Novel Polymorphisms and Genetic Characteristics of the Prion Protein Gene (PRNP) in Dogs—A Resistant Animal of Prion Disease
Abstract
:1. Introduction
2. Results
2.1. Identification of Polymorphisms in Canine PRNP and Genetic Analysis
2.2. Comparison of Tandem Repeat Domains of PrP Among Several Species
2.3. Comparison of the Distribution of the Haplotypes of PRNP Polymorphisms in Eight Dog Breeds
2.4. The Number of Canine PRNP Polymorphisms in Eight Breeds
2.5. Estimation of the Functional Effect of Genetic Polymorphisms of Dog PrP
2.6. Prediction of the Structural Alteration of Dog PrP Induced by Nonsynonymous SNPs
2.7. Evaluation of Polymorphisms on the Aggregation Propensity of Dog PrP
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Genomic DNA Extraction and Genetic Analysis
4.3. Statistical Analysis
4.4. Prediction of Protein Functional Alterations in Dog PrP
4.5. 3D Structure Modeling of Dog PrP
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| PRNP | Prion protein gene |
| TSEs | Transmissible spongiform encephalopathies |
| LD | Linkage disequilibrium |
| PrP | Prion protein |
| SNPs | Single nucleotide polymorphisms |
| BSE | Bovine spongiform encephalopathy |
| MDCK | Madin–Darby canine kidney |
| PMCA | Protein misfolding cyclic amplification |
| CWD | Chronic wasting disease |
| RML | Rocky Mountain Laboratory |
| CJD | Creutzfeldt–Jakob disease |
| ORF | Open reading frame |
| HWE | Hardy-Weinberg equilibrium |
| IC | Intracerebral |
| IP | Intraperitoneal |
| NZW | New Zealand White |
| EtBr | Ethidium bromide |
| FPR | False positive rate |
| NMR | Nuclear magnetic resonance |
References
- McIntyre, K.M.; Gubbins, S.; Goldmann, W.; Hunter, N.; Baylis, M. Epidemiological characteristics of classical scrapie outbreaks in 30 sheep flocks in the United Kingdom. PLoS ONE 2008, 3. [Google Scholar] [CrossRef]
- Curcio, L.; Sebastiani, C.; Di Lorenzo, P.; Lasagna, E.; Biagetti, M. A review on classical and atypical scrapie in caprine: Prion protein gene polymorphisms and their role in the disease. Animal 2016, 10, 1585–1593. [Google Scholar] [CrossRef] [Green Version]
- Sheridan, H.A.; McGrath, G.; White, P.; Fallon, R.; Shoukri, M.M.; Martin, S. A temporal-spatial analysis of bovine spongiform encephalopathy in Irish cattle herds, from 1996 to 2000. Can. J. Vet. Res. 2005, 69, 19–25. [Google Scholar]
- Imran, M.; Mahmood, S. An overview of animal prion diseases. Virol. J. 2011, 8, 493. [Google Scholar] [CrossRef] [Green Version]
