Quantitative Phosphoproteomics Reveals Cell Alignment and Mitochondrial Length Change under Cyclic Stretching in Lung Cells
Abstract
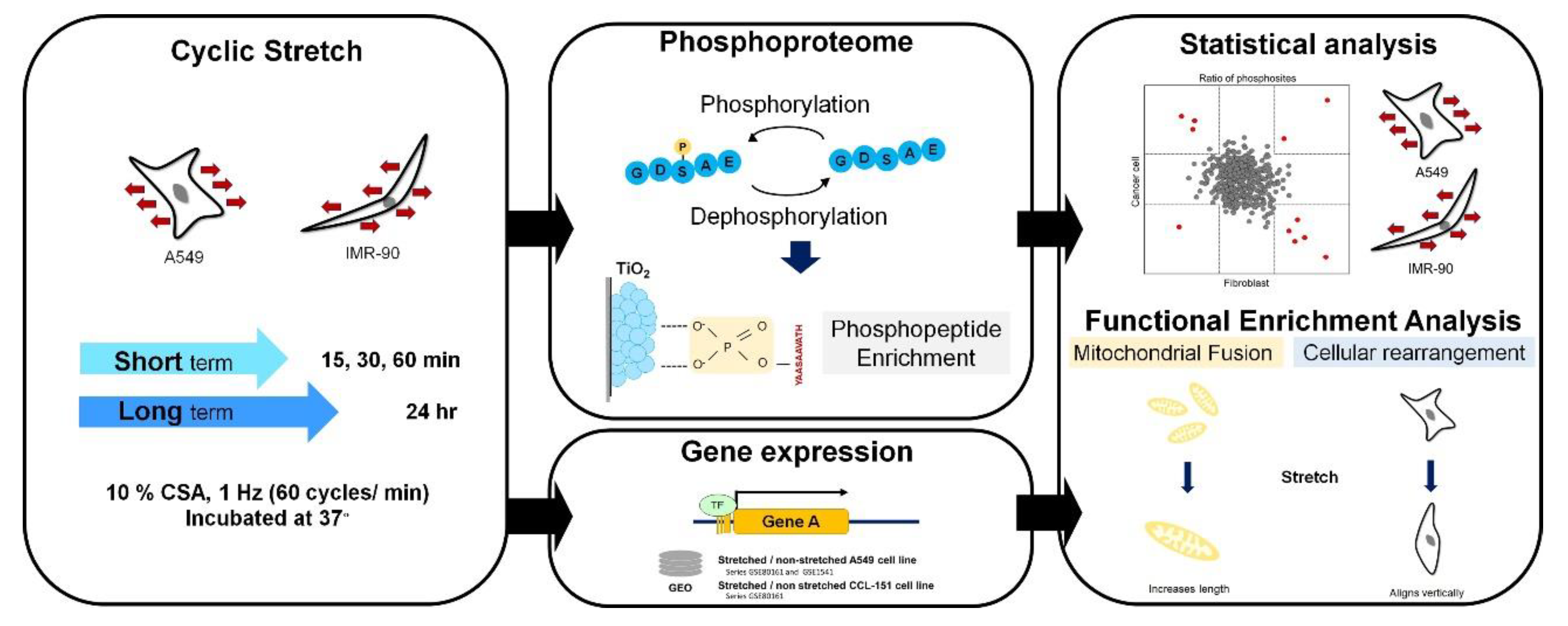
1. Introduction
2. Results
2.1. Cell Surface Area (CSA) Determination
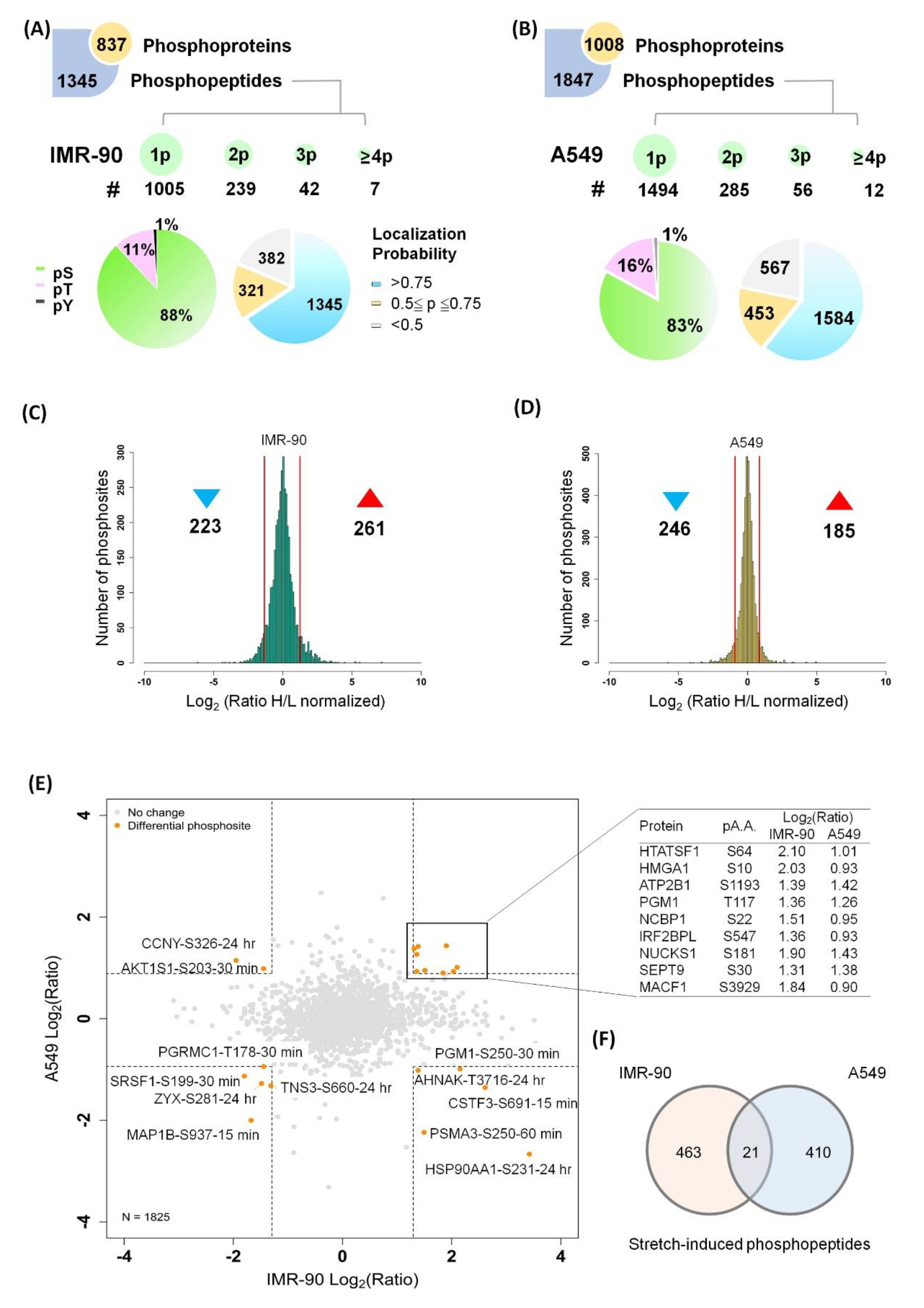
2.2. Quantitative Phosphoproteome of Lung Cells in Response to Cyclic Stretching
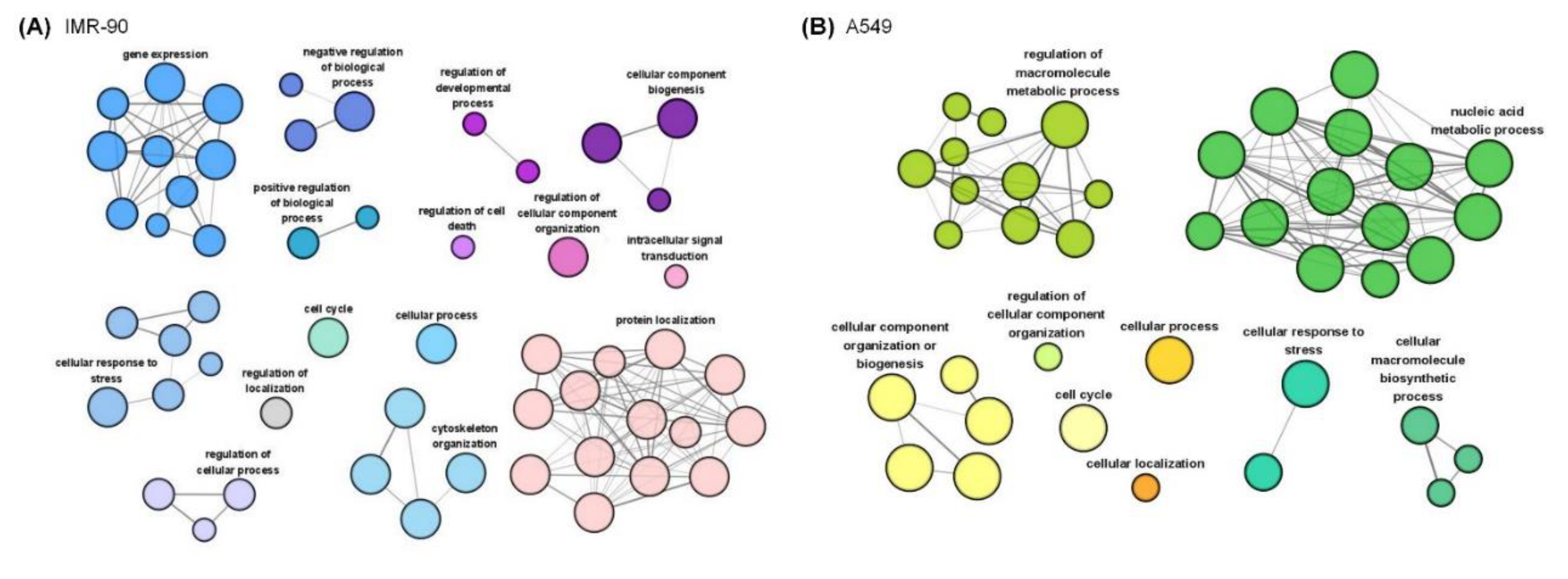
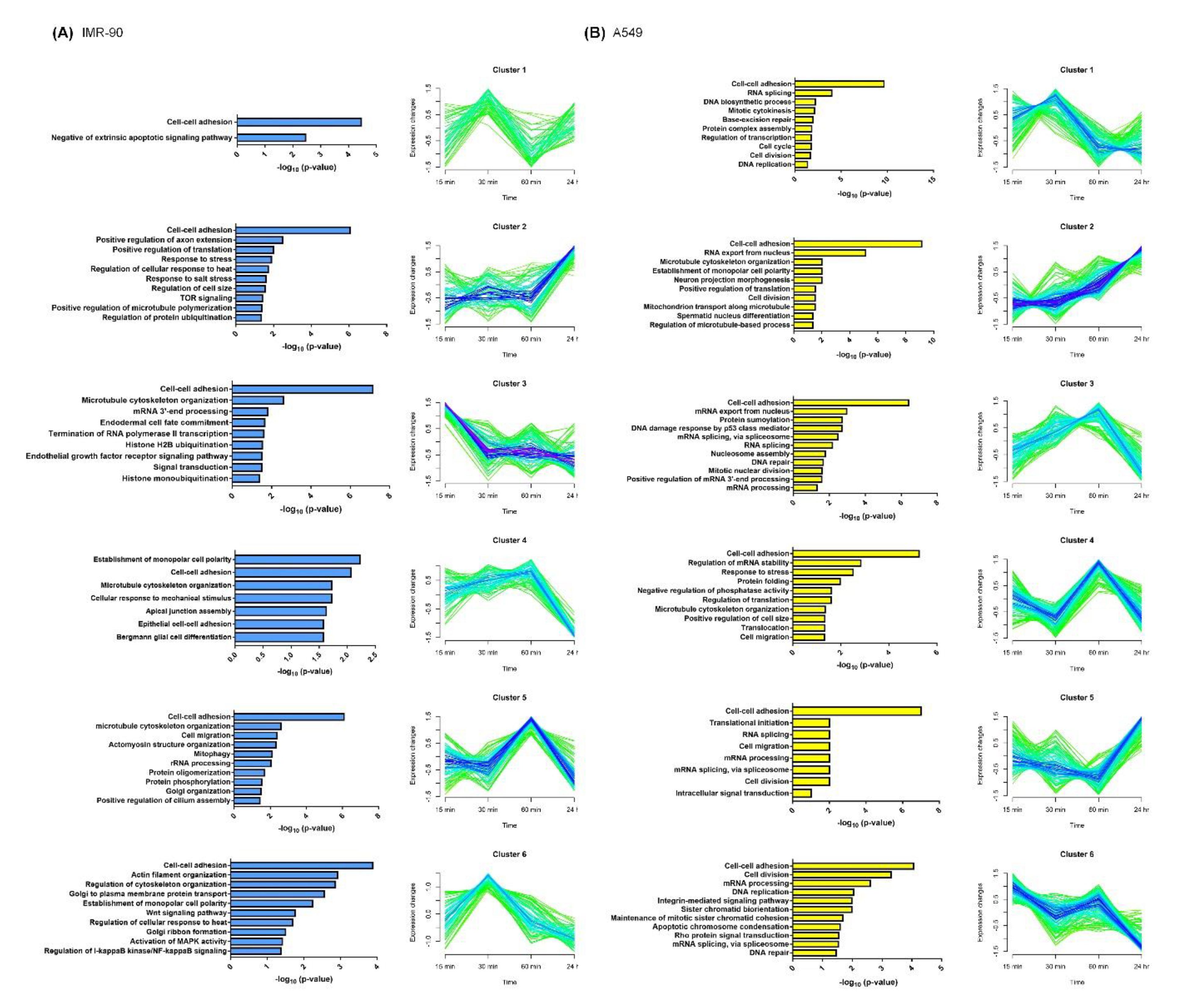
2.3. Functional Enrichment of Stretching-Regulated Phosphoproteins
2.4. Functional Enrichment of Stretching-Regulated Gene Expression
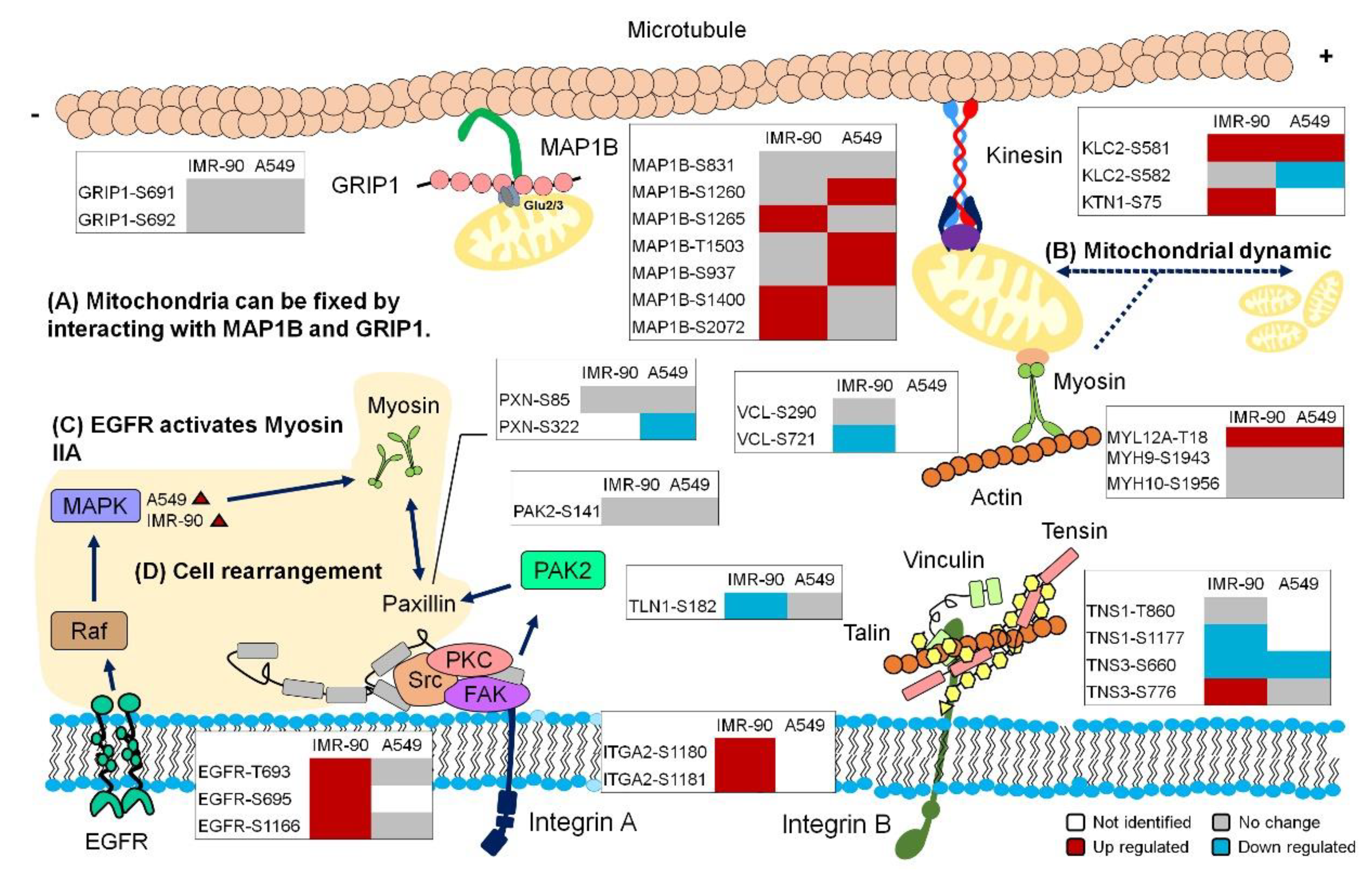
2.5. Uniaxial Cyclic Stretching Resulted in Cell Rearrangement
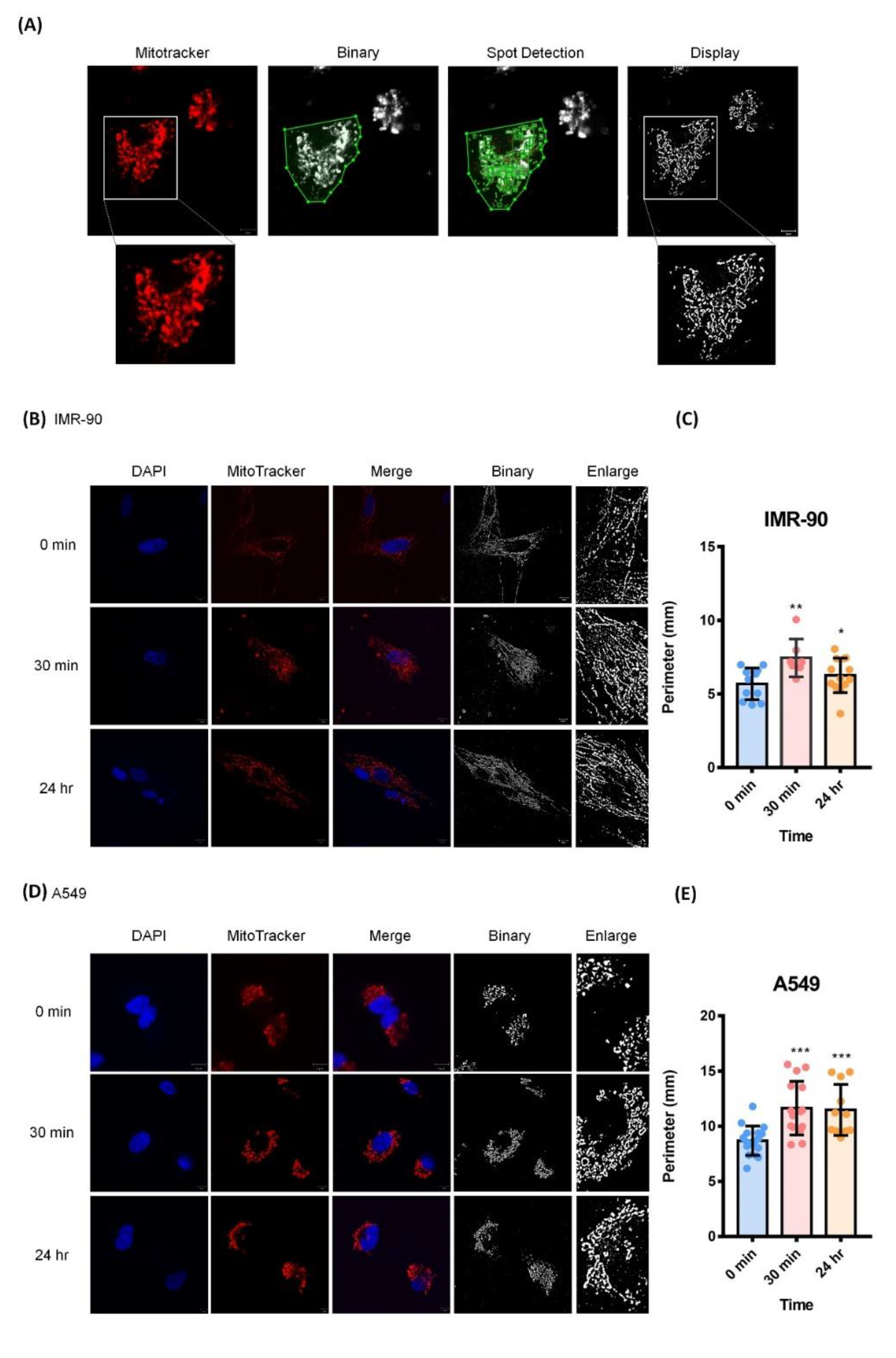
2.6. Cyclic Stretching Enhanced Mitochondrial Length
2.7. Global View of the Phosphorylation Events Induced by Cyclic Stretching
3. Discussion
4. Materials and Methods
4.1. Cell Cultures
4.2. Selection of Cyclic Stretching Conditions
4.3. Cellular Orientation Measurement
4.4. Protein Extraction
4.5. Phosphoproteome Experiments
4.6. Nano LC-MS/MS Analysis
4.7. Phosphoproteomics Data Processing and Analysis
4.8. Functional Enrichment Analysis of Differential Phosphoproteins
4.9. Transcriptomics Data Analysis
4.10. Immunofluorescence Staining
4.11. Mitochondrial Image Analysis
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wirtz, H.R.; Dobbs, L.G. The effects of mechanical forces on lung functions. Respir. Physiol. 2000, 119, 1–17. [Google Scholar] [CrossRef]
- Matsugaki, A.; Fujiwara, N.; Nakano, T. Continuous cyclic stretch induces osteoblast alignment and formation of anisotropic collagen fiber matrix. Acta Biomater. 2013, 9, 7227–7235. [Google Scholar] [CrossRef] [PubMed]
- Sasai, Y. Cytosystems dynamics in self-organization of tissue architecture. Nature 2013, 493, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Le, H.Q.; Ghatak, S.; Yeung, C.Y.; Tellkamp, F.; Gunschmann, C.; Dieterich, C.; Yeroslaviz, A.; Habermann, B.; Pombo, A.; Niessen, C.M.; et al. Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat. Cell Biol. 2016, 18, 864–875. [Google Scholar] [CrossRef]
- Gudipaty, S.A.; Lindblom, J.; Loftus, P.D.; Redd, M.J.; Edes, K.; Davey, C.F.; Krishnegowda, V.; Rosenblatt, J. Mechanical stretch triggers rapid epithelial cell division through Piezo1. Nature 2017, 543, 118–121. [Google Scholar] [CrossRef]
- Reimand, J.; Wagih, O.; Bader, G.D. The mutational landscape of phosphorylation signaling in cancer. Sci. Rep. 2013, 3, 2651. [Google Scholar] [CrossRef]
