Adaptation of the Marine Bacterium Shewanella baltica to Low Temperature Stress
Abstract
1. Introduction
2. Results and Discussion
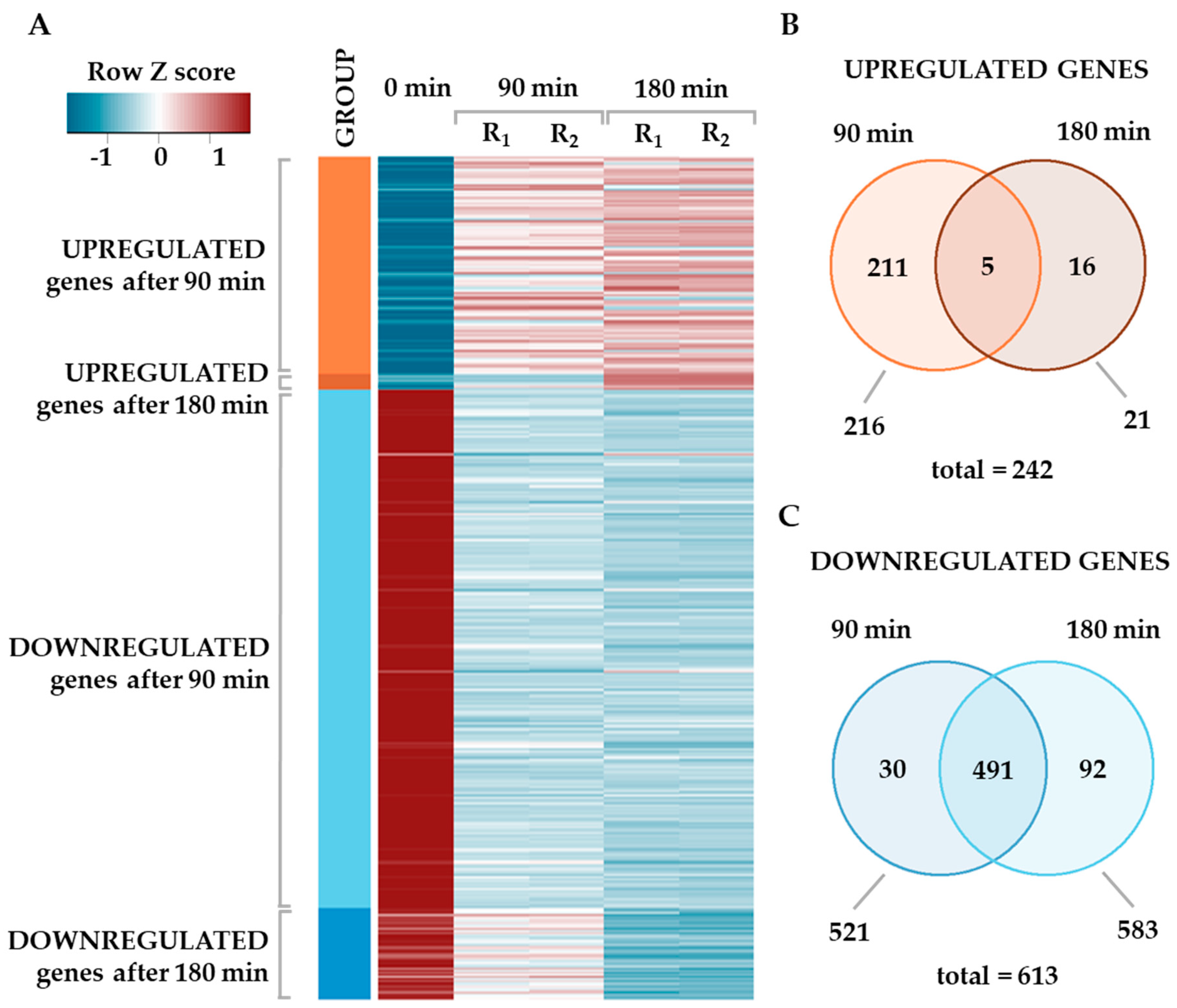
2.1. Global Changes in the Transcriptome of S. baltica in Response to Cold Stress
2.2. Changes in the Metabolic Pathways of S. baltica in the Adaptation and Survival under Cold Stress Conditions
2.3. Transcriptional Reprogramming to Adjust to Cold Stress
2.4. Expression of S. baltica Cold-Induced Proteins in Response to Low Temperatures
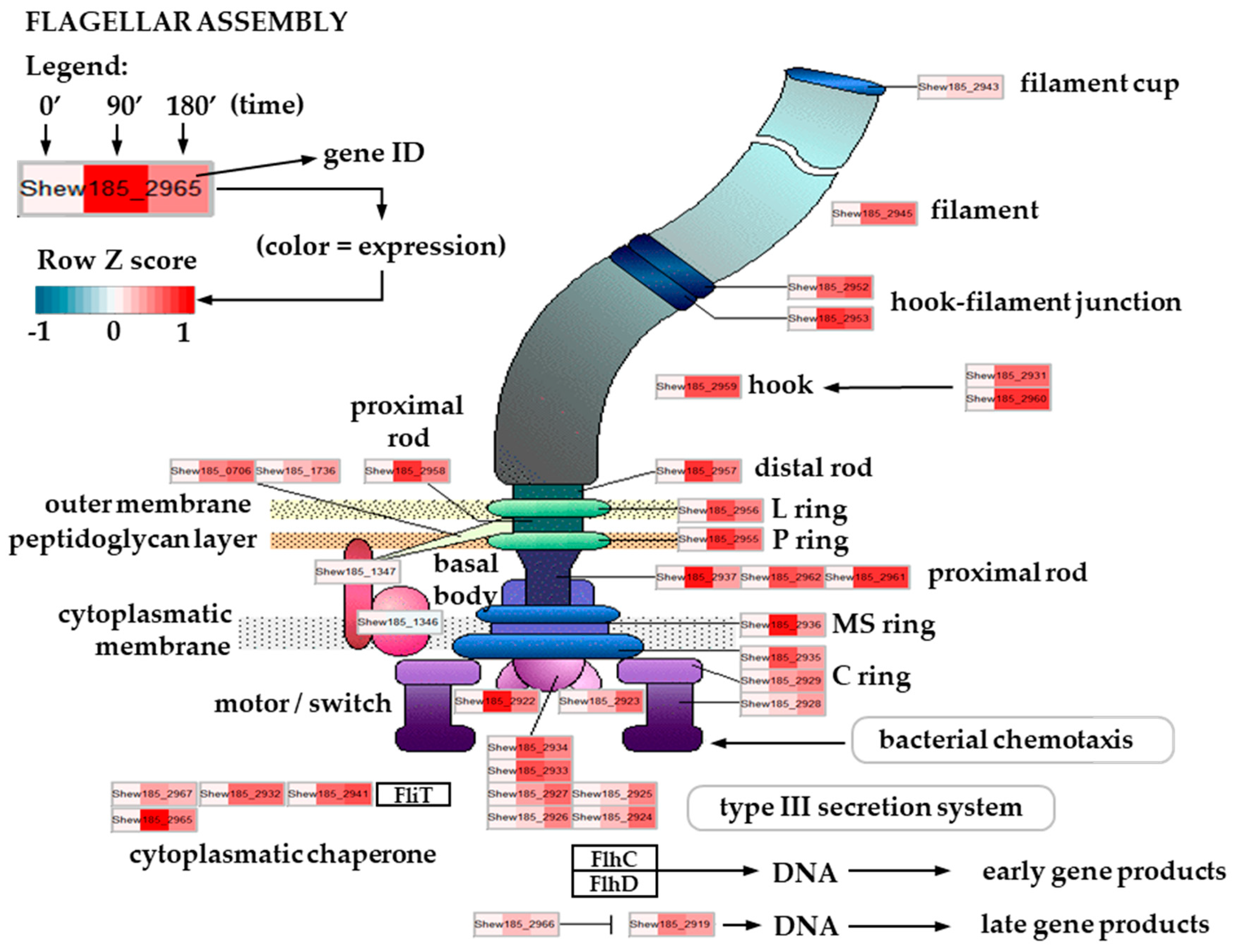
2.5. Expression of Flagellar Assembly-Related Genes in Cold Stress Exposed S. baltica
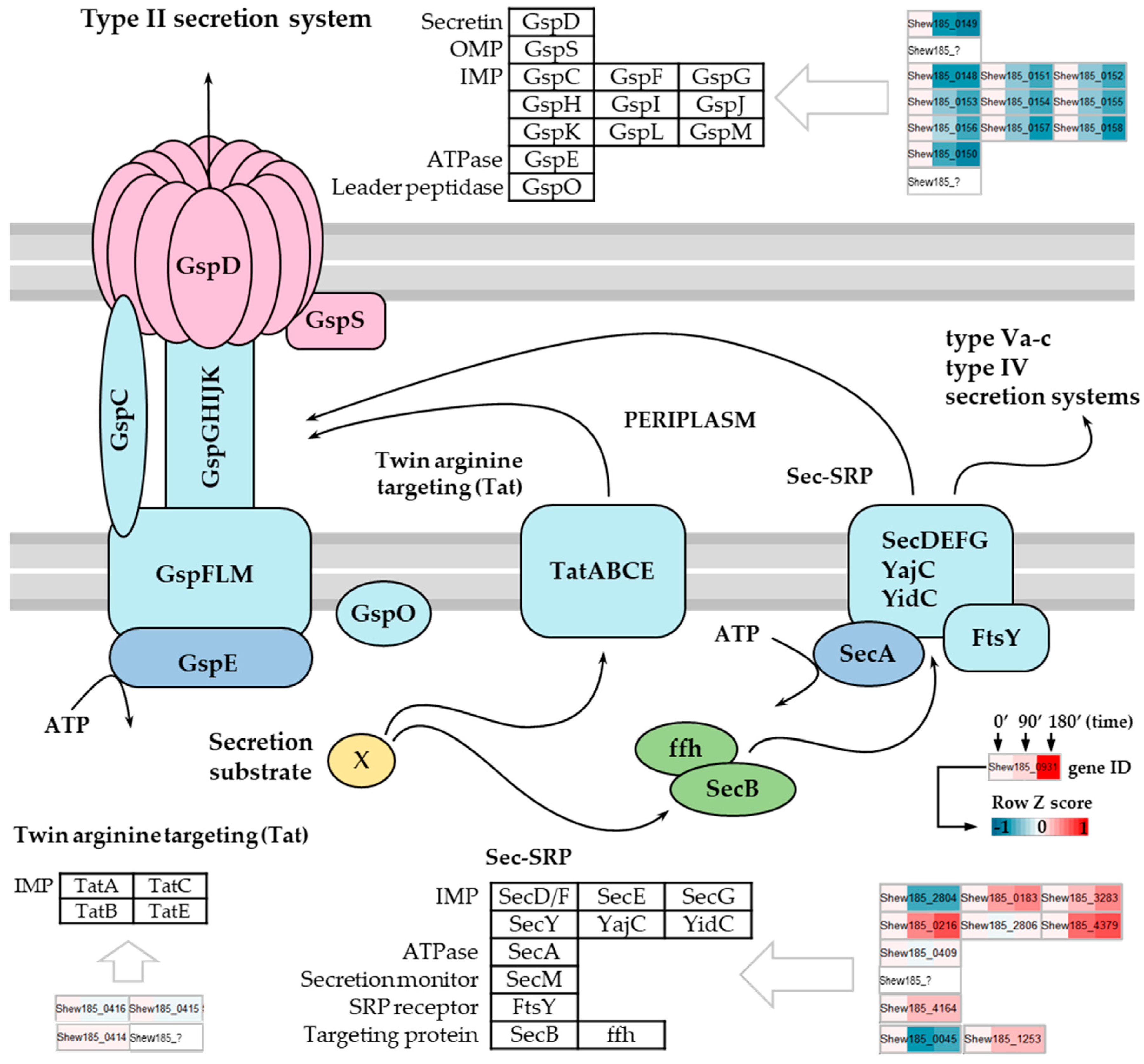
2.6. Downregulation of a Type II Secretion System in Response to Cold Stress
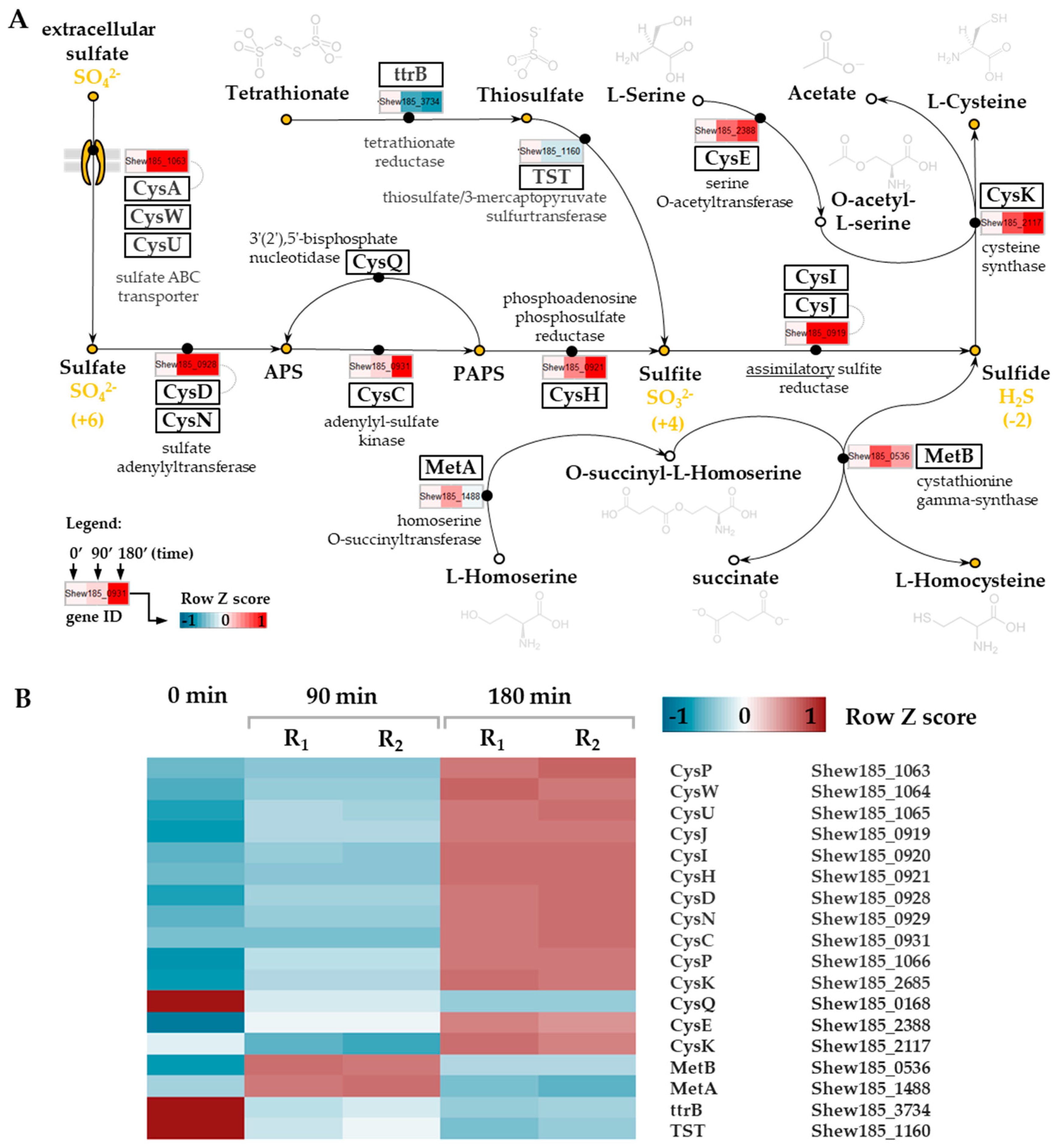
2.7. Upregulation of the Sulfur Assimilatory Pathway upon Cold Stress
2.8. Modified Expression of Genes Responsible for Membrane Integrity and Composition
2.9. Changes in Amino Acid Metabolism and Translation-Related Pathways in Response to Cold Stress
3. Materials and Methods
3.1. Bacteria and Growth Conditions
3.2. RNA Isolation and Sequencing
3.3. Bioinformatics Analysis
3.4. RT-qPCR Assay and Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| APS | adenylyl sulfate |
| Cip | cold induced proteins |
| Csp | cold shock proteins |
| Ct | crossing threshold |
| ECF | extracytoplasmic function |
| FC | fold change |
| Gsp | general secretory pathway |
| IMP | inner membrane proteins |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| OMP | outer membrane proteins |
| PAPS | 3′-phosphoadenylyl sulfate |
| PUFA | polyunsaturated fatty acids |
| RNA-seq | RNA sequencing |
| ROS | reactive oxygen species |
| RT-qPCR | quantitative reverse transcription PCR |
