in barley leads to different phenotypes. BMC Plant Biol. 2012, 12, 206. [Google Scholar] [CrossRef] [PubMed]
- Holubova, K.; Hensel, G.; Vojta, P.; Tarkowski, P.; Bergougnoux, V.; Galuszka, P. Modification of barley plant productivity through regulation of cytokinin content by reverse-genetics approaches. Front. Plant Sci. 2018, 9, 1676. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.L.; Song, G.Q.; Gao, J.; Zhang, S.J.; Zhang, R.Z.; Li, W.; Chen, M.L.; Liu, M.; Xia, X.C.; Risacher, T.; et al. Enhancement of grain number per spike by RNA interference of cytokinin oxidase 2 gene in bread wheat. Hereditas 2018, 155, 33. [Google Scholar] [CrossRef] [PubMed]
- Gasparis, S.; Przyborowski, M.; Kala, M.; Nadolska-Orczyk, A. Knockout of the HvCKX1 or HvCKX3 gene in barley (Hordeum vulgare L.) by RNA-Guided Cas9 Nuclease affects the regulation of cytokinin metabolism and root morphology. Cells 2019, 8, 782. [Google Scholar] [CrossRef]
- Lu, J.; Chang, C.; Zhang, H.P.; Wang, S.X.; Sun, G.; Xiao, S.H.; Ma, C.X. Identification of a Novel Allele of TaCKX6a02 Associated with Grain Size, Filling Rate and Weight of Common Wheat. PLoS ONE 2015, 10, e0144765. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhao, Y.L.; Gao, L.F.; Zhao, G.Y.; Zhou, R.H.; Zhang, B.S.; Jia, J.Z. TaCKX6-D1, the ortholog of rice OsCKX2, is associated with grain weight in hexaploid wheat. New Phytol. 2012, 195, 574–584. [Google Scholar] [CrossRef]
- Chang, C.; Lu, J.; Zhang, H.P.; Ma, C.X.; Sun, G.L. Copy Number Variation of Cytokinin Oxidase Gene Tackx4 Associated with Grain Weight and Chlorophyll Content of Flag Leaf in Common Wheat. PLoS ONE 2015, 10, 15. [Google Scholar] [CrossRef]
