On-Target CRISPR/Cas9 Activity Can Cause Undesigned Large Deletion in Mouse Zygotes
Abstract
1. Introduction
2. Results
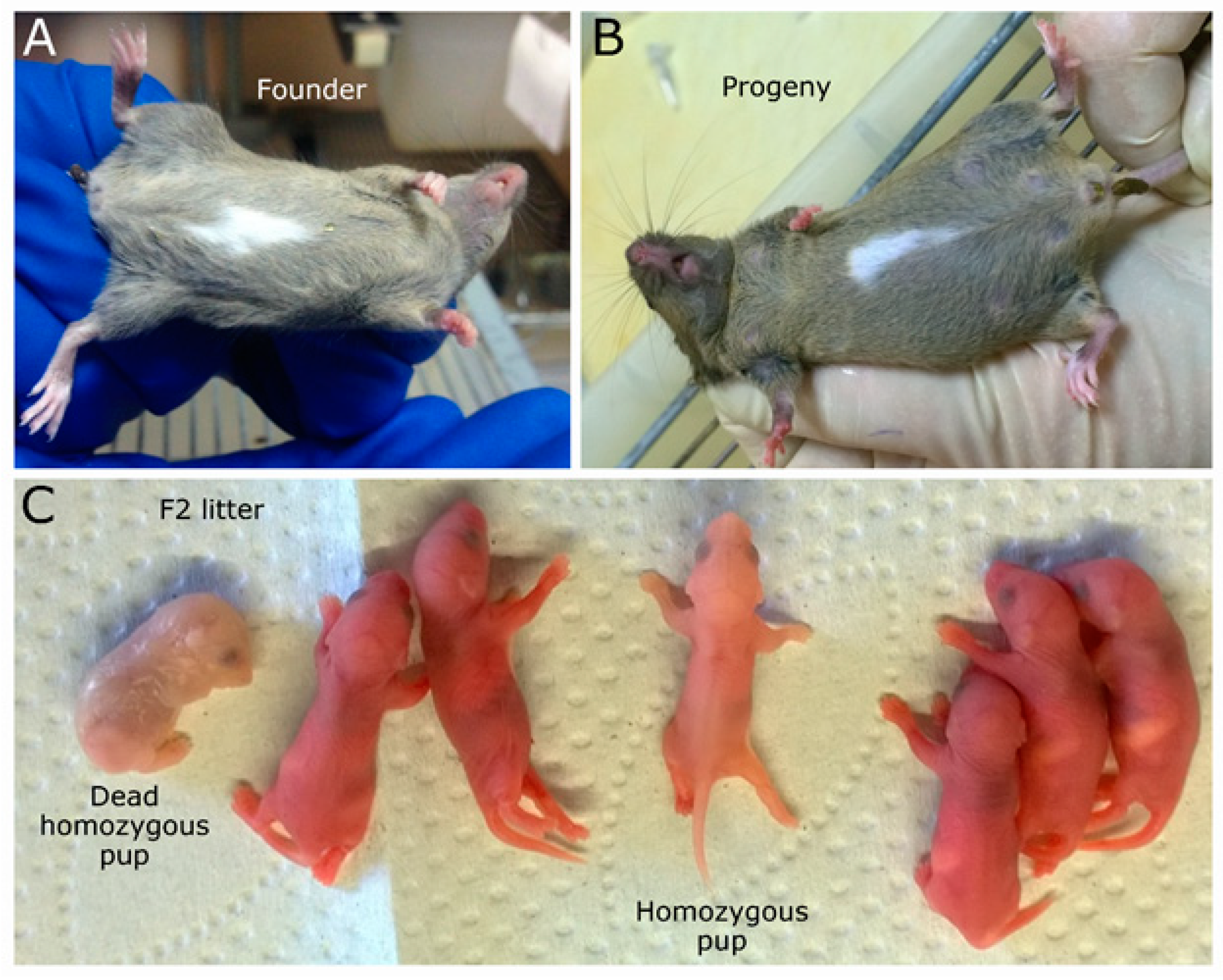
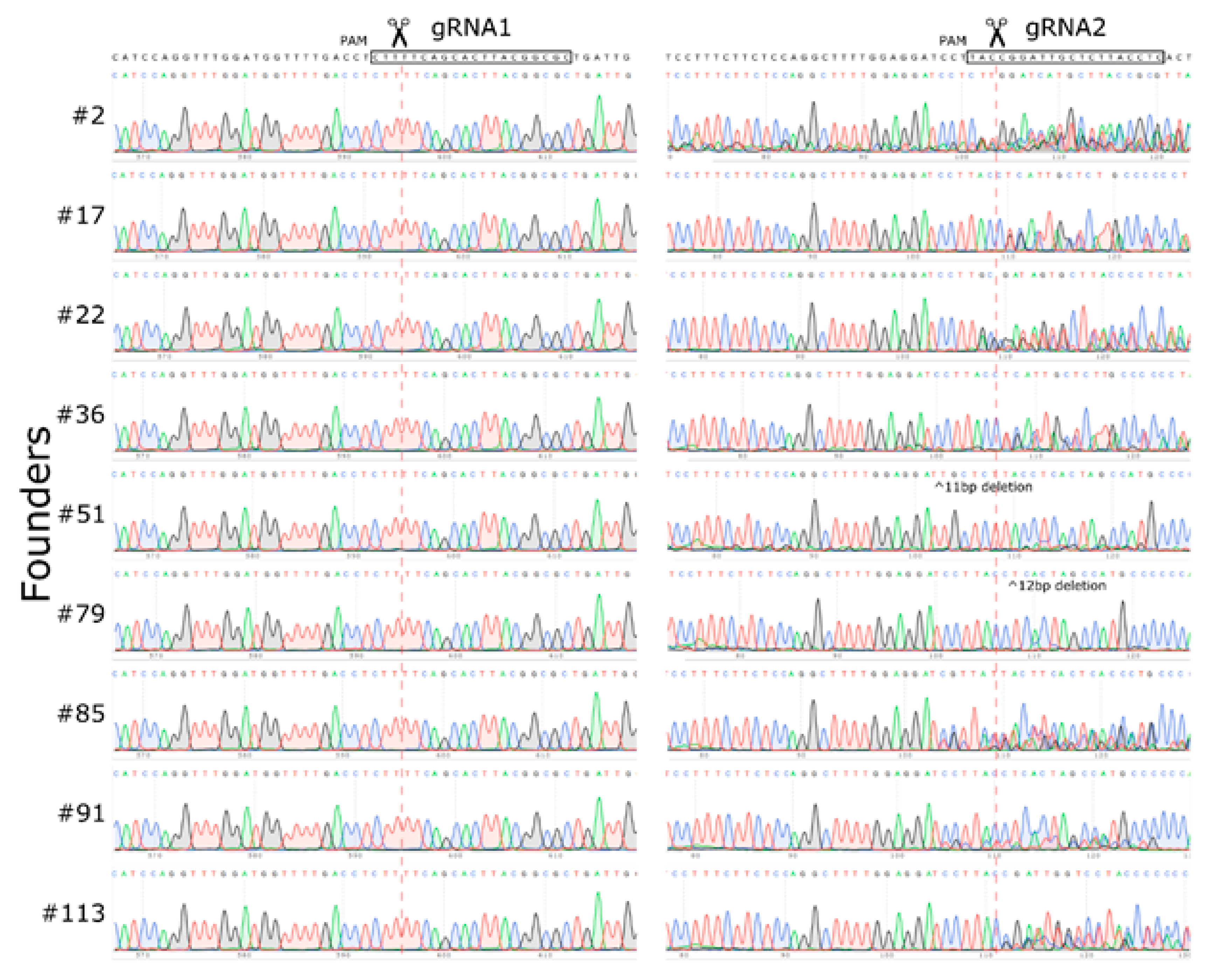
2.1. Mouse Genome Editing
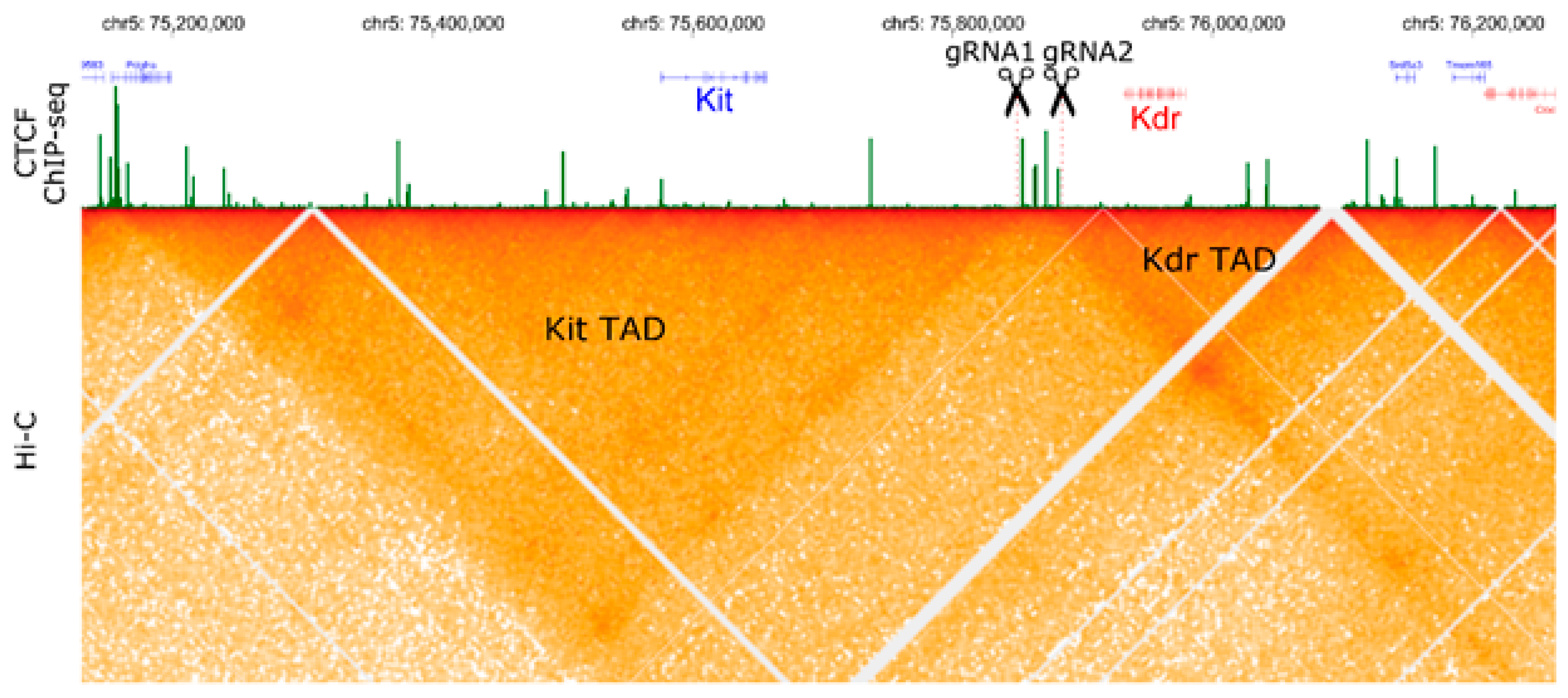
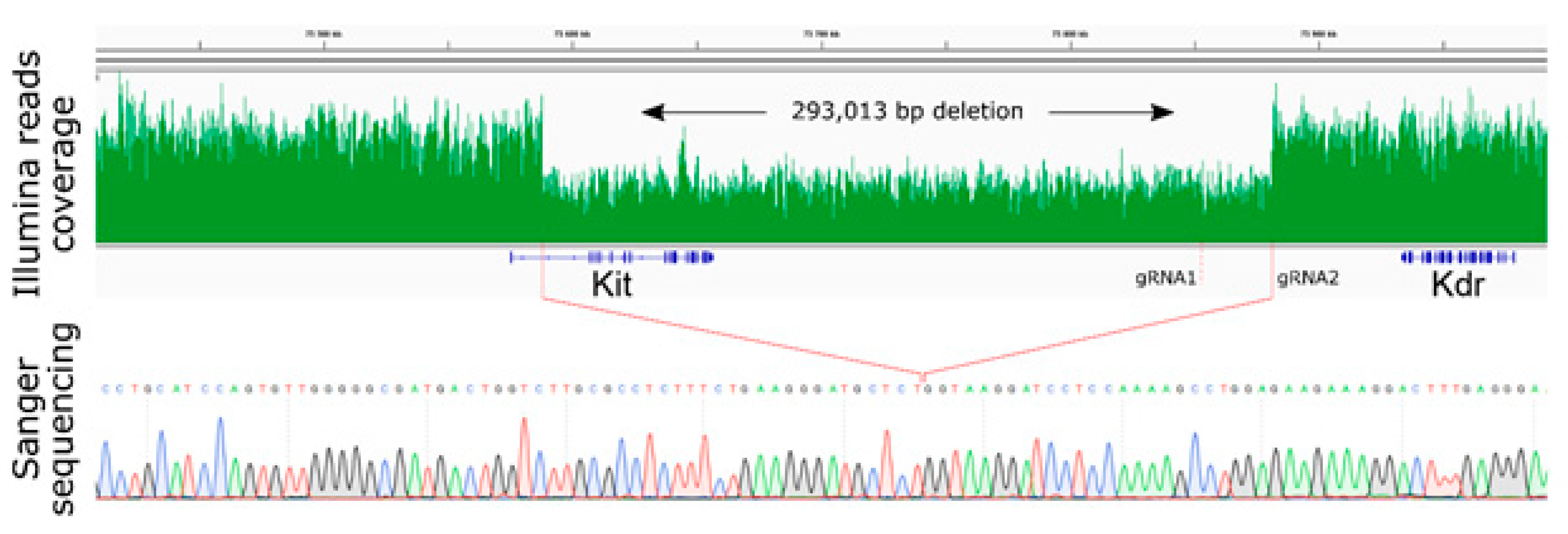
2.2. Large Unexpected Deletion in the Target Region
2.3. Off-Target or Truncation
3. Discussion
4. Materials and Methods
4.1. gRNA Design and gRNA and Cas9 mRNA Preparation
4.2. Preparation of gRNA and Cas9 mRNA
4.3. Animals
4.4. Microinjection
4.5. Genotyping
4.6. Whole Genome Sequencing
4.7. Data Availability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Crowther, M.D.; Dolton, G.; Legut, M.; Caillaud, M.E.; Lloyd, A.; Attaf, M.; Galloway, S.A.E.; Rius, C.; Farrell, C.P.; Szomolay, B.; et al. Genome-wide CRISPR–Cas9 screening reveals ubiquitous T cell cancer targeting via the monomorphic MHC class I-related protein MR1. Nat. Immunol. 2020, 21, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Hart, T.; Chandrashekhar, M.; Aregger, M.; Steinhart, Z.; Brown, K.R.; MacLeod, G.; Mis, M.; Zimmermann, M.; Fradet-Turcotte, A.; Sun, S.; et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell 2015, 163, 1515–1526. [Google Scholar] [CrossRef]
- Picco, G.; Chen, E.D.; Alonso, L.G.; Behan, F.M.; Gonçalves, E.; Bignell, G.; Matchan, A.; Fu, B.; Banerjee, R.; Anderson, E.; et al. Functional linkage of gene fusions to cancer cell fitness assessed by pharmacological and CRISPR-Cas9 screening. Nat. Commun. 2019, 10, 2198. [Google Scholar] [CrossRef] [PubMed]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelsen, T.S.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G.; et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science 2014, 343, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Carroll, K.J.; Makarewich, C.A.; McAnally, J.; Anderson, D.M.; Zentilin, L.; Liu, N.; Giacca, M.; Bassel-Duby, R.; Olson, E.N. A mouse model for adult cardiac-specific gene deletion with CRISPR/Cas9. Proc. Natl. Acad. Sci. USA 2016, 113, 338–343. [Google Scholar] [CrossRef] [PubMed]
- Korablev, A.N.; Serova, I.A.; Serov, O.L. Generation of megabase-scale deletions, inversions and duplications involving the Contactin-6 gene in mice by CRISPR/Cas9 technology. BMC Genet. 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- Pristyazhnyuk, I.E.; Minina, J.; Korablev, A.; Serova, I.; Fishman, V.; Gridina, M.; Rozhdestvensky, T.S.; Gubar, L.; Skryabin, B.V.; Serov, O.L. Time origin and structural analysis of the induced CRISPR/cas9 megabase-sized deletions and duplications involving the Cntn6 gene in mice. Sci. Rep. 2019, 9, 14161. [Google Scholar] [CrossRef] [PubMed]
