Abstract
Centromeres are essential for proper chromosome segregation to the daughter cells during mitosis and meiosis. Chromosomes of most eukaryotes studied so far have regional centromeres that form primary constrictions on metaphase chromosomes. These monocentric chromosomes vary from point centromeres to so-called “meta-polycentromeres”, with multiple centromere domains in an extended primary constriction, as identified in Pisum and Lathyrus species. However, in various animal and plant lineages centromeres are distributed along almost the entire chromosome length. Therefore, they are called holocentromeres. In holocentric plants, centromere-specific proteins, at which spindle fibers usually attach, are arranged contiguously (line-like), in clusters along the chromosomes or in bands. Here, we summarize findings of ultrastructural investigations using immunolabeling with centromere-specific antibodies and super-resolution microscopy to demonstrate the structural diversity of plant centromeres. A classification of the different centromere types has been suggested based on the distribution of spindle attachment sites. Based on these findings we discuss the possible evolution and advantages of holocentricity, and potential strategies to segregate holocentric chromosomes correctly.
1. Introduction
During mitotic and meiotic cell proliferation, correct segregation of the genetic material to the daughter cells is essential. Spindle fibers attach to specific regions, called centromeres, at the highly condensed metaphase chromosomes (reviewed in [1]). Different types of centromeres have been described in yeasts, animals, and plants and are classified into two main categories: monocentromeres and holocentromeres.
The centromere size based on centromere-specific chromatin varies highly among eukaryotes, even in monocentric chromosomes. It ranges from 125 bp in the budding yeast point centromere to several Mbp in regional human centromeres [2,3,4]. At relatively large chromosomes (>2 µm), regional centromeres are localized within a distinct primary constriction cytologically visible at metaphase. Smaller chromosomes (<2 µm) often do not show such constrictions and the localization of their centromeres is difficult, as in duckweed [5] or Genlisea [6,7] species. Nevertheless, spindle fibers attach at a distinct chromosome region. Common bean (Phaseolus vulgaris) chromosomes are ~2 µm, but the primary constrictions are only visible in some chromosomes and cells [8]. The same is true for Citrus, but here the constrictions are visible depending on chromosome condensation and preparation [9]. In contrast to point and regional centromeres, holocentromeres are distributed almost along the entire metaphase chromosome length, and therefore do not exhibit a primary constriction [10]. Interestingly, regional centromeres with elongated constrictions may also occur in legumes [11,12,13], fire ant species [14], muntjacs [15], as well as in marsupial hybrids [16] and cancer cell lines [17].
Proteinaceous kinetochore complexes assemble at centromeres where microtubules attach for chromosome segregation. In most eukaryotes (except in budding yeast, [18]), these assembly sites are not determined by specific DNA sequences. Instead, the centromere-specific histone H3 (CENH3)/centromere protein A (CENP-A) [19] specifies the kinetochore positions in many eukaryotes and recruits additional kinetochore proteins [1]. However, no CENH3/CENP-A has been found in holocentric Lepidoptera and Hemiptera insects [20], kinetoplastids (e.g., trypanosomes, [21]) and some fungi, such as Mucor [22,23], Phycomyces blakesleeanus and Mortierella verticillata [24].
To decipher the structure and organization of centromeres, super-resolution microscopy, beside other microscopy techniques, has been applied. Super-resolution microscopy techniques, such as spatial structured illumination microscopy (3D-SIM), are subdiffraction imaging methods bridging the resolution gap between light and electron microscopy [25,26,27,28,29,30]. 3D-SIM allows, compared to other super-resolution microscopy techniques, fast high throughput multicolor imaging by doubling the resolution of wide-field microscopy and achieving best contrast in thin specimens [31]. Super-resolution microscopy was applied successfully in cell biology [32,33,34] to specimens from both prokaryotes and eukaryotes and allowed discovery of new structures within mammalian [35] and plant chromatin [36].
Here, we summarize findings achieved via investigating the plant ultrastructural centromere variability by 3D-SIM imaging of chromatin after immunostaining with centromere-specific antibodies.
2. Centromere Diversity in Plants
In recent studies with the application of 3D-SIM super-resolution microscopy and immunodetection of CENH3/CENP-A, several other centromere and (peri)centromere-specific proteins and tubulin (Figure 1, Figure 2 and Figure 3) provided the basis for creating detailed models of centromere organization in plants (Figure 4 and Table 1). Most studied plant species with large chromosomes (>2 µm) typically show a distinct primary constriction. They represent a regional monocentromere containing one cluster of CENH3/CENP-A surrounded by (peri)centromeric chromatin marked by cell-cycle dependent post-translational histone modifications, such as H2A phosphorylated at threonine 120 (H2AT120ph) [37] and histone H3 phosphorylated at serines 10 [38] and 28 [39]. However, other less common centromere-specific structures are present in plants.