- Kersey, P.J.; Allen, J.E.; Allot, A.; Barba, M.; Boddu, S.; Bolt, B.J.; Carvalho-Silva, D.; Christensen, M.; Davis, P.; Grabmueller, C.; et al. Ensembl Genomes 2018: An integrated omics infrastructure for non-vertebrate species. Nucleic Acids Res. 2018, 46, D802–D808. [Google Scholar] [CrossRef] [PubMed]
- Uauy, C. The high grain protein content gene Gpc-B1 accelerates senescence and has pleiotropic effects on protein content in wheat. J. Exp. Bot. 2006, 57, 2785–2794. [Google Scholar] [CrossRef] [PubMed]
- Travella, S.; Klimm, T.E.; Keller, B. RNA interference-based gene silencing as an efficient tool for functional genomics in hexaploid bread wheat. Plant Physiol. 2006, 142, 6–20. [Google Scholar] [CrossRef] [PubMed]
- Gasparis, S.; Orczyk, W.; Zalewski, W.; Nadolska-Orczyk, A. The RNA-mediated silencing of one of the Pin genes in allohexaploid wheat simultaneously decreases the expression of the other, and increases grain hardness. J. Exp. Bot. 2011, 62, 4025–4036. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.P.; Francis, D.; Ormrod, J.C.; Bennett, M.D. Changes in cell number and cell-division activity during endosperm development in allohexaploid wheat, Triticum-aestivum L. J. Exp. Bot. 1992, 43, 1603–1609. [Google Scholar] [CrossRef]
- Hess, J.R.; Carman, J.G.; Banowetz, G.M. Hormones in wheat kernels during embryony. J. Plant Physiol. 2002, 159, 379–386. [Google Scholar] [CrossRef]
- Gajdosova, S.; Spichal, L.; Kaminek, M.; Hoyerova, K.; Novak, O.; Dobrev, P.I.; Galuszka, P.; Klima, P.; Gaudinova, A.; Zizkova, E.; et al. Distribution, biological activities, metabolism, and the conceivable function of cis-zeatin-type cytokinins in plants. J. Exp. Bot. 2011, 62, 2827–2840. [Google Scholar] [CrossRef]
- Morris, R.O.; Blevins, D.G.; Dietrich, J.T.; Durley, R.C.; Gelvin, S.B.; Gray, J.; Hommes, N.G.; Kaminek, M.; Mathews, L.J.; Meilan, R.; et al. Cytokinins in plant-pathogenic bacteria and developing cereal-grains. Funct. Plant Physiol. 1993, 20, 621–637. [Google Scholar] [CrossRef]