| Sec-SRP | SecA and signal recognition particle |
| SOX | sulfur oxidation pathway |
| T2SS | type II secretion system |
| Tat | twin-arginine targeting |
| TCA | tricarboxylic acid |
| UFA | unsaturated fatty acids |
References
- Kot-Wasik, A.; Zukowska, B.; Dabrowska, D.; Debska, J.; Pacyna, J.; Namiesnik, J. Physical, chemical, and biological changes in the Gulf of Gdansk ecosystem (southern Baltic sea). Rev. Environ. Contam. Toxicol. 2003, 179, 1–36. [Google Scholar] [PubMed]
- Polissi, A.; De Laurentis, W.; Zangrossi, S.; Briani, F.; Longhi, V.; Pesole, G.; Deho, G. Changes in Escherichia coli transcriptome during acclimatization at low temperature. Res. Microbiol. 2003, 154, 573–580. [Google Scholar] [CrossRef]
- Hoffmann, T.; Bremer, E. Protection of Bacillus subtilis against cold stress via compatible-solute acquisition. J. Bacteriol. 2011, 193, 1552–1562. [Google Scholar] [CrossRef] [PubMed]
- Montanchez, I.; Arana, I.; Parada, C.; Garaizabal, I.; Orruno, M.; Barcina, I.; Kaberdin, V.R. Reprogramming of Vibrio harveyi gene expression during adaptation in cold seawater. FEMS Microbiol. Ecol. 2014, 87, 193–203. [Google Scholar] [CrossRef]
- Carroll, J.W.; Mateescu, M.C.; Chava, K.; Colwell, R.R.; Bej, A.K. Response and tolerance of toxigenic Vibro cholerae O1 to cold temperatures. Antonie Van Leeuwenhoek 2001, 79, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhou, D.; Liu, X.; Han, H.; Zhan, L.; Guo, Z.; Zhang, L.; Qin, C.; Wong, H.; Yang, R. Cold-induced gene expression profiles of Vibrio parahaemolyticus: A time-course analysis. FEMS Microbiol. Lett. 2009, 291, 50–58. [Google Scholar] [CrossRef]
- Wood, R.R.; Arias, C.R. Evaluation of global gene expression during cold shock in the human pathogen Vibrio vulnificus. Mar. Biotechnol. (NY) 2011, 13, 942–954. [Google Scholar] [CrossRef]
