Discovery of the Novel Inhibitor Against New Delhi Metallo-β-Lactamase Based on Virtual Screening and Molecular Modelling
Abstract
1. Introduction
2. Results and Discussion
2.1. Virtual Screening Analysis
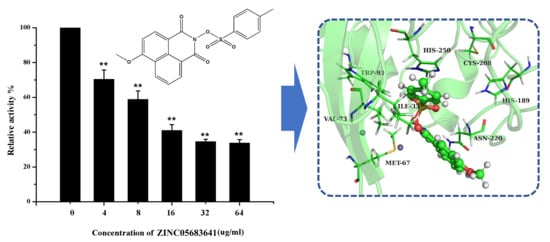
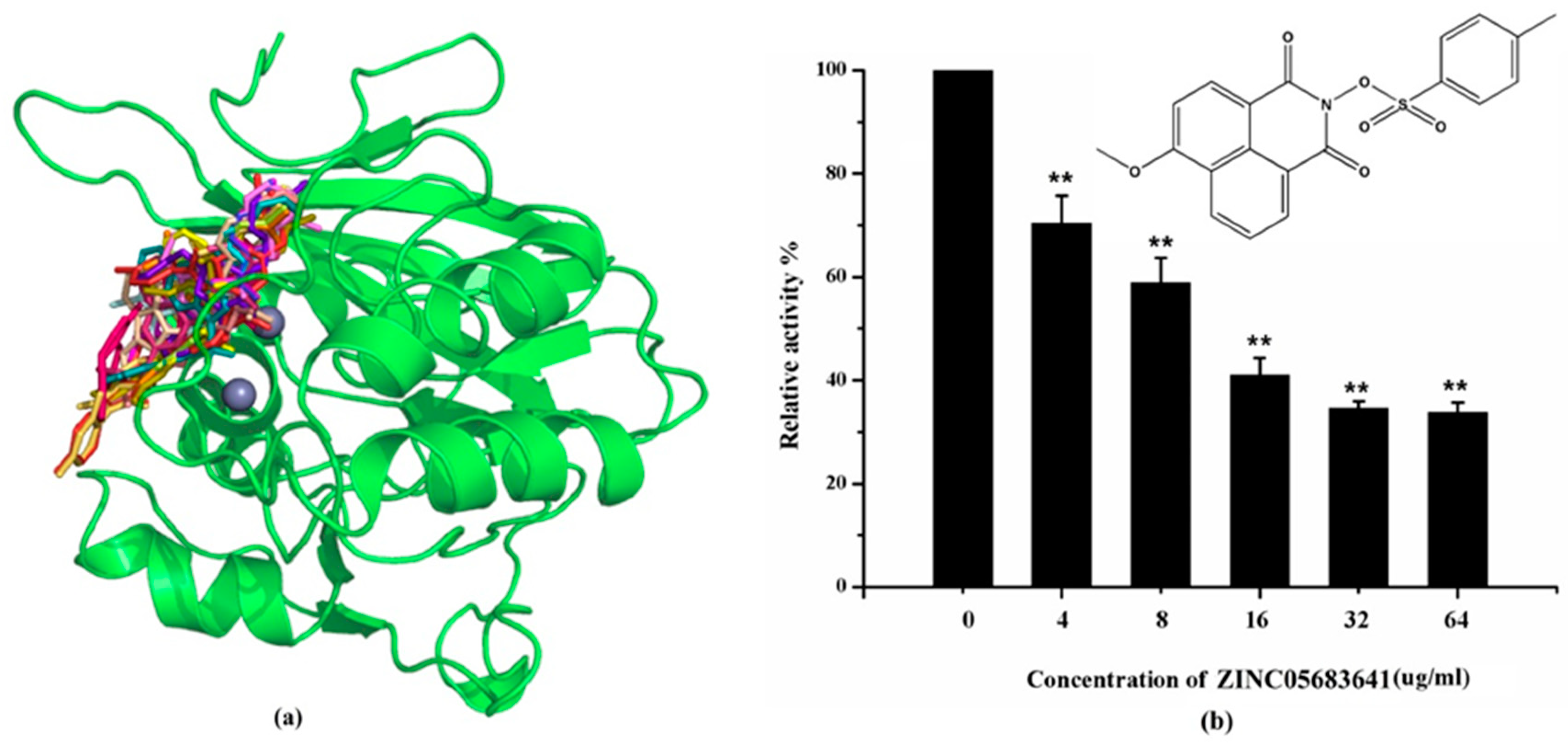
2.2. Nitrocefin Assay
2.3. Antibacterial Activity Assay
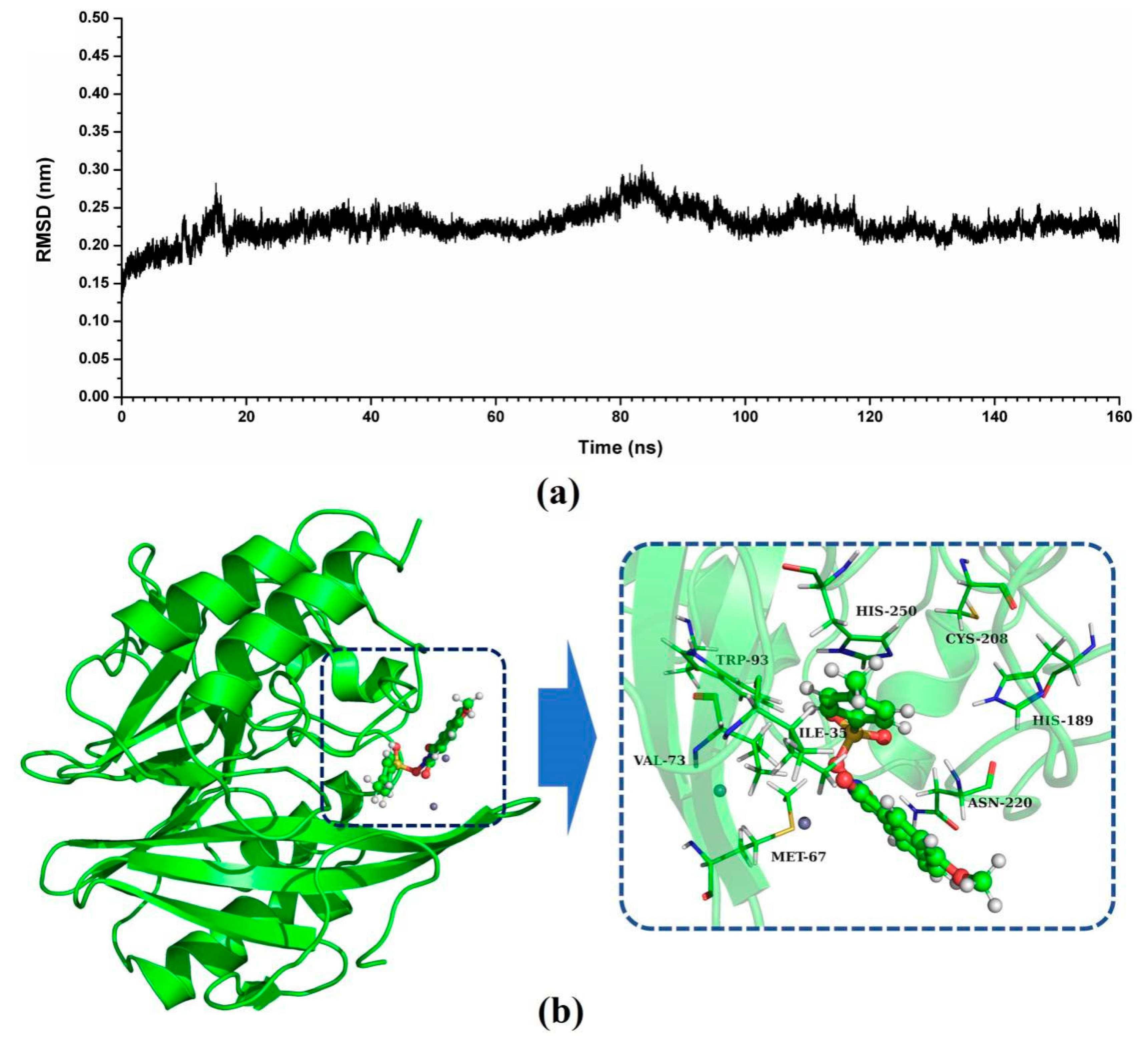
2.4. The Potential Binding Mode of Compound ZINC05683641 with NDM-1
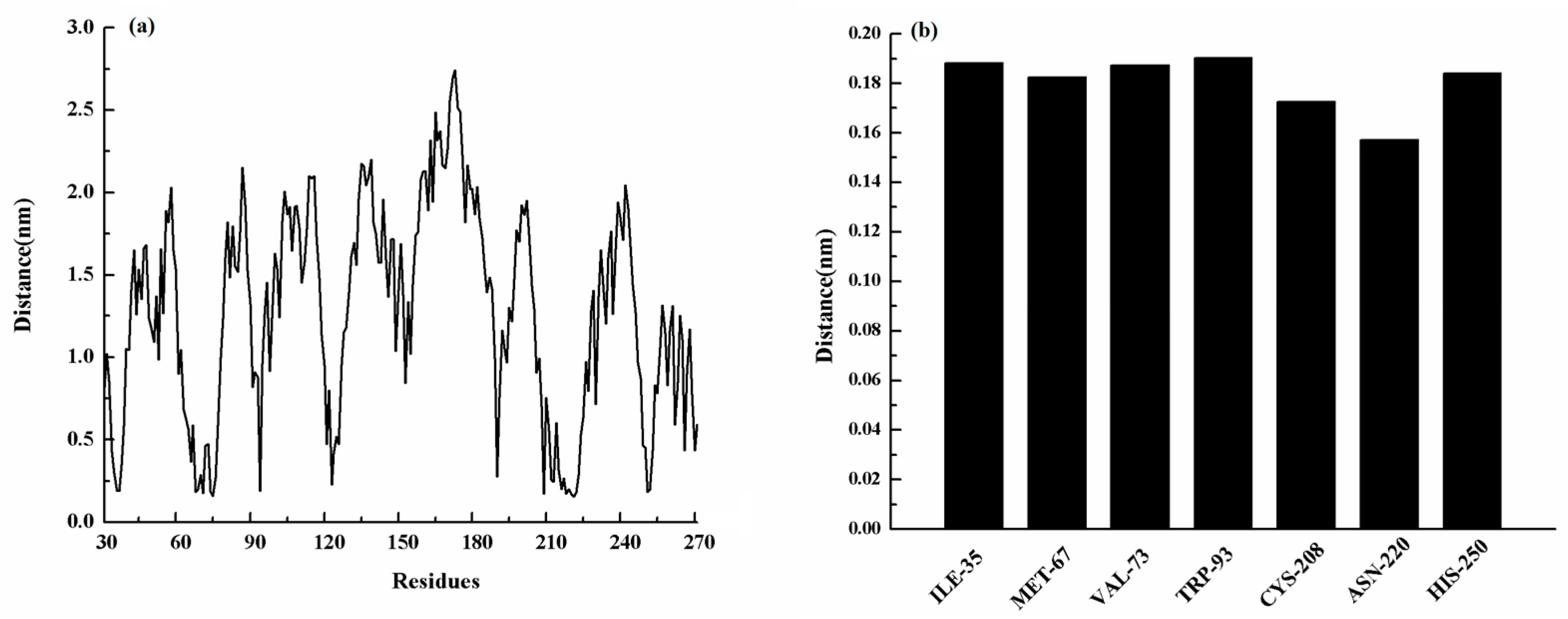
2.5. Confirmation of the Binding Mode in the Complex
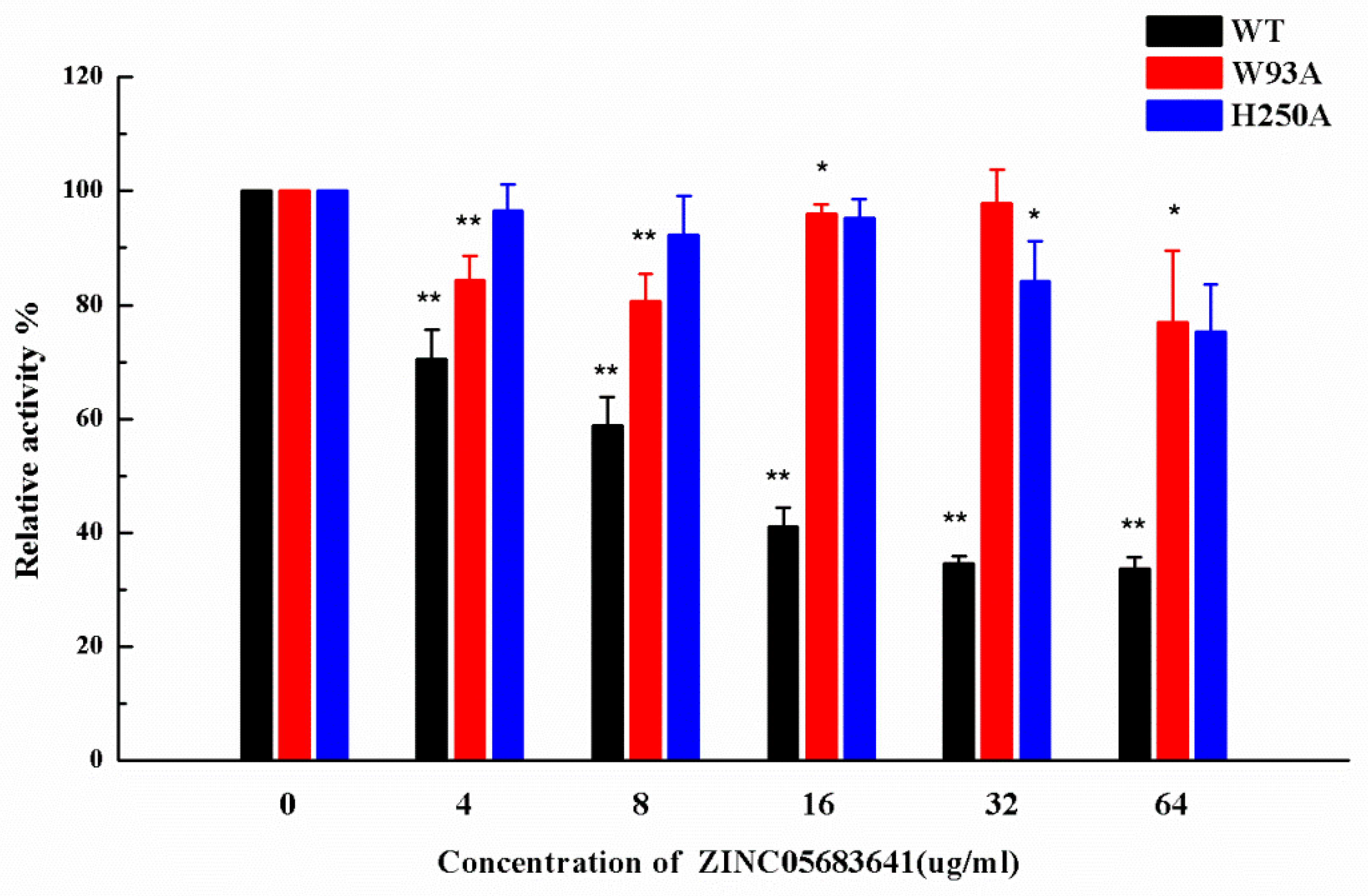
2.6. Identification of the Competitive Inhibition Mechanism
3. Method and Computation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Demain, A.L.; Elander, R.P. The beta-lactam antibiotics: Past, present, and future. Antonie Van Leeuwenhoek 1999, 75, 5–19. [Google Scholar] [CrossRef]
- Drawz, S.M.; Bonomo, R.A. Three decades of beta-lactamase inhibitors. Clin. Microbiol. Rev. 2010, 23, 160–201. [Google Scholar] [CrossRef]
- Spyrakis, F.; Celenza, G.; Marcoccia, F.; Santucci, M.; Cross, S.; Bellio, P.; Cendron, L.; Perilli, M.; Tondi, D. Structure-Based Virtual Screening for the Discovery of Novel Inhibitors of New Delhi Metallo-beta-lactamase-1. ACS Med. Chem. Lett. 2018, 9, 45–50. [Google Scholar] [CrossRef]
- Frere, J.M.; Sauvage, E.; Kerff, F. From “An Enzyme Able to Destroy Penicillin” to Carbapenemases: 70 Years of Beta-lactamase Misbehaviour. Curr. Drug Targets 2016, 17, 974–982. [Google Scholar] [CrossRef]
- Tondi, D.; Cross, S.; Venturelli, A.; Costi, M.P.; Cruciani, G.; Spyrakis, F. Decoding the Structural Basis for Carbapenem Hydrolysis by Class A beta-lactamases: Fishing for a Pharmacophore. Curr. Drug Targets 2016, 17, 983–1005. [Google Scholar] [CrossRef]