- Bartrina, I.; Otto, E.; Strnad, M.; Werner, T.; Schmulling, T. Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 2011, 23, 69–80. [Google Scholar] [CrossRef]
- Schafer, M.; Brutting, C.; Meza-Canales, I.D.; Grosskinsky, D.K.; Vankova, R.; Baldwin, I.T.; Meldau, S. The role of cis-zeatin-type cytokinins in plant growth regulation and mediating responses to environmental interactions. J. Exp. Bot. 2015, 66, 4873–4884. [Google Scholar] [CrossRef]
- Bilyeu, K.D.; Cole, J.L.; Laskey, J.G.; Riekhof, W.R.; Esparza, T.J.; Kramer, M.D.; Morris, R.O. Molecular and biochemical characterization of a cytokinin oxidase from maize. Plant Physiol. 2001, 125, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Saleem, M.; Lamkemeyer, T.; Schutzenmeister, A.; Madlung, J.; Sakai, H.; Piepho, H.P.; Nordheim, A.; Hochholdinger, F. Specification of Cortical Parenchyma and Stele of Maize Primary Roots by Asymmetric Levels of Auxin, Cytokinin, and Cytokinin-Regulated Proteins. Plant Physiol. 2010, 152, 4–18. [Google Scholar] [CrossRef] [PubMed]
- Zalabak, D.; Galuszka, P.; Mrizova, K.; Podlesakova, K.; Gu, R.L.; Frebortova, J. Biochemical characterization of the maize cytokinin dehydrogenase family and cytokinin profiling in developing maize plantlets in relation to the expression of cytokinin dehydrogenase genes. Plant Physiol. Biochem. 2014, 74, 283–293. [Google Scholar] [CrossRef]
- Powell, A.E.; Paleczny, A.R.; Olechowski, H.; Emery, R.J.N. Changes in cytokinin form and concentration in developing kernels correspond with variation in yield among field-grown barley cultivars. Plant Physiol. Biochem. 2013, 64, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Zd’arska, M.; Zatloukalova, P.; Benitez, M.; Sedo, O.; Potesil, D.; Novak, O.; Svacinova, J.; Pesek, B.; Malbeck, J.; Vasickova, J.; et al. Proteome analysis in Arabidopsis reveals shoot- and root-specific targets of cytokinin action and differential regulation of hormonal homeostasis. Plant Physiol. 2013, 161, 918–930. [Google Scholar] [CrossRef]
