Abstract
Melanoma is a deadly skin cancer that becomes especially difficult to treat after it metastasizes. Timely identification of melanoma is critical for effective therapy, but histopathologic diagnosis can frequently pose a significant challenge to this goal. Therefore, auxiliary diagnostic tools are imperative to facilitating prompt recognition of malignant lesions. Melanoma develops as result of a number of genetic mutations, with UV radiation often acting as a mutagenic risk factor. Novel methods of genetic testing have improved detection of these molecular alterations, which subsequently revealed important information for diagnosis and prognosis. Rapid detection of genetic alterations is also significant for choosing appropriate treatment and developing targeted therapies for melanoma. This review will delve into the understanding of various mutations and the implications they may pose for clinical decision making.
1. Introduction
Melanoma represents the most lethal type of skin cancer, with an estimated 7000 deaths in 2019 in the United States [1]. Furthermore, its incidence has steadily risen since the 1960s, with approximately 96,000 new cases in 2019 [2]. Ultraviolet radiation (UVR) represents a major contributor to cutaneous melanomagenesis through its deleterious effects on the skin and direct damage to DNA [3]. These processes trigger the acceleration of tumorigenesis and thus have facilitated the emergence of malignant melanoma as a significant public health problem.
The financial burden of treating melanoma remains cumbersome. In the United States, the estimated annual cost per patient for Stage I melanoma is $2169–$14,499, while the cost for Stage IV melanoma is $34,103–$152,244 [4]. While the advent of new immunotherapies has helped to reduce the mortality rate over the last decade, melanoma remains difficult to treat. Given the expensive costs for treatment of advanced-stage melanoma, diagnosing and treating melanoma at an early stage is crucial. However, the heterogeneous morphological and histopathological appearance of these tumors can make diagnosis challenging.
As per the 2018 WHO classification, melanoma was newly divided into three classes: melanomas associated with cumulative solar damage (CSD), melanomas not associated with CSD, and nodular melanoma [5]. Pathways of melanoma associated with CSD include superficial spreading, lentigo maligna, and desmoplastic melanomas. Superficial spreading melanoma is the most common subtype and is noted for its early radial growth, followed by vertical growth and subsequent invasion into the dermis [6]. Melanomas not associated with CSD are subclassified into spitzoid, acral, mucosal, and uveal melanomas and melanomas arising in congenital and blue nevi. Finally, nodular melanoma is distinctive for its early progression to vertical growth [6,7]. This new classification better delineates the mutagenic changes that arise in melanoma formation.
In addition to the formulation of melanoma on the skin, uveal melanoma has emerged due to its morbidity and mortality. Uveal melanoma originates from melanocytes that are located in the iris, choroid, or ciliary body [8,9]. Uveal melanoma has an estimated annual incidence of 5.1 cases per million and comprises approximately 3%–5% of all melanomas [8,9,10]. Although relatively rare, this malignancy is highly lethal due to its rapidly metastatic nature. Interestingly, uveal melanoma may be associated with similar risk factors to those of cutaneous melanoma, including fair skin and sunlight exposure [10,11,12]. Treatment is limited but has revolved around radiation and possible enucleation [13].
Diagnostic and therapeutic molecular markers have been increasingly used to assist in histopathological assessment of these tumors. These markers are not only helpful for diagnosing melanoma, but also in distinguishing certain subtypes that may otherwise be difficult to identify (Table 1). In addition, therapeutic markers can guide the selection of treatment with the development of novel targeted therapies (Table 2).

Table 1.
Summary of melanoma mutations.

Table 2.
Function of and known clinical therapies targeting melanoma mutations.
2. Molecular Pathways of Melanoma Formation
Cutaneous melanomagenesis can generally be traced to mutations in signaling pathways critical to cell survival. Most notably, the mitogen-activated protein kinase (MAPK) pathway regulates cell growth, proliferation, differentiation, and apoptosis [77,78,79,80]. Mutations along this pathway result in overamplification of signaling, leading to cell cycle dysregulation and uninhibited cell growth. The MAPK pathway is activated by binding of a growth factor to a receptor tyrosine kinase (RTK) on the surface of the cell and stimulates the GTPase activity of RAS. The signal is propagated down through the cascade of RAF, MEK, and ERK, which enters the nucleus to activate transcription factors promoting the cell cycle [81,82,83,84,85,86]. The PI3K/AKT pathway regulates cell growth and proliferation [87,88,89,90]. PI3Ks can be similarly activated by binding of RTK to a growth factor or directly stimulated by RAS [91,92]. PI3K subsequently phosphorylates its substrate PIP2 on the cell membrane into PIP3, helping to recruit and activate AKT [93,94,95]. AKT promotes cell growth and survival through multiple effectors, such as mTOR, Bad, and Mdm2 [87,88,89]. Mutations to these regulatory signals such as the oncogene NRAS or the tumor suppressor PTEN can occur alone or even in addition to other mutations in melanoma [91,96].
In contrast to cutaneous melanoma, uveal melanoma tends to develop from different mutations along the MAPK or PI3K/AKT pathways. The most common mutations are in GNAQ or GNA11, which can lead to overactivation of both pathways. These genes are responsible for encoding the Gα subunit of G proteins, leading to a constitutively active GTP-bound state [70,71,72]. GNAQ and GNA11 mutations may also increase activity through the Hippo pathway. The Hippo pathway has been identified for its role in cell homeostasis and mammalian organ size, including heart, liver, and pancreas [97,98,99,100]. GNAQ and GNA11 mutations result in downstream activation of YAP/TAZ to stimulate melanomagenesis [101]. Uveal melanoma has been thought to result from an initiating GNAQ/GNA11 mutation, followed by a secondary BSE event from mutations in the genes BAP1, SF3B1, and EIF1AX [13,102].
3. Molecular Markers
Melanoma is generally diagnosed by assessment of skin histological and architectural features but can be prone to subjectivity. Further, traditional characteristics of melanomas such as thickness or mitotic rate can be inaccurate in diagnosis and prognosis. For these reasons, there is an ever-present search for novel detection methods. Detecting molecular markers or genetic alterations has emerged as an innovative form of testing that guides therapeutic decisions and aids the diagnosis of histologically challenging cases. Sequencing studies have illuminated the role of UV exposure in different mutations that lead to melanoma. For example, identifying UV signature mutations, such as C T and CC TT substitutions, can provide an idea of the underlying impact of UV radiation [103]. Whole-genome sequencing has revealed the different mutations that contribute to the development of UV-dependent and -independent melanomas [104]. Methods including comparative genomic hybridization (CGH), fluorescence in situ hybridization (FISH), and quantitative gene expression profiling contribute to the detection of genetic mutations and determination of expression levels. Tests in clinical use include DecisionDx-Melanoma (Castle Biosciences), myPath Melanoma (Myriad Genetics), and Pigmented Lesion Assay (DermTech, Inc.), which profile a wide array of genes [105,106,107,108]. As these tests become more refined, the meaning of various markers in diagnosis and therapy of melanoma has expanded as well. These markers can be represented by melanoma mutations, gene polymorphisms, signaling receptors, and melanin pigment. For this review, we will discuss the significance of these in the context of their role in prognostic and diagnostic value, the melanin synthesis pathway, and targeted therapeutics.
3.1. Prognostic or Diagnostic Markers
3.1.1. GNAQ/GNA11
GNAQ and GNA11 mutations result in overamplification of signaling through the MAPK and PI3K pathways via blocking GTPase activity. G proteins become active when bound to GTP and are inactivated by GTPase hydrolysis to GDP. With GNAQ and GNA11 mutations, GTP is persistently bound to the G protein and lead to constitutive downstream signaling [9,70,71,72]. These mutations are mutually exclusive and are detected in approximately 80%–90% of cases of uveal melanoma [29,109]. However, they are known to occur with BAP1 and SF3B1 mutations, with GNAQ/GNA11 mutation representing the initial event [13,102]. Because uveal melanoma rapidly metastasizes, in such advanced stage cases, identification of the primary tumor can be difficult. Analysis of oncogene status showing positive GNAQ or GNA11 expression can be a valuable diagnostic tool to differentiate uveal melanoma from other types of melanoma and cancers [109]. While GNAQ and GNA11 mutations can also be found in cutaneous melanoma, these cases are extremely rare [110].
The evidence for the prognostic value of GNAQ and GNA11 mutations is limited. Multiple studies have shown that the presence of GNAQ or GNA11 mutations is not associated with metastatic progression or patient outcomes [111,112]. In addition, no difference has been found in survival between patients harboring the GNAQ mutation versus patients with the GNA11 mutation [29,111].
3.1.2. CDKN2A
Mutations in the CDKN2A gene are the most common alteration in hereditary melanoma, with presence in 40% of families with strong family history [20,21,113]. This gene encodes the p16 protein, which inhibits cyclin-dependent kinase (CDK) 4 and 6, and the p14ARF protein. Mutations in CDKN2A thus result in hyperphosphorylation of retinoblastoma protein (RB1), releasing the E2F1 transcription factor to promote cell cycle progression from G1 to S. In addition, loss of p14ARF function promotes the ubiquitination of p53, subsequently reducing cell cycle arrest and apoptosis [48,49,50,114].
Those with the CDKN2A mutation have been shown to develop multiple melanomas and significantly more dysplastic nevi, including presentations consistent with dysplastic nevus syndrome [22]. Interestingly, one study found CDKN2A penetrance varied with geographic location, postulating a correlation with UV exposure as highest penetrance by age 80 in families from Australia [20,115]. Histological analysis of CDKN2A-mutated familial melanomas revealed a greater association with the superficial spreading subtype as compared with CDKN2A-wild type familial melanomas [116]. Furthermore, in a Swedish study, familial melanoma cases with the CDKN2A mutation were associated with a younger age at onset and worse survival than those without the mutation. That study suggested that dysregulation of the cell cycle with CDKN2A mutations may exacerbate mutational load and increase tumor aggression [117]. On the other hand, an Italian retrospective cohort study found no association of the mutation with worse survival [118].
3.1.3. BAP1
BAP1 is a tumor suppressor gene with a poorly understood mechanism in melanoma development but has been implicated as a deubiquitinase of cell cycle genes [73,119]. BAP1 mutations are associated with monosomy 3, which is associated with metastatic uveal melanoma [13,33,120]. Multiple other studies since have confirmed the correlation of BAP1 mutations with metastasis, tumor aggression, and worse prognosis in uveal melanoma [30,121,122]. Notably, BAP1 was found to be mutated in early tumorigenesis and not with progression to metastasis [123].
Germline mutations of BAP1 have also been identified, suggesting a hereditary form of uveal melanoma. In fact, BAP1 is the most common mutation found in familial uveal melanoma, with an estimated frequency of 8%–50% of such cases [31]. Genetic testing is helpful on a case-by-case basis, particularly with suggestive family history. Similar to somatic mutations, germline mutation of BAP1 was highly associated with metastasis as compared with uveal melanoma without BAP1 mutation [124]. In another study, somatic mutations were found to have a greater risk of metastasis as compared to germline mutations. As a result, determining BAP1 status in cases of uveal melanoma can be useful to understanding the risk of metastasis [125].
The BAP1 tumor predisposition syndrome caused by germline BAP1 mutations is not only associated with cutaneous melanoma [126,127]. Rarely, somatic mutations may also lead to cutaneous melanoma [126]. Kumar et al. suggested that BAP1 may have differential roles in uveal and cutaneous melanoma cells [128]. In contrast to its role as a tumor suppressor gene, BAP1 expression in cutaneous melanoma was found to promote growth and survival of cells [128]. BAP1-inactivated nevus (BAPoma) is a relatively new entity of an atypical spitzoid tumor, and can serve as an early sign of BAP1 tumor predisposition syndrome [32,129]. The conflicting roles of BAP1 underscore the dearth of understanding into its mechanism. Therapeutic targeting of BAP1 has focused on its role in DNA double-strand break repair via homologous recombination [130]. Mutations in BAP1 thus have been hypothesized to rely on alternative mechanisms of DNA repair with poly (ADP-ribose) polymerase (PARP) emerging as a target for its role in base-excision and nucleotide excision repair [131]. A phase II trial is recruiting to evaluate the effect of niraparib, a PARP inhibitor, in the treatment of uveal melanoma (NCT03207347) [74].
3.1.4. SF3B1
SF3B1 encodes a subunit of splicing factor 3b, and mutations therefore result in aberrant splicing of pre-mRNA into mature mRNA [34,75]. SF3B1 mutations are characterized by disomy 3 and noted to be a marker of good prognosis for uveal melanoma and found in younger patients [13,33,120,122,132]. While tumors bearing the mutation often metastasize, this can take many years and they are thus thought to have intermediate risk for metastasis [35]. Metastasis is thought to occur with the development of additional oncogenic mutations [123].
In one study sequencing melanoma samples, the SF3B1 R625 mutation was found in two out of 231 cutaneous melanoma samples. That report also noted those cases to be metastatic and, similar to BAP1, posed the question of differential roles of SF3B1 depending on the type of melanoma cells [133].
3.1.5. EIF1AX
EIF1AX is important in regulating protein translation, as it encodes for a eukaryotic initiation factor that serves to stabilize the ribosome [34,76]. These mutations are also characterized by disomy 3 [33]. Cases of uveal melanoma with positive EIF1AX mutations rarely metastasize and other genetic alterations are thought to occur when metastasis does occur [35,37,122,123]. It follows that these cases are also generally associated with good prognosis [122].
3.1.6. VDR
The vitamin D receptor (VDR), after binding the active form of vitamin D, 1,25-dihydroxyvitamin D3 (1,25-(OH)2D3), complexes with retinoid X receptor (RXR) to form a trimolecular complex that translocates to the nucleus and binds to VDR response elements (VDRE) on DNA to function as a transcription factor [52,53,54,134]. Further, 1,25(OH)2D3 has been shown to protect against melanoma by inhibiting proliferation, regulating growth factor activity, and promoting apoptosis [135,136,137,138,139,140,141]. Loss of this activity is thought to contribute to the formation and progression of melanoma. In an early study of VDR in melanoma patients, expression was found to be inversely correlated with progression from normal skin to melanocytic nevus to melanoma, suggesting a potential role in aiding differentiation between nevi and early melanoma [23,24]. Reduction in VDR expression was also associated with tumor progression, higher mitotic rates, and shorter survival time [23,24] (Figure 1). Localization of VDR expression is also important as lower expression of cytoplasmic VDR was found more commonly in melanoma as compared to nevi and also associated with tumor size [142]. However, nuclear VDR expression in nevi versus melanoma contrasts with this [23,142].
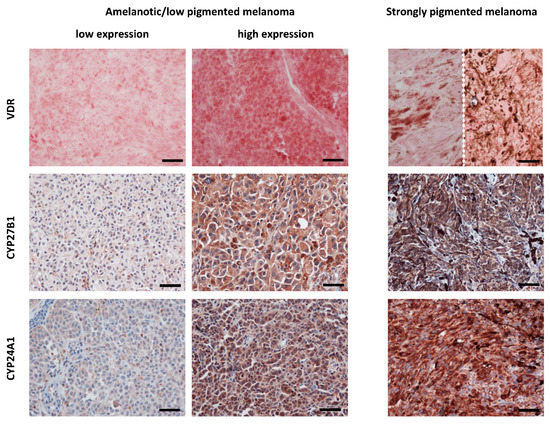
Figure 1.
VDR, CYP27B1 and CYP24A1 immunostaining cutaneous melanomas. Left panel presents lack or low VDR, CYP27B1 and CYP24A1 expression in amelanotic/low pigmented human melanomas obtained from tissues of patients treated in Oncology Center, Bydgoszcz, Poland. Middle panel presents high expression of VDR, CYP27B1 and CYP24A1 expression in amelanotic/low pigmented melanomas. Right panel presents VDR, CYP27B1 and CYP24A1 expression in strongly pigmented melanomas (images of VDR from two different cases are separated with dotted line). VDR was labelled with rat antibody (clone 9A7; Abcam, Cambridge, MA, USA; a dilution 1:75) and visualized with Red AP Substrate (Vector Laboratories, Burlingame, CA, USA). CYP27B1 and CYP24A1 were labelled with rabbit antibody (clone H-90, Santa Cruz Biotechnology, Santa Cruz, CA, USA, a dilution of 1:75) and mouse antibody (Abcam, Cambridge, UK, dilution 1:40), respectively, followed by visualization with ImmPACT NovaRED substrate (Vector Laboratories, Burlingame, CA, USA) and counterstaining with hematoxylin [23,143,144]. Scale bars: 50 μm.
Various polymorphisms of VDR have also been found to affect the risk and prognosis of melanoma, but a consistent pattern has not been identified [141]. Orlow et al. first studied 38 common VDR single-nucleotide polymorphisms (SNPs) and discovered six of these to be associated with increased risk of melanoma development and two with decreased risk [145]. In a later study by the same group, eight SNPs were found to be associated with improved survival in melanoma [146]. Furthermore, the impact of various SNPs on melanoma survival also depends on the amount of sun exposure around time of melanoma diagnosis [147]. However, a 2009 study analyzing six VDR SNPs found no significant change in outcomes with the exception of worse outcome in patients with the BsmI polymorphism and low vitamin D levels [148]. Overall, the understanding of VDR variants in melanoma remains poor [149].