- Bush, K. New beta-lactamases in gram-negative bacteria: Diversity and impact on the selection of antimicrobial therapy. Clin. Infect. Dis. 2001, 32, 1085–1089. [Google Scholar] [CrossRef]
- Ambler, R.P. The structure of beta-lactamases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1980, 289, 321–331. [Google Scholar] [CrossRef]
- Jarlier, V.; Nicolas, M.H.; Fournier, G.; Philippon, A. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: Hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 1988, 10, 867–878. [Google Scholar] [CrossRef]
- Fisher, J.F.; Meroueh, S.O.; Mobashery, S. Bacterial resistance to beta-lactam antibiotics: Compelling opportunism, compelling opportunity. Chem. Rev. 2005, 105, 395–424. [Google Scholar] [CrossRef]
- Palzkill, T. Metallo-beta-lactamase structure and function. Ann. N. Y. Acad. Sci. 2013, 1277, 91–104. [Google Scholar] [CrossRef]
- Farina, D.; Spyrakis, F.; Venturelli, A.; Cross, S.; Tondi, D.; Costi, M.P. The inhibition of extended spectrum beta-lactamases: Hits and leads. Curr. Med. Chem. 2014, 21, 1405–1434. [Google Scholar] [CrossRef]
- Thomas, P.W.; Zheng, M.; Wu, S.; Guo, H.; Liu, D.; Xu, D.; Fast, W. Characterization of purified New Delhi metallo-beta-lactamase-1. Biochemistry 2011, 50, 10102–10113. [Google Scholar] [CrossRef]
- Gibb, A.P.; McCallum, A.K. New Delhi metallo-beta-lactamase 1. Lancet Infect. Dis. 2010, 10, 751–752. [Google Scholar] [CrossRef]
- Fair, R.J.; Tor, Y. Antibiotics and bacterial resistance in the 21st century. Perspect. Medicin. Chem. 2014, 6, 25–64. [Google Scholar] [CrossRef]