- Polanska, L.; Vicankova, A.; Novakova, M.; Malbeck, J.; Dobrev, P.I.; Brzobohaty, B.; Vankova, R.; Machackova, I. Altered cytokinin metabolism affects cytokinin, auxin, and abscisic acid contents in leaves and chloroplasts, and chloroplast ultrastructure in transgenic tobacco. J. Exp. Bot. 2007, 58, 637–649. [Google Scholar] [CrossRef]
- Cerny, M.; Kuklova, A.; Hoehenwarter, W.; Fragner, L.; Novak, O.; Rotkova, G.; Jedelsky, P.L.; Zakova, K.; Smehilova, M.; Strnad, M.; et al. Proteome and metabolome profiling of cytokinin action in Arabidopsis identifying both distinct and similar responses to cytokinin down- and up-regulation. J. Exp. Bot. 2013, 64, 4193–4206. [Google Scholar] [CrossRef]
- Kiba, T.; Kudo, T.; Kojima, M.; Sakakibara, H. Hormonal control of nitrogen acquisition: Roles of auxin, abscisic acid, and cytokinin. J. Exp. Bot. 2011, 62, 1399–1409. [Google Scholar] [CrossRef]
- Abid, M.; Shao, Y.H.; Liu, S.X.; Wang, F.; Gao, J.W.; Jiang, D.; Tian, Z.W.; Dai, T.B. Pre-drought priming sustains grain development under post-anthesis drought stress by regulating the growth hormones in winter wheat (Triticum aestivum L.). Planta 2017, 246, 509–524. [Google Scholar] [CrossRef]
- Perilli, S.; Moubayidin, L.; Sabatini, S. The molecular basis of cytokinin function. Curr. Opin. Plant Biol. 2010, 13, 21–26. [Google Scholar] [CrossRef]
- Miyawaki, K.; Matsumoto-Kitano, M.; Kakimoto, T. Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: Tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J. 2004, 37, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Shimizu-Sato, S.; Tanaka, M.; Mori, H. Auxin-cytokinin interactions in the control of shoot branching. Plant Mol. Biol. 2009, 69, 429–435. [Google Scholar] [CrossRef] [PubMed]