- Kraft, K.; Geuer, S.; Will, A.J.; Chan, W.; Paliou, C.; Borschiwer, M.; Harabula, I.; Wittler, L.; Franke, M.; Ibrahim, D.M.; et al. Deletions, inversions, duplications: Engineering of structural variants using CRISPR/Cas in mice. Cell Rep. 2015, 10, 833–839. [Google Scholar] [CrossRef]
- Artus, J.; Hadjantonakis, A.-K. Generation of Chimeras by Aggregation of Embryonic Stem Cells with Diploid or Tetraploid Mouse Embryos. Methods Mol. Biol. 2011, 693, 37–56. [Google Scholar]
- Lupiáñez, D.G.; Kraft, K.; Heinrich, V.; Krawitz, P.; Brancati, F.; Klopocki, E.; Horn, D.; Kayserili, H.; Opitz, J.M.; Laxova, R.; et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 2015, 161, 1012–1025. [Google Scholar] [CrossRef]
- Yang, H.; Wang, H.; Shivalila, C.S.; Cheng, A.W.; Shi, L.; Jaenisch, R. XOne-step generation of mice carrying reporter and conditional alleles by CRISPR/cas-mediated genome engineering. Cell 2013, 154, 1370–1379. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Lee, A.; Oh, J.H.; Lee, H.W.; Ha, S.J. The R229Q mutation of Rag2 does not characterize severe immunodeficiency in mice. Sci. Rep. 2019, 9, 4415. [Google Scholar] [CrossRef] [PubMed]
- Yoshimi, K.; Kunihiro, Y.; Kaneko, T.; Nagahora, H.; Voigt, B.; Mashimo, T. ssODN-mediated knock-in with CRISPR-Cas for large genomic regions in zygotes. Nat. Commun. 2016, 7, 10431. [Google Scholar] [CrossRef] [PubMed]
- Chu, V.T.; Weber, T.; Graf, R.; Sommermann, T.; Petsch, K.; Sack, U.; Volchkov, P.; Rajewsky, K.; Kühn, R. Efficient generation of Rosa26 knock-in mice using CRISPR/Cas9 in C57BL/6 zygotes. BMC Biotechnol. 2016, 16, 4. [Google Scholar] [CrossRef]
- Kvon, E.Z.; Zhu, Y.; Kelman, G.; Novak, C.S.; Plajzer-Frick, I.; Kato, M.; Garvin, T.H.; Pham, Q.; Harrington, A.N.; Hunter, R.D.; et al. Comprehensive In Vivo Interrogation Reveals Phenotypic Impact of Human Enhancer Variants. Cell 2020, 180, 1262–1271.e15. [Google Scholar] [CrossRef]
- Boroviak, K.; Doe, B.; Banerjee, R.; Yang, F.; Bradley, A. Chromosome engineering in zygotes with CRISPR/Cas9. Genesis 2016, 54, 78–85. [Google Scholar] [CrossRef]
- Saito, R.; Koebis, M.; Nagai, T.; Shimizu, K.; Liao, J.; Wulaer, B.; Sugaya, Y.; Nagahama, K.; Uesaka, N.; Kushima, I.; et al. Comprehensive analysis of a novel mouse model of the 22q11.2 deletion syndrome: A model with the most common 3.0-Mb deletion at the human 22q11.2 locus. Transl. Psychiatry 2020, 10, 35. [Google Scholar] [CrossRef]
- Kato, T.; Hara, S.; Goto, Y.; Ogawa, Y.; Okayasu, H.; Kubota, S.; Tamano, M.; Terao, M.; Takada, S. Creation of mutant mice with megabase-sized deletions containing custom-designed breakpoints by means of the CRISPR/Cas9 system. Sci. Rep. 2017, 7, 59. [Google Scholar] [CrossRef]
- German, D.M.; Mitalipov, S.; Mishra, A.; Kaul, S. Therapeutic Genome Editing in Cardiovascular Diseases. JACC Basic Transl. Sci. 2019, 4, 122–131. [Google Scholar] [CrossRef]