Recently, VDR has also been proven to exist in uveal melanocytes and melanomas as well. While VDR expression was not found to be associated with histopathological characteristics of uveal melanoma, an inverse correlation was shown between expression level and the degree of tumor pigmentation [150]. In light of the positive correlation between uveal tumor pigmentation and metastatic risk, the pattern of VDR expression in uveal melanoma matches closely with that in cutaneous melanoma, implicating similar utility in diagnosis.
VDR expression has been shown to be negatively correlated with overall prognosis in melanoma patients, painting an important picture for vitamin D-based therapy [23,24,140]. In 1981, Colston et al. were the first to discover that 1,25(OH)2D3 inhibits the proliferation of human melanoma cells [151]. Since then, similar results have been elicited by 1,25(OH)2D3 in other human melanoma cell lines and animal models [141,152,153,154,155]. Importantly, VDR mediates the effects of 1,25(OH)2D3 [156]. A recent study by Wasiewicz et al. further confirmed this, as resistance to the anti-proliferative properties of vitamin D was seen in the human melanoma cell line SK-MEL-188b, a VDR-/- subline [157]. In fact, metabolites and analogs of 1,25(OH)2D3 are able to exert similar effects through VDR [152]. This becomes especially important because 1,25(OH)2D3 is limited in its therapeutic use due to its hypercalcemic effects. Analogs with modified side chains, such as calcipotriol, have been demonstrated to inhibit proliferation with low calcemic activity [158,159,160]. Similar effects have been demonstrated for noncalcemic vitamin D3 hydroxyderivatives [159,160,161], which are products of the CYP11A1 action of the side chain of vitamin D3 [63,162]. CYP11A1, in addition to adrenals, placenta and sex organs [163], is also expressed in several peripheral tissue including immune system and skin [164,165,166,167,168,169,170,171]. The anti-melanoma effect of 20-hydroxyvitamin D3 (20(OH)D3) was also reported in an in vivo model of melanoma [153]. It must also be noted that while there was an inverse correlation between CYP27B1 (enzyme activating vitamin D3) and disease progression [143], an inactivating enzyme, CYP24A1, also showed an unexpectedly inverse correlation [144] (Figure 1). This finding was possibly due to alternative hydroxylation pathways forming other anti-tumorigenic dihydroxy-derivatives [144].
In the clinical setting, vitamin D intake has not been associated with melanoma risk; however, its role as adjuvant therapy in melanoma is unknown [172]. A phase 2 trial (Mel-D) is underway in Australia to assess the safety and progression-free survival results of high dose vitamin D therapy in patients who have had surgical excision of melanoma and are at high risk for recurrence [55]. In a Belgian phase 3 trial (ViDMe), relapse-free survival is being assessed in patients treated with cholecalciferol after primary excision of melanoma [56]. The results of these trials will provide key understanding for the therapeutic utility of vitamin D in combination with the assessment of VDR expression.
3.1.7. MC1R
The melanocortin 1 receptor (MC1R) has been identified as a major gene in developing sporadic melanoma [25,57,173]. MC1R responds to melanocyte-stimulating hormone (MSH) to regulate melanogenesis and skin pigmentation. Variants in the MC1R gene have been shown to directly and indirectly induce melanomagenesis. In melanocytes, MC1R activates DNA repair and reduces oxidative stress. Thus, MC1R polymorphisms can exacerbate the UV-induced DNA damage and promote tumor formation [174,175,176,177]. In addition, variants of MC1R upregulate pheomelanin production, which is characterized by a phenotype of fair skin and red hair and susceptibility to UV light [178].
Multiple studies have demonstrated that MC1R variation confers increased risk for melanoma [26,179,180]. While this association was thought to be driven by the predisposition to fair skin, multiple large studies have shown that the presence of a MC1R variant was associated with development of melanoma, independent of all other risk factors including skin type [25,181]. However, in a study stratified by sex, such an association was only discovered in females [181]. While the presence of MC1R variation has not been associated with histopathologic characteristics, it was found to correlate with tumor presentation on the arms, which may provide additional support for its UV-risk independence [182]. Melanomas associated with germline mutations of MC1R have also been shown to have a significantly higher somatic mutational burden, suggesting a higher susceptibility to tumorigenesis in these patients [183]. Notably, MC1R variants have been shown to increase the penetrance of CDKN2A mutations, doubling the risk for melanoma [184].
3.1.8. MITF
Microphthalmia transcription factor (MITF) regulates melanocyte development, differentiation, and function [58,59]. MITF has been shown to serve as a sensitive and specific marker for distinguishing melanoma from histologically similar nonmelanocytic tumors [185,186,187]. MITF has also been found to regulate phenotype switching, wherein low expression leads to increased invasiveness of melanoma cells and high expression leads to decreased invasiveness [188]. Indeed, Cheli et al. showed that MITF silencing in mouse and human melanoma cells enhanced tumorigenicity and metastasis [189]. Investigation of the clinical significance of MITF expression in melanoma samples showed its positive correlation with survival. Higher expression was also associated with negative lymph node status [27].
In addition to its utility as a diagnostic marker for melanoma, MITF has also been found to increase susceptibility to the co-occurrence of melanoma and renal cell carcinoma (RCC). The presence of the Mi-E318K germline mutation in MITF was associated with a fivefold increase in the risk of developing melanoma as compared to patients without the mutation [28,190]. Recently, Ciccarese et al. also reported the association of this variant nodular with the development of nodular melanoma and dysplastic nevi [191]. In cases of familial melanoma, testing for the MITF mutation may be helpful.
3.1.9. HAPLN1
Age represents another risk factor predicting inferior survival in melanoma patients [192]. One explanation is that age-related changes degrade the extracellular matrix (ECM) in the skin and thus promote the growth and migration of melanoma [65]. With age, fibroblasts secrete fewer ECM components with hyaluronan and proteoglycan link protein 1 (HAPLN1) identified as major components [65]. Subsequent study of HAPLN1 revealed its utility as a potential prognostic biomarker. High expression of HAPLN1 in lymphatic tissue was associated with a 56% decrease in death [193]. Thus, determining HAPLN1 expression levels may be particularly helpful in elderly patients.
3.2. Members of the Melanin Synthesis Pathway
3.2.1. Melanin
The synthesis of melanin pigment is characteristic of melanocytes and plays various roles in those cells, including as a free radical scavenger to protect against UV radiation [60,61,62,194,195]. Melanin also serves as a differentiation marker of normal and cancerous melanocytes and thus serves as a suitable marker to distinguish melanoma from other tumors [196].
The degree of melanization has also been shown to impact the aggressiveness and prognosis of melanoma. In one study, patients with stage 3 or 4 melanotic melanoma had poorer survival as compared to patients with amelanotic or hypomelanotic melanomas [197]. Such an association may in part be attributed to the upregulation of HIF-1α by melanogenesis and subsequent downstream stimulation of angiogenesis and cellular metabolism, promoting tumor aggression [198]. Similarly, correlation of melanogenesis to metastasis and death has been shown in uveal melanoma [199,200,201]. On the flip side, a large population-based study showed a twofold increased risk of death for amelanotic compared to pigmented melanoma [202]. To explain this, Sarna et al. have suggested that melanin mechanically reduces the elasticity of melanoma cells and thus inhibits metastasis [203,204].
The anti-oxidative properties of melanin represent an adaptive function of melanocytes to protect the skin from UV radiation. However, the effects of radio-, photo-, or chemotherapy can be unintentionally blunted [205]. One of the early studies investigating this phenomenon by Brozyna et al. showed that inhibition of melanogenesis by N-phenylthiourea (PTU) or D-penicillamine in human melanoma cells increased the sensitivity to killing by gamma rays [206]. Soon after, melanogenesis inhibition was also shown to amplify the cytotoxicity of cyclophosphamide chemotherapy in human melanoma cells [207]. Of note, intermediates of melanogenesis show immunosuppressive effects [207,208,209,210]. In a retrospective melanoma cohort study in Poland, among those treated with radiotherapy, mean survival after therapy and overall survival were more than twofold higher in amelanotic melanomas as compared to pigmented melanomas [205]. Although the diagnosis of an amelanotic melanoma is typically associated with a poorer prognosis, this may be due to its atypical clinical morphology that hinders its diagnosis. This is evidenced by amelanotic melanoma’s higher American Joint Committee on Cancer tumor stage at the time of diagnosis when compared to that of pigmented melanoma [202]. Interestingly, inhibition of melanogenesis also potentiates the efficacy of vitamin D therapy [161]. VDR expression has been shown to be lower in pigmented melanoma cells compared with nonpigmented cells [161]. Accordingly, nonpigmented cells were more sensitive to vitamin D treatment than were pigmented cells [158,161]. Overall, the potential role for melanogenesis inhibition as adjuvant treatment to primary therapy of melanoma has been demonstrated with clinical application standing as the next step.
3.2.2. Melanogenesis Related Proteins (TYR, TRP1, TRP2)
Melanin is synthesized in melanocytes from L-tyrosine through a series of enzymatic reactions leading to production of variety of intermediates of melanogenesis that are biologically active [61,63,64]. Among the proteins involved are tyrosinase (TYR) and tyrosinase-related proteins 1 and 2 (TRP1, TRP2). TYR has become commonplace in the diagnosis of melanoma for its high sensitivity and specificity [211,212,213,214]. Recently, TYR expression was found to correlate with melanocyte differentiation and thus may serve to distinguish between melanoma and benign nevi [215]. TRP1 and TRP2 have also been identified as differentiation markers from the class of melanogenesis-related proteins [216,217]. TRP1 mRNA expression has been correlated with worse survival and depth of invasion; however, TRP1 protein expression was found to inversely correlate with tumor stage and also had no association with patient survival [218,219].
3.3. Therapeutic Targets
3.3.1. B-raf
B-raf is one of the signaling kinases down the MAPK pathway. BRAF mutations comprise the most common genetic alteration in cutaneous melanoma with its presence ranging from 40% to 60% of cases [14,15,220]. Mutations in this oncogene lead to constitutive activation of the MAPK pathway. The most common BRAF mutation is V600E, which represents 80% of alterations in the gene [14]. The V600K and V600R mutations are other known BRAF mutations [14,220]. Studies have shown that V600E expression is associated with the superficial spreading subtype, younger patient age, and skin sites without chronic sun-induced damage, such as the extremities [16,17,221]. In contrast, V600K mutations are correlated with skin sites with CSD, such as the head and neck, and patients of older age [222]. These associations underscore the incompletely understood role of UV radiation in the development of mutations leading to melanoma.
Recently, whole-genome sequencing of benign melanocytic nevi showed the presence of BRAF mutations, in addition to NRAS mutations, with mutational load positively correlated with UV exposure; lower mutational loads were observed in congenital nevi [223]. Similar observations were found in dysplastic nevi with high mutational load as a key distinction between the benign tumors and melanoma. BRAF mutations were thought to be independent of UV exposure due to the absence of UV signature mutations but the consideration of “noninformative” mutations has shifted that belief [41,224]. Of note, Bauer et al. showed that BRAF mutations could not be detected in congenital melanocytic nevi, further suggesting the role of moderate UV exposure in introducing such mutations in the skin [225]. Accordingly, BRAF mutations are associated with melanomas from anatomic locations with intermittent sun exposure, such as the trunk and extremities [226]. Better understanding of the development of these mutations can help to track the rare transformation of benign nevi to malignant melanoma and distinguish the two when histology is equivocal [227]. Refined sequencing technology is also critical for determining the mutational load, which may help evaluate tumor malignancy.
Previously, it had been thought that the presence of BRAF mutations were not associated with worsening prognosis or tumor proliferation [220,228]. Abd Elmageed et al. showed that cellular localization of the BRAF mutation to either the nucleus or cytoplasm was associated with different clinicopathological features. Positive expression of V600E in the nucleus as compared to the cytoplasm was correlated with worse tumor stage, lymph node metastasis, and depth of invasion [229]. Vemurafenib, dabrafenib, and encorafenib are FDA-approved drugs that have been developed as inhibitors of BRAF V600 mutations [38,39,40]. While monotherapy with BRAF inhibitors shows good efficacy against BRAF-mutant melanomas, patients can easily develop resistance through upregulation of RTKs or NRAS [230,231]. As a result, combination therapy with BRAF and MEK inhibitors, including trametinib and cobimetinib, has become standard due to multi-pronged blockade of melanoma growth pathways. The COMBI-v and COMBI-d phase 3 trials showed greater than twofold increases in survival when comparing dabrafenib and trametinib combination to vemurabfenib or dabrafenib monotherapy [232,233]. Newer combinations of encorafenib and binimetinib potentially show even greater efficacy in BRAF-mutant melanomas [38]. Maintaining long-term response is difficult, with just 20% of patients in one study remaining progression free [234]. Ongoing research is investigating the addition of CDK, MAPK, or immune checkpoint inhibitors to combat resistance [231,235,236]. Recently, Misek et al. identified RhoA GTPases as a potential pathway for BRAF resistance and that inhibition of the pathway by Rho kinase (ROCK) inhibitors promotes resensitization to BRAF-targeting therapy [237]. Because of the success of target therapies, identifying patients with BRAF-mutant melanomas is paramount. Vemurafenib had been associated with a paradoxical upregulation of the MAPK signaling pathway in cells with wild-type BRAF [238]. In turn, this paradoxical upregulation had been linked to increased incidence in cutaneous SCCs that harbor RAS mutation, as well as reported cases of dysplastic nevi and wild-type BRAF melanomas, in patients after vemurafenib treatment [239,240].
3.3.2. N-ras
The N-ras GTPase is critical in the transduction of extracellular growth signals via both the MAPK and PI3K/AKT pathways. When the oncogene is mutated, GTPase activity is reduced, resulting in a constitutively active GTP-bound G protein to propagate downstream signals [15,41,85,91,241,242]. Generally, NRAS mutations occur independently of BRAF mutations but dual expression has been reported [243]. In contrast to BRAF mutations, NRAS-mutant melanomas are correlated with the nodular subtype and found in CSD skin [18]. The association of NRAS and skin with CSD suggests that such mutagenesis is induced by UV irradiation [244]. Positive NRAS expression was also associated with a lower grade of tumor-infiltrating lymphocytes and a higher tumor stage [245]. The prognostic value of identification of NRAS mutation is unclear. A large cohort study from the M.D. Anderson Cancer Center revealed shorter survival from metastatic melanoma [17]. Others have reported, however, that overall survival is not different between WT and NRAS-mutated melanomas [246].
Along with BRAF mutations, NRAS mutations are among mutations found in melanocytic and dysplastic nevi and melanomas, with high mutational load as a notable discriminant favoring the latter in diagnosis [223,227]. Notably, NRAS mutations were also commonly found in congenital melanocytic nevi, suggesting mutagenesis independent of UV radiation [225,247]. The contrast between this observation and the association of NRAS mutation with skin with CSD suggests that detecting the UV signature mutations can help diagnose melanoma.
In melanomas bearing NRAS mutations, targeted therapy has shown limited effectiveness and thus immune therapies and chemotherapy are generally used. Direct targeting of N-ras is difficult but most research has focused on inhibiting the farnesylation and subsequent activation of N-ras [248,249]. Subsequently, farnesyl transferase inhibitors (FTIs), such as lonafarnib and tipifarnib, were developed and shown to induce apoptosis in mouse and human melanoma cells [42,43]. However, clinical trials have failed to show efficacy for FTIs, as N-ras becomes activated through alternative post-translational modifications [249,250,251].
Besides N-ras, ongoing research has focused on targeting its downstream signals. MEK inhibitors have been identified as potential therapy for NRAS mutants. In particular, binimetinib has been shown in phase 3 trials to have improved survival and response rate compared to dacarbazine in NRAS-mutant melanoma [252]. Other clinical trials in progress are studying the potential of combining MEK inhibitors with PI3K, RAF, and cell cycle inhibitors [249,253,254,255,256]. Overall, the options to treat NRAS-mutated melanomas remain relatively scarce despite ongoing trials with signaling pathway targets.
3.3.3. C-KIT
C-KIT is a receptor tyrosine kinase (RTK) directly responsible for binding to growth factors as the first signal down the MAPK and PI3K pathways [15,44,85,257]. The majority of c-KIT mutations are found in mucosal and acral melanomas, as well as in melanomas arising from skin with CSD [44,258]. Presence of these mutations has also been associated with worse survival as compared with wild-type melanomas [259].
Because of the relative rarity of c-KIT mutations, the understanding of targeted therapy to treat melanoma is scarce. Imatinib and nilotinib are the main c-KIT inhibitors that have been studied in melanoma. In a pair of phase 2 trials, imatinib was shown to have significant clinical response in tumors bearing c-KIT mutations as compared with wild-type tumors [45,260]. Nilotinib has similarly shown promise in the treatment of patients with c-KIT mutations in phase 2 trials [46,47].