- Osugi, A.; Sakakibara, H. Q&A: How do plants respond to cytokinins and what is their importance? BMC Biol. 2015, 13, 1–10. [Google Scholar]
- Martin, R.C.; Mok, M.C.; Habben, J.E.; Mok, D.W.S. A maize cytokinin gene encoding an O-glucosyltransferase specific to cis-zeatin. Proc. Natl. Acad. Sci. USA 2001, 98, 5922–5926. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Du, Y.Y.; Lu, Q.M.; Chen, H.; Meng, R.S.; Cui, C.G.; Lu, S.; Yang, Y.; Chai, Y.M.; Li, J.; et al. The Photoperiod-Insensitive Allele Ppd-D1a Promotes Earlier Flowering in Rht12 Dwarf Plants of Bread Wheat. Front. Plant Sci. 2018, 9, 1312. [Google Scholar] [CrossRef]
- Guo, Z.F.; Liu, G.Z.; Roder, M.S.; Reif, J.C.; Ganal, M.W.; Schnurbusch, T. Genome-wide association analyses of plant growth traits during the stem elongation phase in wheat. Plant Biotechnol. J. 2018, 16, 2042–2052. [Google Scholar] [CrossRef] [PubMed]
- Fahy, B.; Siddiqui, H.; David, L.C.; Powers, S.J.; Borrill, P.; Uauy, C.; Smith, A.M. Final grain weight is not limited by the activity of key starch-synthesising enzymes during grain filling in wheat. J. Exp. Bot. 2018, 69, 5461–5475. [Google Scholar] [CrossRef] [PubMed]
- Cai, T.; Meng, X.P.; Liu, X.L.; Liu, T.N.; Wang, H.; Jia, Z.K.; Yang, D.Q.; Ren, X.L. Exogenous hormonal application regulates the occurrence of wheat tillers by changing endogenous hormones. Front. Plant Sci. 2018, 9, 1886. [Google Scholar] [CrossRef] [PubMed]
- Pacifici, E.; Polverari, L.; Sabatini, S. Plant hormone cross-talk: The pivot of root growth. J. Exp. Bot. 2015, 66, 1113–1121. [Google Scholar] [CrossRef]