- Jiang, W.; Hou, Y.; Inouye, M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 1997, 272, 196–202. [Google Scholar] [CrossRef]
- Golovlev, E.L. Bacterial cold shock response at the level of DNA transcription, translation and chromosome dynamics. Mikrobiologiia 2003, 72, 5–13. [Google Scholar]
- Tribelli, P.M.; Lopez, N.I. Reporting key features in cold-adapted bacteria. Life 2018, 8, 8. [Google Scholar] [CrossRef]
- Barria, C.; Malecki, M.; Arraiano, C.M. Bacterial adaptation to cold. Microbiology 2013, 159, 2437–2443. [Google Scholar] [CrossRef] [PubMed]
- Mocali, S.; Chiellini, C.; Fabiani, A.; Decuzzi, S.; de Pascale, D.; Parrilli, E.; Tutino, M.L.; Perrin, E.; Bosi, E.; Fondi, M.; et al. Ecology of cold environments: New insights of bacterial metabolic adaptation through an integrated genomic-phenomic approach. Sci. Rep. 2017, 7, 839. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, S.; Collins, T.; Marx, J.-C.; Feller, G.; Gerday, C. Psychrophilic microorganisms: Challenges for life. EMBO Rep. 2006, 7, 385–389. [Google Scholar] [CrossRef]
- Siddiqui, K.S.; Cavicchioli, R. Cold-adapted enzymes. Annu. Rev. Biochem. 2006, 75, 403–433. [Google Scholar] [CrossRef] [PubMed]
- Moreno, R.; Rojo, F. Features of pseudomonads growing at low temperatures: Another facet of their versatility. Environ. Microbiol. Rep. 2014, 6, 417–426. [Google Scholar] [CrossRef]
- Wang, F.; Xiao, X.; Ou, H.-Y.; Gai, Y.; Wang, F. Role and regulation of fatty acid biosynthesis in the response of Shewanella piezotolerans WP3 to different temperatures and pressures. J. Bacteriol. 2009, 191, 2574–2584. [Google Scholar] [CrossRef]
- Trevors, J.T.; Bej, A.K.; Mojib, N.; van Elsas, J.D.; Van Overbeek, L. Bacterial gene expression at low temperatures. Extremophiles 2012, 16, 167–176. [Google Scholar] [CrossRef]