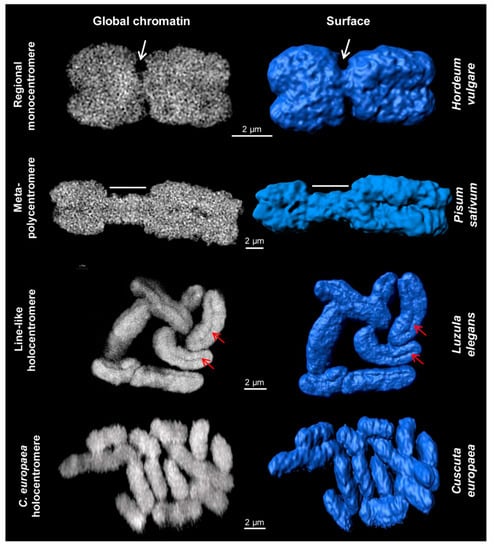
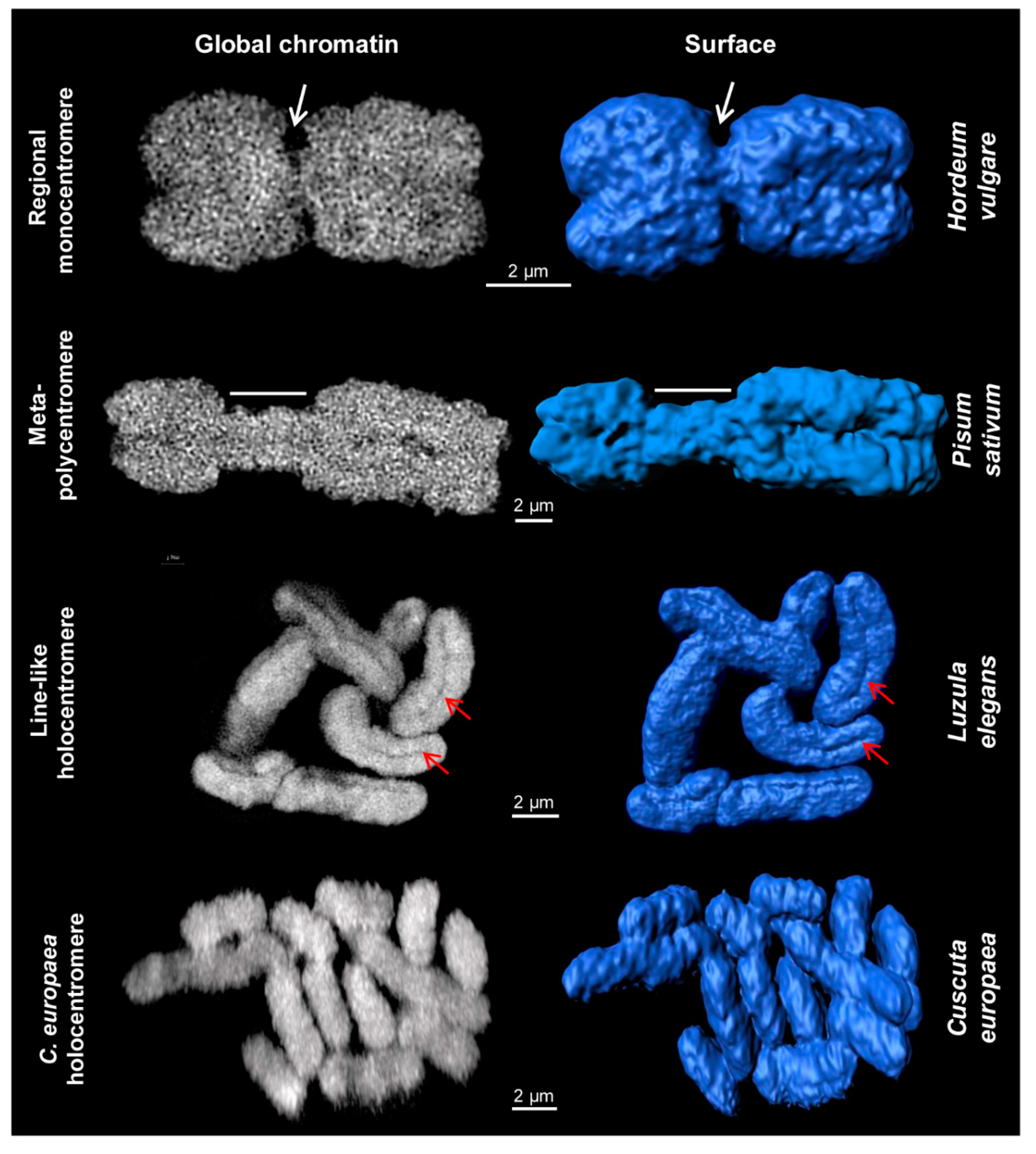
Figure 1.
Different centromere types of somatic plant metaphase chromosomes. Images were obtained via global chromatin labelling by DAPI. After surface rendering of structured illumination microscopy (SIM) image stacks [40] using the Imaris 8.0 software, the centromere structure variability of different plant species becomes visible. Regional monocentromeres are characterized by a district primary constriction (white arrows). Meta-polycentromeres represent an elongated primary constriction (region indicated by dashes). Line-like holocentromeres are characterized by the arrangement of centromere-specific proteins in a distinct line within a groove (red arrows), as found in Luzula and Rhynchospora (Figure 3). Holocentromeres in Cuscuta europaea are structures where spindle fibres attach along the whole chromosome at centromere-specific histone H3 (CENH3)/centromere protein A (CENP-A)-chromatin as well as at CENH3/CENP-A-free regions (see also Figure 2 and Figure 4), but the surface is relatively smooth without a specific constriction.