- Porteus, M.H. Towards a new era in medicine: Therapeutic genome editing. Genome Biol. 2015, 16, 286. [Google Scholar] [CrossRef]
- Kim, D.; Kim, J.-S. DIG-seq: A genome-wide CRISPR off-target profiling method using chromatin DNA. Genome Res. 2018, 28, 1894–1900. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.; Park, J.; Kim, J.-S. Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 2014, 30, 1473–1475. [Google Scholar] [CrossRef] [PubMed]
- Cradick, T.J.; Qiu, P.; Lee, C.M.; Fine, E.J.; Bao, G. COSMID: A Web-based Tool for Identifying and Validating CRISPR/Cas Off-target Sites. Mol. Ther. Nucleic Acids 2014, 3, e214. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef]
- Labuhn, M.; Adams, F.F.; Ng, M.; Knoess, S.; Schambach, A.; Charpentier, E.M.; Schwarzer, A.; Mateo, J.L.; Klusmann, J.-H.; Heckl, D. Refined sgRNA efficacy prediction improves large- and small-scale CRISPR–Cas9 applications. Nucleic Acids Res. 2018, 46, 1375–1385. [Google Scholar] [CrossRef]
- Havlicek, S.; Shen, Y.; Alpagu, Y.; Bruntraeger, M.B.; Zufir, N.B.M.; Phuah, Z.Y.; Fu, Z.; Dunn, N.R.; Stanton, L.W. Re-engineered RNA-Guided FokI-Nucleases for Improved Genome Editing in Human Cells. Mol. Ther. 2017, 25, 342–355. [Google Scholar] [CrossRef]
- Tsai, S.Q.; Wyvekens, N.; Khayter, C.; Foden, J.A.; Thapar, V.; Reyon, D.; Goodwin, M.J.; Aryee, M.J.; Joung, J.K. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 2014, 32, 569–576. [Google Scholar] [CrossRef]
- Fagerlund, R.D.; Staals, R.H.J.; Fineran, P.C. The Cpf1 CRISPR-Cas protein expands genome-editing tools. Genome Biol. 2015, 16, 251. [Google Scholar] [CrossRef]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; Van Der Oost, J.; Regev, A.; et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef]
- Kim, E.; Koo, T.; Park, S.W.; Kim, D.; Kim, K.; Cho, H.-Y.; Song, D.W.; Lee, K.J.; Jung, M.H.; Kim, S.; et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017, 8, 14500. [Google Scholar] [CrossRef]
- Liu, J.-J.; Orlova, N.; Oakes, B.L.; Ma, E.; Spinner, H.B.; Baney, K.L.M.; Chuck, J.; Tan, D.; Knott, G.J.; Harrington, L.B.; et al. CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature 2019, 566, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Prew, M.S.; Tsai, S.Q.; Topkar, V.V.; Nguyen, N.T.; Zheng, Z.; Gonzales, A.P.W.; Li, Z.; Peterson, R.T.; Yeh, J.-R.J.; et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 2015, 523, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Pattanayak, V.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Zheng, Z.; Keith Joung, J. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016. [Google Scholar] [CrossRef] [PubMed]
- Lau, C.-H.; Suh, Y. In vivo genome editing in animals using AAV-CRISPR system: Applications to translational research of human disease. F1000Research 2017, 6, 2153. [Google Scholar] [CrossRef]