- Jeong, B.H.; Kim, Y.S. Genetic studies in human prion diseases. J. Korean. Med. Sci. 2014, 29, 623–632. [Google Scholar] [CrossRef] [Green Version]
- Mathiason, C.K.; Nalls, A.V.; Seelig, D.M.; Kraft, S.L.; Carnes, K.; Anderson, K.R.; Hayes-Klug, J.; Hoover, E.A. Susceptibility of domestic cats to chronic wasting disease. J. Virol. 2013, 87, 1947–1956. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.C.; Kim, S.K.; Jeong, B.H. Scrapie susceptibility-associated indel polymorphism of shadow of prion protein gene (SPRN) in Korean native black goats. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef] [Green Version]
- Jeong, B.H.; Jin, H.T.; Carp, R.I.; Kim, Y.S. Bovine spongiform encephalopathy (BSE)-associated polymorphisms of the prion protein (PRNP) gene in Korean native cattle. Anim. Genet. 2013, 44, 356–357. [Google Scholar] [CrossRef]
- Kim, Y.C.; Jeong, B.H. Bovine spongiform encephalopathy (BSE) associated polymorphisms of the prion-like protein gene (PRND) in Korean dairy cattle and Hanwoo. J. Dairy Res. 2018, 85, 7–11. [Google Scholar] [CrossRef]
- Kim, Y.C.; Jeong, B.H. The first report of prion-related protein gene (PRNT) polymorphisms in goat. Acta Vet. Hung. 2017, 65, 291–300. [Google Scholar] [CrossRef]
- Baylis, M.; Goldmann, W. The genetics of scrapie in sheep and goats. Curr. Mol. Med. 2004, 4, 385–396. [Google Scholar] [CrossRef]
- Collee, J.G.; Bradley, R. BSE: A decade on--Part 2. Lancet 1997, 349, 715–721. [Google Scholar] [CrossRef]
- Collee, J.G.; Bradley, R. BSE: A decade on--Part I. Lancet 1997, 349, 636–641. [Google Scholar] [CrossRef]
- Lysek, D.A.; Schorn, C.; Nivon, L.G.; Esteve-Moya, V.; Christen, B.; Calzolai, L.; von Schroetter, C.; Fiorito, F.; Herrmann, T.; Güntert, p. Prion protein NMR structures of cats, dogs, pigs, and sheep. Proc. Natl. Acad. Sci. USA 2005, 102, 640–645. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Garcia, J.; Fernandez-Funez, P. D159 and S167 are protective residues in the prion protein from dog and horse, two prion-resistant animals. Neurobiol. Dis. 2018, 119, 1–12. [Google Scholar] [CrossRef]
- Li-Li, Q.; Hui, Z.; Lin-Lin, L. Progress on low susceptibility mechanisms of transmissible spongiform encephalopathies. Zool. Res. 2014, 35. [Google Scholar] [CrossRef]