- Corbesier, L.; Prinsen, E.; Jacqmard, A.; Lejeune, P.; Van Onckelen, H.; Perilleux, C.; Bernier, G. Cytokinin levels in leaves, leaf exudate and shoot apical meristem of Arabidopsis thaliana during floral transition. J. Exp. Bot. 2003, 54, 2511–2517. [Google Scholar] [CrossRef]
- Hirose, N.; Takei, K.; Kuroha, T.; Kamada-Nobusada, T.; Hayashi, H.; Sakakibara, H. Regulation of cytokinin biosynthesis, compartmentalization and translocation. J. Exp. Bot. 2008, 59, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Reid, D.E.; Heckmann, A.B.; Novak, O.; Kelly, S.; Stougaard, J. CYTOKININ OXIDASE/DEHYDROGENASE3 Maintains Cytokinin Homeostasis during Root and Nodule Development in Lotus japonicus. Plant Physiol. 2016, 170, 1060–1074. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.J.; He, J.M.; Liu, L.N.; Deng, Q.M.; Yao, X.F.; Liu, C.M.; Qiao, Y.L.; Li, P.; Ming, F. OsNAC2 integrates auxin and cytokinin pathways to modulate rice root development. Plant Biotechnol. J. 2019, 18, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Bishopp, A.; Lehesranta, S.; Vaten, A.; Help, H.; El-Showk, S.; Scheres, B.; Helariutta, K.; Mahonen, A.P.; Sakakibara, H.; Helariutta, Y. Phloem-transported cytokinin regulates polar auxin transport and maintains vascular pattern in the root meristem. Curr. Biol. 2011, 21, 927–932. [Google Scholar] [CrossRef] [PubMed]
- Brenner, W.G.; Schmulling, T. Transcript profiling of cytokinin action in Arabidopsis roots and shoots discovers largely similar but also organ-specific responses. BMC Plant Biol. 2012, 12, 112. [Google Scholar] [CrossRef]
- Kollmer, I.; Novak, O.; Strnad, M.; Schmulling, T.; Werner, T. Overexpression of the cytosolic cytokinin oxidase/dehydrogenase (CKX7) from Arabidopsis causes specific changes in root growth and xylem differentiation. Plant J. 2014, 78, 359–371. [Google Scholar] [CrossRef]