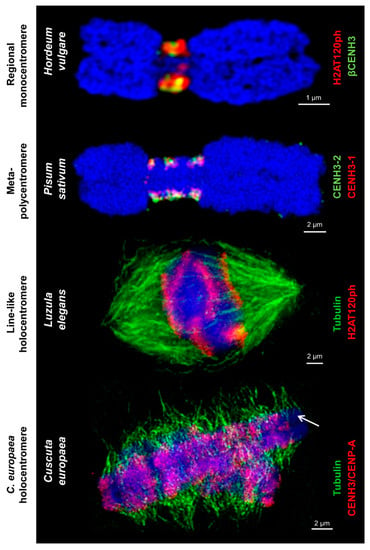
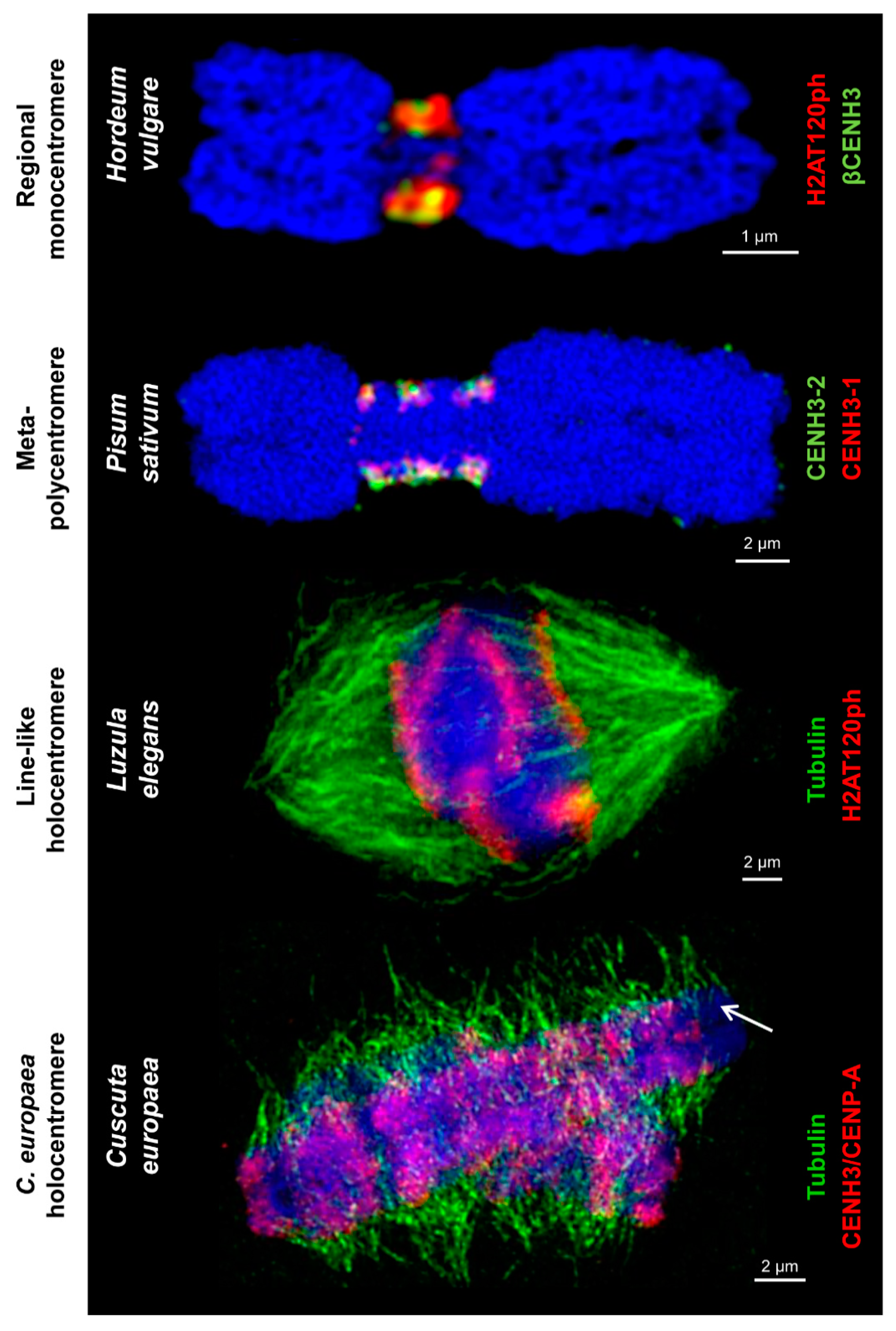
Figure 2.
Different centromere types labeled by centromere-specific histone markers and tubulin. These markers, such as different CENH3/CENP-A variants and H2A phosphorylated at threonine 120 H2AT120ph, intermingle in regional monocentromeres. Spindle fibers attach to H2AT120ph-containing regions of line-like Luzula holocentromeres and CENH3/CENP-A-containing and CENH3/CENP-A-free regions of C. europaea holocentromeres, respectively, along the entire chromosomes. The arrow marks chromosome 1 of C. europaea with a chromosome-wide distribution of tubulin and restricted amount of CENH3/CENP-A. Chromosomes are counterstained with DAPI (in blue).