- Macek, B.; Mann, M.; Olsen, J.V. Global and site-specific quantitative phosphoproteomics: Principles and applications. Annu. Rev. Pharmacol. Toxicol. 2009, 49, 199–221. [Google Scholar] [CrossRef]
- Palumbo, A.M.; Smith, S.A.; Kalcic, C.L.; Dantus, M.; Stemmer, P.M.; Reid, G.E. Tandem mass spectrometry strategies for phosphoproteome analysis. Mass Spectrom. Rev. 2011, 30, 600–625. [Google Scholar] [CrossRef]
- Chen, Z.; Lei, C.; Wang, C.; Li, N.; Srivastava, M.; Tang, M.; Zhang, H.; Choi, J.M.; Jung, S.Y.; Qin, J.; et al. Global phosphoproteomic analysis reveals ARMC10 as an AMPK substrate that regulates mitochondrial dynamics. Nat. Commun. 2019, 10, 104. [Google Scholar] [CrossRef]
- Cheung, C.H.Y.; Hsu, C.L.; Chen, K.P.; Chong, S.T.; Wu, C.H.; Huang, H.C.; Juan, H.F. MCM2-regulated functional networks in lung cancer by multi-dimensional proteomic approach. Sci. Rep. 2017, 7, 13302. [Google Scholar] [CrossRef]
- Montenegro, M.F.; Valdivia, A.; Smolensky, A.; Verma, K.; Taylor, W.R.; San Martin, A. Nox4-dependent activation of cofilin mediates VSMC reorientation in response to cyclic stretching. Free Radic. Biol. Med. 2015, 85, 288–294. [Google Scholar] [CrossRef] [PubMed]
- Richard, M.N.; Deniset, J.F.; Kneesh, A.L.; Blackwood, D.; Pierce, G.N. Mechanical stretching stimulates smooth muscle cell growth, nuclear protein import, and nuclear pore expression through mitogen-activated protein kinase activation. J. Biol. Chem. 2007, 282, 23081–23088. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L. Role of FAK-ERK1/2 Signaling Pathway in roliferation of Rat Bone-marrow Mesenchymal Stem Cells Stimulated by Cyclic Stretching. J. Med. Biol. Eng. 2013, 33, 229. [Google Scholar] [CrossRef]
- Cohen, T.S.; Gray Lawrence, G.; Khasgiwala, A.; Margulies, S.S. MAPK activation modulates permeability of isolated rat alveolar epithelial cell monolayers following cyclic stretch. PLoS ONE 2010, 5, e10385. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.H.; Mungai, P.T.; Schumacker, P.T. Stretch-induced phosphorylation of focal adhesion kinase in endothelial cells: Role of mitochondrial oxidants. Am. J. Physiol. Lung Cell. Mol. Physiol. 2006, 291, L38–L45. [Google Scholar] [CrossRef]
- Pan, J.; Singh, U.S.; Takahashi, T.; Oka, Y.; Palm-Leis, A.; Herbelin, B.S.; Baker, K.M. PKC mediates cyclic stretch-induced cardiac hypertrophy through Rho family GTPases and mitogen-activated protein kinases in cardiomyocytes. J. Cell. Physiol. 2005, 202, 536–553. [Google Scholar] [CrossRef]