- Poirel, L.; Lagrutta, E.; Taylor, P.; Pham, J.; Nordmann, P. Emergence of metallo-beta-lactamase NDM-1-producing multidrug-resistant Escherichia coli in Australia. Antimicrob. Agents Chemother. 2010, 54, 4914–4916. [Google Scholar] [CrossRef]
- Walsh, T.R.; Toleman, M.A.; Poirel, L.; Nordmann, P. Metallo-beta-lactamases: The quiet before the storm? Clin. Microbiol. Rev. 2005, 18, 306–325. [Google Scholar] [CrossRef]
- Thakur, P.K.; Kumar, J.; Ray, D.; Anjum, F.; Hassan, M.I. Search of potential inhibitor against New Delhi metallo-beta-lactamase 1 from a series of antibacterial natural compounds. J. Nat. Sci. Biol. Med. 2013, 4, 51–56. [Google Scholar] [CrossRef]
- Yang, H.; Aitha, M.; Marts, A.R.; Hetrick, A.; Bennett, B.; Crowder, M.W.; Tierney, D.L. Spectroscopic and mechanistic studies of heterodimetallic forms of metallo-beta-lactamase NDM-1. J. Am. Chem. Soc. 2014, 136, 7273–7285. [Google Scholar] [CrossRef]
- King, D.T.; Worrall, L.J.; Gruninger, R.; Strynadka, N.C. New Delhi metallo-beta-lactamase: Structural insights into beta-lactam recognition and inhibition. J. Am. Chem. Soc. 2012, 134, 11362–11365. [Google Scholar] [CrossRef]
- Ma, J.; McLeod, S.; MacCormack, K.; Sriram, S.; Gao, N.; Breeze, A.L.; Hu, J. Real-time monitoring of New Delhi metallo-beta-lactamase activity in living bacterial cells by 1H NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 2014, 53, 2130–2133. [Google Scholar] [CrossRef]