- Mendell, J.R.; Al-Zaidy, S.; Shell, R.; Arnold, W.D.; Rodino-Klapac, L.R.; Prior, T.W.; Lowes, L.; Alfano, L.; Berry, K.; Church, K.; et al. Single-Dose Gene-Replacement Therapy for Spinal Muscular Atrophy. N. Engl. J. Med. 2017, 377, 1713–1722. [Google Scholar] [CrossRef]
- Ding, Y.; Jiang, Z.; Saha, K.; Kim, C.S.; Kim, S.T.; Landis, R.F.; Rotello, V.M. Gold Nanoparticles for Nucleic Acid Delivery. Mol. Ther. 2014, 22, 1075–1083. [Google Scholar] [CrossRef]
- Glass, Z.; Li, Y.; Xu, Q. Nanoparticles for CRISPR–Cas9 delivery. Nat. Biomed. Eng. 2017, 1, 854–855. [Google Scholar] [CrossRef]
- Lee, K.; Conboy, M.; Park, H.M.; Jiang, F.; Kim, H.J.; Dewitt, M.A.; Mackley, V.A.; Chang, K.; Rao, A.; Skinner, C.; et al. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat. Biomed. Eng. 2017, 1, 889–901. [Google Scholar] [CrossRef]
- Sago, C.D.; Lokugamage, M.P.; Paunovska, K.; Vanover, D.A.; Monaco, C.M.; Shah, N.N.; Gamboa Castro, M.; Anderson, S.E.; Rudoltz, T.G.; Lando, G.N.; et al. High-throughput in vivo screen of functional mRNA delivery identifies nanoparticles for endothelial cell gene editing. Proc. Natl. Acad. Sci. USA 2018, 115, E9944–E9952. [Google Scholar] [CrossRef]
- Staahl, B.T.; Benekareddy, M.; Coulon-Bainier, C.; Banfal, A.A.; Floor, S.N.; Sabo, J.K.; Urnes, C.; Munares, G.A.; Ghosh, A.; Doudna, J.A. Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes. Nat. Biotechnol. 2017, 35, 431–434. [Google Scholar] [CrossRef]
- Cameron, P.; Fuller, C.K.; Donohoue, P.D.; Jones, B.N.; Thompson, M.S.; Carter, M.M.; Gradia, S.; Vidal, B.; Garner, E.; Slorach, E.M.; et al. Mapping the genomic landscape of CRISPR–Cas9 cleavage. Nat. Methods 2017, 14, 600–606. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishna, S.; Kwaku Dad, A.-B.; Beloor, J.; Gopalappa, R.; Lee, S.-K.; Kim, H. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014, 24, 1020–1027. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Sander, J.D.; Reyon, D.; Cascio, V.M.; Joung, J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014, 32, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Lin, C.-Y.; Gootenberg, J.S.; Konermann, S.; Trevino, A.E.; Scott, D.A.; Inoue, A.; Matoba, S.; Zhang, Y.; et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013, 154, 1380–1389. [Google Scholar] [CrossRef]
- van Overbeek, M.; Capurso, D.; Carter, M.M.; Thompson, M.S.; Frias, E.; Russ, C.; Reece-Hoyes, J.S.; Nye, C.; Gradia, S.; Vidal, B.; et al. DNA Repair Profiling Reveals Nonrandom Outcomes at Cas9-Mediated Breaks. Mol. Cell 2016. [Google Scholar] [CrossRef]
- Ghezraoui, H.; Piganeau, M.; Renouf, B.; Renaud, J.-B.; Sallmyr, A.; Ruis, B.; Oh, S.; Tomkinson, A.E.; Hendrickson, E.A.; Giovannangeli, C.; et al. Chromosomal Translocations in Human Cells Are Generated by Canonical Nonhomologous End-Joining. Mol. Cell 2014, 55, 829–842. [Google Scholar] [CrossRef]