- Polymenidou, M.; Trusheim, H.; Stallmach, L.; Moos, R.; Julius, C.; Miele, G.; Lenz-Bauer, C.; Aguzzi, A. Canine MDCK cell lines are refractory to infection with human and mouse prions. Vaccine 2008, 26, 2601–2614. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Borges, N.; Parra, B.; Vidal, E.; Eraña, H.; Sánchez-Martín, M.A.; de Castro, J.; Elezgarai, S.R.; Pumarola, M.; Mayoral, T.; Castilla, J. Unraveling the key to the resistance of canids to prion diseases. PLoS Pathog. 2017, 13, e1006716. [Google Scholar] [CrossRef] [Green Version]
- Vidal, E.; Fernández-Borges, N.; Pintado, B.; Ordóñez, M.; Márquez, M.; Fondevila, D.; Torres, J.M.; Pumarola, M.; Castilla, J. Bovine spongiform encephalopathy induces misfolding of alleged prion-resistant species cellular prion protein without altering its pathobiological features. J. Neurosci. 2013, 33, 7778–7786. [Google Scholar] [CrossRef] [Green Version]
- Eiden, M.; Soto, E.O.; Mettenleiter, T.C.; Groschup, M.H. Effects of polymorphisms in ovine and caprine prion protein alleles on cell-free conversion. Vet. Res. 2011, 42. [Google Scholar] [CrossRef] [Green Version]
- Hunter, N. PrP genetics in sheep and the implications for scrapie and BSE. Trends Microbiol. 1997, 5, 331–334. [Google Scholar] [CrossRef]
- Kim, S.K.; Kim, Y.C.; Won, S.Y.; Jeong, B.-H. Potential scrapie-associated polymorphisms of the prion protein gene (PRNP) in Korean native black goats. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Fragkiadaki, E.G.; Vaccari, G.; Ekateriniadou, L.V.; Agrimi, U.; Giadinis, N.D.; Chiappini, B.; Esposito, E.; Conte, M.; Nonno, R. PRNP genetic variability and molecular typing of natural goat scrapie isolates in a high number of infected flocks. Vet. Res. 2011, 42. [Google Scholar] [CrossRef] [Green Version]
- Corbiere, F.; Perrin-Chauvineau, C.; Lacroux, C.; Costes, P.; Thomas, M.; Bremaud, I.; Martin, S.; Lugan, S.; Chartier, C.; Schelcher, F. PrP-associated resistance to scrapie in five highly infected goat herds. J. Gen. Virol. 2013, 94, 241–245. [Google Scholar] [CrossRef]
- Goldmann, W.; Martin, T.; Foster, J.; Hughes, S.; Smith, G.; Hughes, K.; Dawson, M.; Hunter, N. Novel polymorphisms in the caprine PrP gene: A codon 142 mutation associated with scrapie incubation period. J. Gen. Virol. 1996, 77, 2885–2891. [Google Scholar] [CrossRef]
- Jeong, B.H.; Lee, K.H.; Kim, N.H.; Jin, J.K.; Kim, J.I.; Carp, R.I.; Kim, Y.S. Association of sporadic Creutzfeldt–Jakob disease with homozygous genotypes at PRNP codons 129 and 219 in the Korean population. Neurogenetics 2005, 6, 229–232. [Google Scholar] [CrossRef]