3.3.4. Immune Checkpoint (CTLA-4, PD-1, PD-L1)
Immune checkpoint inhibitors represent a novel class of drugs that have increasingly been used in melanoma therapy. By targeting the inactivation signals of the immune system, immunotherapy acts to stimulate the intrinsic immune response to cancer. The main targeted signals are cytotoxic T-lymphocyte antigen-4 (CTLA-4) and programmed death-1 (PD-1). Normally, antigen-presenting cells (APCs) co-stimulate T cells through both binding of the antigen-major histocompatibility complex (MHC) to the T cell receptor (TCR) and of CD80/86 to the CD28 receptor on T cells. As T cells become activated, CTLA-4 is upregulated and binds to the CD28 receptor with greater affinity than does CD80/86. Through this negative feedback, the T cell response is downregulated [66,261]. In peripheral tissue, binding of PD-1 on T cells to PD-L1 on normal or tumor cells reduces the activity and proliferation of T cells [262,263]. Monotherapy and combined therapy with CTLA-4 antibodies (ipilimumab) and PD-1 antibodies (pembrolizumab, nivolumab) have been shown to elicit a positive response against melanoma [67,68,69,264].
While immunotherapy shows promising potential, predicting therapeutic response has proven difficult. PD-L1 has been studied as a potential marker to predict response to immunotherapy without much avail. PD-L1 is estimated to be overexpressed in 45% of melanoma tumor samples [265]. In these samples, PD-L1 expression was associated with improved response rates to checkpoint therapy; however, absence of expression did not rule out the rate or degree of response to combined or monotherapy [265]. Other studies have shown similar patterns of response depending on positive PD-L1 expression [266,267]. Recently, a pair of studies has shown that melanoma cells release vesicles with PD-L1 on the surface and that such exosomal PD-L1 levels correlates with response to anti-PD-1 therapy [268,269]. Thus, identification of PD-L1 expression in tumors may serve as an effective first step in determining treatment with checkpoint therapy serving as a first-line option in tumors with positive expression and as reserve treatment in tumors with negative expression [270].
The expression of PD-1 and CTLA-4 on melanoma cells is not well studied. PD-1 has generally been studied for its expression on immune cells, such as T cells, but receptors have been identified on melanoma cells as well. In fact, the PD-1 receptor was shown to promote growth of melanoma and, as such, bears significance for choosing proper therapy [271]. Similarly, CTLA-4 has been found to be expressed on melanoma cells and to drive tumor formation [272]. An Italian study also found that serum CTLA-4 may serve as a novel biomarker in predicting favorable response to ipilimumab [273]. Timely identification of these markers is crucial for determining treatment for melanoma patients.
4. Summary and Conclusions
The incidence of melanoma continues to rapidly increase in the United States, representing a serious public health problem. Early detection of both cutaneous and uveal melanoma represents one of the most crucial ways of reducing the clinical and financial burden of the disease [274]. Understanding the molecular pathology of melanoma marks a significant stride towards that end. Detecting molecular markers aids diagnosis when histological assessment is challenging. Identification of common mutations, such as BRAF or NRAS, not only helps to narrow down diagnosis and paint a prognostic picture, but also to guide treatment. The expanding knowledge into vitamin D and melanogenesis represents an untapped reservoir for new clinical therapies of melanoma. Overall, analysis of these markers should become reflexive when melanoma is suspected or after diagnosis to contribute to improving outcomes for patients.
Funding
Writing of this review was in part supported by the NIH grants 1R01AR073004 and R01AR071189 to ATS and VA merit 1I01BX004293-01A1 to ATS.
Acknowledgments
Partial support from NIH and VA is acknowledged. In compliance with journal regulations, many original papers on the topic are cited in the referenced reviews.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cancer Facts & Figures; American Cancer Society: Atlanta, GA, USA, 2019.
- Rastrelli, M.; Tropea, S.; Rossi, C.R.; Alaibac, M. Melanoma: Epidemiology, Risk Factors, Pathogenesis, Diagnosis and Classification. In Vivo 2014, 28, 1005–1011. [Google Scholar] [PubMed]
- Sample, A.; He, Y.-Y. Mechanisms and Prevention of Uv-Induced Melanoma. Photodermatol. Photoimmunol. Photomed. 2018, 34, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Guy, G.P., Jr.; Ekwueme, D.U.; Tangka, F.K.; Richardson, L.C. Melanoma Treatment Costs: A Systematic Review of the Literature, 1990–2011. Am. J. Prev. Med. 2012, 43, 537–545. [Google Scholar] [CrossRef]
- Elder, D.E.; Bastian, B.C.; Cree, I.A.; Massi, D.; Scolyer, R.A. The 2018 World Health Organization Classification of Cutaneous, Mucosal, and Uveal Melanoma: Detailed Analysis of 9 Distinct Subtypes Defined by Their Evolutionary Pathway. Arch. Pathol. Lab. Med. 2020, 144, 500–522. [Google Scholar] [CrossRef]
- Pan, Y.; Adler, N.R.; Wolfe, R.; McLean, C.A.; Kelly, J.W. Nodular Melanoma Is Less Likely Than Superficial Spreading Melanoma to Be Histologically Associated with a Naevus. Med. J. Aust. 2017, 207, 333–338. [Google Scholar] [CrossRef]
- Bolognia, J.L.S.; Julie, V.; Duncan, K.O.; Ko, C.J. Cutaneous Melanoma. In Dermatology Essentials; Elsevier Inc.: Amsterdam, The Netherlands, 2014. [Google Scholar]
- McLaughlin, C.C.; Wu, X.C.; Jemal, A.; Martin, H.J.; Roche, L.M.; Chen, V.W. Incidence of Noncutaneous Melanomas in the U.S. Cancer 2005, 103, 1000–1007. [Google Scholar] [CrossRef]
- Krantz, B.A.; Dave, N.; Komatsubara, K.M.; Marr, B.P.; Carvajal, R.D. Uveal Melanoma: Epidemiology, Etiology, and Treatment of Primary Disease. Clin. Ophthalmol. 2017, 11, 279–289. [Google Scholar] [CrossRef]
- Kaliki, S.; Shields, C.L. Uveal Melanoma: Relatively Rare but Deadly Cancer. Eye (London) 2017, 31, 241–257. [Google Scholar] [CrossRef]
- Shah, C.P.; Weis, E.; Lajous, M.; Shields, J.A.; Shields, C.L. Intermittent and Chronic Ultraviolet Light Exposure and Uveal Melanoma: A Meta-Analysis. Ophthalmology 2005, 112, 1599–1607. [Google Scholar] [CrossRef]
- Weis, E.; Shah, C.P.; Lajous, M.; Shields, J.A.; Shields, C.L. The Association between Host Susceptibility Factors and Uveal Melanoma: A Meta-Analysis. Arch. Ophthalmol. 2006, 124, 54–60. [Google Scholar] [CrossRef]
- Jager, M.J.; Shields, C.L.; Cebulla, C.M.; Abdel-Rahman, M.H.; Grossniklaus, H.E.; Stern, M.-H.; Carvajal, R.D.; Belfort, R.N.; Jia, R.; Shields, J.A.; et al. Uveal Melanoma. Nat. Rev. Dis. Prim. 2020, 6, 24. [Google Scholar] [CrossRef] [PubMed]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the Braf Gene in Human Cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef] [PubMed]
- Melis, C.; Rogiers, A.; Bechter, O.; van den Oord, J.J. Molecular Genetic and Immunotherapeutic Targets in Metastatic Melanoma. Virchows Arch. 2017, 471, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Pracht, M.; Mogha, A.; Lespagnol, A.; Fautrel, A.; Mouchet, N.; Le Gall, F.; Paumier, V.; Lefeuvre-Plesse, C.; Rioux-Leclerc, N.; Mosser, J.; et al. Prognostic and Predictive Values of Oncogenic Braf, Nras, C-Kit and Mitf in Cutaneous and Mucous Melanoma. J. Eur. Acad. Dermatol. Venereol. 2015, 29, 1530–1538. [Google Scholar] [CrossRef]
- Jakob, J.A.; Bassett, R.L., Jr.; Ng, C.S.; Curry, J.L.; Joseph, R.W.; Alvarado, G.C.; Rohlfs, M.L.; Richard, J.; Gershenwald, J.E.; Kim, K.B.; et al. Nras Mutation Status Is an Independent Prognostic Factor in Metastatic Melanoma. Cancer 2012, 118, 4014–4023. [Google Scholar] [CrossRef]
- Lee, J.H.; Choi, J.W.; Kim, Y.S. Frequencies of Braf and Nras Mutations Are Different in Histological Types and Sites of Origin of Cutaneous Melanoma: A Meta-Analysis. Br. J. Dermatol. 2011, 164, 776–784. [Google Scholar] [CrossRef]
- Beadling, C.; Jacobson-Dunlop, E.; Hodi, F.S.; Le, C.; Warrick, A.; Patterson, J.; Town, A.; Harlow, A.; Cruz, F., 3rd; Azar, S.; et al. Kit Gene Mutations and Copy Number in Melanoma Subtypes. Clin. Cancer Res. 2008, 14, 6821–6828. [Google Scholar] [CrossRef]
- Goldstein, A.M.; Chan, M.; Harland, M.; Hayward, N.K.; Demenais, F.; Timothy Bishop, D.; Azizi, E.; Bergman, W.; Bianchi-Scarra, G.; Bruno, W.; et al. Features Associated with Germline Cdkn2a Mutations: A Genomel Study of Melanoma-Prone Families from Three Continents. J. Med. Genet. 2007, 44, 99–106. [Google Scholar] [CrossRef]
- Aoude, L.G.; Wadt, K.A.W.; Pritchard, A.L.; Hayward, N.K. Genetics of Familial Melanoma: 20 Years after Cdkn2a. Pigment Cell Melanoma Res. 2015, 28, 148–160. [Google Scholar] [CrossRef]
- Florell, S.R.; Meyer, L.J.; Boucher, K.M.; Porter-Gill, P.A.; Hart, M.; Erickson, J.; Cannon-Albright, L.A.; Pershing, L.K.; Harris, R.M.; Samlowski, W.E.; et al. Longitudinal Assessment of the Nevus Phenotype in a Melanoma Kindred. J. Investig. Dermatol. 2004, 123, 576–582. [Google Scholar] [CrossRef]
- Brożyna, A.A.; Jozwicki, W.; Janjetovic, Z.; Slominski, A.T. Expression of Vitamin D Receptor Decreases During Progression of Pigmented Skin Lesions. Hum. Pathol. 2011, 42, 618–631. [Google Scholar] [CrossRef] [PubMed]
- Brożyna, A.A.; Jóźwicki, W.; Slominski, A.T. Decreased Vdr Expression in Cutaneous Melanomas as Marker of Tumor Progression: New Data and Analyses. Anticancer Res. 2014, 34, 2735–2743. [Google Scholar] [PubMed]
- Tagliabue, E.; Gandini, S.; Bellocco, R.; Maisonneuve, P.; Newton-Bishop, J.; Polsky, D.; Lazovich, D.; Kanetsky, P.A.; Ghiorzo, P.; Gruis, N.A.; et al. Mc1r Variants as Melanoma Risk Factors Independent of at-Risk Phenotypic Characteristics: A Pooled Analysis from the M-Skip Project. Cancer Manag. Res. 2018, 10, 1143–1154. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, S.; Sera, F.; Gandini, S.; Iodice, S.; Caini, S.; Maisonneuve, P.; Fargnoli, M.C. Mc1r Variants, Melanoma and Red Hair Color Phenotype: A Meta-Analysis. Int. J. Cancer 2008, 122, 2753–2760. [Google Scholar] [CrossRef]
- Naffouje, S.; Naffouje, R.; Bhagwandin, S.; Salti, G.I. Microphthalmia Transcription Factor in Malignant Melanoma Predicts Occult Sentinel Lymph Node Metastases and Survival. Melanoma Res. 2015, 25, 496–502. [Google Scholar] [CrossRef]
- Yokoyama, S.; Woods, S.L.; Boyle, G.M.; Aoude, L.G.; MacGregor, S.; Zismann, V.; Gartside, M.; Cust, A.E.; Haq, R.; Harland, M.; et al. A Novel Recurrent Mutation in Mitf Predisposes to Familial and Sporadic Melanoma. Nature 2011, 480, 99–103. [Google Scholar] [CrossRef]
- Van Raamsdonk, C.D.; Griewank, K.G.; Crosby, M.B.; Garrido, M.C.; Vemula, S.; Wiesner, T.; Obenauf, A.C.; Wackernagel, W.; Green, G.; Bouvier, N.; et al. Mutations in Gna11 in Uveal Melanoma. N. Engl. J. Med. 2010, 363, 2191–2199. [Google Scholar] [CrossRef]
- Kalirai, H.; Dodson, A.; Faqir, S.; Damato, B.E.; Coupland, S.E. Lack of Bap1 Protein Expression in Uveal Melanoma Is Associated with Increased Metastatic Risk and Has Utility in Routine Prognostic Testing. Br. J. Cancer 2014, 111, 1373–1380. [Google Scholar] [CrossRef]
- Rai, K.; Pilarski, R.; Boru, G.; Rehman, M.; Saqr, A.H.; Massengill, J.B.; Singh, A.; Marino, M.J.; Davidorf, F.H.; Cebulla, C.M.; et al. Germline Bap1 Alterations in Familial Uveal Melanoma. Genes Chromosom. Cancer 2017, 56, 168–174. [Google Scholar] [CrossRef]
- Garfield, E.M.; Walton, K.E.; Quan, V.L.; VandenBoom, T.; Zhang, B.; Kong, B.Y.; Isales, M.C.; Panah, E.; Kim, G.; Gerami, P. Histomorphologic Spectrum of Germline-Related and Sporadic Bap1-Inactivated Melanocytic Tumors. J. Am. Acad. Dermatol. 2018, 79, 525–534. [Google Scholar] [CrossRef]
- Robertson, A.G.; Shih, J.; Yau, C.; Gibb, E.A.; Oba, J.; Mungall, K.L.; Hess, J.M.; Uzunangelov, V.; Walter, V.; Danilova, L.; et al. Integrative Analysis Identifies Four Molecular and Clinical Subsets in Uveal Melanoma. Cancer Cell 2017, 32, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Smit, K.N.; Jager, M.J.; de Klein, A.; Kiliҫ, E. Uveal Melanoma: Towards a Molecular Understanding. Prog. Retin. Eye Res. 2019, 75, 100800. [Google Scholar] [CrossRef] [PubMed]
- Yavuzyigitoglu, S.; Koopmans, A.E.; Verdijk, R.M.; Vaarwater, J.; Eussen, B.; van Bodegom, A.; Paridaens, D.; Kiliç, E.; de Klein, A.; Rotterdam Ocular Melanoma Study Group. Uveal Melanomas with Sf3b1 Mutations: A Distinct Subclass Associated with Late-Onset Metastases. Ophthalmology 2016, 123, 1118–1128. [Google Scholar] [CrossRef]