- Zhu, K.; Lu, J.; Liang, Z.; Kong, X.; Ye, F.; Jin, L.; Geng, H.; Chen, Y.; Zheng, M.; Jiang, H.; et al. A quantum mechanics/molecular mechanics study on the hydrolysis mechanism of New Delhi metallo-beta-lactamase-1. J. Comput. Aided Mol. Des. 2013, 27, 247–256. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, J.; Niu, G.; Shui, W.; Sun, Y.; Zhou, H.; Zhang, Y.; Yang, C.; Lou, Z.; Rao, Z. A structural view of the antibiotic degradation enzyme NDM-1 from a superbug. Protein Cell 2011, 2, 384–394. [Google Scholar] [CrossRef]
- Xiang, Y.; Chen, C.; Wang, W.M.; Xu, L.W.; Yang, K.W.; Oelschlaeger, P.; He, Y. Rhodanine as a Potent Scaffold for the Development of Broad-Spectrum Metallo-beta-lactamase Inhibitors. ACS Med. Chem. Lett. 2018, 9, 359–364. [Google Scholar] [CrossRef]
- Chen, C.; Xiang, Y.; Yang, K.W.; Zhang, Y.; Wang, W.M.; Su, J.P.; Ge, Y.; Liu, Y. A protein structure-guided covalent scaffold selectively targets the B1 and B2 subclass metallo-beta-lactamases. Chem. Commun. 2018, 54, 4802–4805. [Google Scholar] [CrossRef]
- Kang, J.S.; Zhang, A.L.; Faheem, M.; Zhang, C.J.; Ai, N.; Buynak, J.D.; Welsh, W.J.; Oelschlaeger, P. Virtual Screening and Experimental Testing of B1 Metallo-beta-lactamase Inhibitors. J. Chem. Inf. Model 2018, 58, 1902–1914. [Google Scholar] [CrossRef]
- Chen, A.Y.; Thomas, P.W.; Stewart, A.C.; Bergstrom, A.; Cheng, Z.; Miller, C.; Bethel, C.R.; Marshall, S.H.; Credille, C.V.; Riley, C.L.; et al. Dipicolinic Acid Derivatives as Inhibitors of New Delhi Metallo-beta-lactamase-1. J. Med. Chem. 2017, 60, 7267–7283. [Google Scholar] [CrossRef]
- Hecker, S.J.; Reddy, K.R.; Totrov, M.; Hirst, G.C.; Lomovskaya, O.; Griffith, D.C.; King, P.; Tsivkovski, R.; Sun, D.; Sabet, M.; et al. Discovery of a Cyclic Boronic Acid beta-Lactamase Inhibitor (RPX7009) with Utility vs Class A Serine Carbapenemases. J. Med. Chem. 2015, 58, 3682–3692. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Forli, S.; Huey, R.; Pique, M.E.; Sanner, M.F.; Goodsell, D.S.; Olson, A.J. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef]