- Phadtare, S. Unwinding activity of cold shock proteins and RNA metabolism. RNA Biol. 2011, 8, 394–397. [Google Scholar] [CrossRef]
- Hau, H.H.; Gralnick, J.A. Ecology and biotechnology of the genus Shewanella. Annu. Rev. Microbiol. 2007, 61, 237–258. [Google Scholar] [CrossRef]
- Fredrickson, J.K.; Romine, M.F.; Beliaev, A.S.; Auchtung, J.M.; Driscoll, M.E.; Gardner, T.S.; Nealson, K.H.; Osterman, A.L.; Pinchuk, G.; Reed, J.L.; et al. Towards environmental systems biology of Shewanella. Nat. Rev. Microbiol. 2008, 6, 592–603. [Google Scholar] [CrossRef]
- Fonnesbech Vogel, B.; Venkateswaran, K.; Satomi, M.; Gram, L. Identification of Shewanella baltica as the most important H2S-producing species during iced storage of Danish marine fish. Appl. Environ. Microbiol. 2005, 71, 6689–6697. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Xiao, X.; Sun, P.; Wang, F. Screening of genes regulated by cold shock in Shewanella piezotolerans WP3 and time course expression of cold-regulated genes. Arch. Microbiol. 2008, 189, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Yang, Z.K.; Wu, L.; Thompson, D.K.; Zhou, J. Global transcriptome analysis of the cold shock response of Shewanella oneidensis MR-1 and mutational analysis of its classical cold shock proteins. J. Bacteriol. 2006, 188, 4560–4569. [Google Scholar] [CrossRef] [PubMed]
- Kawamoto, J.; Kurihara, T.; Kitagawa, M.; Kato, I.; Esaki, N. Proteomic studies of an Antarctic cold-adapted bacterium, Shewanella livingstonensis Ac10, for global identification of cold-inducible proteins. Extremophiles 2007, 11, 819–826. [Google Scholar] [CrossRef]
- Jian, H.; Li, S.; Tang, X.; Xiao, X. A transcriptome resource for the deep-sea bacterium Shewanella piezotolerans WP3 under cold and high hydrostatic pressure shock stress. Mar. Genomics 2016, 30, 87–91. [Google Scholar] [CrossRef]