- Tan, E.-P.; Li, Y.; Del Castillo Velasco-Herrera, M.; Yusa, K.; Bradley, A. Off-target assessment of CRISPR-Cas9 guiding RNAs in human iPS and mouse ES cells. Genesis 2015, 53, 225–236. [Google Scholar] [CrossRef]
- Birling, M.-C.; Schaeffer, L.; André, P.; Lindner, L.; Maréchal, D.; Ayadi, A.; Sorg, T.; Pavlovic, G.; Hérault, Y. Efficient and rapid generation of large genomic variants in rats and mice using CRISMERE. Sci. Rep. 2017, 7, 43331. [Google Scholar] [CrossRef]
- Li, D.; Qiu, Z.; Shao, Y.; Chen, Y.; Guan, Y.; Liu, M.; Li, Y.; Gao, N.; Wang, L.; Lu, X.; et al. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 681–683. [Google Scholar] [CrossRef]
- Wang, H.; Yang, H.; Shivalila, C.S.; Dawlaty, M.M.; Cheng, A.W.; Zhang, F.; Jaenisch, R. One-step generation of mice carrying mutations in multiple genes by CRISPR/cas-mediated genome engineering. Cell 2013, 153, 910–918. [Google Scholar] [CrossRef]
- Kosicki, M.; Tomberg, K.; Bradley, A. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018, 36, 765–771. [Google Scholar] [CrossRef] [PubMed]
- Boroviak, K.; Fu, B.; Yang, F.; Doe, B.; Bradley, A. Revealing hidden complexities of genomic rearrangements generated with Cas9. Sci. Rep. 2017, 7, 12867. [Google Scholar] [CrossRef] [PubMed]
- Pattanayak, V.; Lin, S.; Guilinger, J.P.; Ma, E.; Doudna, J.A.; Liu, D.R. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 2013, 31, 839–843. [Google Scholar] [CrossRef] [PubMed]
- Frock, R.L.; Hu, J.; Meyers, R.M.; Ho, Y.-J.; Kii, E.; Alt, F.W. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 2015, 33, 179–186. [Google Scholar] [CrossRef]
- Tsai, S.Q.; Zheng, Z.; Nguyen, N.T.; Liebers, M.; Topkar, V.V.; Thapar, V.; Wyvekens, N.; Khayter, C.; Iafrate, A.J.; Le, L.P.; et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015, 33, 187–197. [Google Scholar] [CrossRef]
- Cullot, G.; Boutin, J.; Toutain, J.; Prat, F.; Pennamen, P.; Rooryck, C.; Teichmann, M.; Rousseau, E.; Lamrissi-Garcia, I.; Guyonnet-Duperat, V.; et al. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat. Commun. 2019. [Google Scholar] [CrossRef]
- Bonev, B.; Mendelson Cohen, N.; Szabo, Q.; Fritsch, L.; Papadopoulos, G.L.; Lubling, Y.; Xu, X.; Lv, X.; Hugnot, J.P.; Tanay, A.; et al. Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell 2017, 171, 557–572.e24. [Google Scholar] [CrossRef]
- Jackling, F.C.; Johnson, W.E.; Appleton, B.R. The Genetic Inheritance of the Blue-eyed White Phenotype in Alpacas (Vicugna pacos). J. Hered. 2014, 105, 941–951. [Google Scholar] [CrossRef]
- Holl, H.; Isaza, R.; Mohamoud, Y.; Ahmed, A.; Almathen, F.; Youcef, C.; Gaouar, S.; Antczak, D.; Brooks, S. A Frameshift Mutation in KIT is Associated with White Spotting in the Arabian Camel. Genes 2017, 8, 102. [Google Scholar] [CrossRef]