- Sugiyama, N.; Masuda, T.; Shinoda, K.; Nakamura, A.; Tomita, M.; Ishihama, Y. Phosphopeptide enrichment by aliphatic hydroxy acid-modified metal oxide chromatography for nano-LC-MS/MS in proteomics applications. Mol. Cell. Proteom. 2007, 6, 1103–1109. [Google Scholar] [CrossRef]
- Chapman, K.E.; Sinclair, S.E.; Zhuang, D.; Hassid, A.; Desai, L.P.; Waters, C.M. Cyclic mechanical strain increases reactive oxygen species production in pulmonary epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2005, 289, L834–L841. [Google Scholar] [CrossRef]
- Tschumperlin, D.J.; Margulies, S.S. Alveolar epithelial surface area-volume relationship in isolated rat lungs. J. Appl. Physiol. 1999, 86, 2026–2033. [Google Scholar] [CrossRef]
- Chen, J.; Liu, Z.; Zhong, G.; Li, Z.; Qian, L.; Li, X.; Chen, B.; Lao, L.; Wang, H. Cyclic stretch enhances apoptosis in human lumbar ligamentum fl avum cells via the induction of reactive oxygen species generation. J. Spinal Cord Med. 2016, 39, 450–454. [Google Scholar] [CrossRef]
- Liao, X.D.; Wang, X.H.; Jin, H.J.; Chen, L.Y.; Chen, Q. Mechanical stretch induces mitochondria-dependent apoptosis in neonatal rat cardiomyocytes and G2 /M accumulation in cardiac fibroblasts. Cell Res. 2004, 14, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Murata, N.; Ito, S.; Furuya, K.; Takahara, N.; Naruse, K.; Aso, H.; Kondo, M.; Sokabe, M.; Hasegawa, Y. Ca2+ influx and ATP release mediated by mechanical stretch in human lung fibroblasts. Biochem. Biophys. Res. Commun. 2014, 453, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Kaunas, R.; Nguyen, P.; Usami, S.; Chien, S. Cooperative effects of Rho and mechanical stretch on stress fiber organization. Proc. Natl. Acad. Sci. USA 2005, 102, 15895–15900. [Google Scholar] [CrossRef] [PubMed]
- Geiger, R.C.; Taylor, W.; Glucksberg, M.R.; Dean, D.A. Cyclic stretch-induced reorganization of the cytoskeleton and its role in enhanced gene transfer. Gene Ther. 2006, 13, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Waters, C.M.; Ridge, K.M.; Sunio, G.; Venetsanou, K.; Sznajder, J.I. Mechanical stretching of alveolar epithelial cells increases Na(+)-K(+)-ATPase activity. J. Appl. Physiol. 1999, 87, 715–721. [Google Scholar] [CrossRef] [PubMed]
- Abiko, H.; Fujiwara, S.; Ohashi, K.; Hiatari, R.; Mashiko, T.; Sakamoto, N.; Sato, M.; Mizuno, K. Rho guanine nucleotide exchange factors involved in cyclic-stretch-induced reorientation of vascular endothelial cells. J. Cell Sci. 2015, 128, 1683–1695. [Google Scholar] [CrossRef]