- Cosconati, S.; Forli, S.; Perryman, A.L.; Harris, R.; Goodsell, D.S.; Olson, A.J. Virtual Screening with AutoDock: Theory and Practice. Expert Opin. Drug Discov. 2010, 5, 597–607. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Huey, R.; Morris, G.M.; Olson, A.J.; Goodsell, D.S. A semiempirical free energy force field with charge-based desolvation. J. Comput. Chem. 2007, 28, 1145–1152. [Google Scholar] [CrossRef]
- Kwofie, S.K.; Broni, E.; Teye, J.; Quansah, E.; Issah, I.; Wilson, M.D.; Miller, W.A., 3rd; Tiburu, E.K.; Bonney, J.H.K. Pharmacoinformatics-based identification of potential bioactive compounds against Ebola virus protein VP24. Comput. Biol. Med. 2019, 113, 103414. [Google Scholar] [CrossRef]
- Labbe, C.M.; Rey, J.; Lagorce, D.; Vavrusa, M.; Becot, J.; Sperandio, O.; Villoutreix, B.O.; Tuffery, P.; Miteva, M.A. MTiOpenScreen: A web server for structure-based virtual screening. Nucleic Acids Res. 2015, 43, W448–W454. [Google Scholar] [CrossRef]
- Montes-Grajales, D.; Olivero-Verbel, J. Structure-based Identification of Endocrine Disrupting Pesticides Targeting Breast Cancer Proteins. Toxicology 2020, 9, 152459. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. Swiss. ADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Lipinski, C.A. Drug-like properties and the causes of poor solubility and poor permeability. Pharmacol. Toxicol. Methods. 2000, 44, 235–249. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Odds, F.C. Synergy, antagonism, and what the chequerboard puts between them. J. Antimicrob. Chemother. 2003, 52, 1. [Google Scholar] [CrossRef]