- Haase, B.; Doherr, M.G.; Seuberlich, T.; Drogemuller, C.; Dolf, G.; Nicken, p.; Schiebel, K.; Ziegler, U.; Groschup, M.H.; Zurbriggen, A.; et al. PRNP promoter polymorphisms are associated with BSE susceptibility in Swiss and German cattle. BMC Genetics 2007, 8. [Google Scholar] [CrossRef] [Green Version]
- Geldermann, H.; He, H.; Bobal, P.; Bartenschlager, H.; Preuss, S. Comparison of DNA variants in the PRNP and NF1 regions between bovine spongiform encephalopathy and control cattle. Anim. Genet. 2006, 37, 469–474. [Google Scholar] [CrossRef]
- Kashkevich, K.; Humeny, A.; Ziegler, U.; Groschup, M.H.; Nicken, P.; Leeb, T.; Fischer, C.; Becker, C.M.; Schiebel, K. Functional relevance of DNA polymorphisms within the promoter region of the prion protein gene and their association to BSE infection. FASEB J. 2007, 21, 1547–1555. [Google Scholar] [CrossRef]
- Bowie, J.U.; Reidhaar-Olson, J.F.; Lim, W.A.; Sauer, R.T. Deciphering the message in protein sequences: Tolerance to amino acid substitutions. Science 1990, 247, 1306–1310. [Google Scholar] [CrossRef]
- Singh, J.; Kumar, H.; Sabareesan, A.T.; Udgaonkar, J.B. Rational stabilization of helix 2 of the prion protein prevents its misfolding and oligomerization. J. Am. Chem. Soc. 2014, 136, 16704–16707. [Google Scholar] [CrossRef]
- Stewart, P.; Campbell, L.; Skogtvedt, S.; Griffin, K.A.; Arnemo, J.M.; Tryland, M.; Girling, S.; Miller, M.W.; Tranulis, M.A.; Goldmann, W. Genetic predictions of prion disease susceptibility in carnivore species based on variability of the prion gene coding region. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [Green Version]
- Iglesias, V.; Conchillo-Sole, O.; Batlle, C.; Ventura, S. AMYCO: Evaluation of mutational impact on prion-like proteins aggregation propensity. BMC Bioinformatics 2019, 20. [Google Scholar] [CrossRef]
- Tang, H.; Thomas, P.D. PANTHER-PSEP: Predicting disease-causing genetic variants using position-specific evolutionary preservation. Bioinformatics 2016, 32, 2230–2232. [Google Scholar] [CrossRef]
- Adzhubei, I.; Jordan, D.M.; Sunyaev, S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013, 7. [Google Scholar] [CrossRef] [Green Version]
- Won, S.Y.; Kim, Y.C.; Kim, K.; Kim, A.D.; Jeong, B.H. The First Report of Polymorphisms and Genetic Features of the prion-like Protein Gene (PRND) in a Prion Disease-Resistant Animal, Dog. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.C.; Jeong, B.H. In Silico Evaluation of Acetylation Mimics in the 27 Lysine Residues of Human Tau Protein. Curr. Alzheimer Res. 2019, 16, 379–387. [Google Scholar] [CrossRef]