- Gan, S.S.; Amasino, R.M. Inhibition of Leaf senescence by autoregulated production of cytokinin. Science 1995, 270, 1986–1988. [Google Scholar] [CrossRef]
- Gregersen, P.L.; Holm, P.B.; Krupinska, K. Leaf senescence and nutrient remobilisation in barley and wheat. Plant Biol. 2008, 10, 37–49. [Google Scholar] [CrossRef]
- Behr, M.; Motyka, V.; Weihmann, F.; Malbeck, J.; Deising, H.B.; Wirsel, S.G.R. Remodeling of Cytokinin Metabolism at Infection Sites of Colletotrichum graminicola on Maize Leaves. Mol. Plant-Microbe Interact. 2012, 25, 1073–1082. [Google Scholar] [CrossRef]
- Wang, W.Q.; Hao, Q.Q.; Wang, W.L.; Li, Q.X.; Chen, F.J.; Ni, F.; Wang, Y.; Fu, D.L.; Wu, J.J.; Wang, W. The involvement of cytokinin and nitrogen metabolism in delayed flag leaf senescence in a wheat stay-green mutant, tasg1. Plant Sci. 2019, 278, 70–79. [Google Scholar] [CrossRef]
- Przetakiewicz, A.; Orczyk, W.; Nadolska-Orczyk, A. The effect of auxin on plant regeneration of wheat, barley and triticale. Plant Cell Tissue Organ Cult. 2003, 73, 245–256. [Google Scholar] [CrossRef]
- Przetakiewicz, A.; Karas, A.; Orczyk, W.; Nadolska-Orczyk, A. Agrobacterium-mediated transformation of polyploid cereals. The efficiency of selection and transgene expression in wheat. Cell. Mol. Biol. Lett. 2004, 9, 903–917. [Google Scholar] [PubMed]
- Murray, M.G.; Thompson, W.F. Rapid Isolation of High Molecular-Weight Plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef] [PubMed]
- Simura, J.; Antoniadi, I.; Siroka, J.; Tarkowska, D.; Strnad, M.; Ljung, K.; Novak, O. Plant hormonomics: Multiple phytohormone profiling by targeted metabolomics. Plant Physiol. 2018, 177, 476–489. [Google Scholar] [CrossRef] [PubMed]

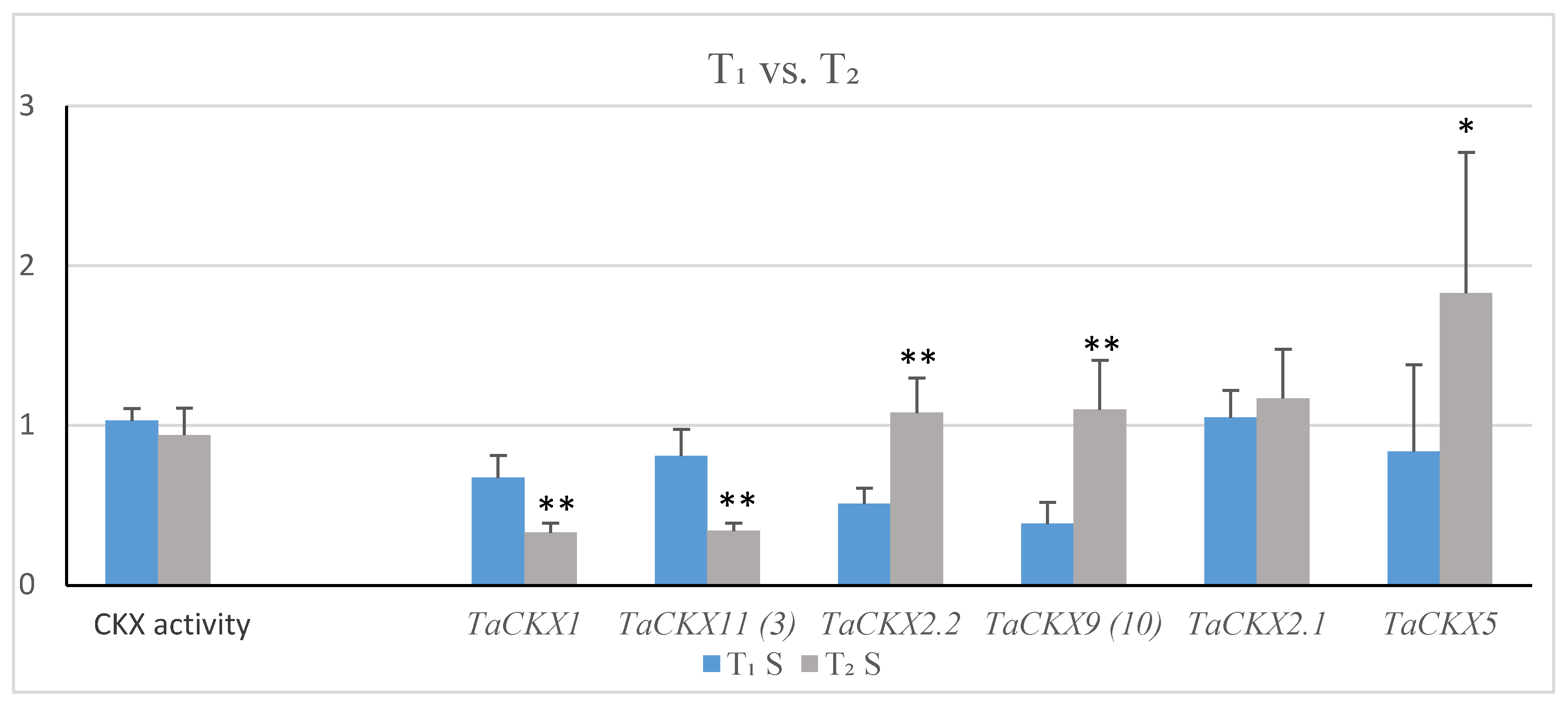
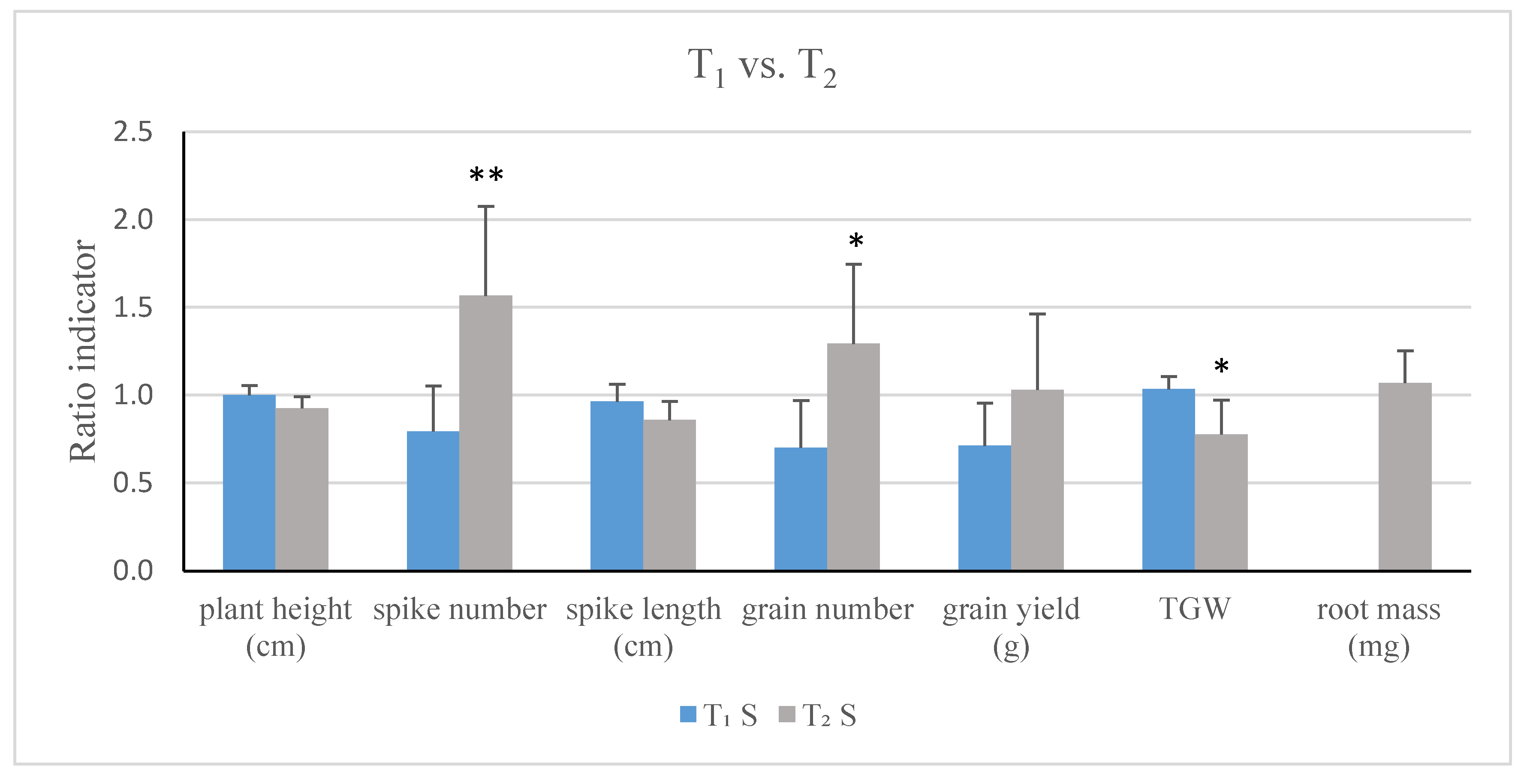
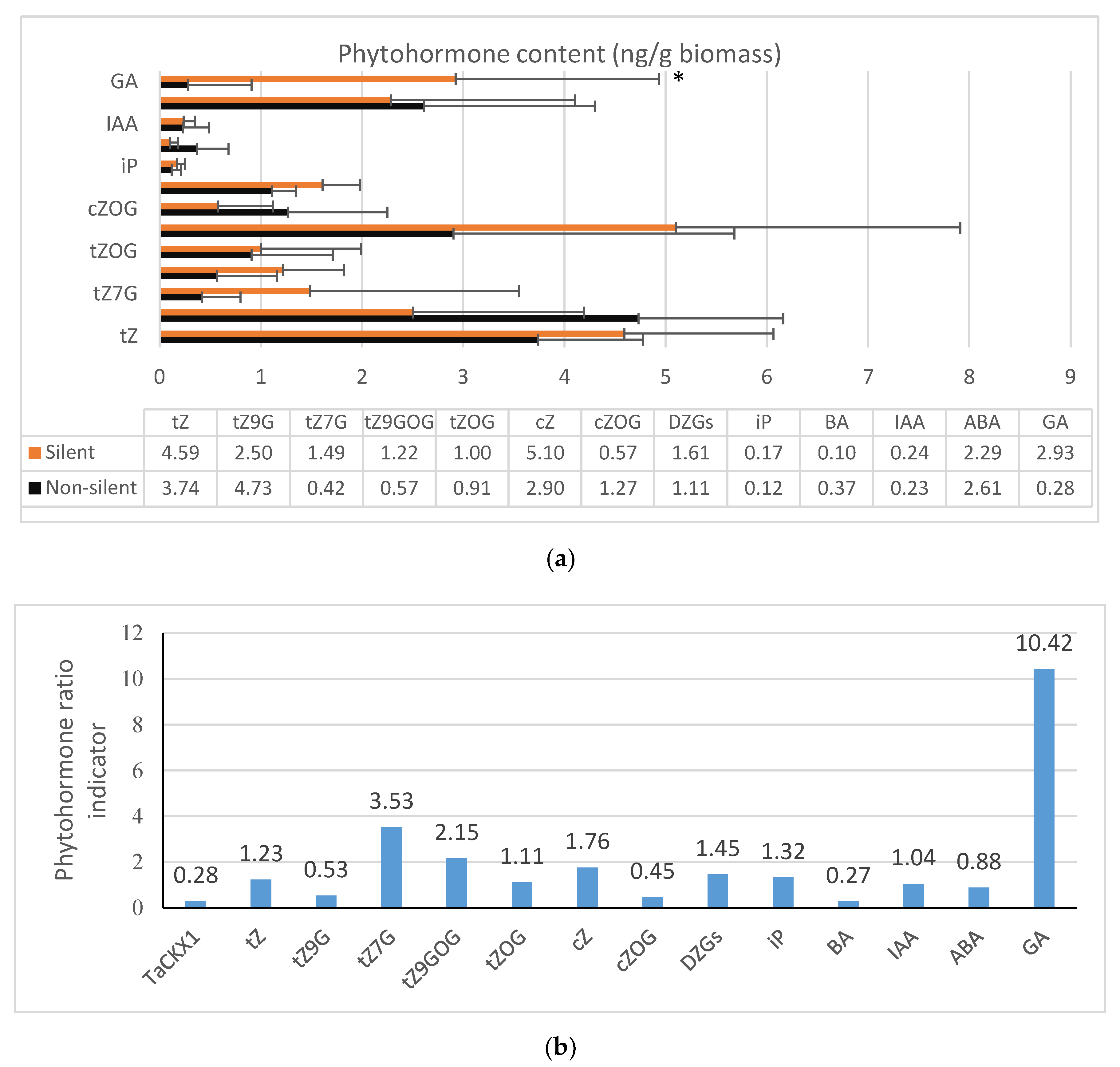