- Mustafa, S.B.; Isaac, J.; Seidner, S.R.; Dixon, P.S.; Henson, B.M.; DiGeronimo, R.J. Mechanical stretch induces lung alpha-epithelial Na(+) channel expression. Exp. Lung Res. 2014, 40, 380–391. [Google Scholar] [CrossRef]
- Livne, A.; Bouchbinder, E.; Geiger, B. Cell reorientation under cyclic stretching. Nat. Commun. 2014, 5, 3938. [Google Scholar] [CrossRef]
- Hoffman, B.D.; Grashoff, C.; Schwartz, M.A. Dynamic molecular processes mediate cellular mechanotransduction. Nature 2011, 475, 316–323. [Google Scholar] [CrossRef]
- Basson, M.A. Signaling in cell differentiation and morphogenesis. Cold Spring Harb. Perspect. Biol. 2012, 4, a008151. [Google Scholar] [CrossRef]
- Heisenberg, C.P.; Bellaiche, Y. Forces in tissue morphogenesis and patterning. Cell 2013, 153, 948–962. [Google Scholar] [CrossRef] [PubMed]
- van Helvert, S.; Storm, C.; Friedl, P. Mechanoreciprocity in cell migration. Nat. Cell Biol. 2018, 20, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Kumar, L.; Futschik, M.E. Mfuzz: A software package for soft clustering of microarray data. Bioinformation 2007, 2, 5–7. [Google Scholar] [CrossRef]
- Da Huang, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Ravandi, F.; Talpaz, M.; Estrov, Z. Modulation of cellular signaling pathways: Prospects for targeted therapy in hematological malignancies. Clin. Cancer Res. 2003, 9, 535–550. [Google Scholar]
- Yordy, J.S.; Muise-Helmericks, R.C. Signal transduction and the Ets family of transcription factors. Oncogene 2000, 19, 6503–6513. [Google Scholar] [CrossRef]
- Nardone, G.; Oliver-De La Cruz, J.; Vrbsky, J.; Martini, C.; Pribyl, J.; Skladal, P.; Pesl, M.; Caluori, G.; Pagliari, S.; Martino, F.; et al. YAP regulates cell mechanics by controlling focal adhesion assembly. Nat. Commun. 2017, 8, 15321. [Google Scholar] [CrossRef]
- Smoot, R.L.; Werneburg, N.W.; Sugihara, T.; Hernandez, M.C.; Yang, L.; Mehner, C.; Graham, R.P.; Bronk, S.F.; Truty, M.J.; Gores, G.J. Platelet-derived growth factor regulates YAP transcriptional activity via Src family kinase dependent tyrosine phosphorylation. J. Cell Biochem. 2018, 119, 824–836. [Google Scholar] [CrossRef]
- dos Santos, C.C.; Han, B.; Andrade, C.F.; Bai, X.; Uhlig, S.; Hubmayr, R.; Tsang, M.; Lodyga, M.; Keshavjee, S.; Slutsky, A.S.; et al. DNA microarray analysis of gene expression in alveolar epithelial cells in response to TNFalpha, LPS, and cyclic stretch. Physiol. Genom. 2004, 19, 331–342. [Google Scholar] [CrossRef]