- Dono, M.; Angelini, G.; Cecconi, M.; Amaro, A.; Esposito, A.I.; Mirisola, V.; Maric, I.; Lanza, F.; Nasciuti, F.; Viaggi, S.; et al. Mutation Frequencies of Gnaq, Gna11, Bap1, Sf3b1, Eif1ax and Tert in Uveal Melanoma: Detection of an Activating Mutation in the Tert Gene Promoter in a Single Case of Uveal Melanoma. Br. J. Cancer 2014, 110, 1058–1065. [Google Scholar] [CrossRef]
- Martin, M.; Maßhöfer, L.; Temming, P.; Rahmann, S.; Metz, C.; Bornfeld, N.; van de Nes, J.; Klein-Hitpass, L.; Hinnebusch, A.G.; Horsthemke, B.; et al. Exome Sequencing Identifies Recurrent Somatic Mutations in Eif1ax and Sf3b1 in Uveal Melanoma with Disomy 3. Nat. Genet. 2013, 45, 933–936. [Google Scholar] [CrossRef]
- Dummer, R.; Ascierto, P.A.; Gogas, H.J.; Arance, A.; Mandala, M.; Liszkay, G.; Garbe, C.; Schadendorf, D.; Krajsova, I.; Gutzmer, R.; et al. Encorafenib Plus Binimetinib Versus Vemurafenib or Encorafenib in Patients with Braf-Mutant Melanoma (Columbus): A Multicentre, Open-Label, Randomised Phase 3 Trial. Lancet Oncol. 2018, 19, 603–615. [Google Scholar] [CrossRef]
- Hauschild, A.; Grob, J.J.; Demidov, L.V.; Jouary, T.; Gutzmer, R.; Millward, M.; Rutkowski, P.; Blank, C.U.; Miller, W.H., Jr.; Kaempgen, E.; et al. Dabrafenib in Braf-Mutated Metastatic Melanoma: A Multicentre, Open-Label, Phase 3 Randomised Controlled Trial. Lancet 2012, 380, 358–365. [Google Scholar] [CrossRef]
- Sarkisian, S.; Davar, D. Mek Inhibitors for the Treatment of Nras Mutant Melanoma. Drug Des. Dev. Ther. 2018, 12, 2553–2565. [Google Scholar] [CrossRef]
- Hodis, E.; Watson, I.R.; Kryukov, G.V.; Arold, S.T.; Imielinski, M.; Theurillat, J.P.; Nickerson, E.; Auclair, D.; Li, L.; Place, C.; et al. A Landscape of Driver Mutations in Melanoma. Cell 2012, 150, 251–263. [Google Scholar] [CrossRef]
- Smalley, K.S.M.; Eisen, T.G. Farnesyl Transferase Inhibitor Sch66336 Is Cytostatic, Pro-Apoptotic and Enhances Chemosensitivity to Cisplatin in Melanoma Cells. Int. J. Cancer 2003, 105, 165–175. [Google Scholar] [CrossRef]
- Niessner, H.; Beck, D.; Sinnberg, T.; Lasithiotakis, K.; Maczey, E.; Gogel, J.; Venturelli, S.; Berger, A.; Mauthe, M.; Toulany, M.; et al. The Farnesyl Transferase Inhibitor Lonafarnib Inhibits Mtor Signaling and Enforces Sorafenib-Induced Apoptosis in Melanoma Cells. J. Investig. Dermatol. 2011, 131, 468–479. [Google Scholar] [CrossRef] [PubMed]
- Curtin, J.A.; Busam, K.; Pinkel, D.; Bastian, B.C. Somatic Activation of Kit in Distinct Subtypes of Melanoma. J. Clin. Oncol. 2006, 24, 4340–4346. [Google Scholar] [CrossRef] [PubMed]
- Carvajal, R.D.; Antonescu, C.R.; Wolchok, J.D.; Chapman, P.B.; Roman, R.-A.; Teitcher, J.; Panageas, K.S.; Busam, K.J.; Chmielowski, B.; Lutzky, J.; et al. Kit as a Therapeutic Target in Metastatic Melanoma. JAMA 2011, 305, 2327–2334. [Google Scholar] [CrossRef] [PubMed]
- Carvajal, R.D.; Lawrence, D.P.; Weber, J.S.; Gajewski, T.F.; Gonzalez, R.; Lutzky, J.; O’Day, S.J.; Hamid, O.; Wolchok, J.D.; Chapman, P.B.; et al. Phase Ii Study of Nilotinib in Melanoma Harboring Kit Alterations Following Progression to Prior Kit Inhibition. Clin. Cancer Res. 2015, 21, 2289–2296. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Carvajal, R.D.; Dummer, R.; Hauschild, A.; Daud, A.; Bastian, B.C.; Markovic, S.N.; Queirolo, P.; Arance, A.; Berking, C.; et al. Efficacy and Safety of Nilotinib in Patients with Kit-Mutated Metastatic or Inoperable Melanoma: Final Results from the Global, Single-Arm, Phase Ii Team Trial. Ann. Oncol. 2017, 28, 1380–1387. [Google Scholar] [CrossRef] [PubMed]
- Serrano, M.; Hannon, G.J.; Beach, D. A New Regulatory Motif in Cell-Cycle Control Causing Specific Inhibition of Cyclin D/Cdk4. Nature 1993, 366, 704–707. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiong, Y.; Yarbrough, W.G. Arf Promotes Mdm2 Degradation and Stabilizes P53: Arf-Ink4a Locus Deletion Impairs Both the Rb and P53 Tumor Suppression Pathways. Cell 1998, 92, 725–734. [Google Scholar] [CrossRef]
- Soura, E.; Eliades, P.J.; Shannon, K.; Stratigos, A.J.; Tsao, H. Hereditary Melanoma: Update on Syndromes and Management: Genetics of Familial Atypical Multiple Mole Melanoma Syndrome. J. Am. Acad. Dermatol. 2016, 74, 395–410. [Google Scholar] [CrossRef]
- Burdette-Radoux, S.; Tozer, R.G.; Lohmann, R.C.; Quirt, I.; Ernst, D.S.; Walsh, W.; Wainman, N.; Colevas, A.D.; Eisenhauer, E.A. Phase Ii Trial of Flavopiridol, a Cyclin Dependent Kinase Inhibitor, in Untreated Metastatic Malignant Melanoma. Investig. New Drugs 2004, 22, 315–322. [Google Scholar] [CrossRef]
- Baker, A.R.; McDonnell, D.P.; Hughes, M.; Crisp, T.M.; Mangelsdorf, D.J.; Haussler, M.R.; Pike, J.W.; Shine, J.; O’Malley, B.W. Cloning and Expression of Full-Length Cdna Encoding Human Vitamin D Receptor. Proc. Natl. Acad. Sci. USA 1988, 85, 3294–3298. [Google Scholar] [CrossRef]
- Demay, M.B.; Kiernan, M.S.; DeLuca, H.F.; Kronenberg, H.M. Sequences in the Human Parathyroid Hormone Gene That Bind the 1,25-Dihydroxyvitamin D3 Receptor and Mediate Transcriptional Repression in Response to 1,25-Dihydroxyvitamin D3. Proc. Natl. Acad. Sci. USA 1992, 89, 8097–8101. [Google Scholar] [CrossRef] [PubMed]
- Colnot, S.; Lambert, M.; Blin, C.; Thomasset, M.; Perret, C. Identification of DNA Sequences That Bind Retinoid X Receptor-1,25(Oh)2d3-Receptor Heterodimers with High Affinity. Mol. Cell. Endocrinol. 1995, 113, 89–98. [Google Scholar] [CrossRef]
- Saw, R.P.M.; Armstrong, B.K.; Mason, R.S.; Morton, R.L.; Shannon, K.F.; Spillane, A.J.; Stretch, J.R.; Thompson, J.F. Adjuvant Therapy with High Dose Vitamin D Following Primary Treatment of Melanoma at High Risk of Recurrence: A Placebo Controlled Randomised Phase Ii Trial (Anzmtg 02.09 Mel-D). BMC Cancer 2014, 14, 780. [Google Scholar] [CrossRef] [PubMed]
- De Smedt, J.; Van Kelst, S.; Boecxstaens, V.; Stas, M.; Bogaerts, K.; Vanderschueren, D.; Aura, C.; Vandenberghe, K.; Lambrechts, D.; Wolter, P.; et al. Vitamin D Supplementation in Cutaneous Malignant Melanoma Outcome (Vidme): A Randomized Controlled Trial. BMC Cancer 2017, 17, 562. [Google Scholar] [CrossRef]
- Levy, C.; Khaled, M.; Fisher, D.E. Mitf: Master Regulator of Melanocyte Development and Melanoma Oncogene. Trends Mol. Med. 2006, 12, 406–414. [Google Scholar] [CrossRef]
- Price, E.R.; Horstmann, M.A.; Wells, A.G.; Weilbaecher, K.N.; Takemoto, C.M.; Landis, M.W.; Fisher, D.E. Alpha-Melanocyte-Stimulating Hormone Signaling Regulates Expression of Microphthalmia, a Gene Deficient in Waardenburg Syndrome. J. Biol. Chem. 1998, 273, 33042–33047. [Google Scholar] [CrossRef]
- Bertolotto, C.; Abbe, P.; Hemesath, T.J.; Bille, K.; Fisher, D.E.; Ortonne, J.P.; Ballotti, R. Microphthalmia Gene Product as a Signal Transducer in Camp-Induced Differentiation of Melanocytes. J. Cell Biol. 1998, 142, 827–835. [Google Scholar] [CrossRef]
- Rózanowska, M.; Sarna, T.; Land, E.J.; Truscott, T.G. Free Radical Scavenging Properties of Melanin Interaction of Eu- and Pheo-Melanin Models with Reducing and Oxidising Radicals. Free Radic. Biol. Med. 1999, 26, 518–525. [Google Scholar]
- Slominski, A.; Tobin, D.J.; Shibahara, S.; Wortsman, J. Melanin Pigmentation in Mammalian Skin and Its Hormonal Regulation. Physiol. Rev. 2004, 84, 1155–1228. [Google Scholar] [CrossRef]
- Slominski, R.M.; Zmijewski, M.A.; Slominski, A.T. The Role of Melanin Pigment in Melanoma. Exp. Dermatol. 2015, 24, 258–259. [Google Scholar] [CrossRef]
- Slominski, A.; Zmijewski, M.A.; Pawelek, J. L-Tyrosine and L-Dihydroxyphenylalanine as Hormone-Like Regulators of Melanocyte Functions. Pigment Cell Melanoma Res. 2012, 25, 14–27. [Google Scholar] [CrossRef] [PubMed]
- Ito, S. The Ifpcs Presidential Lecture: A Chemist’s View of Melanogenesis. Pigment Cell Res. 2003, 16, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Kaur, A.; Ecker, B.L.; Douglass, S.M.; Kugel, C.H.; Webster, M.R.; Almeida, F.V.; Somasundaram, R.; Hayden, J.; Ban, E.; Ahmadzadeh, H.; et al. Remodeling of the Collagen Matrix in Aging Skin Promotes Melanoma Metastasis and Affects Immune Cell Motility. Cancer Discov. 2019, 9, 64–81. [Google Scholar] [CrossRef] [PubMed]
- Fong, L.; Small, E.J. Anti-Cytotoxic T-Lymphocyte Antigen-4 Antibody: The First in an Emerging Class of Immunomodulatory Antibodies for Cancer Treatment. J. Clin. Oncol. 2008, 26, 5275–5283. [Google Scholar] [CrossRef] [PubMed]
- Hodi, F.S.; O’Day, S.J.; McDermott, D.F.; Weber, R.W.; Sosman, J.A.; Haanen, J.B.; Gonzalez, R.; Robert, C.; Schadendorf, D.; Hassel, J.C.; et al. Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. N. Engl. J. Med. 2010, 363, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Topalian, S.L.; Sznol, M.; McDermott, D.F.; Kluger, H.M.; Carvajal, R.D.; Sharfman, W.H.; Brahmer, J.R.; Lawrence, D.P.; Atkins, M.B.; Powderly, J.D.; et al. Survival, Durable Tumor Remission, and Long-Term Safety in Patients with Advanced Melanoma Receiving Nivolumab. J. Clin. Oncol. 2014, 32, 1020–1030. [Google Scholar] [CrossRef] [PubMed]
- Postow, M.A.; Callahan, M.K.; Wolchok, J.D. Immune Checkpoint Blockade in Cancer Therapy. J. Clin. Oncol. 2015, 33, 1974–1982. [Google Scholar] [CrossRef] [PubMed]
- Landis, C.A.; Masters, S.B.; Spada, A.; Pace, A.M.; Bourne, H.R.; Vallar, L. Gtpase Inhibiting Mutations Activate the Alpha Chain of Gs and Stimulate Adenylyl Cyclase in Human Pituitary Tumours. Nature 1989, 340, 692–696. [Google Scholar] [CrossRef]
- Kalinec, G.; Nazarali, A.J.; Hermouet, S.; Xu, N.; Gutkind, J.S. Mutated Alpha Subunit of the Gq Protein Induces Malignant Transformation in Nih 3t3 Cells. Mol. Cell. Biol. 1992, 12, 4687–4693. [Google Scholar] [CrossRef]
- Amaro, A.; Gangemi, R.; Piaggio, F.; Angelini, G.; Barisione, G.; Ferrini, S.; Pfeffer, U. The Biology of Uveal Melanoma. Cancer Metastasis Rev. 2017, 36, 109–140. [Google Scholar] [CrossRef]
- Pan, H.; Jia, R.; Zhang, L.; Xu, S.; Wu, Q.; Song, X.; Zhang, H.; Ge, S.; Leon Xu, X.; Fan, X. Bap1 Regulates Cell Cycle Progression through E2f1 Target Genes and Mediates Transcriptional Silencing Via H2a Monoubiquitination in Uveal Melanoma Cells. Int. J. Biochem. Cell Biol. 2015, 60, 176–184. [Google Scholar] [CrossRef]
- Vivet-Noguer, R.; Tarin, M.; Roman-Roman, S.; Alsafadi, S. Emerging Therapeutic Opportunities Based on Current Knowledge of Uveal Melanoma Biology. Cancers 2019, 11, 1019. [Google Scholar] [CrossRef] [PubMed]
- Alsafadi, S.; Houy, A.; Battistella, A.; Popova, T.; Wassef, M.; Henry, E.; Tirode, F.; Constantinou, A.; Piperno-Neumann, S.; Roman-Roman, S.; et al. Cancer-Associated Sf3b1 Mutations Affect Alternative Splicing by Promoting Alternative Branchpoint Usage. Nat. Commun. 2016, 7, 10615. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, J.; Si, K.; Maitra, U. Function of Eukaryotic Translation Initiation Factor 1a (Eif1a) (Formerly Called Eif-4c) in Initiation of Protein Synthesis. J. Biol. Chem. 1997, 272, 7883–7891. [Google Scholar] [CrossRef]
- Blanchard, D.A.; Mouhamad, S.; Auffredou, M.-T.; Pesty, A.; Bertoglio, J.; Leca, G.; Vazquez, A. Cdk2 Associates with Map Kinase in Vivo and Its Nuclear Translocation Is Dependent on Map Kinase Activation in Il-2-Dependent Kit 225 T Lymphocytes. Oncogene 2000, 19, 4184–4189. [Google Scholar] [CrossRef][Green Version]
- Seger, R.; Seger, D.; Reszka, A.A.; Munar, E.S.; Eldar-Finkelman, H.; Dobrowolska, G.; Jensen, A.M.; Campbell, J.S.; Fischer, E.H.; Krebs, E.G. Overexpression of Mitogen-Activated Protein Kinase Kinase (Mapkk) and Its Mutants in Nih 3t3 Cells. Evidence That Mapkk Involvement in Cellular Proliferation Is Regulated by Phosphorylation of Serine Residues in Its Kinase Subdomains Vii and Viii. J. Biol. Chem. 1994, 269, 25699–25709. [Google Scholar]
- Lavoie, J.N.; L’Allemain, G.; Brunet, A.; Müller, R.; Pouysségur, J. Cyclin D1 Expression Is Regulated Positively by the P42/P44mapk and Negatively by the P38/Hogmapk Pathway. J. Biol. Chem. 1996, 271, 20608–20616. [Google Scholar] [CrossRef]
- Raman, M.; Chen, W.; Cobb, M.H. Differential Regulation and Properties of Mapks. Oncogene 2007, 26, 3100–3112. [Google Scholar] [CrossRef]
- Dent, P.; Haser, W.; Haystead, T.A.; Vincent, L.A.; Roberts, T.M.; Sturgill, T.W. Activation of Mitogen-Activated Protein Kinase Kinase by V-Raf in Nih 3t3 Cells and in Vitro. Science 1992, 257, 1404–1407. [Google Scholar] [CrossRef]
- Kyriakis, J.M.; App, H.; Zhang, X.F.; Banerjee, P.; Brautigan, D.L.; Rapp, U.R.; Avruch, J. Raf-1 Activates Map Kinase-Kinase. Nature 1992, 358, 417–421. [Google Scholar] [CrossRef]