- Volpato, G.; Dioli, M.; Di Nardo, A. Piebald Camels. Pastoralism 2017, 7, 3. [Google Scholar] [CrossRef]
- David, V.A.; Menotti-Raymond, M.; Wallace, A.C.; Roelke, M.; Kehler, J.; Leighty, R.; Eizirik, E.; Hannah, S.S.; Nelson, G.; Schäffer, A.A.; et al. Endogenous Retrovirus Insertion in the KIT Oncogene Determines White and White spotting in Domestic Cats. G3 Genes Genomes Genet. 2014, 4, 1881–1891. [Google Scholar] [CrossRef]
- Fontanesi, L.; Tazzoli, M.; Russo, V.; Beever, J. Genetic heterogeneity at the bovine KIT gene in cattle breeds carrying different putative alleles at the spotting locus. Anim. Genet. 2010, 41, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Häfliger, I.M.; Hirter, N.; Paris, J.M.; Wolf Hofstetter, S.; Seefried, F.R.; Drögemüller, C. A de novo germline mutation of KIT in a white-spotted Brown Swiss cow. Anim. Genet. 2020, 51, 449–452. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.K.; Ruhe, A.L.; Robertson, K.R.; Loew, E.R.; Williams, D.C.; Neff, M.W. A de novo mutation in KIT causes white spotting in a subpopulation of German Shepherd dogs. Anim. Genet. 2013, 44, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Haase, B.; Rieder, S.; Leeb, T. Two variants in the KIT gene as candidate causative mutations for a dominant white and a white spotting phenotype in the donkey. Anim. Genet. 2015, 46, 321–324. [Google Scholar] [CrossRef]
- Nazari-Ghadikolaei, A.; Mehrabani-Yeganeh, H.; Miarei-Aashtiani, S.R.; Staiger, E.A.; Rashidi, A.; Huson, H.J. Genome-Wide Association Studies Identify Candidate Genes for Coat Color and Mohair Traits in the Iranian Markhoz Goat. Front. Genet. 2018, 9. [Google Scholar] [CrossRef]
- Hauswirth, R.; Jude, R.; Haase, B.; Bellone, R.R.; Archer, S.; Holl, H.; Brooks, S.A.; Tozaki, T.; Penedo, M.C.T.; Rieder, S.; et al. Novel variants in the KIT and PAX3 genes in horses with white-spotted coat colour phenotypes. Anim. Genet. 2013, 44, 763–765. [Google Scholar] [CrossRef]
- Chabot, B.; Stephenson, D.A.; Chapman, V.M.; Besmer, P.; Bernstein, A. The proto-oncogene c-kit encoding a transmembrane tyrosine kinase receptor maps to the mouse W locus. Nature 1988, 335, 88–89. [Google Scholar] [CrossRef]
- Geissler, E.N.; Ryan, M.A.; Housman, D.E. The dominant-white spotting (W) locus of the mouse encodes the c-kit proto-oncogene. Cell 1988, 55, 185–192. [Google Scholar] [CrossRef]
- Nocka, K.; Majumder, S.; Chabot, B.; Ray, P.; Cervone, M.; Bernstein, A.; Besmer, P. Expression of c-kit gene products in known cellular targets of W mutations in normal and W mutant mice--evidence for an impaired c-kit kinase in mutant mice. Genes Dev. 1989, 3, 816–826. [Google Scholar] [CrossRef]