- Benard, G.; Karbowski, M. Mitochondrial fusion and division: Regulation and role in cell viability. Semin. Cell Dev. Biol. 2009, 20, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Wai, T.; Langer, T. Mitochondrial Dynamics and Metabolic Regulation. Trends Endocrinol. Metab. 2016, 27, 105–117. [Google Scholar] [CrossRef] [PubMed]
- de Chaumont, F.; Dallongeville, S.; Chenouard, N.; Hervé, N.; Pop, S.; Provoost, T.; Meas-Yedid, V.; Pankajakshan, P.; Lecomte, T.; Le Montagner, Y.; et al. Icy: An open bioimage informatics platform for extended reproducible research. Nat. Methods 2012, 9, 690–696. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lu, M.; Xiong, L.; Fan, J.; Zhou, Y.; Li, H.; Peng, X.; Zhong, Z.; Wang, Y.; Huang, F.; et al. Drp1-mediated mitochondrial fission promotes renal fibroblast activation and fibrogenesis. Cell Death Dis. 2020, 11, 29. [Google Scholar] [CrossRef] [PubMed]
- Westrate, L.M.; Drocco, J.A.; Martin, K.R.; Hlavacek, W.S.; MacKeigan, J.P. Mitochondrial morphological features are associated with fission and fusion events. PLoS ONE 2014, 9, e95265. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Zhang, P.; Song, I.; Petralia, R.S.; Liao, D.; Huganir, R.L. Characterization of the glutamate receptor-interacting proteins GRIP1 and GRIP2. J. Neurosci. 1999, 19, 6930–6941. [Google Scholar] [CrossRef]
- Palenzuela, R.; Gutierrez, Y.; Draffin, J.E.; Lario, A.; Benoist, M.; Esteban, J.A. MAP1B Light Chain Modulates Synaptic Transmission via AMPA Receptor Intracellular Trapping. J. Neurosci. 2017, 37, 9945–9963. [Google Scholar] [CrossRef]
- Moore, A.S.; Wong, Y.C.; Simpson, C.L.; Holzbaur, E.L. Dynamic actin cycling through mitochondrial subpopulations locally regulates the fission-fusion balance within mitochondrial networks. Nat. Commun. 2016, 7, 12886. [Google Scholar] [CrossRef]
- Picca, A.; Mankowski, R.T.; Burman, J.L.; Donisi, L.; Kim, J.S.; Marzetti, E.; Leeuwenburgh, C. Mitochondrial quality control mechanisms as molecular targets in cardiac ageing. Nat. Rev. Cardiol. 2018, 15, 543–554. [Google Scholar] [CrossRef]
- Reyes, A.; He, J.; Mao, C.C.; Bailey, L.J.; Di Re, M.; Sembongi, H.; Kazak, L.; Dzionek, K.; Holmes, J.B.; Cluett, T.J.; et al. Actin and myosin contribute to mammalian mitochondrial DNA maintenance. Nucleic Acids Res. 2011, 39, 5098–5108. [Google Scholar] [CrossRef]