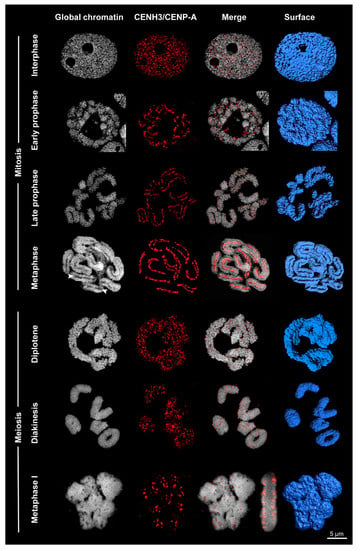
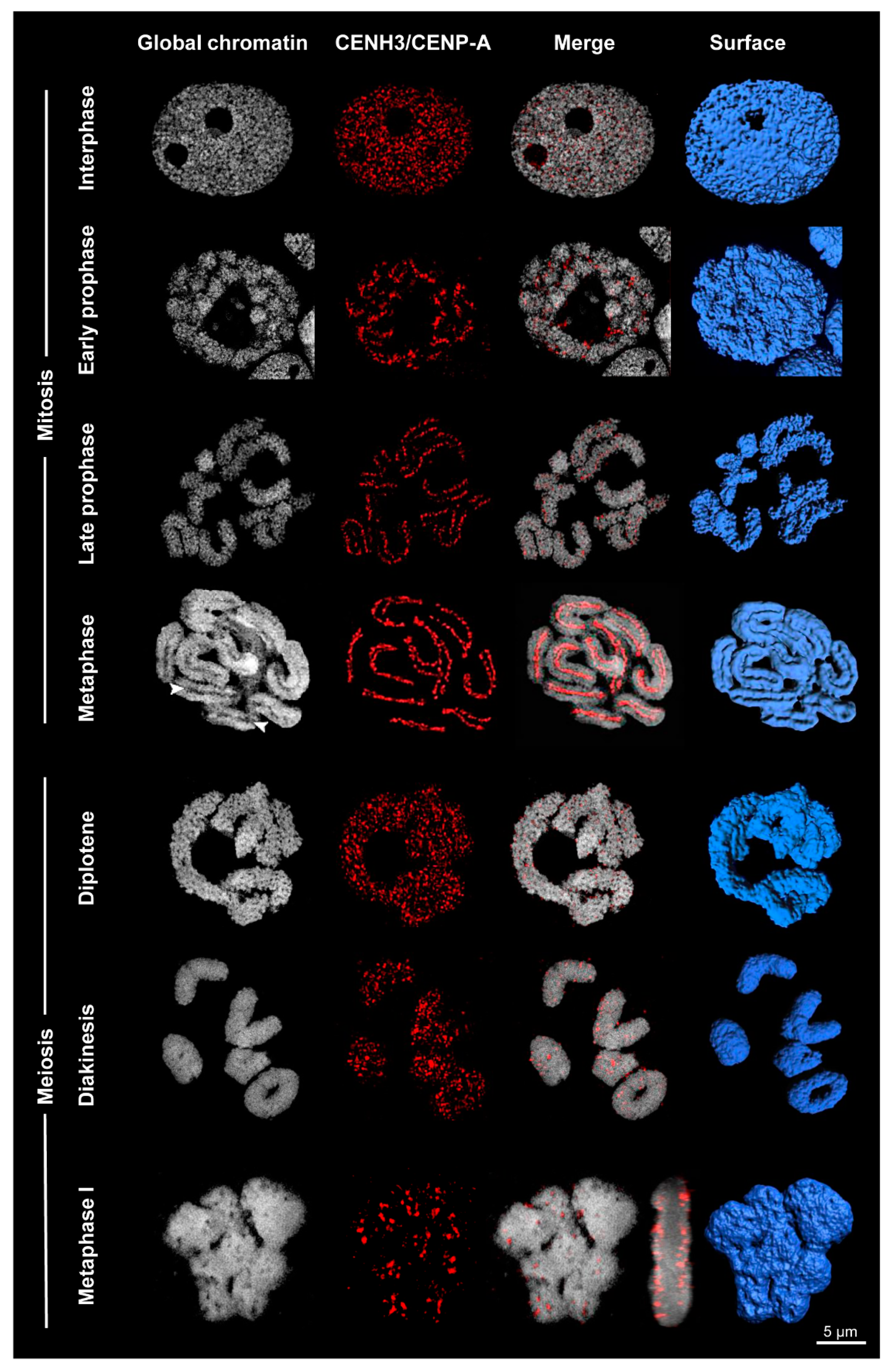
Figure 3.
Centromere formation differs between mitosis and meiosis of Rhynchospora pubera. Whereas line-like holocentromeres appear in mitosis, cluster-like holocentromeres become established in meiosis. The process of global chromatin condensation and the dynamics of CENH3/CENP-A arrangement is visualized by DAPI staining and immunolabeling with CENH3/CENP-A-specific antibodies. Surface rendering of SIM image stacks clearly indicates the presence of grooves (arrowheads) at somatic metaphase chromosomes, but their absence at metaphase I bivalents. The merged side-view of the metaphase I cell reveals CENH3/CENP-A at the surface, but not inside the bivalents.
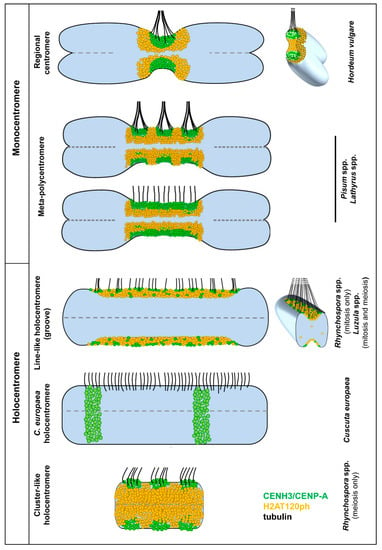
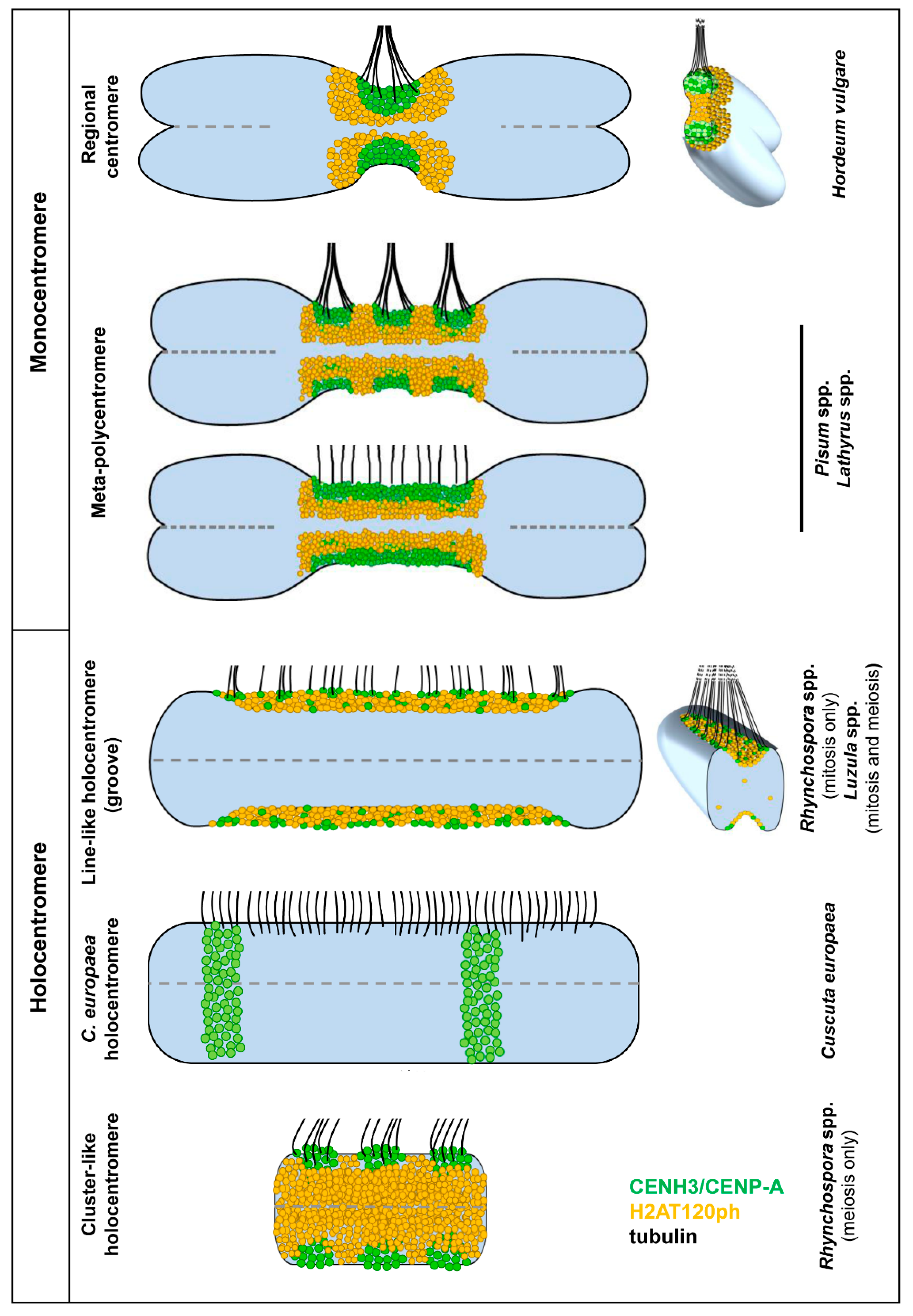
Figure 4.
Models of the different mono- and holocentromere types appearing in different plant species indicate the possible centromere plasticity during mitosis and meiosis. The classification is based on the distribution of the spindle fibre attachment sites. In mono- and meta-polycentromeres, the microtubules (tubulin) form branching bundles and attach mainly at the flanks of the CENH3/CENP-A clusters, but not at H2AT120ph. The bundle formation is less pronounced at holocentromeres. In line-like holocentromeres, spindle fibres attach mainly as single microtubules at the rim along the entire groove containing CENH3/CENP-A and H2AT120ph, as is clearly visible in the cross-section [41]. The CENH3/CENP-A-containing domains in meta-polycentromeres are usually well discernible (the upper model), but may also fuse into one line-like domain (bottom model). In C. europaea holocentromeres, the spindle fibres also attach to CENH3/CENP-A-free chromatin.

Table 1.
Centromere types present in yeast and plant species classified based on the distribution of spindle fiber attachment sites.