- Moore, R.A.; Herzog, C.; Errett, J.; Kocisko, D.A.; Arnold, K.M.; Hayes, S.F.; Priola, S.A. Octapeptide repeat insertions increase the rate of protease-resistant prion protein formation. Protein Sci. 2006, 15, 609–619. [Google Scholar] [CrossRef]
- Beck, J.A.; Mead, S.; Campbell, T.A.; Dickinson, A.; Wientjens, D.P.; Croes, E.A.; Van Duijn, C.M.; Collinge, J. Two-octapeptide repeat deletion of prion protein associated with rapidly progressive dementia. Neurology 2001, 57, 354–356. [Google Scholar] [CrossRef]
- Stanczak, P.; Luczkowski, M.; Juszczyk, P.; Grzonka, Z.; Kozlowski, H. Interactions of Cu2+ ions with chicken prion tandem repeats. Dalton Trans. 2004, 2102–2107. [Google Scholar] [CrossRef]
- Di Natale, G.; Pappalardo, G.; Milardi, D.; Sciacca, M.F.; Attanasio, F.; La Mendola, D.; Rizzarelli, E. Membrane interactions and conformational preferences of human and avian prion N-terminal tandem repeats: The role of copper(II) ions, pH, and membrane mimicking environments. J. Phys. Chem. B. 2010, 114, 13830–13838. [Google Scholar] [CrossRef]
- Hornshaw, M.P.; McDermott, J.R.; Candy, J.M.; Lakey, J.H. Copper binding to the N-terminal tandem repeat region of mammalian and avian prion protein: Structural studies using synthetic peptides. Biochem. Biophys. Res. Commun. 1995, 214, 993–999. [Google Scholar] [CrossRef]
- Gralka, E.; Valensin, D.; Gajda, K.; Bacco, D.; Szyrwiel, L.; Remelli, M.; Valensin, G.; Kamasz, W.; Baranska-Rybak, W.; Kozlowski, H. Copper(II) coordination outside the tandem repeat region of an unstructured domain of chicken prion protein. Mol. Biosyst. 2009, 5, 497–510. [Google Scholar] [CrossRef]
- Seabury, C.M.; Honeycutt, R.L.; Rooney, A.P.; Halbert, N.D.; Derr, J.N. Prion protein gene (PRNP) variants and evidence for strong purifying selection in functionally important regions of bovine exon 3. Proc. Natl. Acad. Sci. USA 2004, 101, 15142–15147. [Google Scholar] [CrossRef] [Green Version]
- Castle, A.R.; Gill, A.C. Physiological Functions of the Cellular Prion Protein. Front. Mol. Biosci. 2017, 4. [Google Scholar] [CrossRef] [Green Version]
- Konold, T.; Spiropoulos, J.; Chaplin, M.J.; Thorne, L.; Spencer, Y.I.; Wells, G.A.; Hawkins, S.A. Transmissibility studies of vacuolar changes in the rostral colliculus of pigs. BMC Vet. Res. 2009, 5. [Google Scholar] [CrossRef] [Green Version]
- Ryder, S.; Hawkins, S.; Dawson, M.; Wells, G. The neuropathology of experimental bovine spongiform encephalopathy in the pig. J. Comp. Pathol. 2000, 122, 131–143. [Google Scholar] [CrossRef]