- Sigoillot, F.D.; Evans, D.R.; Guy, H.I. Growth-Dependent Regulation of Mammalian Pyrimidine Biosynthesis by the Protein Kinase a and Mapk Signaling Cascades. J. Biol. Chem. 2002, 277, 15745–15751. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Harrison, J.K.; Vincent, L.A.; Haystead, C.; Haystead, T.A.; Michel, H.; Hunt, D.F.; Lynch, K.R.; Sturgill, T.W. Molecular Structure of a Protein-Tyrosine/Threonine Kinase Activating P42 Mitogen-Activated Protein (Map) Kinase: Map Kinase Kinase. Proc. Natl. Acad. Sci. USA 1993, 90, 173–177. [Google Scholar] [CrossRef] [PubMed]
- Leonardi, G.C.; Falzone, L.; Salemi, R.; Zanghì, A.; Spandidos, D.A.; McCubrey, J.A.; Candido, S.; Libra, M. Cutaneous Melanoma: From Pathogenesis to Therapy (Review). Int. J. Oncol. 2018, 52, 1071–1080. [Google Scholar] [CrossRef] [PubMed]
- Burotto, M.; Chiou, V.L.; Lee, J.-M.; Kohn, E.C. The Mapk Pathway across Different Malignancies: A New Perspective. Cancer 2014, 120, 3446–3456. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.R.; Dudek, H.; Tao, X.; Masters, S.; Fu, H.; Gotoh, Y.; Greenberg, M.E. Akt Phosphorylation of Bad Couples Survival Signals to the Cell-Intrinsic Death Machinery. Cell 1997, 91, 231–241. [Google Scholar] [CrossRef]
- Mayo, L.D.; Donner, D.B. A Phosphatidylinositol 3-Kinase/Akt Pathway Promotes Translocation of Mdm2 from the Cytoplasm to the Nucleus. Proc. Natl. Acad. Sci. USA 2001, 98, 11598–11603. [Google Scholar] [CrossRef]
- Inoki, K.; Li, Y.; Zhu, T.; Wu, J.; Guan, K.L. Tsc2 Is Phosphorylated and Inhibited by Akt and Suppresses Mtor Signalling. Nat. Cell Biol. 2002, 4, 648–657. [Google Scholar] [CrossRef]
- Yuan, T.L.; Cantley, L.C. Pi3k Pathway Alterations in Cancer: Variations on a Theme. Oncogene 2008, 27, 5497–5510. [Google Scholar] [CrossRef]
- Rodriguez-Viciana, P.; Warne, P.H.; Dhand, R.; Vanhaesebroeck, B.; Gout, I.; Fry, M.J.; Waterfield, M.D.; Downward, J. Phosphatidylinositol-3-Oh Kinase as a Direct Target of Ras. Nature 1994, 370, 527–532. [Google Scholar] [CrossRef]
- Carpenter, C.L.; Auger, K.R.; Chanudhuri, M.; Yoakim, M.; Schaffhausen, B.; Shoelson, S.; Cantley, L.C. Phosphoinositide 3-Kinase Is Activated by Phosphopeptides That Bind to the Sh2 Domains of the 85-Kda Subunit. J. Biol. Chem. 1993, 268, 9478–9483. [Google Scholar]
- Klippel, A.; Kavanaugh, W.M.; Pot, D.; Williams, L.T. A Specific Product of Phosphatidylinositol 3-Kinase Directly Activates the Protein Kinase Akt through Its Pleckstrin Homology Domain. Mol. Cell. Biol. 1997, 17, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Rameh, L.E.; Tolias, K.F.; Duckworth, B.C.; Cantley, L.C. A New Pathway for Synthesis of Phosphatidylinositol-4,5-Bisphosphate. Nature 1997, 390, 192–196. [Google Scholar] [CrossRef] [PubMed]
- De Luca, A.; Maiello, M.R.; D’Alessio, A.; Pergameno, M.; Normanno, N. The Ras/Raf/Mek/Erk and the Pi3k/Akt Signalling Pathways: Role in Cancer Pathogenesis and Implications for Therapeutic Approaches. Expert Opin. Ther. Targets 2012, 16 (Suppl. 2), S17–S27. [Google Scholar] [CrossRef] [PubMed]
- Maehama, T.; Dixon, J.E. The Tumor Suppressor, Pten/Mmac1, Dephosphorylates the Lipid Second Messenger, Phosphatidylinositol 3,4,5-Trisphosphate. J. Biol. Chem. 1998, 273, 13375–13378. [Google Scholar] [CrossRef]
- Camargo, F.D.; Gokhale, S.; Johnnidis, J.B.; Fu, D.; Bell, G.W.; Jaenisch, R.; Brummelkamp, T.R. Yap1 Increases Organ Size and Expands Undifferentiated Progenitor Cells. Curr. Biol. 2007, 17, 2054–2060. [Google Scholar] [CrossRef]
- Xin, M.; Kim, Y.; Sutherland, L.B.; Qi, X.; McAnally, J.; Schwartz, R.J.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. Regulation of Insulin-Like Growth Factor Signaling by Yap Governs Cardiomyocyte Proliferation and Embryonic Heart Size. Sci. Signal. 2011, 4, ra70. [Google Scholar] [CrossRef]
- George, N.M.; Day, C.E.; Boerner, B.P.; Johnson, R.L.; Sarvetnick, N.E. Hippo Signaling Regulates Pancreas Development through Inactivation of Yap. Mol. Cell. Biol. 2012, 32, 5116–5128. [Google Scholar] [CrossRef]
- Hossain, Z.; Ali, S.M.; Ko, H.L.; Xu, J.; Ng, C.P.; Guo, K.; Qi, Z.; Ponniah, S.; Hong, W.; Hunziker, W. Glomerulocystic Kidney Disease in Mice with a Targeted Inactivation of Wwtr1. Proc. Natl. Acad. Sci. USA 2007, 104, 1631–1636. [Google Scholar] [CrossRef]
- Yu, F.X.; Luo, J.; Mo, J.S.; Liu, G.; Kim, Y.C.; Meng, Z.; Zhao, L.; Peyman, G.; Ouyang, H.; Jiang, W.; et al. Mutant Gq/11 Promote Uveal Melanoma Tumorigenesis by Activating Yap. Cancer Cell 2014, 25, 822–830. [Google Scholar] [CrossRef]
- Field, M.G.; Durante, M.A.; Anbunathan, H.; Cai, L.Z.; Decatur, C.L.; Bowcock, A.M.; Kurtenbach, S.; Harbour, J.W. Punctuated Evolution of Canonical Genomic Aberrations in Uveal Melanoma. Nat. Commun. 2018, 9, 116. [Google Scholar] [CrossRef]
- Pleasance, E.D.; Cheetham, R.K.; Stephens, P.J.; McBride, D.J.; Humphray, S.J.; Greenman, C.D.; Varela, I.; Lin, M.L.; Ordóñez, G.R.; Bignell, G.R.; et al. A Comprehensive Catalogue of Somatic Mutations from a Human Cancer Genome. Nature 2010, 463, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Hayward, N.K.; Wilmott, J.S.; Waddell, N.; Johansson, P.A.; Field, M.A.; Nones, K.; Patch, A.M.; Kakavand, H.; Alexandrov, L.B.; Burke, H.; et al. Whole-Genome Landscapes of Major Melanoma Subtypes. Nature 2017, 545, 175–180. [Google Scholar] [CrossRef]
- Berger, A.C.; Davidson, R.S.; Poitras, J.K.; Chabra, I.; Hope, R.; Brackeen, A.; Johnson, C.E.; Maetzold, D.J.; Middlebrook, B.; Oelschlager, K.M.; et al. Clinical Impact of a 31-Gene Expression Profile Test for Cutaneous Melanoma in 156 Prospectively and Consecutively Tested Patients. Curr. Med. Res. Opin. 2016, 32, 1599–1604. [Google Scholar] [CrossRef] [PubMed]
- Cassarino, D.S.; Lewine, N.; Cole, D.; Wade, B.; Gustavsen, G. Budget Impact Analysis of a Novel Gene Expression Assay for the Diagnosis of Malignant Melanoma. J. Med. Econ. 2014, 17, 782–791. [Google Scholar] [CrossRef] [PubMed]
- Ferris, L.K.; Jansen, B.; Ho, J.; Busam, K.J.; Gross, K.; Hansen, D.D.; Alsobrook, J.P., 2nd; Yao, Z.; Peck, G.L.; Gerami, P. Utility of a Noninvasive 2-Gene Molecular Assay for Cutaneous Melanoma and Effect on the Decision to Biopsy. JAMA Dermatol. 2017, 153, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.J.; Lian, C.G. Molecular Testing for Cutaneous Melanoma: An Update and Review. Arch. Pathol. Lab. Med. 2019, 143, 811–820. [Google Scholar] [CrossRef]
- Griewank, K.G.; Schilling, B.; Scholz, S.L.; Metz, C.H.; Livingstone, E.; Sucker, A.; Moller, I.; Reis, H.; Franklin, C.; Cosgarea, I.; et al. Oncogene Status as a Diagnostic Tool in Ocular and Cutaneous Melanoma. Eur. J. Cancer 2016, 57, 112–117. [Google Scholar] [CrossRef]
- Patel, S.P.; Kim, D.W.; Lacey, C.L.; Hwu, P. Gna11 Mutation in a Patient with Cutaneous Origin Melanoma: A Case Report. Medicine 2016, 95, e2336. [Google Scholar] [CrossRef]
- Koopmans, A.E.; Vaarwater, J.; Paridaens, D.; Naus, N.C.; Kilic, E.; de Klein, A. Patient Survival in Uveal Melanoma Is Not Affected by Oncogenic Mutations in Gnaq and Gna11. Br. J. Cancer 2013, 109, 493–496. [Google Scholar] [CrossRef]
- Staby, K.M.; Gravdal, K.; Mork, S.J.; Heegaard, S.; Vintermyr, O.K.; Krohn, J. Prognostic Impact of Chromosomal Aberrations and Gnaq, Gna11 and Bap1 Mutations in Uveal Melanoma. Acta Ophthalmol. 2018, 96, 31–38. [Google Scholar] [CrossRef]
- Nobori, T.; Miura, K.; Wu, D.J.; Lois, A.; Takabayashi, K.; Carson, D.A. Deletions of the Cyclin-Dependent Kinase-4 Inhibitor Gene in Multiple Human Cancers. Nature 1994, 368, 753–756. [Google Scholar] [CrossRef] [PubMed]
- Sharpless, N.E.; Kannan, K.; Xu, J.; Bosenberg, M.W.; Chin, L. Both Products of the Mouse Ink4a/Arf Locus Suppress Melanoma Formation in Vivo. Oncogene 2003, 22, 5055–5059. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Read, J.; Wadt, K.A.; Hayward, N.K. Melanoma Genetics. J. Med. Genet. 2016, 53, 1–14. [Google Scholar] [CrossRef]
- Sargen, M.R.; Kanetsky, P.A.; Newton-Bishop, J.; Hayward, N.K.; Mann, G.J.; Gruis, N.A.; Tucker, M.A.; Goldstein, A.M.; Bianchi-Scarra, G.; Puig, S.; et al. Histologic Features of Melanoma Associated with Cdkn2a Genotype. J. Am. Acad. Dermatol. 2015, 72, 496–507.e7. [Google Scholar] [CrossRef]
- Helgadottir, H.; Höiom, V.; Tuominen, R.; Nielsen, K.; Jönsson, G.; Olsson, H.; Hansson, J. Germline Cdkn2a Mutation Status and Survival in Familial Melanoma Cases. J. Natl. Cancer Inst. 2016, 108. [Google Scholar] [CrossRef]
- Dalmasso, B.; Pastorino, L.; Ciccarese, G.; Andreotti, V.; Grillo, F.; Mastracci, L.; Spagnolo, F.; Ballestrero, A.; Queirolo, P.; Bruno, W.; et al. Cdkn2a Germline Mutations Are Not Associated with Poor Survival in an Italian Cohort of Melanoma Patients. J. Am. Acad. Dermatol. 2019, 80, 1263–1271. [Google Scholar] [CrossRef]
- Yu, H.; Mashtalir, N.; Daou, S.; Hammond-Martel, I.; Ross, J.; Sui, G.; Hart, G.W.; Rauscher, F.J., 3rd; Drobetsky, E.; Milot, E.; et al. The Ubiquitin Carboxyl Hydrolase Bap1 Forms a Ternary Complex with Yy1 and Hcf-1 and Is a Critical Regulator of Gene Expression. Mol. Cell. Biol. 2010, 30, 5071–5085. [Google Scholar] [CrossRef]
- Harbour, J.W.; Onken, M.D.; Roberson, E.D.O.; Duan, S.; Cao, L.; Worley, L.A.; Council, M.L.; Matatall, K.A.; Helms, C.; Bowcock, A.M. Frequent Mutation of Bap1 in Metastasizing Uveal Melanomas. Science 2010, 330, 1410–1413. [Google Scholar] [CrossRef]
- Koopmans, A.E.; Verdijk, R.M.; Brouwer, R.W.W.; van den Bosch, T.P.P.; van den Berg, M.M.P.; Vaarwater, J.; Kockx, C.E.M.; Paridaens, D.; Naus, N.C.; Nellist, M.; et al. Clinical Significance of Immunohistochemistry for Detection of Bap1 Mutations in Uveal Melanoma. Mod. Pathol. 2014, 27, 1321–1330. [Google Scholar] [CrossRef]
- Decatur, C.L.; Ong, E.; Garg, N.; Anbunathan, H.; Bowcock, A.M.; Field, M.G.; Harbour, J.W. Driver Mutations in Uveal Melanoma: Associations with Gene Expression Profile and Patient Outcomes. JAMA Ophthalmol. 2016, 134, 728–733. [Google Scholar] [CrossRef]
- Shain, A.H.; Bagger, M.M.; Yu, R.; Chang, D.; Liu, S.; Vemula, S.; Weier, J.F.; Wadt, K.; Heegaard, S.; Bastian, B.C.; et al. The Genetic Evolution of Metastatic Uveal Melanoma. Nat. Genet. 2019, 51, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.P.; Lane, A.M.; DeAngelis, M.M.; Mayne, K.; Crabtree, M.; Gragoudas, E.S.; Kim, I.K. Clinical Characteristics of Uveal Melanoma in Patients with Germline Bap1 Mutations. JAMA Ophthalmol. 2015, 133, 881–887. [Google Scholar] [CrossRef] [PubMed]
- Ewens, K.G.; Lalonde, E.; Richards-Yutz, J.; Shields, C.L.; Ganguly, A. Comparison of Germline Versus Somatic Bap1 Mutations for Risk of Metastasis in Uveal Melanoma. BMC Cancer 2018, 18, 1172. [Google Scholar] [CrossRef] [PubMed]
- Wiesner, T.; Obenauf, A.C.; Murali, R.; Fried, I.; Griewank, K.G.; Ulz, P.; Windpassinger, C.; Wackernagel, W.; Loy, S.; Wolf, I.; et al. Germline Mutations in Bap1 Predispose to Melanocytic Tumors. Nat. Genet. 2011, 43, 1018–1021. [Google Scholar] [CrossRef]
- Njauw, C.N.; Kim, I.; Piris, A.; Gabree, M.; Taylor, M.; Lane, A.M.; DeAngelis, M.M.; Gragoudas, E.; Duncan, L.M.; Tsao, H. Germline Bap1 Inactivation Is Preferentially Associated with Metastatic Ocular Melanoma and Cutaneous-Ocular Melanoma Families. PLoS ONE 2012, 7, e35295. [Google Scholar] [CrossRef]
- Kumar, R.; Taylor, M.; Miao, B.; Ji, Z.; Njauw, J.C.N.; Jönsson, G.; Frederick, D.T.; Tsao, H. Bap1 Has a Survival Role in Cutaneous Melanoma. J. Investig. Dermatol. 2015, 135, 1089–1097. [Google Scholar] [CrossRef]
- Atkinson, A.; Linos, K.; Yan, S.; Tsongalis, G.; Lefferts, J. Molecular Diagnostic Techniques Used to Confirm an Unusual Case of Bapoma. FASEB J. 2017, 31 (Suppl. 1), 807.17. [Google Scholar]
- Yu, H.; Pak, H.; Hammond-Martel, I.; Ghram, M.; Rodrigue, A.; Daou, S.; Barbour, H.; Corbeil, L.; Hébert, J.; Drobetsky, E.; et al. Tumor Suppressor and Deubiquitinase Bap1 Promotes DNA Double-Strand Break Repair. Proc. Natl. Acad. Sci. USA 2014, 111, 285–290. [Google Scholar] [CrossRef]
- de Koning, L.; Decaudin, D.; El Botty, R.; Nicolas, A.; Carita, G.; Schuller, M.; Ouine, B.; Cartier, A.; Naguez, A.; Fleury, J.; et al. Parp Inhibition Increases the Response to Chemotherapy in Uveal Melanoma. Cancers 2019, 11, 751. [Google Scholar] [CrossRef]
- Harbour, J.W.; Roberson, E.D.; Anbunathan, H.; Onken, M.D.; Worley, L.A.; Bowcock, A.M. Recurrent Mutations at Codon 625 of the Splicing Factor Sf3b1 in Uveal Melanoma. Nat. Genet. 2013, 45, 133–135. [Google Scholar] [CrossRef]
- Kong, Y.; Krauthammer, M.; Halaban, R. Rare Sf3b1 R625 Mutations in Cutaneous Melanoma. Melanoma Res. 2014, 24, 332–334. [Google Scholar] [CrossRef]
- Bikle, D.D. Vitamin D Metabolism and Function in the Skin. Mol. Cell. Endocrinol. 2011, 347, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Verlinden, L.; Verstuyf, A.; Convents, R.; Marcelis, S.; Van Camp, M.; Bouillon, R. Action of 1,25(Oh)2d3 on the Cell Cycle Genes, Cyclin D1, P21 and P27 in Mcf-7 Cells. Mol. Cell. Endocrinol. 1998, 142, 57–65. [Google Scholar] [CrossRef]