- Wang, Y.T.; Cheng, T.L. Refined models of New Delhi metallo-beta-lactamase-1 with inhibitors: An QM/MM modeling study. J. Biomol. Struct. Dyn. 2016, 34, 2214–2223. [Google Scholar] [CrossRef]
- Zhu, K.; Lu, J.; Ye, F.; Jin, L.; Kong, X.; Liang, Z.; Chen, Y.; Yu, K.; Jiang, H.; Li, J.Q.; et al. Structure-based computational study of the hydrolysis of New Delhi metallo-beta-lactmase-1. Biochem. Biophys. Res. Commun. 2013, 431, 2–7. [Google Scholar] [CrossRef]
- Eftink, M.R.; Ghiron, C.A. Fluorescence quenching studies with proteins. Anal. Biochem. 1981, 114, 199–227. [Google Scholar] [CrossRef]
- Bandyopadhyay, S.; Valder, C.R.; Huynh, H.G.; Ren, H.; Allison, W.S. The beta G156C substitution in the F1-ATPase from the thermophilic Bacillus PS3 affects catalytic site cooperativity by destabilizing the closed conformation of the catalytic site. Biochemistry 2003, 41, 14421–14429. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, J.; Zhou, Y.; Hu, N.; Li, J.; Wang, Y.; Niu, X.; Deng, X.; Wang, J. Pterostilbene restores carbapenem susceptibility in New Delhi metallo-beta-lactamase-producing isolates by inhibiting the activity of New Delhi metallo-beta-lactamases. Br. J. Pharmacol. 2019, 176, 4548–4557. [Google Scholar] [CrossRef]
- Hess, B.; Kutzner, C.; van der Spoel, D.; Lindahl, E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef]
- Schaffner-Barbero, C.; Laura, R.G.-R.; Ruiz-Avila, B.; Huecas, S.; Lppchen, T.; den Blaauwen, t.; Diaz, J.F.; Morreale, A.; Andreu, J.M. Insights into Nucleotide Recognition by Cell Division Protein FtsZ from a mant-GTP Competition Assay and Molecular Dynamics. Biochemistry 2010, 49, 10458–10472. [Google Scholar] [CrossRef][Green Version]