- Cristobal, H.A.; Breccia, J.D.; Abate, C.M. Isolation and molecular characterization of Shewanella sp. G5, a producer of cold-active beta-d-glucosidases. J. Basic Microbiol. 2008, 48, 16–24. [Google Scholar] [CrossRef]
- Tsuruta, H.; Mikami, B.; Higashi, T.; Aizono, Y. Crystal structure of cold-active alkaline phosphatase from the psychrophile Shewanella sp. Biosci. Biotechnol. Biochem. 2010, 74, 69–74. [Google Scholar] [CrossRef]
- Brettar, I.; Moore, E.R.B.; Hofle, M.G. Phylogeny and abundance of novel denitrifying bacteria isolated from the water column of the Central Baltic Sea. Microb. Ecol. 2001, 42, 295–305. [Google Scholar] [CrossRef]
- Deng, J.; Brettar, I.; Luo, C.; Auchtung, J.; Konstantinidis, K.T.; Rodrigues, J.L.M.; Hofle, M.; Tiedje, J.M. Stability, genotypic and phenotypic diversity of Shewanella baltica in the redox transition zone of the Baltic Sea. Environ. Microbiol. 2014, 16, 1854–1866. [Google Scholar] [CrossRef]
- Milewska, K.; Krause, K.; Szalewska-Palasz, A. The stringent response of marine bacteria—Assessment of (p)ppGpp accumulation upon stress conditions. J. Appl. Genet. 2020, 61, 123–130. [Google Scholar] [CrossRef]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szczesniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X.; et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016, 17, 13. [Google Scholar] [CrossRef] [PubMed]
- Therneau, T.; Hart, S.; Kocher, J. Calculating Samplessize Estimates for RNA Seq Studies, R Package Version 1.28.0. 2020.
- Zhao, S.; Guo, Y.; Shyr, Y. KEGG Profile: An Annotation and Visualization Package for Multi-Types and Multi-Groups Expression Data in KEGG Pathway, R Package Version 1.30.0. 2020.