- Lim, H.T.; Zhong, T.; Cho, I.C.; Seo, B.Y.; Kim, J.H.; Lee, S.S.; Ko, M.S.; Park, H.B.; Kim, B.W.; Lee, J.H.; et al. Novel alternative splicing by exon skipping in KIT associated with whole-body roan in an intercrossed population of Landrace and Korean Native pigs. Anim. Genet. 2011, 42, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Fontanesi, L.; Vargiolu, M.; Scotti, E.; Latorre, R.; Faussone Pellegrini, M.S.; Mazzoni, M.; Asti, M.; Chiocchetti, R.; Romeo, G.; Clavenzani, P.; et al. The KIT Gene Is Associated with the English Spotting Coat Color Locus and Congenital Megacolon in Checkered Giant Rabbits (Oryctolagus cuniculus). PLoS ONE 2014, 9, e93750. [Google Scholar] [CrossRef] [PubMed]
- Tsujimura, T.; Hirota, S.; Nomura, S.; Niwa, Y.; Yamazaki, M.; Tono, T.; Morii, E.; Kim, H.; Kondo, K.; Nishimune, Y. Characterization of Ws mutant allele of rats: A 12-base deletion in tyrosine kinase domain of c-kit gene. Blood 1991, 78, 1942–1946. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.Q.; Xu, X.; Luo, S.J. The genetics of brown coat color and white spotting in domestic yaks (Bos grunniens). Anim. Genet. 2014. [Google Scholar] [CrossRef]
- Ezoe, K.; Holmes, S.A.; Ho, L.; Bennett, C.P.; Bolognia, J.L.; Brueton, L.; Burn, J.; Falabella, R.; Gatto, E.M.; Ishii, N. Novel mutations and deletions of the KIT (steel factor receptor) gene in human piebaldism. Am. J. Hum. Genet. 1995, 56, 58–66. [Google Scholar]
- Giebel, L.B.; Spritz, R.A. Mutation of the KIT (mast/stem cell growth factor receptor) protooncogene in human piebaldism. Proc. Natl. Acad. Sci. USA 1991, 88, 8696–8699. [Google Scholar] [CrossRef]
- Parichy, D.M.; Mellgren, E.M.; Rawls, J.F.; Lopes, S.S.; Kelsh, R.N.; Johnson, S.L. Mutational Analysis of Endothelin Receptor b1 (rose) during Neural Crest and Pigment Pattern Development in the Zebrafish Danio rerio. Dev. Biol. 2000, 227, 294–306. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Rees, H.A.; Liu, D.R. Base editing: Precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018, 19, 770–788. [Google Scholar] [CrossRef]
- Shin, H.Y.; Wang, C.; Lee, H.K.; Yoo, K.H.; Zeng, X.; Kuhns, T.; Yang, C.M.; Mohr, T.; Liu, C.; Hennighausen, L. CRISPR/Cas9 targeting events cause complex deletions and insertions at 17 sites in the mouse genome. Nat. Commun. 2017. [Google Scholar] [CrossRef]
- Adikusuma, F.; Piltz, S.; Corbett, M.A.; Turvey, M.; McColl, S.R.; Helbig, K.J.; Beard, M.R.; Hughes, J.; Pomerantz, R.T.; Thomas, P.Q. Large deletions induced by Cas9 cleavage. Nature 2018. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, S.; Aoto, K.; Hiraide, T.; Nakashima, M.; Takabayashi, S.; Saitsu, H. Nanopore sequencing reveals a structural alteration of mirror-image duplicated genes in a genome-editing mouse line. Congenit. Anom. 2019. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Marti-Gutierrez, N.; Park, S.-W.; Wu, J.; Hayama, T.; Darby, H.; Van Dyken, C.; Li, Y.; Koski, A.; Liang, D.; et al. Ma et al. reply. Nature 2018, 560, E10–E23. [Google Scholar] [CrossRef] [PubMed]