- Punkvang, A.; Saparpakorn, P.; Hannongbua, S.; Wolschann, P.; Beyer, A.; Pungpo, P. Investigating the structural basis of arylamides to improve potency against M. tuberculosis strain through molecular dynamics simulations. Eur. J. Med. Chem. 2010, 45, 5585–5593. [Google Scholar] [CrossRef]

| Ligands | Structure | Binding Affinity (kcal/mol) | IC50(μM) |
|---|---|---|---|
| ZINC00639379 |  | −10.0 | 207.28 ± 15.51 |
| ZINC03198432 | 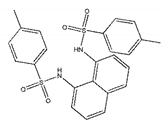 | −9.4 | >300 |
| ZINC25558269 | 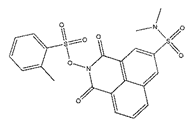 | −9.2 | N/A a |
| ZINC02938448 | 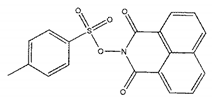 | −9.2 | >300 |
| ZINC02079077 |  | −9.2 | 107.68 ± 5.64 |
| ZINC12524283 |  | −9.1 | >300 |
| ZINC02953923 | 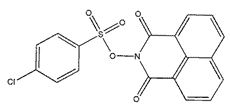 | −9.1 | >300 |
| ZINC06166781 | 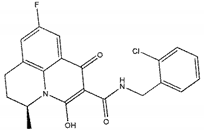 | −9.1 | >300 |
| ZINC25558273 | 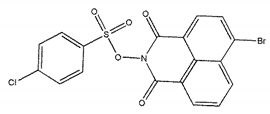 | −9.0 | >300 |
| ZINC11865709 |  | −9.0 | >300 |
| ZINC05683641 | 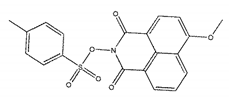 | −9.0 | 13.59 ± 0.52 |
| ZINC02577071 | 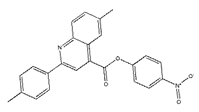 | −9.0 | >300 |
| ZINC06421319 | 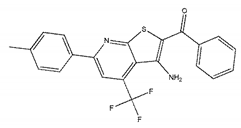 | −8.9 | N/A |
| ZINC04218138 | 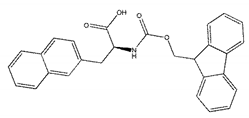 | −8.9 | N/A |
| ZINC20115378 |  | −8.8 | >300 |
| ZINC25558691 |  | −8.8 | N/A |
| ZINC25694564 |  | −8.8 | N/A |
| ZINC01799114 | 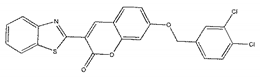 | −8.8 | >300 |