- Gruber, T.M.; Gross, C.A. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 2003, 57, 441–466. [Google Scholar] [CrossRef] [PubMed]
- Walsh, N.P.; Alba, B.M.; Bose, B.; Gross, C.A.; Sauer, R.T. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 2003, 113, 61–71. [Google Scholar] [CrossRef]
- Lima, S.; Guo, M.S.; Chaba, R.; Gross, C.A.; Sauer, R.T. Dual molecular signals mediate the bacterial response to outer-membrane stress. Science 2013, 340, 837–841. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.S.; Updegrove, T.B.; Gogol, E.B.; Shabalina, S.A.; Gross, C.A.; Storz, G. MicL, a new sigmaE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev. 2014, 28, 1620–1634. [Google Scholar] [CrossRef] [PubMed]
- Helmann, J.D. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 2002, 46, 47–110. [Google Scholar] [PubMed]
- Staron, A.; Sofia, H.J.; Dietrich, S.; Ulrich, L.E.; Liesegang, H.; Mascher, T. The third pillar of bacterial signal transduction: Classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol. Microbiol. 2009, 74, 557–581. [Google Scholar] [CrossRef]
- Dai, J.; Wei, H.; Tian, C.; Damron, F.H.; Zhou, J.; Qiu, D. An extracytoplasmic function sigma factor-dependent periplasmic glutathione peroxidase is involved in oxidative stress response of Shewanella oneidensis. BMC Microbiol. 2015, 15, 34. [Google Scholar] [CrossRef]
- Chattopadhyay, M.K.; Raghu, G.; Sharma, Y.V.R.K.; Biju, A.R.; Rajasekharan, M.V.; Shivaji, S. Increase in oxidative stress at low temperature in an antarctic bacterium. Curr. Microbiol. 2011, 62, 544–546. [Google Scholar] [CrossRef]
- Piette, F.; Leprince, P.; Feller, G. Is there a cold shock response in the Antarctic psychrophile Pseudoalteromonas haloplanktis? Extremophiles 2012, 16, 681–683. [Google Scholar] [CrossRef]
- Shatalin, K.; Shatalina, E.; Mironov, A.; Nudler, E. H2S: A universal defense against antibiotics in bacteria. Science 2011, 334, 986–990. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Li, N.; Mao, Y.; Zhou, G.; Gao, H. Endogenous generation of hydrogen sulfide and its regulation in Shewanella oneidensis. Front. Microbiol. 2015, 6, 374. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.H.; Marahiel, M.A. Bacterial cold shock responses. Sci. Prog. 2003, 86, 9–75. [Google Scholar] [CrossRef] [PubMed]
- Phadtare, S.; Alsina, J.; Inouye, M. Cold-shock response and cold-shock proteins. Curr. Opin. Microbiol. 1999, 2, 175–180. [Google Scholar] [CrossRef]
- Woodson, S.A.; Panja, S.; Santiago-Frangos, A. Proteins that chaperone RNA regulation. Microbiol. Spectr. 2018, 6, 4. [Google Scholar] [CrossRef]
- Moens, S.; Vanderleyden, J. Functions of bacterial flagella. Crit. Rev. Microbiol. 1996, 22, 67–100. [Google Scholar] [CrossRef]
- Pratt, L.A.; Kolter, R. Genetic analysis of Escherichia coli biofilm formation: Roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 1998, 30, 285–293. [Google Scholar] [CrossRef]
- Young, G.M.; Schmiel, D.H.; Miller, V.L. A new pathway for the secretion of virulence factors by bacteria: The flagellar export apparatus functions as a protein-secretion system. Proc. Natl. Acad. Sci. USA 1999, 96, 6456–6461. [Google Scholar] [CrossRef]
- Childers, S.E.; Ciufo, S.; Lovley, D.R. Geobacter metallireducens accesses insoluble Fe(III) oxide by chemotaxis. Nature 2002, 416, 767–769. [Google Scholar] [CrossRef]
- Liu, R.; Ochman, H. Stepwise formation of the bacterial flagellar system. Proc. Natl. Acad. Sci. USA 2007, 104, 11507. [Google Scholar] [CrossRef]
- Macnab, R.M. How Bacteria Assemble Flagella. Annu. Rev. Microbiol. 2003, 57, 77–100. [Google Scholar] [CrossRef] [PubMed]
- Aldridge, P.; Hughes, K.T. Regulation of flagellar assembly. Curr. Opin. Microbiol. 2002, 5, 160–165. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, Y.; Zhang, W. RNA-seq-based analysis of cold shock response in Thermoanaerobacter tengcongensis, a bacterium harboring a single cold shock protein encoding gene. PLoS ONE 2014, 9, e93289. [Google Scholar] [CrossRef] [PubMed]
- McCarter, L.L. Regulation of flagella. Curr. Opin. Microbiol. 2006, 9, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wang, J.; Tang, P.; Chen, H.; Gao, H. Genetic and molecular characterization of flagellar assembly in Shewanella oneidensis. PLoS ONE 2011, 6, e21479. [Google Scholar] [CrossRef]
- Mattila, M.; Lindström, M.; Somervuo, P.; Markkula, A.; Korkeala, H. Role of flhA and motA in growth of Listeria monocytogenes at low temperatures. Int. J. Food Microbiol. 2011, 148, 177–183. [Google Scholar] [CrossRef]
- Eshwar, A.K.; Guldimann, C.; Oevermann, A.; Tasara, T. Cold-shock domain family proteins (Csps) are involved in regulation of virulence, cellular aggregation, and flagella-based motility in Listeria monocytogenes. Front. Cell. Infect. Microbiol. 2017, 7, 453. [Google Scholar] [CrossRef]