- Smirnov, A.; Fishman, V.; Yunusova, A.; Korablev, A.; Serova, I.; Skryabin, B.V.; Rozhdestvensky, T.S.; Battulin, N. DNA barcoding reveals that injected transgenes are predominantly processed by homologous recombination in mouse zygote. Nucleic Acids Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Skryabin, B.V.; Kummerfeld, D.-M.; Gubar, L.; Seeger, B.; Kaiser, H.; Stegemann, A.; Roth, J.; Meuth, S.G.; Pavenstädt, H.; Sherwood, J.; et al. Pervasive head-to-tail insertions of DNA templates mask desired CRISPR-Cas9–mediated genome editing events. Sci. Adv. 2020, 6, eaax2941. [Google Scholar] [CrossRef] [PubMed]
- Kerpedjiev, P.; Abdennur, N.; Lekschas, F.; McCallum, C.; Dinkla, K.; Strobelt, H.; Luber, J.M.; Ouellette, S.B.; Azhir, A.; Kumar, N.; et al. HiGlass: Web-based visual exploration and analysis of genome interaction maps. Genome Biol. 2018, 19, 125. [Google Scholar] [CrossRef] [PubMed]
| Name | Sequence |
|---|---|
| T7-gRNA1-FWD | GTTAATACGACTCACTATAGCGCCGTAAGTGCTGAAAAGGTTTTAGAGCTAGAAATAGCAAGTTAA |
| T7-gRNA2-FWD | GTTAATACGACTCACTATAGAGGTAAGAGCAATCCGGTAGTTTTAGAGCTAGAAATAGCAAGTTAA |
| gRNA-REV | AAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTA |
| REV | AAAAGCACCGACTCGGTGCCACTTTTTCAAG |
| 86 | TCTTTGTCCTGTTACCGCCC |
| 87 | TTGACATGACTCCATGCCCC |
| 88 | CCTACGAGCCTTCACGTTGT |
| 89 | TGAGGACCGCTGATAGGGAA |
| 90 | AAGGCTGTTGTACTGCGTGA |
| 91 | ATGATCTCGTGGCGTCATCC |
| 93 | GTGCTATGGGAGCCGAAAGA |
| 94 | CATAGCCTCTTGCCTTCCGT |
| 179 | ATGTTTGGCTGACGCTGAGA |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Korablev, A.; Lukyanchikova, V.; Serova, I.; Battulin, N. On-Target CRISPR/Cas9 Activity Can Cause Undesigned Large Deletion in Mouse Zygotes. Int. J. Mol. Sci. 2020, 21, 3604. https://doi.org/10.3390/ijms21103604
Korablev A, Lukyanchikova V, Serova I, Battulin N. On-Target CRISPR/Cas9 Activity Can Cause Undesigned Large Deletion in Mouse Zygotes. International Journal of Molecular Sciences. 2020; 21(10):3604. https://doi.org/10.3390/ijms21103604
Chicago/Turabian StyleKorablev, Alexey, Varvara Lukyanchikova, Irina Serova, and Nariman Battulin. 2020. "On-Target CRISPR/Cas9 Activity Can Cause Undesigned Large Deletion in Mouse Zygotes" International Journal of Molecular Sciences 21, no. 10: 3604. https://doi.org/10.3390/ijms21103604
APA StyleKorablev, A., Lukyanchikova, V., Serova, I., & Battulin, N. (2020). On-Target CRISPR/Cas9 Activity Can Cause Undesigned Large Deletion in Mouse Zygotes. International Journal of Molecular Sciences, 21(10), 3604. https://doi.org/10.3390/ijms21103604