- Korobova, F.; Gauvin, T.J.; Higgs, H.N. A role for myosin II in mammalian mitochondrial fission. Curr. Biol. 2014, 24, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Yao, M.; Goult, B.T.; Klapholz, B.; Hu, X.; Toseland, C.P.; Guo, Y.; Cong, P.; Sheetz, M.P.; Yan, J. The mechanical response of talin. Nat. Commun. 2016, 7, 11966. [Google Scholar] [CrossRef] [PubMed]
- Stutchbury, B.; Atherton, P.; Tsang, R.; Wang, D.-Y.; Ballestrem, C. Distinct focal adhesion protein modules control different aspects of mechanotransduction. J. Cell Sci. 2017, 130, 1612–1624. [Google Scholar] [CrossRef] [PubMed]
- Rubsam, M.; Mertz, A.F.; Kubo, A.; Marg, S.; Jungst, C.; Goranci-Buzhala, G.; Schauss, A.C.; Horsley, V.; Dufresne, E.R.; Moser, M.; et al. E-cadherin integrates mechanotransduction and EGFR signaling to control junctional tissue polarization and tight junction positioning. Nat. Commun. 2017, 8, 1250. [Google Scholar] [CrossRef]
- Kippenberger, S.; Loitsch, S.; Guschel, M.; Muller, J.; Knies, Y.; Kaufmann, R.; Bernd, A. Mechanical stretch stimulates protein kinase B/Akt phosphorylation in epidermal cells via angiotensin II type 1 receptor and epidermal growth factor receptor. J. Biol. Chem. 2005, 280, 3060–3067. [Google Scholar] [CrossRef]
- Wozniak, M.A.; Modzelewska, K.; Kwong, L.; Keely, P.J. Focal adhesion regulation of cell behavior. Biochim. Biophys. Acta 2004, 1692, 103–119. [Google Scholar] [CrossRef]
- Wu, J.C.; Chen, Y.C.; Kuo, C.T.; Yu, H.W.; Chen, Y.Q.; Chiou, A.; Kuo, J.C. Focal adhesion kinase-dependent focal adhesion recruitment of SH2 domains directs SRC into focal adhesions to regulate cell adhesion and migration. Sci. Rep. 2015, 5, 18476. [Google Scholar] [CrossRef]
- Kuo, J.C. Mechanotransduction at focal adhesions: Integrating cytoskeletal mechanics in migrating cells. J. Cell Mol. Med. 2013, 17, 704–712. [Google Scholar] [CrossRef]
- Morioka, M.; Parameswaran, H.; Naruse, K.; Kondo, M.; Sokabe, M.; Hasegawa, Y.; Suki, B.; Ito, S. Microtubule dynamics regulate cyclic stretch-induced cell alignment in human airway smooth muscle cells. PLoS ONE 2011, 6, e26384. [Google Scholar] [CrossRef]
- Aoki, M.S.; Miyabara, E.H.; Soares, A.G.; Saito, E.T.; Moriscot, A.S. mTOR pathway inhibition attenuates skeletal muscle growth induced by stretching. Cell Tissue Res. 2006, 324, 149–156. [Google Scholar] [CrossRef]