A specialized monocentric organization has been found for mitotic chromosomes in Pisum and Lathyrus species [11,12,13]. In these species, primary constrictions span up to a third of the chromosome length, corresponding to 263 Mbp, and contain up to five explicit CENH3/CENP-A-containing domains. All these domains contain both CENH3/CENP-A variants (CENH3-1 and CENH3-2) identified in these species and assemble on arrays of satellite DNA with a length varying from hundreds of kilobases to almost 3 Mbp. Thus, each of these domains can be viewed as one regional centromere. Individual CENH3/CENP-A-containing domains are particularly well discernible from prophase to prometaphase, but when condensation proceeds towards metaphase, they attach to each other or even merge into line-like structures similar to holocentromeres. H2AT120ph chromatin surrounds the CENH3/CENP-A regions, but does not intermingle with them. Microtubules of the mitotic spindle attach to chromosomes at each CENH3/CENP-A-containing domain or along the entire CENH3/CENP-A-containing line-like region, indicating that CENH3/CENP-A is a faithful marker of functional kinetochores in these species (Figure 4). Although these centromeres are functionally similar to monocentromeres, they were designated as “meta-polycentromeres” to reflect the unique organization of CENH3/CENP-A-containing chromatin domains.
Line-like contiguous holocentromeres within a groove were first identified in Luzula (Juncaceae) species during mitosis and meiosis [41,48,49,50,53]. Later on, a similar line-like holocentromere structure and arrangement was found during mitosis in related species of the genus Rhynchospora (Cyperaceae) [51,54]. In both cases, line-like holocentromeres show similar localization of CENH3/CENP-A and H2AT120ph and complete association to spindle microtubules during mitotic cell divisions [41,55]. Interestingly, holocentromeres in Rhynchospora were also associated with centromere-specific DNA sequences (tandem repeats and centromeric retrotransposons of maize) [55], while in Luzula, no centromere-specific repeats have been found.