- Hershberger, P.A.; Modzelewski, R.A.; Shurin, Z.R.; Rueger, R.M.; Trump, D.L.; Johnson, C.S. 1,25-Dihydroxycholecalciferol (1,25-D3) Inhibits the Growth of Squamous Cell Carcinoma and Down-Modulates P21(Waf1/Cip1) in Vitro and in Vivo. Cancer Res. 1999, 59, 2644–2649. [Google Scholar]
- Danielsson, C.; Fehsel, K.; Polly, P.; Carlberg, C. Differential Apoptotic Response of Human Melanoma Cells to 1 Alpha,25-Dihydroxyvitamin D3 and Its Analogues. Cell Death Differ. 1998, 5, 946–952. [Google Scholar] [CrossRef]
- Cordero, J.B.; Cozzolino, M.; Lu, Y.; Vidal, M.; Slatopolsky, E.; Stahl, P.D.; Barbieri, M.A.; Dusso, A. 1,25-Dihydroxyvitamin D Down-Regulates Cell Membrane Growth- and Nuclear Growth-Promoting Signals by the Epidermal Growth Factor Receptor. J. Biol. Chem. 2002, 277, 38965–38971. [Google Scholar] [CrossRef]
- Pinczewski, J.; Slominski, A. The Potential Role of Vitamin D in the Progression of Benign and Malignant Melanocytic Neoplasms. Exp. Dermatol. 2010, 19, 860–864. [Google Scholar] [CrossRef]
- Slominski, A.T.; Brożyna, A.A.; Skobowiat, C.; Zmijewski, M.A.; Kim, T.K.; Janjetovic, Z.; Oak, A.S.; Jozwicki, W.; Jetten, A.M.; Mason, R.S.; et al. On the Role of Classical and Novel Forms of Vitamin D in Melanoma Progression and Management. J. Steroid Biochem. Mol. Biol. 2018, 177, 159–170. [Google Scholar] [CrossRef]
- Slominski, A.T.; Brożyna, A.A.; Zmijewski, M.A.; Jóźwicki, W.; Jetten, A.M.; Mason, R.S.; Tuckey, R.C.; Elmets, C.A. Vitamin D Signaling and Melanoma: Role of Vitamin D and Its Receptors in Melanoma Progression and Management. Lab. Investig. 2017, 97, 706–724. [Google Scholar] [CrossRef]
- Del Puerto, C.; Navarrete-Dechent, C.; Molgó, M.; Camargo, C.A., Jr.; Borzutzky, A.; González, S. Immunohistochemical Expression of Vitamin D Receptor in Melanocytic Naevi and Cutaneous Melanoma: A Case-Control Study. Br. J. Dermatol. 2018, 179, 95–100. [Google Scholar] [CrossRef]
- Brozyna, A.A.; Jozwicki, W.; Janjetovic, Z.; Slominski, A.T. Expression of the Vitamin D-Activating Enzyme 1alpha-Hydroxylase (Cyp27b1) Decreases During Melanoma Progression. Hum. Pathol. 2013, 44, 374–387. [Google Scholar] [CrossRef] [PubMed]
- Brozyna, A.A.; Jochymski, C.; Janjetovic, Z.; Jozwicki, W.; Tuckey, R.C.; Slominski, A.T. Cyp24a1 Expression Inversely Correlates with Melanoma Progression: Clinic-Pathological Studies. Int. J. Mol. Sci. 2014, 15, 19000–19017. [Google Scholar] [CrossRef] [PubMed]
- Orlow, I.; Roy, P.; Reiner, A.S.; Yoo, S.; Patel, H.; Paine, S.; Armstrong, B.K.; Kricker, A.; Marrett, L.D.; Millikan, R.C.; et al. Vitamin D Receptor Polymorphisms in Patients with Cutaneous Melanoma. Int. J. Cancer 2012, 130, 405–418. [Google Scholar] [CrossRef] [PubMed]
- Orlow, I.; Reiner, A.S.; Thomas, N.E.; Roy, P.; Kanetsky, P.A.; Luo, L.; Paine, S.; Armstrong, B.K.; Kricker, A.; Marrett, L.D.; et al. Vitamin D Receptor Polymorphisms and Survival in Patients with Cutaneous Melanoma: A Population-Based Study. Carcinogenesis 2016, 37, 30–38. [Google Scholar] [CrossRef]
- Orlow, I.; Shi, Y.; Kanetsky, P.A.; Thomas, N.E.; Luo, L.; Corrales-Guerrero, S.; Cust, A.E.; Sacchetto, L.; Zanetti, R.; Rosso, S.; et al. The Interaction between Vitamin D Receptor Polymorphisms and Sun Exposure around Time of Diagnosis Influences Melanoma Survival. Pigment Cell Melanoma Res. 2018, 31, 287–296. [Google Scholar] [CrossRef]
- Newton-Bishop, J.A.; Beswick, S.; Randerson-Moor, J.; Chang, Y.M.; Affleck, P.; Elliott, F.; Chan, M.; Leake, S.; Karpavicius, B.; Haynes, S.; et al. Serum 25-Hydroxyvitamin D3 Levels Are Associated with Breslow Thickness at Presentation and Survival from Melanoma. J. Clin. Oncol. 2009, 27, 5439–5444. [Google Scholar] [CrossRef]
- Vasilovici, A.F.; Grigore, L.E.; Ungureanu, L.; Fechete, O.; Candrea, E.; Trifa, A.P.; Vișan, S.; Șenilă, S.; Cosgarea, R. Vitamin D Receptor Polymorphisms and Melanoma. Oncol. Lett. 2019, 17, 4162–4169. [Google Scholar] [CrossRef]
- Markiewicz, A.; Brożyna, A.A.; Podgórska, E.; Elas, M.; Urbańska, K.; Jetten, A.M.; Slominski, A.T.; Jóźwicki, W.; Orłowska-Heitzman, J.; Dyduch, G.; et al. Vitamin D Receptors (Vdr), Hydroxylases Cyp27b1 and Cyp24a1 and Retinoid-Related Orphan Receptors (Ror) Level in Human Uveal Tract and Ocular Melanoma with Different Melanization Levels. Sci. Rep. 2019, 9, 9142. [Google Scholar] [CrossRef]
- Colston, K.; Colston, M.J.; Feldman, D. 1,25-Dihydroxyvitamin D3 and Malignant Melanoma: The Presence of Receptors and Inhibition of Cell Growth in Culture. Endocrinology 1981, 108, 1083–1086. [Google Scholar] [CrossRef]
- Frampton, R.J.; Omond, S.A.; Eisman, J.A. Inhibition of Human Cancer Cell Growth by 1,25-Dihydroxyvitamin D3 Metabolites. Cancer Res. 1983, 43, 4443. [Google Scholar]
- Skobowiat, C.; Oak, A.S.; Kim, T.K.; Yang, C.H.; Pfeffer, L.M.; Tuckey, R.C.; Slominski, A.T. Noncalcemic 20-Hydroxyvitamin D3 Inhibits Human Melanoma Growth in in Vitro and in Vivo Models. Oncotarget 2017, 8, 9823–9834. [Google Scholar] [CrossRef] [PubMed]
- Piotrowska, A.; Wierzbicka, J.; Nadkarni, S.; Brown, G.; Kutner, A.; Żmijewski, M.A. Antiproliferative Activity of Double Point Modified Analogs of 1,25-Dihydroxyvitamin D2 against Human Malignant Melanoma Cell Lines. Int. J. Mol. Sci. 2016, 17, 76. [Google Scholar] [CrossRef] [PubMed]
- Brożyna, A.A.; Hoffman, R.M.; Slominski, A.T. Relevance of Vitamin D in Melanoma Development, Progression and Therapy. Anticancer Res. 2020, 40, 473–489. [Google Scholar] [CrossRef]
- Harant, H.; Spinner, D.; Reddy, G.S.; Lindley, I.J. Natural Metabolites of 1alpha,25-Dihydroxyvitamin D(3) Retain Biologic Activity Mediated through the Vitamin D Receptor. J. Cell. Biochem. 2000, 78, 112–120. [Google Scholar] [CrossRef]
- Wasiewicz, T.; Piotrowska, A.; Wierzbicka, J.; Slominski, A.T.; Zmijewski, M.A. Antiproliferative Activity of Non-Calcemic Vitamin D Analogs on Human Melanoma Lines in Relation to Vdr and Pdia3 Receptors. Int. J. Mol. Sci. 2018, 19, 2583. [Google Scholar] [CrossRef]
- Wasiewicz, T.; Szyszka, P.; Cichorek, M.; Janjetovic, Z.; Tuckey, R.C.; Slominski, A.T.; Zmijewski, M.A. Antitumor Effects of Vitamin D Analogs on Hamster and Mouse Melanoma Cell Lines in Relation to Melanin Pigmentation. Int. J. Mol. Sci. 2015, 16, 6645–6667. [Google Scholar] [CrossRef]
- Slominski, A.T.; Kim, T.-K.; Janjetovic, Z.; Tuckey, R.C.; Bieniek, R.; Yue, J.; Li, W.; Chen, J.; Nguyen, M.N.; Tang, E.K.Y.; et al. 20-Hydroxyvitamin D2 Is a Noncalcemic Analog of Vitamin D with Potent Antiproliferative and Prodifferentiation Activities in Normal and Malignant Cells. Am. J. Physiol. Cell Physiol. 2011, 300, C526–C541. [Google Scholar] [CrossRef]
- Slominski, A.T.; Janjetovic, Z.; Kim, T.-K.; Wright, A.C.; Grese, L.N.; Riney, S.J.; Nguyen, M.N.; Tuckey, R.C. Novel Vitamin D Hydroxyderivatives Inhibit Melanoma Growth and Show Differential Effects on Normal Melanocytes. Anticancer Res. 2012, 32, 3733–3742. [Google Scholar]
- Janjetovic, Z.; Brozyna, A.A.; Tuckey, R.C.; Kim, T.K.; Nguyen, M.N.; Jozwicki, W.; Pfeffer, S.R.; Pfeffer, L.M.; Slominski, A.T. High Basal Nf-Κb Activity in Nonpigmented Melanoma Cells Is Associated with an Enhanced Sensitivity to Vitamin D3 Derivatives. Br. J. Cancer 2011, 105, 1874–1884. [Google Scholar] [CrossRef]
- Slominski, A.T.; Kim, T.K.; Li, W.; Postlethwaite, A.; Tieu, E.W.; Tang, E.K.Y.; Tuckey, R.C. Detection of Novel Cyp11a1-Derived Secosteroids in the Human Epidermis and Serum and Pig Adrenal Gland. Sci. Rep. 2015, 5, 14875. [Google Scholar] [CrossRef]
- Tuckey, R.C.; Cheng, C.Y.S.; Slominski, A.T. The Serum Vitamin D Metabolome: What We Know and What Is Still to Discover. J. Steroid Biochem. Mol. Biol. 2019, 186, 4–21. [Google Scholar] [CrossRef] [PubMed]
- He, G.; Xu, W.; Chen, Y.; Liu, X.; Xi, M. Abnormal Apoptosis of Trophoblastic Cells Is Related to the up-Regulation of Cyp11a Gene in Placenta of Preeclampsia Patients. PLoS ONE 2013, 8, e59609. [Google Scholar] [CrossRef] [PubMed]
- Slominski, A.; Zjawiony, J.; Wortsman, J.; Semak, I.; Stewart, J.; Pisarchik, A.; Sweatman, T.; Marcos, J.; Dunbar, C.; C Tuckey, R. A Novel Pathway for Sequential Transformation of 7-Dehydrocholesterol and Expression of the P450scc System in Mammalian Skin. Eur. J. Biochem. 2004, 271, 4178–4188. [Google Scholar] [CrossRef] [PubMed]
- Slominski, A.; Semak, I.; Zjawiony, J.; Wortsman, J.; Li, W.; Szczesniewski, A.; Tuckey, R.C. The Cytochrome P450scc System Opens an Alternate Pathway of Vitamin D3 Metabolism. FEBS J. 2005, 272, 4080–4090. [Google Scholar] [CrossRef] [PubMed]
- Manna, P.R.; Sennoune, S.R.; Martinez-Zaguilan, R.; Slominski, A.T.; Pruitt, K. Regulation of Retinoid Mediated Cholesterol Efflux Involves Liver X Receptor Activation in Mouse Macrophages. Biochem. Biophys. Res. Commun. 2015, 464, 312–317. [Google Scholar] [CrossRef]
- Schedel, M.; Jia, Y.; Michel, S.; Takeda, K.; Domenico, J.; Joetham, A.; Ning, F.; Strand, M.; Han, J.; Wang, M.; et al. 1,25d3 Prevents Cd8(+)Tc2 Skewing and Asthma Development through Vdr Binding Changes to the Cyp11a1 Promoter. Nat. Commun. 2016, 7, 10213. [Google Scholar] [CrossRef]
- Manna, P.R.; Stetson, C.L.; Daugherty, C.; Shimizu, I.; Syapin, P.J.; Garrel, G.; Cohen-Tannoudji, J.; Huhtaniemi, I.; Slominski, A.T.; Pruitt, K.; et al. Up-Regulation of Steroid Biosynthesis by Retinoid Signaling: Implications for Aging. Mech. Ageing Dev. 2015, 150, 74–82. [Google Scholar] [CrossRef]
- Slominski, R.M.; Tuckey, R.C.; Manna, P.R.; Jetten, A.M.; Postlethwaite, A.; Raman, C.; Slominski, A.T. Extra-Adrenal Glucocorticoid Biosynthesis: Implications for Autoimmune and Inflammatory Disorders. Genes Immun. 2020. [Google Scholar] [CrossRef]
- Slominski, A.T.; Manna, P.R.; Tuckey, R.C. On the Role of Skin in the Regulation of Local and Systemic Steroidogenic Activities. Steroids 2015, 103, 72–88. [Google Scholar] [CrossRef]
- Asgari, M.M.; Maruti, S.S.; Kushi, L.H.; White, E. A Cohort Study of Vitamin D Intake and Melanoma Risk. J. Investig. Dermatol. 2009, 129, 1675–1680. [Google Scholar] [CrossRef]
- Weinstein, D.; Leininger, J.; Hamby, C.; Safai, B. Diagnostic and Prognostic Biomarkers in Melanoma. J. Clin. Aesthet. Dermatol. 2014, 7, 13–24. [Google Scholar] [PubMed]
- Abdel-Malek, Z.A.; Ruwe, A.; Kavanagh-Starner, R.; Kadekaro, A.L.; Swope, V.; Haskell-Luevano, C.; Koikov, L.; Knittel, J.J. Alpha-Msh Tripeptide Analogs Activate the Melanocortin 1 Receptor and Reduce Uv-Induced DNA Damage in Human Melanocytes. Pigment Cell Melanoma Res. 2009, 22, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Scherer, D.; Bermejo, J.L.; Rudnai, P.; Gurzau, E.; Koppova, K.; Hemminki, K.; Kumar, R. Mc1r Variants Associated Susceptibility to Basal Cell Carcinoma of Skin: Interaction with Host Factors and Xrcc3 Polymorphism. Int. J. Cancer 2008, 122, 1787–1793. [Google Scholar] [CrossRef] [PubMed]
- Hauser, J.E.; Kadekaro, A.L.; Kavanagh, R.J.; Wakamatsu, K.; Terzieva, S.; Schwemberger, S.; Babcock, G.; Rao, M.B.; Ito, S.; Abdel-Malek, Z.A. Melanin Content and Mc1r Function Independently Affect Uvr-Induced DNA Damage in Cultured Human Melanocytes. Pigment Cell Res. 2006, 19, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Feller, L.; Khammissa, R.A.G.; Kramer, B.; Altini, M.; Lemmer, J. Basal Cell Carcinoma, Squamous Cell Carcinoma and Melanoma of the Head and Face. Head Face Med. 2016, 12, 11. [Google Scholar] [CrossRef] [PubMed]
- Nan, H.; Kraft, P.; Hunter, D.J.; Han, J. Genetic Variants in Pigmentation Genes, Pigmentary Phenotypes, and Risk of Skin Cancer in Caucasians. Int. J. Cancer 2009, 125, 909–917. [Google Scholar] [CrossRef]
- Kanetsky, P.A.; Panossian, S.; Elder, D.E.; Guerry, D.; Ming, M.E.; Schuchter, L.; Rebbeck, T.R. Does Mc1r Genotype Convey Information About Melanoma Risk Beyond Risk Phenotypes? Cancer 2010, 116, 2416–2428. [Google Scholar]
- Williams, P.F.; Olsen, C.M.; Hayward, N.K.; Whiteman, D.C. Melanocortin 1 Receptor and Risk of Cutaneous Melanoma: A Meta-Analysis and Estimates of Population Burden. Int. J. Cancer 2011, 129, 1730–1740. [Google Scholar] [CrossRef]
- Wendt, J.; Rauscher, S.; Burgstaller-Muehlbacher, S.; Fae, I.; Fischer, G.; Pehamberger, H.; Okamoto, I. Human Determinants and the Role of Melanocortin-1 Receptor Variants in Melanoma Risk Independent of Uv Radiation Exposure. JAMA Dermatol. 2016, 152, 776–782. [Google Scholar] [CrossRef]
- Taylor, N.J.; Busam, K.J.; From, L.; Groben, P.A.; Anton-Culver, H.; Cust, A.E.; Begg, C.B.; Dwyer, T.; Gallagher, R.P.; Gruber, S.B.; et al. Inherited Variation at Mc1r and Histological Characteristics of Primary Melanoma. PLoS ONE 2015, 10, e0119920. [Google Scholar] [CrossRef]