- Bresolin, G.; Neuhaus, K.; Scherer, S.; Fuchs, T.M. Transcriptional analysis of long-term adaptation of Yersinia enterocolitica to low-temperature growth. J. Bacteriol. 2006, 188, 2945–2958. [Google Scholar] [CrossRef]
- Phadtare, S.; Inouye, M. Genome-wide transcriptional analysis of the cold shock response in wild-type and cold-sensitive, quadruple-csp-deletion strains of Escherichia coli. J. Bacteriol. 2004, 186, 7007–7014. [Google Scholar] [CrossRef]
- Humphries, S. A physical explanation of the temperature dependence of physiological processes mediated by cilia and flagella. Proc. Natl. Acad. Sci. USA 2013, 110, 14693–14698. [Google Scholar] [CrossRef]
- Yang, R.S.; Chen, Y.T. Flagellation of Shewanella oneidensis impacts bacterial fitness in different environments. Curr. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Costa, T.R.D.; Felisberto-Rodrigues, C.; Meir, A.; Prevost, M.S.; Redzej, A.; Trokter, M.; Waksman, G. Secretion systems in Gram-negative bacteria: Structural and mechanistic insights. Nat. Rev. Microbiol. 2015, 13, 343–359. [Google Scholar] [CrossRef] [PubMed]
- Nivaskumar, M.; Francetic, O. Type II secretion system: A magic beanstalk or a protein escalator. Biochim. Biophys. Acta 2014, 1843, 1568–1577. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Deng, S.; Marshall, M.J.; Wang, Z.; Kennedy, D.W.; Dohnalkova, A.C.; Mottaz, H.M.; Hill, E.A.; Gorby, Y.A.; Beliaev, A.S.; et al. Direct involvement of type II secretion system in extracellular translocation of Shewanella oneidensis outer membrane cytochromes MtrC and OmcA. J. Bacteriol. 2008, 190, 5512–5516. [Google Scholar] [CrossRef]
- DiChristina, T.J.; Moore, C.M.; Haller, C.A. Dissimilatory Fe(III) and Mn(IV) reduction by Shewanella putrefaciens requires ferE, a homolog of the pulE (gspE) type II protein secretion gene. J. Bacteriol. 2002, 184, 142–151. [Google Scholar] [CrossRef]
- Korotkov, K.V.; Sandkvist, M. Architecture, Function, and Substrates of the Type II Secretion System. EcoSal Plus 2019, 8. [Google Scholar] [CrossRef]
- Douzi, B.; Filloux, A.; Voulhoux, R. On the path to uncover the bacterial type II secretion system. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2012, 367, 1059–1072. [Google Scholar] [CrossRef]
- Keenan, R.J.; Freymann, D.M.; Stroud, R.M.; Walter, P. The signal recognition particle. Annu. Rev. Biochem. 2001, 70, 755–775. [Google Scholar] [CrossRef]
- Ghosh, W.; Dam, B. Biochemistry and molecular biology of lithotrophic sulfur oxidation by taxonomically and ecologically diverse bacteria and archaea. FEMS Microbiol. Rev. 2009, 33, 999–1043. [Google Scholar] [CrossRef]
- Pronk, J.T.; Meulenberg, R.; Hazeu, W.; Bos, P.; Kuenen, J.G. Oxidation of reduced inorganic sulphur compounds by acidophilic thiobacilli. FEMS Microbiol. Rev. 1990, 6, 293–306. [Google Scholar] [CrossRef]
- Grein, F.; Ramos, A.R.; Venceslau, S.S.; Pereira, I.A.C. Unifying concepts in anaerobic respiration: Insights from dissimilatory sulfur metabolism. Biochim. Biophys. Acta Bioenerg. 2013, 1827, 145–160. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, B.B.; Findlay, A.J.; Pellerin, A. The biogeochemical sulfur cycle of marine sediments. Front. Microbiol. 2019, 10, 849. [Google Scholar] [CrossRef]
- Tiedje, J.M. Shewanella—The environmentally versatile genome. Nat. Biotechnol. 2002, 20, 1093–1094. [Google Scholar] [CrossRef] [PubMed]
- Burns, J.L.; DiChristina, T.J. Anaerobic Respiration of Elemental Sulfur and Thiosulfate by Shewanella oneidensis MR-1 Requires psrA, a Homolog of the phsA Gene of Salmonella enterica Serovar Typhimurium LT2. Appl. Environ. Microbiol. 2009, 75, 5209–5217. [Google Scholar] [CrossRef] [PubMed]
- Rabus, R.; Venceslau, S.S.; Wohlbrand, L.; Voordouw, G.; Wall, J.D.; Pereira, I.A.C. A Post-genomic view of the ecophysiology, catabolism and biotechnological relevance of sulphate-reducing prokaryotes. Adv. Microb. Physiol. 2015, 66, 55–321. [Google Scholar]
- Shewan, J.M.; HOBBS, G.; Hodgkiss, W. A determinative scheme for the identification of certain genera of Gram-negative bacteria, with special reference to the Pseudomonadaceae. J. Appl. Bacteriol. 1960, 23, 379–390. [Google Scholar] [CrossRef]
- Fu, X.; Wang, D.; Yin, X.; Du, P.; Kan, B. Time course transcriptome changes in Shewanella algae in response to salt stress. PLoS ONE 2014, 9, e96001. [Google Scholar] [CrossRef]
- Markus Meier, H.E.; Feistel, R.; Piechura, J.; Arneborg, L.; Burchard, H.; Fiekas, V.; Golenko, N.; Kuzmina, N.; Mohrholz, V.; Nohr, C. Ventilation of the Baltic Sea deep water: A brief review of present knowledge from observations and models. Oceanologia 2006, 48, 133–164. [Google Scholar]
- Strahl, H.; Errington, J. Bacterial membranes: Structure, domains, and function. Annu. Rev. Microbiol. 2017, 71, 519–538. [Google Scholar] [CrossRef]