- Ma, J.; Zhao, D.; Wu, Y.; Xu, C.; Zhang, F. Cyclic stretch induced gene expression of extracellular matrix and adhesion molecules in human periodontal ligament cells. Arch. Oral Biol. 2015, 60, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Nakai, N.; Kawano, F.; Nakata, K. Mechanical stretch activates mammalian target of rapamycin and AMP-activated protein kinase pathways in skeletal muscle cells. Mol. Cell Biochem. 2015, 406, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.G.; Miyazu, M.; Xiang, P.; Li, S.N.; Sokabe, M.; Naruse, K. Stretch-induced cell proliferation is mediated by FAK-MAPK pathway. Life Sci. 2005, 76, 2817–2825. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Fukuda, K.; Saito, M.; Matsuzaki, J.; Kodama, H.; Sano, M.; Takahashi, T.; Kato, T.; Ogawa, S. Mechanical Stretch Activates the JAK/STAT Pathway in Rat Cardiomyocytes. Circ. Res. 1999, 84, 1127–1136. [Google Scholar] [CrossRef]
- Scholz, R.; Imami, K.; Scott, N.E.; Trimble, W.S.; Foster, L.J.; Finlay, B.B. Novel Host Proteins and Signaling Pathways in Enteropathogenic E. coli Pathogenesis Identified by Global Phosphoproteome Analysis. Mol. Cell. Proteom. 2015, 14, 1927–1945. [Google Scholar] [CrossRef]
- Cavanaugh, K.J.; Oswari, J.; Margulies, S.S. Role of stretch on tight junction structure in alveolar epithelial cells. Am. J. Respir. Cell Mol. 2001, 25, 584–591. [Google Scholar] [CrossRef]
- Zhou, J.; Zeng, Y.; Cui, L.; Chen, X.; Stauffer, S.; Wang, Z.; Yu, F.; Lele, S.M.; Talmon, G.A.; Black, A.R.; et al. Zyxin promotes colon cancer tumorigenesis in a mitotic phosphorylation-dependent manner and through CDK8-mediated YAP activation. Proc. Natl. Acad. Sci. USA 2018, 115, E6760–E6769. [Google Scholar] [CrossRef]
- Ben-Porath, I.; Weinberg, R.A. When cells get stressed: An integrative view of cellular senescence. J. Clin. Investig. 2004, 113, 8–13. [Google Scholar] [CrossRef]
- He, C.L.; Bian, Y.Y.; Xue, Y.; Liu, Z.X.; Zhou, K.Q.; Yao, C.F.; Lin, Y.; Zou, H.F.; Luo, F.X.; Qu, Y.Y.; et al. Pyruvate Kinase M2 Activates mTORC1 by Phosphorylating AKT1S1. Sci. Rep. 2016, 6, 21524. [Google Scholar] [CrossRef]
- Mishra, P.; Chan, D.C. Metabolic regulation of mitochondrial dynamics. J. Cell Biol. 2016, 212, 379–387. [Google Scholar] [CrossRef]
- Yu, S.B.; Pekkurnaz, G. Mechanisms Orchestrating Mitochondrial Dynamics for Energy Homeostasis. J. Mol. Biol. 2018, 430, 3922–3941. [Google Scholar] [CrossRef] [PubMed]