- Robles-Espinoza, C.D.; Roberts, N.D.; Chen, S.; Leacy, F.P.; Alexandrov, L.B.; Pornputtapong, N.; Halaban, R.; Krauthammer, M.; Cui, R.; Timothy Bishop, D.; et al. Germline Mc1r Status Influences Somatic Mutation Burden in Melanoma. Nat. Commun. 2016, 7, 12064. [Google Scholar] [CrossRef] [PubMed]
- Fargnoli, M.C.; Gandini, S.; Peris, K.; Maisonneuve, P.; Raimondi, S. Mc1r Variants Increase Melanoma Risk in Families with Cdkn2a Mutations: A Meta-Analysis. Eur. J. Cancer 2010, 46, 1413–1420. [Google Scholar] [CrossRef] [PubMed]
- King, R.; Googe, P.B.; Weilbaecher, K.N.; Mihm, M.C., Jr.; Fisher, D.E. Microphthalmia Transcription Factor Expression in Cutaneous Benign, Malignant Melanocytic, and Nonmelanocytic Tumors. Am. J. Surg. Pathol. 2001, 25, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Franco-Palacios, M.; Russell, M.; Goddard, L.; Hassell, L.; Gillies, E.; Fung, K.-M. Micropthalmia Transcription Factor (Mitf) as a Diagnostic Marker for Metastatic Melanomas Negative for Other Melanoma Markers. Int. J. Clin. Exp. Pathol. 2013, 6, 1658–1664. [Google Scholar] [PubMed]
- Miettinen, M.; Fernandez, M.; Franssila, K.; Gatalica, Z.; Lasota, J.; Sarlomo-Rikala, M. Microphthalmia Transcription Factor in the Immunohistochemical Diagnosis of Metastatic Melanoma: Comparison with Four Other Melanoma Markers. Am. J. Surg. Pathol. 2001, 25, 205–211. [Google Scholar] [CrossRef]
- Carreira, S.; Goodall, J.; Denat, L.; Rodriguez, M.; Nuciforo, P.; Hoek, K.S.; Testori, A.; Larue, L.; Goding, C.R. Mitf Regulation of Dia1 Controls Melanoma Proliferation and Invasiveness. Genes Dev. 2006, 20, 3426–3439. [Google Scholar] [CrossRef]
- Cheli, Y.; Giuliano, S.; Fenouille, N.; Allegra, M.; Hofman, V.; Hofman, P.; Bahadoran, P.; Lacour, J.P.; Tartare-Deckert, S.; Bertolotto, C.; et al. Hypoxia and Mitf Control Metastatic Behaviour in Mouse and Human Melanoma Cells. Oncogene 2012, 31, 2461–2470. [Google Scholar] [CrossRef]
- Bertolotto, C.; Lesueur, F.; Giuliano, S.; Strub, T.; de Lichy, M.; Bille, K.; Dessen, P.; d’Hayer, B.; Mohamdi, H.; Remenieras, A.; et al. A Sumoylation-Defective Mitf Germline Mutation Predisposes to Melanoma and Renal Carcinoma. Nature 2011, 480, 94–98. [Google Scholar] [CrossRef]
- Ciccarese, G.; Dalmasso, B.; Bruno, W.; Queirolo, P.; Pastorino, L.; Andreotti, V.; Spagnolo, F.; Tanda, E.; Ponti, G.; Massone, C.; et al. Clinical, Pathological and Dermoscopic Phenotype of Mitf P.E318k Carrier Cutaneous Melanoma Patients. J. Transl. Med. 2020, 18, 78. [Google Scholar] [CrossRef]
- Balch, C.M.; Soong, S.J.; Gershenwald, J.E.; Thompson, J.F.; Coit, D.G.; Atkins, M.B.; Ding, S.; Cochran, A.J.; Eggermont, A.M.; Flaherty, K.T.; et al. Age as a Prognostic Factor in Patients with Localized Melanoma and Regional Metastases. Ann. Surg. Oncol. 2013, 20, 3961–3968. [Google Scholar] [CrossRef]
- Ecker, B.L.; Kaur, A.; Douglass, S.M.; Webster, M.R.; Almeida, F.V.; Marino, G.E.; Sinnamon, A.J.; Neuwirth, M.G.; Alicea, G.M.; Ndoye, A.; et al. Age-Related Changes in Hapln1 Increase Lymphatic Permeability and Affect Routes of Melanoma Metastasis. Cancer Discov. 2019, 9, 82–95. [Google Scholar] [CrossRef] [PubMed]
- Herrling, T.; Jung, K.; Fuchs, J. The Role of Melanin as Protector against Free Radicals in Skin and Its Role as Free Radical Indicator in Hair. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2008, 69, 1429–1435. [Google Scholar] [CrossRef] [PubMed]
- Slominski, A.T.; Zmijewski, M.A.; Plonka, P.M.; Szaflarski, J.P.; Paus, R. How Uv Light Touches the Brain and Endocrine System through Skin, and Why. Endocrinology 2018, 159, 1992–2007. [Google Scholar] [CrossRef] [PubMed]
- Jimbow, K.; Lee, S.K.; King, M.G.; Hara, H.; Chen, H.; Dakour, J.; Marusyk, H. Melanin Pigments and Melanosomal Proteins as Differentiation Markers Unique to Normal and Neoplastic Melanocytes. J. Investig. Dermatol. 1993, 100, 259S–268S. [Google Scholar] [CrossRef] [PubMed]
- Brożyna, A.A.; Jóźwicki, W.; Carlson, J.A.; Slominski, A.T. Melanogenesis Affects Overall and Disease-Free Survival in Patients with Stage Iii and Iv Melanoma. Hum. Pathol. 2013, 44, 2071–2074. [Google Scholar] [CrossRef]
- Slominski, A.; Kim, T.K.; Brożyna, A.A.; Janjetovic, Z.; Brooks, D.L.P.; Schwab, L.P.; Skobowiat, C.; Jóźwicki, W.; Seagroves, T.N. The Role of Melanogenesis in Regulation of Melanoma Behavior: Melanogenesis Leads to Stimulation of Hif-1α Expression and Hif-Dependent Attendant Pathways. Arch. Biochem. Biophys. 2014, 563, 79–93. [Google Scholar] [CrossRef]
- Urbanska, K.; Romanowska-Dixon, B.; Elas, M.; Pajak, S.; Paziewski, E.; Bryk, J.; Kukielczak, B.; Slominski, A.; Zygulska-Mach, H.; Lukiewicz, S. Experimental Ruthenium Plaque Therapy of Amelanotic and Melanotic Melanomas in the Hamster Eye. Melanoma Res. 2000, 10, 26–35. [Google Scholar] [CrossRef]
- Shields, C.L.; Kaliki, S.; Cohen, M.N.; Shields, P.W.; Furuta, M.; Shields, J.A. Prognosis of Uveal Melanoma Based on Race in 8100 Patients: The 2015 Doyne Lecture. Eye (London) 2015, 29, 1027–1035. [Google Scholar] [CrossRef]
- Shields, C.L.; Kaliki, S.; Furuta, M.; Fulco, E.; Alarcon, C.; Shields, J.A. American Joint Committee on Cancer Classification of Uveal Melanoma (Anatomic Stage) Predicts Prognosis in 7731 Patients: The 2013 Zimmerman Lecture. Ophthalmology 2015, 122, 1180–1186. [Google Scholar] [CrossRef]
- Thomas, N.E.; Kricker, A.; Waxweiler, W.T.; Dillon, P.M.; Busman, K.J.; From, L.; Groben, P.A.; Armstrong, B.K.; Anton-Culver, H.; Gruber, S.B.; et al. Comparison of Clinicopathologic Features and Survival of Histopathologically Amelanotic and Pigmented Melanomas: A Population-Based Study. JAMA Dermatol. 2014, 150, 1306–1314. [Google Scholar] [CrossRef]
- Sarna, M.; Krzykawska-Serda, M.; Jakubowska, M.; Zadlo, A.; Urbanska, K. Melanin Presence Inhibits Melanoma Cell Spread in Mice in a Unique Mechanical Fashion. Sci. Rep. 2019, 9, 9280. [Google Scholar] [CrossRef] [PubMed]
- Sarna, M.; Zadlo, A.; Czuba-Pelech, B.; Urbanska, K. Nanomechanical Phenotype of Melanoma Cells Depends Solely on the Amount of Endogenous Pigment in the Cells. Int. J. Mol. Sci. 2018, 19, 607. [Google Scholar] [CrossRef] [PubMed]
- Brożyna, A.A.; Jóźwicki, W.; Roszkowski, K.; Filipiak, J.; Slominski, A.T. Melanin Content in Melanoma Metastases Affects the Outcome of Radiotherapy. Oncotarget 2016, 7, 17844–17853. [Google Scholar] [CrossRef] [PubMed]
- Brozyna, A.A.; VanMiddlesworth, L.; Slominski, A.T. Inhibition of Melanogenesis as a Radiation Sensitizer for Melanoma Therapy. Int. J. Cancer 2008, 123, 1448–1456. [Google Scholar] [CrossRef]
- Slominski, A.; Zbytek, B.; Slominski, R. Inhibitors of Melanogenesis Increase Toxicity of Cyclophosphamide and Lymphocytes against Melanoma Cells. Int. J. Cancer 2009, 124, 1470–1477. [Google Scholar] [CrossRef]
- Slominski, A.; Goodman-Snitkoff, G.G. Dopa Inhibits Induced Proliferative Activity of Murine and Human Lymphocytes. Anticancer Res. 1992, 12, 753–756. [Google Scholar]
- Slominski, A.; Paus, R.; Mihm, M.C. Inhibition of Melanogenesis as an Adjuvant Strategy in the Treatment of Melanotic Melanomas: Selective Review and Hypothesis. Anticancer Res. 1998, 18, 3709–3715. [Google Scholar]
- Slominski, A.T.; Carlson, J.A. Melanoma Resistance: A Bright Future for Academicians and a Challenge for Patient Advocates. Mayo Clin. Proc. 2014, 89, 429–433. [Google Scholar] [CrossRef]
- Hofbauer, G.F.; Kamarashev, J.; Geertsen, R.; Böni, R.; Dummer, R. Tyrosinase Immunoreactivity in Formalin-Fixed, Paraffin-Embedded Primary and Metastatic Melanoma: Frequency and Distribution. J. Cutan. Pathol. 1998, 25, 204–209. [Google Scholar] [CrossRef]
- Kaufmann, O.; Koch, S.; Burghardt, J.; Audring, H.; Dietel, M. Tyrosinase, Melan-a, and Kba62 as Markers for the Immunohistochemical Identification of Metastatic Amelanotic Melanomas on Paraffin Sections. Mod. Pathol. 1998, 11, 740–746. [Google Scholar]
- Clarkson, K.S.; Sturdgess, I.C.; Molyneux, A.J. The Usefulness of Tyrosinase in the Immunohistochemical Assessment of Melanocytic Lesions: A Comparison of the Novel T311 Antibody (Anti-Tyrosinase) with S-100, Hmb45, and A103 (Anti-Melan-a). J. Clin. Pathol. 2001, 54, 196–200. [Google Scholar] [CrossRef] [PubMed]
- Reinke, S.; Königer, P.; Herberth, G.; Audring, H.; Wang, H.; Ma, J.; Guo, Y.; Sterry, W.; Trefzer, U. Differential Expression of Mart-1, Tyrosinase, and Sm5-1 in Primary and Metastatic Melanoma. Am. J. Dermatopathol. 2005, 27, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Murtas, D.; Pilloni, L.; Diana, A.; Casula, L.; Tomei, S.; Piras, F.; Ferreli, C.; Maxia, C.; Perra, M.T. Tyrosinase and Nestin Immunohistochemical Expression in Melanocytic Nevi as a Histopathologic Pattern to Trace Melanocyte Differentiation and Nevogenesis. Histochem. Cell Biol. 2019, 151, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Bloom, M.B.; Perry-Lalley, D.; Robbins, P.F.; Li, Y.; el-Gamil, M.; Rosenberg, S.A.; Yang, J.C. Identification of Tyrosinase-Related Protein 2 as a Tumor Rejection Antigen for the B16 Melanoma. J. Exp. Med. 1997, 185, 453–459. [Google Scholar] [CrossRef]
- Kameyama, K.; Sakai, C.; Kuge, S.; Nishiyama, S.; Tomita, Y.; Ito, S.; Wakamatsu, K.; Hearing, V.J. The Expression of Tyrosinase, Tyrosinase-Related Proteins 1 and 2 (Trp1 and Trp2), the Silver Protein, and a Melanogenic Inhibitor in Human Melanoma Cells of Differing Melanogenic Activities. Pigment Cell Res. 1995, 8, 97–104. [Google Scholar] [CrossRef]
- Journe, F.; Id Boufker, H.; Van Kempen, L.; Galibert, M.D.; Wiedig, M.; Salès, F.; Theunis, A.; Nonclercq, D.; Frau, A.; Laurent, G.; et al. Tyrp1 Mrna Expression in Melanoma Metastases Correlates with Clinical Outcome. Br. J. Cancer 2011, 105, 1726–1732. [Google Scholar] [CrossRef]
- Bolander, A.; Agnarsdóttir, M.; Strömberg, S.; Ponten, F.; Hesselius, P.; Uhlen, M.; Bergqvist, M. The Protein Expression of Trp-1 and Galectin-1 in Cutaneous Malignant Melanomas. Cancer Genomics Proteomics 2008, 5, 293–300. [Google Scholar]
- Long, G.V.; Menzies, A.M.; Nagrial, A.M.; Haydu, L.E.; Hamilton, A.L.; Mann, G.J.; Hughes, T.M.; Thompson, J.F.; Scolyer, R.A.; Kefford, R.F. Prognostic and Clinicopathologic Associations of Oncogenic Braf in Metastatic Melanoma. J. Clin. Oncol. 2011, 29, 1239–1246. [Google Scholar] [CrossRef]
- Kim, S.Y.; Kim, S.N.; Hahn, H.J.; Lee, Y.W.; Choe, Y.B.; Ahn, K.J. Metaanalysis of Braf Mutations and Clinicopathologic Characteristics in Primary Melanoma. J. Am. Acad. Dermatol. 2015, 72, 1036–1046.e2. [Google Scholar] [CrossRef]
- Menzies, A.M.; Haydu, L.E.; Visintin, L.; Carlino, M.S.; Howle, J.R.; Thompson, J.F.; Kefford, R.F.; Scolyer, R.A.; Long, G.V. Distinguishing Clinicopathologic Features of Patients with V600e and V600k Braf-Mutant Metastatic Melanoma. Clin. Cancer Res. 2012, 18, 3242–3249. [Google Scholar] [CrossRef]
- Colebatch, A.J.; Ferguson, P.; Newell, F.; Kazakoff, S.H.; Witkowski, T.; Dobrovic, A.; Johansson, P.A.; Saw, R.P.M.; Stretch, J.R.; McArthur, G.A.; et al. Molecular Genomic Profiling of Melanocytic nevi. J. Investig. Dermatol. 2019, 139, 1762–1768. [Google Scholar] [CrossRef] [PubMed]
- Brash, D.E. Uv Signature Mutations. Photochem. Photobiol. 2015, 91, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Bauer, J.; Curtin, J.A.; Pinkel, D.; Bastian, B.C. Congenital Melanocytic Nevi Frequently Harbor Nras Mutations but No Braf Mutations. J. Investig. Dermatol. 2007, 127, 179–182. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, J.L.; Fridlyand, J.; Patel, H.; Jain, A.N.; Busam, K.; Kageshita, T.; Ono, T.; Albertson, D.G.; Pinkel, D.; Bastian, B.C. Determinants of Braf Mutations in Primary Melanomas. J. Natl. Cancer Inst. 2003, 95, 1878–1890. [Google Scholar] [CrossRef]
- Melamed, R.D.; Aydin, I.T.; Rajan, G.S.; Phelps, R.; Silvers, D.N.; Emmett, K.J.; Brunner, G.; Rabadan, R.; Celebi, J.T. Genomic Characterization of Dysplastic Nevi Unveils Implications for Diagnosis Of melanoma. J. Investig. Dermatol. 2017, 137, 905–909. [Google Scholar] [CrossRef]
- Akslen, L.A.; Angelini, S.; Straume, O.; Bachmann, I.M.; Molven, A.; Hemminki, K.; Kumar, R. Braf and Nras Mutations Are Frequent in Nodular Melanoma but Are Not Associated with Tumor Cell Proliferation or Patient Survival. J. Investig. Dermatol. 2005, 125, 312–317. [Google Scholar] [CrossRef]
- Abd Elmageed, Z.Y.; Moore, R.F.; Tsumagari, K.; Lee, M.M.; Sholl, A.B.; Friedlander, P.; Al-Qurayshi, Z.; Hassan, M.; Wang, A.R.; Boulares, H.A.; et al. Prognostic Role of Braf(V600e) Cellular Localization in Melanoma. J. Am. Coll. Surg. 2018, 226, 526–537. [Google Scholar] [CrossRef]