Recently, an atypical holocentromere type was identified in mitotic chromosomes of Cuscuta europaea [46]. In this species, the mitotic spindle microtubules attach at uniform density along the entire chromosome length, clearly indicating the holocentric nature of the chromosomes. However, CENH3/CENP-A is restricted to only one to three discrete heterochromatin bands per chromosome, and H2AT120ph is not detectable (P. Neumann, unpublished). Additionally, CENH3/CENP-A is not confined to the chromosome periphery as would be expected for a kinetochore-specific protein. Instead, it homogenously fills the entire heterochromatin band. These data suggest that CENH3/CENP-A either lost its function or acts in parallel to an additional CENH3/CENP-A-independent mechanism of kinetochore positioning [46]. Interestingly, in fire ants, the centromere expansion occurred via a mechanism independent of CENH3/CENP-A duplication [14].
3. Variation at Different Stages of the Cell Cycle
Centromere organization on meiotic chromosomes differs between Rhynchospora and Luzula. In Luzula, meiotic chromosomes maintain the CENH3/CENP-A-containing line-like holocentromeres within a groove [49]. However, in contrast to Luzula, R. pubera shows a different meiotic chromosome structure. No linear holocentromeres, but a dispersed distribution of H2AT120ph, are present [47,55] (Figure 3). CENH3/CENP-A- and CENP-C-positive centromere domains are localized exclusively at the bivalent surface. The surface of the bivalents has a similar rough structure after surface rendering as somatic prophase chromosomes and interphase nuclei. Several CENH3/CENP-A domains are present along the entire meiotic chromosomes, but as distinct clusters that specifically interact with spindle fibers. Thus, during meiosis in Rhynchospora, the chromosomes are holocentric according to the definition that a chromosome is holocentric when kinetochore proteins and the spindle fibers are distributed along almost the entire poleward surface of the chromatids [10,56,57]. To distinguish the centromere arrangement between mitotic and meiotic chromosomes of R. pubera, we name these subtypes line-like and cluster-like holocentromeres, respectively. Interestingly, the line-like holocentromere becomes reestablished during first pollen mitosis [47,58]. Similar centromere structures in both mitotic and meiotic chromosomes have been observed in other Rhynchospora species (A. Marques, unpublished).
During interphase in L. elegans and R. pubera, the line-like holocentromeres disperse and a high number of CENH3/CENP-A-containing centromeric units are formed [48,51,53]. The line-like metaphase holocentromeres may be the result of merging various centromeric subunits caused by chromosome condensation (Figure 3). In L. elegans, the centromere-specific colocalization of α-kleisin cohesin subunits with CENH3/CENP-A along the holocentromeres was proven during mitosis and meiosis [59]. This suggests that centromere-specific cohesin could act to connect the centromere units along the centromere groove.
4. Centromere Evolution
Holocentric chromosomes are present in green algae, protozoans, invertebrates, and various plants [10,60]. They developed repeatedly via convergent evolution [20,56,60,61,62,63,64]. All known holocentric plants are monocot (species of Melanthiaceae, Juncaceae, and Cyperaceae) or eudicot (species of Drosera and Cuscuta subg. Cuscuta) angiosperms [20,60,65]. Even within the same genus, such as Cuscuta, mono- (C. japonica) and holocentric (C. europaea) species may appear [46]. Such closely related species offer the possibility for comparative studies to analyze the molecular basis of centromere type variability. However, the factors that induced the transition from mono- to holocentromeres and the underlying mechanisms are still under debate. The surprisingly high centromere diversity in independent eukaryotic lineages raises the questions of whether this variability offers evolutionary advantages and how holocentromeres evolved from monocentric ones or vice versa. Additionally, it is of interest to investigate how the different centromere types guarantee proper chromosome segregation during mitosis and meiosis.