- Rog-Zielinska, E.A.; O’Toole, E.T.; Hoenger, A.; Kohl, P. Mitochondrial Deformation During the Cardiac Mechanical Cycle. Anat. Rec. (Hoboken) 2019, 302, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Bartolak-Suki, E.; Suki, B. Tuning mitochondrial structure and function to criticality by fluctuation-driven mechanotransduction. Sci. Rep. 2020, 10, 407. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.W.; Lin, M.H.; Huang, H.C.; Ku, W.C.; Yi, T.H.; Tsai, C.F.; Chen, Y.J.; Sugiyama, N.; Ishihama, Y.; Juan, H.F.; et al. Phosphoproteomic analysis of Rhodopseudomonas palustris reveals the role of pyruvate phosphate dikinase phosphorylation in lipid production. J. Proteome Res. 2012, 11, 5362–5375. [Google Scholar] [CrossRef] [PubMed]
- Kyono, Y.; Sugiyama, N.; Imami, K.; Tomita, M.; Ishihama, Y. Successive and selective release of phosphorylated peptides captured by hydroxy acid-modified metal oxide chromatography. J. Proteome Res. 2008, 7, 4585–4593. [Google Scholar] [CrossRef] [PubMed]
- Rappsilber, J.; Mann, M.; Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007, 2, 1896–1906. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Supek, F.; Bosnjak, M.; Skunca, N.; Smuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef]
- Pagliuso, A.; Tham, T.N.; Stevens, J.K.; Lagache, T.; Persson, R.; Salles, A.; Olivo-Marin, J.C.; Oddos, S.; Spang, A.; Cossart, P.; et al. A role for septin 2 in Drp1-mediated mitochondrial fission. EMBO Rep. 2016, 17, 858–873. [Google Scholar] [CrossRef] [PubMed]
- Mitra, K.; Lippincott-Schwartz, J. Analysis of Mitochondrial Dynamics and Functions Using Imaging Approaches. Curr. Protoc. Cell Biol. 2010, 46, 4–25. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.-H.; Hsu, C.-L.; Huang, H.-C.; Juan, H.-F. Quantitative Phosphoproteomics Reveals Cell Alignment and Mitochondrial Length Change under Cyclic Stretching in Lung Cells. Int. J. Mol. Sci. 2020, 21, 4074. https://doi.org/10.3390/ijms21114074
Wang W-H, Hsu C-L, Huang H-C, Juan H-F. Quantitative Phosphoproteomics Reveals Cell Alignment and Mitochondrial Length Change under Cyclic Stretching in Lung Cells. International Journal of Molecular Sciences. 2020; 21(11):4074. https://doi.org/10.3390/ijms21114074
Chicago/Turabian StyleWang, Wei-Hsuan, Chia-Lang Hsu, Hsuan-Cheng Huang, and Hsueh-Fen Juan. 2020. "Quantitative Phosphoproteomics Reveals Cell Alignment and Mitochondrial Length Change under Cyclic Stretching in Lung Cells" International Journal of Molecular Sciences 21, no. 11: 4074. https://doi.org/10.3390/ijms21114074
APA StyleWang, W.-H., Hsu, C.-L., Huang, H.-C., & Juan, H.-F. (2020). Quantitative Phosphoproteomics Reveals Cell Alignment and Mitochondrial Length Change under Cyclic Stretching in Lung Cells. International Journal of Molecular Sciences, 21(11), 4074. https://doi.org/10.3390/ijms21114074