- Dawson, M.; Wells, G.; Parker, B.; Scott, A. Primary parenteral transmission of bovine spongiform encephalopathy to the pig. Vet. Rec. 1990, 127, 338. [Google Scholar]
- Espinosa, J.-C.; Herva, M.-E.; Andréoletti, O.; Padilla, D.; Lacroux, C.; Cassard, H.; Lantier, I.; Castilla, J.; Torres, J.-M. Transgenic mice expressing porcine prion protein resistant to classical scrapie but susceptible to sheep bovine spongiform encephalopathy and atypical scrapie. Emerg. Infect. Dis. 2009, 15. [Google Scholar] [CrossRef]
- Chianini, F.; Fernandez-Borges, N.; Vidal, E.; Gibbard, L.; Pintado, B.; de Castro, J.; Priola, S.A.; Hamilton, S.; Eaton, S.L.; Finlayson, J.; et al. Rabbits are not resistant to prion infection. Proc. Natl. Acad. Sci. USA 2012, 109, 5080–5085. [Google Scholar] [CrossRef] [Green Version]
- Murdoch, B.M.; Murdoch, G.K. Genetics of Prion Disease in Cattle. Bioinform. Biol. Insights 2015, 9, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Lloyd, S.; Mead, S.; Collinge, J. Genetics of prion disease. Top. Curr. Chem. 2011, 305, 1–22. [Google Scholar] [CrossRef]
- Vidal, E.; Fernandez-Borges, N.; Erana, H.; Parra, B.; Pintado, B.; Sanchez-Martin, M.A.; Charco, J.M.; Ordonez, M.; Perez-Castro, M.A.; Pumarola, M.; et al. Dogs are resistant to prion infection, due to the presence of aspartic or glutamic acid at position 163 of their prion protein. FASEB J. 2020, 34, 3969–3982. [Google Scholar] [CrossRef]
- Myers, J.K.; Pace, C.N. Hydrogen bonding stabilizes globular proteins. Biophys. J. 1996, 71, 2033–2039. [Google Scholar] [CrossRef] [Green Version]
- Marqusee, S.; Sauer, R.T. Contributions of a hydrogen bond/salt bridge network to the stability of secondary and tertiary structure in λ repressor. Protein Sci. 1994, 3, 2217–2225. [Google Scholar] [CrossRef]
- Alber, T.; Dao-Pin, S.; Wilson, K.; Wozniak, J.A.; Cook, S.p.; Matthews, B.W. Contributions of hydrogen bonds of Thr 157 to the thermodynamic stability of phage T4 lysozyme. Nature 1987, 330. [Google Scholar] [CrossRef]
- Fung, K.L.; Pan, J.; Ohnuma, S.; Lund, P.E.; Pixley, J.N.; Kimchi-Sarfaty, C.; Ambudkar, S.V.; Gottesman, M.M. MDR1 synonymous polymorphisms alter transporter specificity and protein stability in a stable epithelial monolayer. Cancer Res. 2014, 74, 598–608. [Google Scholar] [CrossRef] [Green Version]
- Im, E.H.; Choi, S.S. Synonymous Codon Usage Controls Various Molecular Aspects. Genom. Inform. 2017, 15, 123–127. [Google Scholar] [CrossRef] [Green Version]