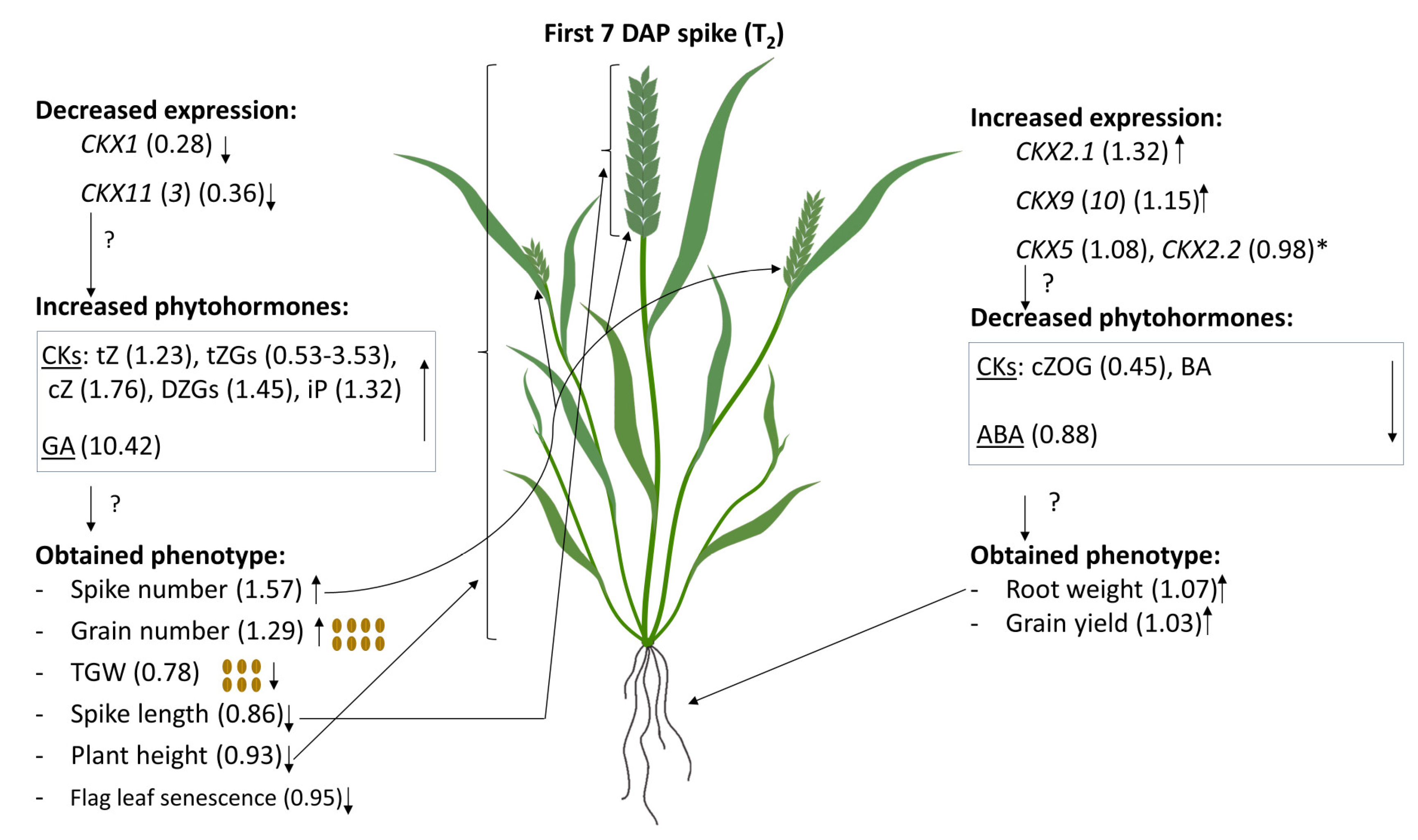
| T1 (SD) | T2 (SD) | Effect of TaCKX1 Silencing T1/T2 | |
|---|---|---|---|
| TaCKX1 * | 0.58 (0.12) | 0.28 (0.05) | decreased/strongly decreased |
| TaCKX11 (3) | 0.80 (0.16) | 0.36 (0.05) | decreased/strongly decreased |
| TaCKX2.2 | 1.08 (0.22) | 0.98 (0.18) | slightly increased/similar |
| TaCKX9 (10) | 0.59 (0.20) | 1.15 (0.32) | strongly decreased/slightly increased |
| TaCKX2.1 | 1.22 (0.19) | 1.32 (0.35) | increased/increased |
| TaCKX5 | 1.00 (0.65) | 1.08 (0.52) | the same/similar |
| CKX activity | 1.01 (0.07) | 0.99 (0.18) | the same/the same |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jabłoński, B.; Ogonowska, H.; Szala, K.; Bajguz, A.; Orczyk, W.; Nadolska-Orczyk, A. Silencing of TaCKX1 Mediates Expression of Other TaCKX Genes to Increase Yield Parameters in Wheat. Int. J. Mol. Sci. 2020, 21, 4809. https://doi.org/10.3390/ijms21134809
Jabłoński B, Ogonowska H, Szala K, Bajguz A, Orczyk W, Nadolska-Orczyk A. Silencing of TaCKX1 Mediates Expression of Other TaCKX Genes to Increase Yield Parameters in Wheat. International Journal of Molecular Sciences. 2020; 21(13):4809. https://doi.org/10.3390/ijms21134809
Chicago/Turabian StyleJabłoński, Bartosz, Hanna Ogonowska, Karolina Szala, Andrzej Bajguz, Wacław Orczyk, and Anna Nadolska-Orczyk. 2020. "Silencing of TaCKX1 Mediates Expression of Other TaCKX Genes to Increase Yield Parameters in Wheat" International Journal of Molecular Sciences 21, no. 13: 4809. https://doi.org/10.3390/ijms21134809
APA StyleJabłoński, B., Ogonowska, H., Szala, K., Bajguz, A., Orczyk, W., & Nadolska-Orczyk, A. (2020). Silencing of TaCKX1 Mediates Expression of Other TaCKX Genes to Increase Yield Parameters in Wheat. International Journal of Molecular Sciences, 21(13), 4809. https://doi.org/10.3390/ijms21134809