4.1. Is Holocentricity Original or Derived?
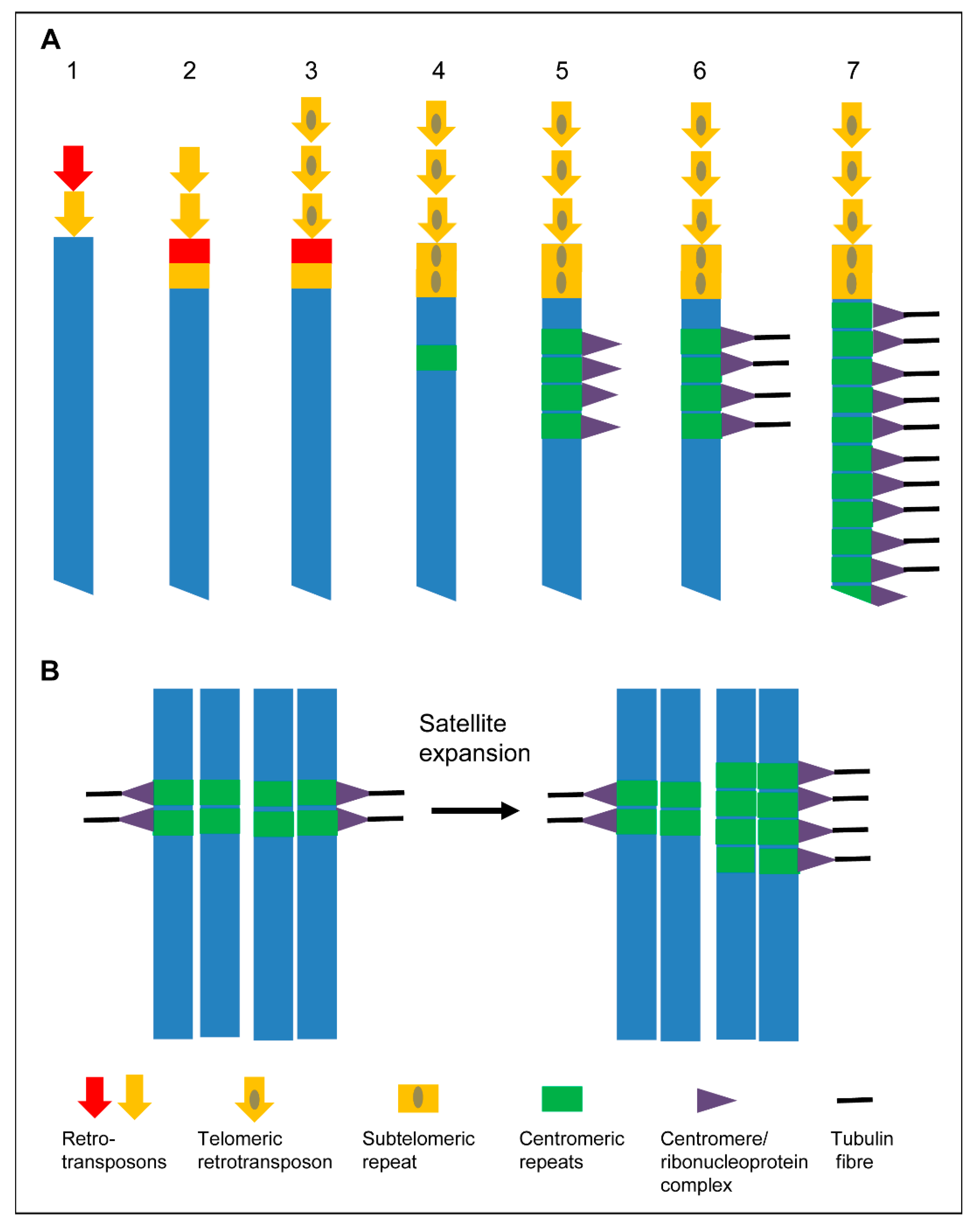
Lima de Faria [66] postulated that mono- and holocentromeres may have derived from distinct groups of special chromomeres that are either restricted to a particular site or uniformly dispersed on mitotic chromosomes. Other authors proposed that holocentricity is a primitive feature, because it has been found in phylogenetically basal plant and animal taxa and offers fast evolution combined with the high tolerance of chromosomal fragmentation and rearrangements [67,68,69,70,71,72,73]. Sybenga [71] suggested that monocentricity developed from holocentricity. In contrast, due to the presence of holocentricity in phylogenetically distant and derived taxa, such as holocentric hemipterans and rushes (Juncaceae), which descended from more primitive monocentric species, Swanson [74] hypothesized that holocentricity has been derived from monocentricity. Greilhuber [75] and Melters et al. [60] supported this hypothesis by arguing that holocentric chromosomes developed four times independently in angiosperms. Nagaki et al. [53] suggested that in Luzula holocentricity may have arisen from monocentricity via subsequent centromere extension, and Neumann et al. [11] proposed that centromere-competent satellite(s) caused the transition from mono- to holocentric chromosomes based on the findings in extended Pisum centromeres. The “telomere to centromere” model of Villasante et al. [76,77] and the “centromere drive” hypothesis of Malik and Henikoff [78] agree with the hypothesis that holocentricity evolved from monocentric chromosomes (Figure 5).
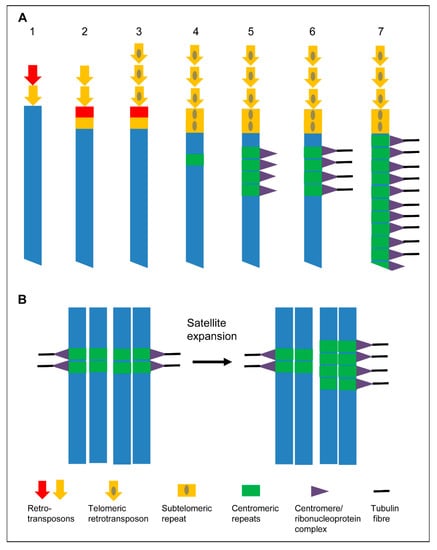
Figure 5.
Models to expand centromeres. (A) “Telomere to centromere” model based on Villasante et al. [76,77]. (1) Different retroelements are mobilized to heal the DNA ends of a broken chromosome. (2) The most effective telomeric retrotransposons become selected. (3) Retrotransposons with a more effective capping capability are selected. (4) Subtelomeric repeats appear. (5) Subtelomeric repeats are amplified and become centromeric repeats. Ribonucleoprotein complexes are formed after transcription of theses repeats. (6) Centromeric repeats become a protocentromere after being recognized by microtubules. (7) Chromosome wide extension of the centromere to form a holocentromere. (B) “Centromere drive model” of Henikoff et al. [2]. The expansion of a satellite that binds CENH3/CENP-A provides more microtubule attachment sites. This stronger centromere drives in female meiosis into the egg cell.
Similar to Pisum and Lathyrus species [11,12,13], the red fire ant, Solenopsis invicta, and closely related species possess elongated centromeres. Consequently, they are regarded as evolutionary intermediates via runaway expansion of their centromeres towards the development of holocentromeres [14]. Our recent ultrastructural centromere investigations, which highlight the variation of centromere organization in holocentric plants, support the view that holocentricity may have been derived from monocentricity independently multiple times.
The occurrence of increased exposure to clastogens (factors inducing chromosome fragmentation), such as cosmic irradiation during terrestrialization (land colonization), may have caused the convergent evolution of holocentricity in different eukaryotic taxa [79]. Král et al. [80] found that holocentricity in spiders is an autapomorphy (a derived trait unique to a given taxon) of the superfamily Dysderoidea. It may have developed via multiple subsequent chromosome fusions of monocentric chromosomes combined with genome size reduction. But, considering the unstable nature of dicentric chromosomes [81], simple fusion of monocentric chromosomes is less likely to be the route towards holocentric chromosomes.