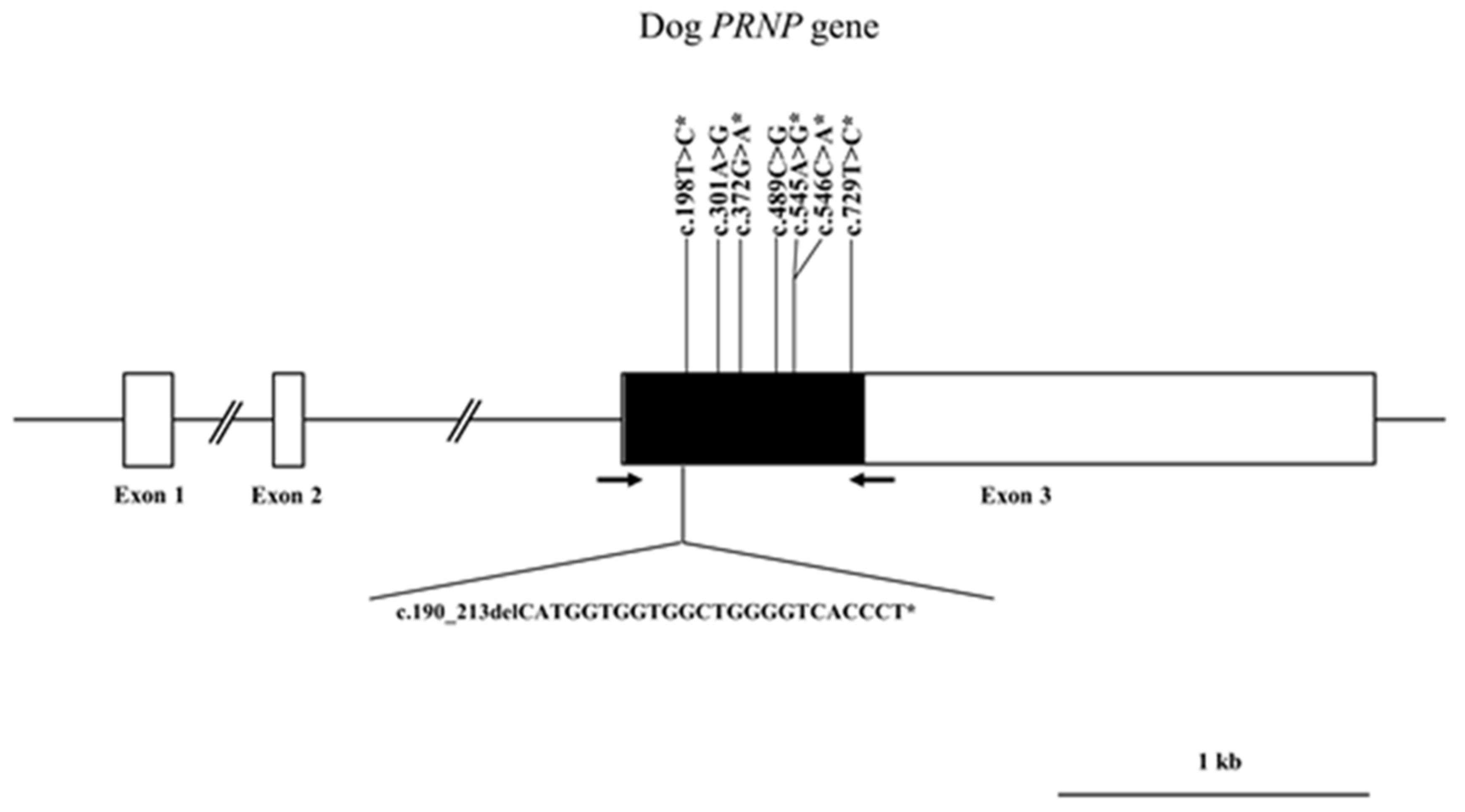

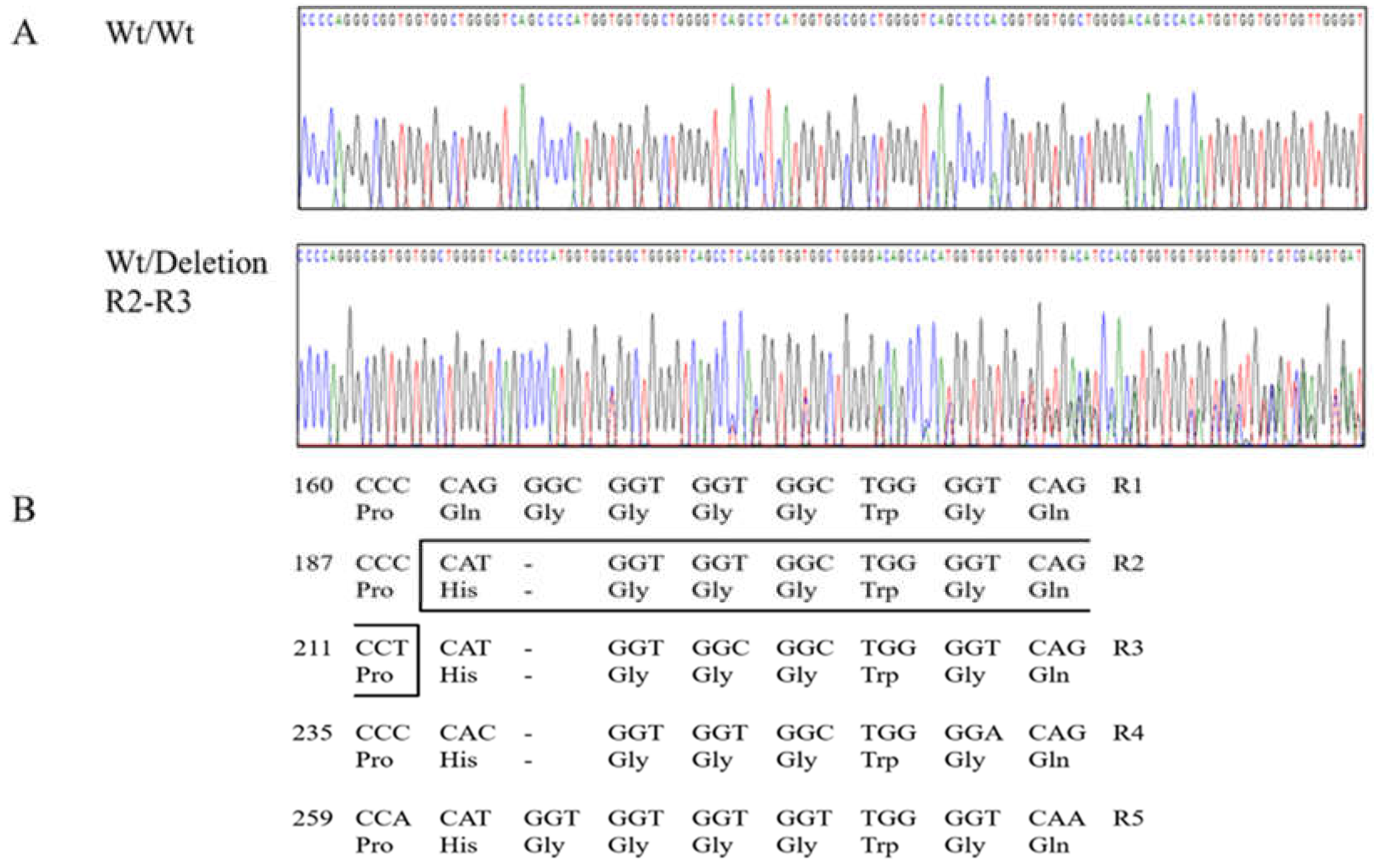
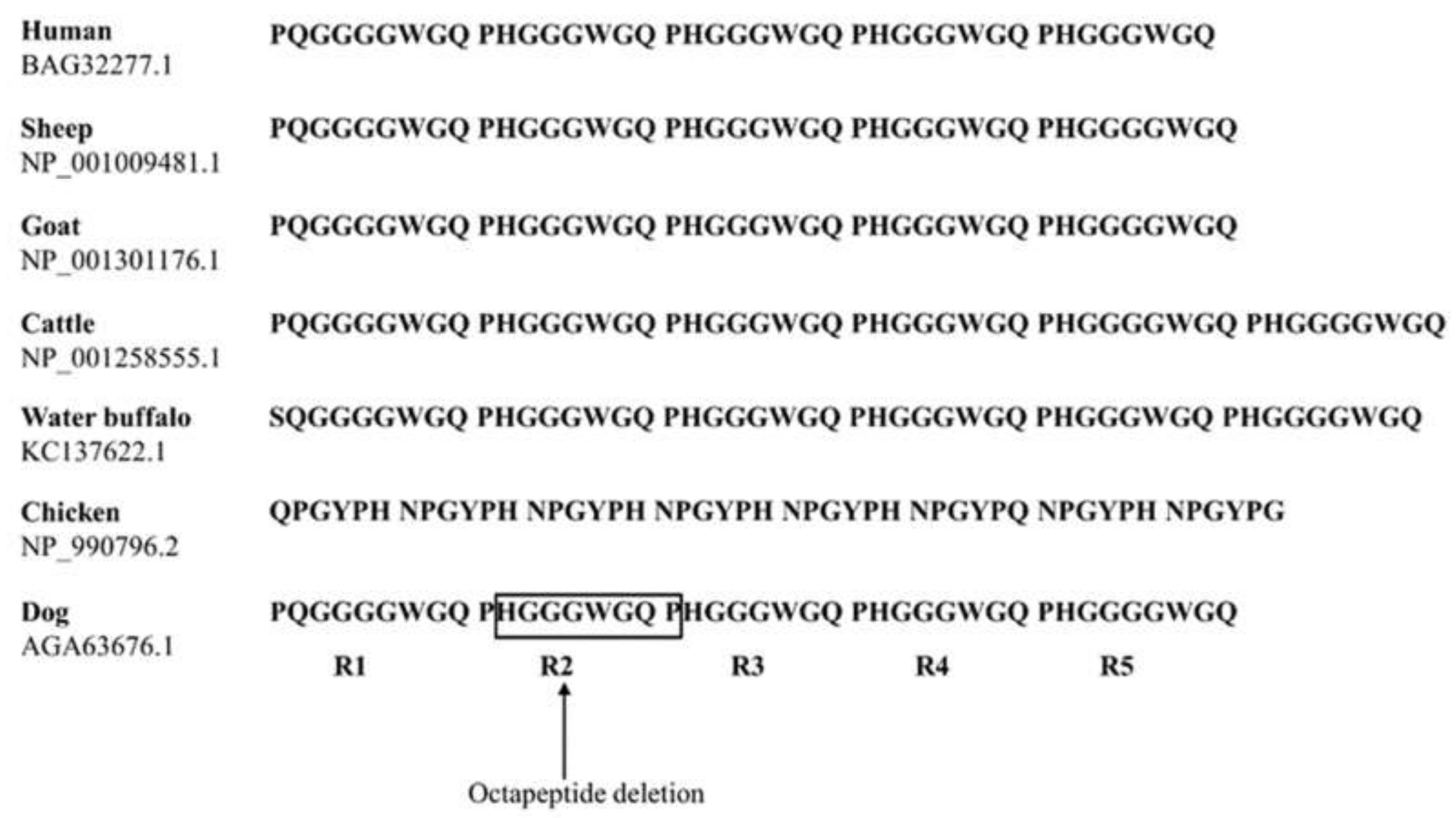



| Polymorphisms | Genotype Frequency, n (%) | Allele frequency, n (%) | HWE | ||||
|---|---|---|---|---|---|---|---|
| c.190_213del | 64_71del HGGGWGQP | Wt/Wt 201 (98.5) | Wt/Del 3 (1.5) | Del/Del 0 (0) | Wt 405 (99.3) | Del 3 (0.7) | 1.0 |
| c.198T>C | Gly66Gly | T/T 159 (77.9) | T/C 33 (16.2) | C/C 12 (5.9) | T 351 (86.0) | C 57 (14.0) | <0.001 |
| c.301A>G | Ser101Gly | A/A 112 (54.9) | A/G 73 (35.8) | G/G 19 (9.3) | A 297 (72.8) | G 111 (27.2) | 0.2168 |
| c.372G>A | Ala124Ala | G/G 196 (96.1) | G/A 8 (3.9) | A/A 0 (0) | G 400 (98.0) | A 8 (2.0) | 1.0 |
| c.489C>G | Asp163Glu | C/C 125 (61.3) | C/G 62 (30.4) | G/G 17 (8.3) | C 312 (76.5) | G 96 (23.5) | 0.0432 |
| c.545A>G | Asp182Gly | A/A 202 (99.0) | A/G 2 (1.0) | G/G 0 (0) | A 406 (99.5) | G 2 (0.5) | 1.0 |
| c.546C>A | Asp182Glu | C/C 200 (98.0) | C/A 2 (1.0) | A/A 2 (1.0) | C 402 (98.5) | A 6 (1.5) | <0.001 |
| c.729T>C | Pro243Pro | T/T 120 (58.8) | T/C 63 (30.9) | C/C 21 (10.3) | T 303 (74.3) | C 105 (25.7) | 0.011 |
| Haplotypes | c.190_213del | c.198T>C | c.301A>G | c.372G>A | c.489C>G | c.545A>G | c.546C>A | c.729T>C | Frequency (n = 204) |
|---|---|---|---|---|---|---|---|---|---|
| ht1 | Wt | T | A | G | C | A | C | T | 284 (0.696) |
| ht2 | Wt | T | G | G | G | A | C | C | 56 (0.137) |
| ht3 | Wt | C | G | G | G | A | C | C | 36 (0.088) |
| ht4 | Wt | C | G | G | C | A | C | T | 7 (0.017) |
| ht5 | Wt | C | G | A | C | A | C | C | 6 (0.015) |
| ht6 | Wt | T | A | G | C | A | A | T | 5 (0.012) |
| others a | 14 (0.035) |
| ∣D’∣ | ||||||||
|---|---|---|---|---|---|---|---|---|
| r2 | c.190_213del | c.198T>C | c.301A>G | c.372G>A | c.489C>G | c.545A>G | c.546C>A | c.729T>C |
| c.190_213del | - | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| c.198T>C | 0.046 | - | 0.919 | 1.0 | 0.55 | 0.308 | 1.0 | 0.688 |
| c.301A>G | 0.002 | 0.367 | - | 1.0 | 0.985 | 0.263 | 0.387 | 0.96 |
| c.372G>A | 0 | 0.123 | 0.054 | - | 1.0 | 1.0 | 1.0 | 0.574 |
| c.489C>G | 0.024 | 0.16 | 0.799 | 0.006 | - | 0.056 | 0.292 | 1.0 |
| c.545A>G | 0 | 0.003 | 0.0 | 0 | 0 | - | 1.0 | 0.089 |
| c.546C>A | 0 | 0.002 | 0.001 | 0 | 0.0 | 0.0 | - | 0.352 |
| c.729T>C | 0.021 | 0.222 | 0.855 | 0.019 | 0.888 | 0.0 | 0.001 | - |
| Breeds (n) | Polymorphisms | Total Number | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Maltese (77) | 64_71delHGGGWGQP | Gly66Gly | Ser101Gly | Ala124Ala | Asp163Glu | Asp182Gly | Asp182Glu | Pro243Pro | 8 |