- De Maayer, P.; Anderson, D.; Cary, C.; Cowan, D.A. Some like it cold: Understanding the survival strategies of psychrophiles. EMBO Rep. 2014, 15, 508–517. [Google Scholar] [CrossRef]
- Mendoza, D. de Temperature Sensing by Membranes. Annu. Rev. Microbiol. 2014, 68, 101–116. [Google Scholar] [CrossRef] [PubMed]
- Diaz, A.R.; Mansilla, M.C.; Vila, A.J.; de Mendoza, D. Membrane topology of the Acyl-Lipid desaturase from Bacillus subtilis. J. Biol. Chem. 2002, 277, 48099–48106. [Google Scholar] [CrossRef] [PubMed]
- Carty, S.M.; Sreekumar, K.R.; Raetz, C.R.H. Effect of cold shock on lipid a biosynthesis in Escherichia coli. J. Biol. Chem. 1999, 274, 9677–9685. [Google Scholar] [CrossRef] [PubMed]
- Klein, W.; Weber, M.H.W.; Marahiel, M.A. Cold shock response of Bacillus subtilis: Isoleucine-dependent switch in the fatty acid branching pattern for membrane adaptation to low temperatures. J. Bacteriol. 1999, 181, 5341–5349. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, P.S.; Lopez, P.; de Mendoza, D. Transcriptional control of the low-temperature-inducible des gene, encoding the delta5 desaturase of Bacillus subtilis. J. Bacteriol. 1999, 181, 7028–7033. [Google Scholar] [CrossRef]
- Campbell, J.W.; Cronan, J.E. Bacterial fatty acid biosynthesis: Targets for antibacterial drug discovery. Annu. Rev. Microbiol. 2001, 55, 305–332. [Google Scholar] [CrossRef]
- Zhu, K.; Zhang, Y.-M.; Rock, C.O. Transcriptional regulation of membrane lipid homeostasis in Escherichia coli. J. Biol. Chem. 2009, 284, 34880–34888. [Google Scholar] [CrossRef]
- Metz, J.G. Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science 2001, 293, 290–293. [Google Scholar] [CrossRef]
- Luo, Q.; Shi, M.; Ren, Y.; Gao, H. Transcription factors FabR and FadR regulate both aerobic and anaerobic pathways for unsaturated fatty acid biosynthesis in Shewanella oneidensis. Front. Microbiol. 2014, 5, 736. [Google Scholar] [CrossRef]
- Yang, S.P.; Xie, J.; Cheng, Y.; Zhang, Z.; Zhao, Y.; Qian, Y.F. Response of Shewanella putrefaciens to low temperature regulated by membrane fluidity and fatty acid metabolism. LWT 2020, 117, 108638. [Google Scholar] [CrossRef]
- Abboud, R.; Popa, R.; Souza-Egipsy, V.; Giometti, C.S.; Tollaksen, S.; Mosher, J.J.; Findlay, R.H.; Nealson, K.H. Low-temperature growth of Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 2005, 71, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Jian, H.; Xiong, L.; He, Y.; Xiao, X. The regulatory function of LexA is temperature-dependent in the deep-sea bacterium Shewanella piezotolerans WP3. Front. Microbiol. 2015, 6, 627. [Google Scholar] [CrossRef] [PubMed]
- Beckering, C.L.; Steil, L.; Weber, M.H.W.; Völker, U.; Marahiel, M.A. Genomewide transcriptional analysis of the cold shock response in Bacillus subtilis. J. Bacteriol. 2002, 184, 6395–6402. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.-S.; Deng, Y.; Manno, D.; Hawari, J. Shewanella spp. Genomic evolution for a cold marine lifestyle and in-situ explosive biodegradation. PLoS ONE 2010, 5, e9109. [Google Scholar] [CrossRef] [PubMed]
- Karczewska-Golec, J.; Strapagiel, D.; Sadowska, M.; Szalewska-Pałasz, A.; Golec, P. Draft genome sequence of Shewanella baltica M1 Isolated from brackish surface water of the Gulf of Gdańsk. Genome Announc. 2016, 4, e00611-16. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- McClure, R.; Balasubramanian, D.; Sun, Y.; Bobrovskyy, M.; Sumby, P.; Genco, C.A.; Vanderpool, C.K.; Tjaden, B. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 2013, 41, e140. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Contchart Software ControlFreak. Outlier Tests. Available online: https://contchart.com/outliers.aspx (accessed on 6 March 2020).
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kloska, A.; Cech, G.M.; Sadowska, M.; Krause, K.; Szalewska-Pałasz, A.; Olszewski, P. Adaptation of the Marine Bacterium Shewanella baltica to Low Temperature Stress. Int. J. Mol. Sci. 2020, 21, 4338. https://doi.org/10.3390/ijms21124338
Kloska A, Cech GM, Sadowska M, Krause K, Szalewska-Pałasz A, Olszewski P. Adaptation of the Marine Bacterium Shewanella baltica to Low Temperature Stress. International Journal of Molecular Sciences. 2020; 21(12):4338. https://doi.org/10.3390/ijms21124338
Chicago/Turabian StyleKloska, Anna, Grzegorz M. Cech, Marta Sadowska, Klaudyna Krause, Agnieszka Szalewska-Pałasz, and Paweł Olszewski. 2020. "Adaptation of the Marine Bacterium Shewanella baltica to Low Temperature Stress" International Journal of Molecular Sciences 21, no. 12: 4338. https://doi.org/10.3390/ijms21124338
APA StyleKloska, A., Cech, G. M., Sadowska, M., Krause, K., Szalewska-Pałasz, A., & Olszewski, P. (2020). Adaptation of the Marine Bacterium Shewanella baltica to Low Temperature Stress. International Journal of Molecular Sciences, 21(12), 4338. https://doi.org/10.3390/ijms21124338