| Meropenem | ZINC05683641 | Meropenem (+64 μg/mL ZINC05683641) | Meropenem (+128 μg/mL ZINC05683641) | |
|---|---|---|---|---|
| MIC (μg/mL) | 32 | >512 | 8 | 8 |
| FICI | - | - | 0.375 | 0.50 |
| Energy Components (kcal/mol) | WT | W93A | H250A |
|---|---|---|---|
| ΔEele | −12.12 ± 0.96 | −14.11 ± 0.75 | −7.23 ± 1.12 |
| ΔEvdw | −38.05 ± 1.15 | −35.43 ± 0.54 | −34.34 ± 1.18 |
| ΔEMM | −50.17 ± 1.20 | −49.54 ± 1.12 | −41.57 ± 1.34 |
| ΔGsol | 23.21 ± 0.92 | 24.53 ± 0.85 | 19.27 ± 1.13 |
| ΔGbind | −26.96 ± 0.88 | −25.00 ± 0.56 | −22.30 ± 0.99 |
| KA | 4.17 ± 1.02 | 3.05 ± 0.83 * | 2.15 ± 0.88 * |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Yang, Y.; Gao, Y.; Niu, X. Discovery of the Novel Inhibitor Against New Delhi Metallo-β-Lactamase Based on Virtual Screening and Molecular Modelling. Int. J. Mol. Sci. 2020, 21, 3567. https://doi.org/10.3390/ijms21103567
Wang X, Yang Y, Gao Y, Niu X. Discovery of the Novel Inhibitor Against New Delhi Metallo-β-Lactamase Based on Virtual Screening and Molecular Modelling. International Journal of Molecular Sciences. 2020; 21(10):3567. https://doi.org/10.3390/ijms21103567
Chicago/Turabian StyleWang, Xiyan, Yanan Yang, Yawen Gao, and Xiaodi Niu. 2020. "Discovery of the Novel Inhibitor Against New Delhi Metallo-β-Lactamase Based on Virtual Screening and Molecular Modelling" International Journal of Molecular Sciences 21, no. 10: 3567. https://doi.org/10.3390/ijms21103567
APA StyleWang, X., Yang, Y., Gao, Y., & Niu, X. (2020). Discovery of the Novel Inhibitor Against New Delhi Metallo-β-Lactamase Based on Virtual Screening and Molecular Modelling. International Journal of Molecular Sciences, 21(10), 3567. https://doi.org/10.3390/ijms21103567