| Shih Tzu (29) | Gly66Gly | Ser101Gly | Ala124Ala | Asp163Glu | Pro243Pro | 5 | |||
| Toy Poodle (25) | 64_71delHGGGWGQP | Gly66Gly | Ser101Gly | Ala 124Ala | Asp163Glu | Pro243Pro | 6 | ||
| Yorkshire Terrier (19) | Asp182Glu | 1 | |||||||
| Pomeranian (15) | Gly66Gly | Ser101Gly | Asp163Glu | Pro243Pro | 4 | ||||
| Chihuahua (11) | Gly66Gly | Ser101Gly | Asp163Glu | Asp182Gly | Asp182Glu | Pro243Pro | 6 | ||
| Schnauzer (7) | Gly66Gly | Ser101Gly | Asp163Glu | Pro243Pro | 4 | ||||
| Bichon Frise (5) | Ser101Gly | Asp163Glu | Pro243Pro | 3 | |||||
| Mixed dog (16) | Gly66Gly | Ser101Gly | Ala 124Ala | Asp163Glu | Asp182Glu | Pro243Pro | 6 | ||
| Polymorphisms | PolyPhen-2 | PROVEAN | PANTHER | ||||
|---|---|---|---|---|---|---|---|
| Score | Prediction | Score | Prediction | Score | Prediction | ||
| c.190_213del | p.64_71del HGGGWGQP | * NA | −13.135 | Deleterious | * NA | ||
| c.301A>G | Ser101Gly | 0.000 | Benign | −0.260 | Neutral | 85 | Probably benign |
| c.489C>G | Asp163Glu | 0.001 | Benign | −0.194 | Neutral | 85 | Probably benign |
| c.545A>G | Asp182Glu | 0.999 | Probably damaging | −1.452 | Neutral | 361 | Possibly damaging |
| c.546C>A | Asp182Gly | 1 | Probably damaging | −1.909 | Neutral | 361 | Possibly damaging |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, D.-J.; Kim, Y.-C.; Kim, A.-D.; Jeong, B.-H. Novel Polymorphisms and Genetic Characteristics of the Prion Protein Gene (PRNP) in Dogs—A Resistant Animal of Prion Disease. Int. J. Mol. Sci. 2020, 21, 4160. https://doi.org/10.3390/ijms21114160
Kim D-J, Kim Y-C, Kim A-D, Jeong B-H. Novel Polymorphisms and Genetic Characteristics of the Prion Protein Gene (PRNP) in Dogs—A Resistant Animal of Prion Disease. International Journal of Molecular Sciences. 2020; 21(11):4160. https://doi.org/10.3390/ijms21114160
Chicago/Turabian StyleKim, Dong-Ju, Yong-Chan Kim, An-Dang Kim, and Byung-Hoon Jeong. 2020. "Novel Polymorphisms and Genetic Characteristics of the Prion Protein Gene (PRNP) in Dogs—A Resistant Animal of Prion Disease" International Journal of Molecular Sciences 21, no. 11: 4160. https://doi.org/10.3390/ijms21114160
APA StyleKim, D.-J., Kim, Y.-C., Kim, A.-D., & Jeong, B.-H. (2020). Novel Polymorphisms and Genetic Characteristics of the Prion Protein Gene (PRNP) in Dogs—A Resistant Animal of Prion Disease. International Journal of Molecular Sciences, 21(11), 4160. https://doi.org/10.3390/ijms21114160