- Nazarian, R.; Shi, H.; Wang, Q.; Kong, X.; Koya, R.C.; Lee, H.; Chen, Z.; Lee, M.-K.; Attar, N.; Sazegar, H.; et al. Melanomas Acquire Resistance to B-Raf(V600e) Inhibition by Rtk or N-Ras Upregulation. Nature 2010, 468, 973–977. [Google Scholar] [CrossRef]
- Amann, V.C.; Ramelyte, E.; Thurneysen, S.; Pitocco, R.; Bentele-Jaberg, N.; Goldinger, S.M.; Dummer, R.; Mangana, J. Developments in Targeted Therapy in Melanoma. Eur. J. Surg. Oncol. 2017, 43, 581–593. [Google Scholar] [CrossRef]
- Robert, C.; Karaszewska, B.; Schachter, J.; Rutkowski, P.; Mackiewicz, A.; Stroyakovskiy, D.; Dummer, R.; Grange, F.; Mortier, L.; Chiarion-Sileni, V.; et al. Three-Year Estimate of Overall Survival in Combi-V, a Randomized Phase 3 Study Evaluating First-Line Dabrafenib (D) + Trametinib (T) in Patients (Pts) with Unresectable or Metastatic Braf V600e/K–Mutant Cutaneous Melanoma. Ann. Oncol. 2016, 27 (Suppl. 6). [Google Scholar] [CrossRef]
- Long, G.V.; Flaherty, K.T.; Stroyakovskiy, D.; Gogas, H.; Levchenko, E.; de Braud, F.; Larkin, J.; Garbe, C.; Jouary, T.; Hauschild, A.; et al. Dabrafenib Plus Trametinib Versus Dabrafenib Monotherapy in Patients with Metastatic Braf V600e/K-Mutant Melanoma: Long-Term Survival and Safety Analysis of a Phase 3 Study. Ann. Oncol. 2017, 28, 1631–1639. [Google Scholar] [CrossRef] [PubMed]
- Long, G.V.; Weber, J.S.; Infante, J.R.; Kim, K.B.; Daud, A.; Gonzalez, R.; Sosman, J.A.; Hamid, O.; Schuchter, L.; Cebon, J.; et al. Overall Survival and Durable Responses in Patients with Braf V600-Mutant Metastatic Melanoma Receiving Dabrafenib Combined with Trametinib. J. Clin. Oncol. 2016, 34, 871–878. [Google Scholar] [CrossRef] [PubMed]
- Pavlick, A.C.; Fecher, L.; Ascierto, P.A.; Sullivan, R.J. Frontline Therapy for Braf-Mutated Metastatic Melanoma: How Do You Choose, and Is There One Correct Answer? Am. Soc. Clin. Oncol. Educ. Book 2019, 39, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.A.; Cullinane, C.; Kirby, L.; Abuhammad, S.; Lelliott, E.J.; Waldeck, K.; Young, R.J.; Brajanovski, N.; Cameron, D.P.; Walker, R.; et al. Palbociclib Synergizes with Braf and Mek Inhibitors in Treatment Naive Melanoma but Not after the Development of Braf Inhibitor Resistance. Int. J. Cancer 2018, 142, 2139–2152. [Google Scholar] [CrossRef]
- Misek, S.A.; Appleton, K.M.; Dexheimer, T.S.; Lisabeth, E.M.; Lo, R.S.; Larsen, S.D.; Gallo, K.A.; Neubig, R.R. Rho-Mediated Signaling Promotes Braf Inhibitor Resistance in De-Differentiated Melanoma Cells. Oncogene 2020, 39, 1466–1483. [Google Scholar] [CrossRef]
- Hatzivassiliou, G.; Song, K.; Yen, I.; Brandhuber, B.J.; Anderson, D.J.; Alvarado, R.; Ludlam, M.J.; Stokoe, D.; Gloor, S.L.; Vigers, G.; et al. Raf Inhibitors Prime Wild-Type Raf to Activate the Mapk Pathway and Enhance Growth. Nature 2010, 464, 431–435. [Google Scholar] [CrossRef]
- Su, F.; Viros, A.; Milagre, C.; Trunzer, K.; Bollag, G.; Spleiss, O.; Reis-Filho, J.S.; Kong, X.; Koya, R.C.; Flaherty, K.T.; et al. Ras Mutations in Cutaneous Squamous-Cell Carcinomas in Patients Treated with Braf Inhibitors. N. Engl. J. Med. 2012, 366, 207–215. [Google Scholar] [CrossRef]
- Göppner, D.; Müller, J.; Krüger, S.; Franke, I.; Gollnick, H.; Quist, S.R. High Incidence of Naevi-Associated Braf Wild-Type Melanoma and Dysplastic Naevi under Treatment with the Class I Braf Inhibitor Vemurafenib. Acta Derm. Venereol. 2014, 94, 517–520. [Google Scholar] [CrossRef]
- Shimizu, K.; Goldfarb, M.; Perucho, M.; Wigler, M. Isolation and Preliminary Characterization of the Transforming Gene of a Human Neuroblastoma Cell Line. Proc. Natl. Acad. Sci. USA 1983, 80, 383–387. [Google Scholar] [CrossRef]
- Hall, A.; Marshall, C.J.; Spurr, N.K.; Weiss, R.A. Identification of Transforming Gene in Two Human Sarcoma Cell Lines as a New Member of the Ras Gene Family Located on Chromosome 1. Nature 1983, 303, 396–400. [Google Scholar] [CrossRef]
- Chiappetta, C.; Proietti, I.; Soccodato, V.; Puggioni, C.; Zaralli, R.; Pacini, L.; Porta, N.; Skroza, N.; Petrozza, V.; Potenza, C.; et al. Braf and Nras Mutations Are Heterogeneous and Not Mutually Exclusive in Nodular Melanoma. Appl. Immunohistochem. Mol. Morphol. 2015, 23, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Van’t Veer, L.J.; Burgering, B.M.; Versteeg, R.; Boot, A.J.; Ruiter, D.J.; Osanto, S.; Schrier, P.I.; Bos, J.L. N-Ras Mutations in Human Cutaneous Melanoma from Sun-Exposed Body Sites. Mol. Cell. Biol. 1989, 9, 3114–3116. [Google Scholar] [CrossRef] [PubMed]
- Thomas, N.E.; Edmiston, S.N.; Alexander, A.; Groben, P.A.; Parrish, E.; Kricker, A.; Armstrong, B.K.; Anton-Culver, H.; Gruber, S.B.; From, L.; et al. Association between Nras and Braf Mutational Status and Melanoma-Specific Survival among Patients with Higher-Risk Primary Melanoma. JAMA Oncol. 2015, 1, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Ellerhorst, J.A.; Greene, V.R.; Ekmekcioglu, S.; Warneke, C.L.; Johnson, M.M.; Cooke, C.P.; Wang, L.E.; Prieto, V.G.; Gershenwald, J.E.; Wei, Q.; et al. Clinical Correlates of Nras and Braf Mutations in Primary Human Melanoma. Clin. Cancer Res. 2011, 17, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Charbel, C.; Fontaine, R.H.; Malouf, G.G.; Picard, A.; Kadlub, N.; El-Murr, N.; How-Kit, A.; Su, X.; Coulomb-L’Hermine, A.; Tost, J.; et al. Nras Mutation Is the Sole Recurrent Somatic Mutation in Large Congenital Melanocytic Nevi. J. Investig. Dermatol. 2014, 134, 1067–1074. [Google Scholar] [CrossRef]
- Hancock, J.F.; Magee, A.I.; Childs, J.E.; Marshall, C.J. All Ras Proteins Are Polyisoprenylated but Only Some Are Palmitoylated. Cell 1989, 57, 1167–1177. [Google Scholar] [CrossRef]
- Boespflug, A.; Caramel, J.; Dalle, S.; Thomas, L. Treatment of Nras-Mutated Advanced or Metastatic Melanoma: Rationale, Current Trials and Evidence to Date. Ther. Adv. Med. Oncol. 2017, 9, 481–492. [Google Scholar] [CrossRef]
- Margolin, K.A.; Moon, J.; Flaherty, L.E.; Lao, C.D.; Akerley, W.L., 3rd; Othus, M.; Sosman, J.A.; Kirkwood, J.M.; Sondak, V.K. Randomized Phase Ii Trial of Sorafenib with Temsirolimus or Tipifarnib in Untreated Metastatic Melanoma (S0438). Clin. Cancer Res. 2012, 18, 1129–1137. [Google Scholar] [CrossRef]
- Whyte, D.B.; Kirschmeier, P.; Hockenberry, T.N.; Nunez-Oliva, I.; James, L.; Catino, J.J.; Bishop, W.R.; Pai, J.K. K- and N-Ras Are Geranylgeranylated in Cells Treated with Farnesyl Protein Transferase Inhibitors. J. Biol. Chem. 1997, 272, 14459–14464. [Google Scholar] [CrossRef]
- Dummer, R.; Schadendorf, D.; Ascierto, P.A.; Arance, A.; Dutriaux, C.; Di Giacomo, A.M.; Rutkowski, P.; Del Vecchio, M.; Gutzmer, R.; Mandala, M.; et al. Binimetinib Versus Dacarbazine in Patients with Advanced Nras-Mutant Melanoma (Nemo): A Multicentre, Open-Label, Randomised, Phase 3 Trial. Lancet Oncol. 2017, 18, 435–445. [Google Scholar] [CrossRef]
- Posch, C.; Moslehi, H.; Feeney, L.; Green, G.A.; Ebaee, A.; Feichtenschlager, V.; Chong, K.; Peng, L.; Dimon, M.T.; Phillips, T.; et al. Combined Targeting of Mek and Pi3k/Mtor Effector Pathways Is Necessary to Effectively Inhibit Nras Mutant Melanoma in Vitro and in Vivo. Proc. Natl. Acad. Sci. USA 2013, 110, 4015–4020. [Google Scholar] [CrossRef] [PubMed]
- Weisberg, E.; Nonami, A.; Chen, Z.; Liu, F.; Zhang, J.; Sattler, M.; Nelson, E.; Cowens, K.; Christie, A.L.; Mitsiades, C.; et al. Identification of Wee1 as a Novel Therapeutic Target for Mutant Ras-Driven Acute Leukemia and Other Malignancies. Leukemia 2015, 29, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, N.; Kikuchi, K.; Yagyu, S.; Miyachi, M.; Iehara, T.; Tajiri, T.; Sakai, T.; Hosoi, H. Mutations in the Ras Pathway as Potential Precision Medicine Targets in Treatment of Rhabdomyosarcoma. Biochem. Biophys. Res. Commun. 2019, 512, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Vu, H.L.; Aplin, A.E. Targeting Mutant Nras Signaling Pathways in Melanoma. Pharmacol. Res. 2016, 107, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Duensing, A.; Medeiros, F.; McConarty, B.; Joseph, N.E.; Panigrahy, D.; Singer, S.; Fletcher, C.D.; Demetri, G.D.; Fletcher, J.A. Mechanisms of Oncogenic Kit Signal Transduction in Primary Gastrointestinal Stromal Tumors (Gists). Oncogene 2004, 23, 3999–4006. [Google Scholar] [CrossRef] [PubMed]
- Slipicevic, A.; Herlyn, M. Kit in Melanoma: Many Shades of Gray. J. Investig. Dermatol. 2015, 135, 337–338. [Google Scholar] [CrossRef]
- Ma, X.; Wu, Y.; Zhang, T.; Song, H.; Jv, H.; Guo, W.; Ren, G. The Clinical Significance of C-Kit Mutations in Metastatic Oral Mucosal Melanoma in China. Oncotarget 2017, 8, 82661–82673. [Google Scholar] [CrossRef]
- Guo, J.; Si, L.; Kong, Y.; Flaherty, K.T.; Xu, X.; Zhu, Y.; Corless, C.L.; Li, L.; Li, H.; Sheng, X.; et al. Phase Ii, Open-Label, Single-Arm Trial of Imatinib Mesylate in Patients with Metastatic Melanoma Harboring C-Kit Mutation or Amplification. J. Clin. Oncol. 2011, 29, 2904–2909. [Google Scholar] [CrossRef]
- O’Day, S.J.; Hamid, O.; Urba, W.J. Targeting Cytotoxic T-Lymphocyte Antigen-4 (Ctla-4): A Novel Strategy for the Treatment of Melanoma and Other Malignancies. Cancer 2007, 110, 2614–2627. [Google Scholar] [CrossRef]
- Freeman, G.J.; Long, A.J.; Iwai, Y.; Bourque, K.; Chernova, T.; Nishimura, H.; Fitz, L.J.; Malenkovich, N.; Okazaki, T.; Byrne, M.C.; et al. Engagement of the Pd-1 Immunoinhibitory Receptor by a Novel B7 Family Member Leads to Negative Regulation of Lymphocyte Activation. J. Exp. Med. 2000, 192, 1027–1034. [Google Scholar] [CrossRef]
- Zitvogel, L.; Kroemer, G. Targeting Pd-1/Pd-L1 Interactions for Cancer Immunotherapy. Oncoimmunology 2012, 1, 1223–1225. [Google Scholar] [CrossRef] [PubMed]
- Wolchok, J.D.; Kluger, H.; Callahan, M.K.; Postow, M.A.; Rizvi, N.A.; Lesokhin, A.M.; Segal, N.H.; Ariyan, C.E.; Gordon, R.A.; Reed, K.; et al. Nivolumab Plus Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2013, 369, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Postow, M.A.; Cardona, D.M.; Taube, J.M.; Anders, R.A.; Taylor, C.R.; Wolchok, J.D.; Callahan, M.K.; Curran, M.A.; Lesokhin, A.M.; Grosso, J.F.; et al. Peripheral and Tumor Immune Correlates in Patients with Advanced Melanoma Treated with Nivolumab (Anti-Pd-1, Bms-936558, Ono-4538) Monotherapy or in Combination with Ipilimumab. J. Transl. Med. 2014, 12, O8. [Google Scholar] [CrossRef]
- Robert, C.; Long, G.V.; Brady, B.; Dutriaux, C.; Maio, M.; Mortier, L.; Hassel, J.C.; Rutkowski, P.; McNeil, C.; Kalinka-Warzocha, E.; et al. Nivolumab in Previously Untreated Melanoma without Braf Mutation. N. Engl. J. Med. 2015, 372, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.S.; Kudchadkar, R.R.; Yu, B.; Gallenstein, D.; Horak, C.E.; Inzunza, H.D.; Zhao, X.; Martinez, A.J.; Wang, W.; Gibney, G.; et al. Safety, Efficacy, and Biomarkers of Nivolumab with Vaccine in Ipilimumab-Refractory or -Naive Melanoma. J. Clin. Oncol. 2013, 31, 4311–4318. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Huang, A.C.; Zhang, W.; Zhang, G.; Wu, M.; Xu, W.; Yu, Z.; Yang, J.; Wang, B.; Sun, H.; et al. Exosomal Pd-L1 Contributes to Immunosuppression and Is Associated with Anti-Pd-1 Response. Nature 2018, 560, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Del Re, M.; Marconcini, R.; Pasquini, G.; Rofi, E.; Vivaldi, C.; Bloise, F.; Restante, G.; Arrigoni, E.; Caparello, C.; Bianco, M.G.; et al. Pd-L1 Mrna Expression in Plasma-Derived Exosomes Is Associated with Response to Anti-Pd-1 Antibodies in Melanoma and Nsclc. Br. J. Cancer 2018, 118, 820–824. [Google Scholar] [CrossRef]
- Topalian, S.L.; Taube, J.M.; Anders, R.A.; Pardoll, D.M. Mechanism-Driven Biomarkers to Guide Immune Checkpoint Blockade in Cancer Therapy. Nat. Rev. Cancer 2016, 16, 275–287. [Google Scholar] [CrossRef]
- Kleffel, S.; Posch, C.; Barthel, S.R.; Mueller, H.; Schlapbach, C.; Guenova, E.; Elco, C.P.; Lee, N.; Juneja, V.R.; Zhan, Q.; et al. Melanoma Cell-Intrinsic Pd-1 Receptor Functions Promote Tumor Growth. Cell 2015, 162, 1242–1256. [Google Scholar] [CrossRef]
- Zhang, B.; Dang, J.; Ba, D.; Wang, C.; Han, J.; Zheng, F. Potential Function of Ctla-4 in the Tumourigenic Capacity of Melanoma Stem Cells. Oncol. Lett. 2018, 16, 6163–6170. [Google Scholar] [CrossRef]
- Pistillo, M.P.; Fontana, V.; Morabito, A.; Dozin, B.; Laurent, S.; Carosio, R.; Banelli, B.; Ferrero, F.; Spano, L.; Tanda, E.; et al. Soluble Ctla-4 as a Favorable Predictive Biomarker in Metastatic Melanoma Patients Treated with Ipilimumab: An Italian Melanoma Intergroup Study. Cancer Immunol. Immunother. 2019, 68, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Petrie, T.; Samatham, R.; Witkowski, A.M.; Esteva, A.; Leachman, S.A. Melanoma Early Detection: Big Data, Bigger Picture. J. Investig. Dermatol. 2019, 139, 25–30. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).