4.2. Is Holocentricity Related to CENH3/CENP-A Loss?
Spindle fibers commonly attach to kinetochores organized by CENH3/CENP-A-containing regions of centromeres. However, recent studies showed that a number of insects have recurrently lost CENH3/CENP-A, implying that they evolved a CENH3/CENP-A-independent mechanism of kinetochore assembly. Interestingly, all of them have holocentric chromosomes, derived independently from monocentric ones. Despite the loss of CENH3/CENP-A in holocentric insects, these holocentromeres still comprise kinetochore proteins that can force apart the chromosomes [20]. Thus, the transition to holocentricity may facilitate the loss of CENH3/CENP-A, at least in some species, or vice versa [20,82]. Since the mitotic spindle microtubuli attach to chromosomes at sites devoid of CENH3/CENP-A in C. europaea, it is likely that a CENH3/CENP-A-independent pathway of kinetochore assembly evolved also in this species [46].
A hybrid-type centromere has been discovered in the pathogenic fungus Mucor circinelloides. This CENH3/CENP-A-lacking centromere type displays a mosaic of point and regional centromeres [22,23].
It can be speculated that the loss of CENH3/CENP-A and the variation of the CENH3/CENP-A content might cause the observed centromere plasticity. Alternatively, structural changes in the centromere architecture might have enabled CENH3/CENP-A-independent centromere activity [57].
5. Advantages and Challenges of Holocentricity
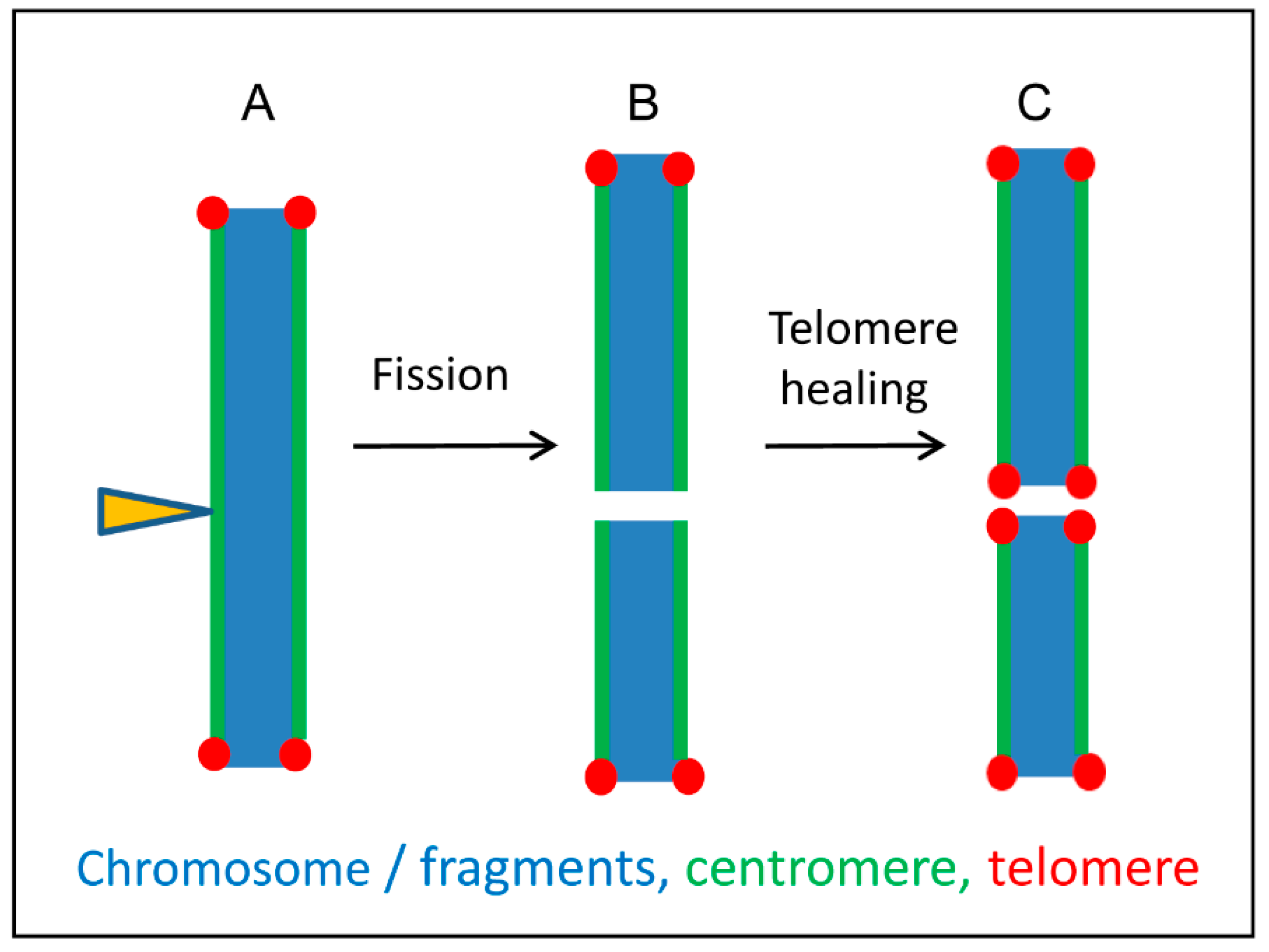
One reason that holocentricity evolved independently in different taxa of eukaryotes could be that chromosome fragmentation by DNA breakage seems to be more easily tolerated in species with holocentric chromosomes [79,83]. Contrary to monocentric species where acentric fragments usually become lost during cell division, the breakage of holocentric chromosomes creates fragments with normal spindle fiber attachment sites [84,85]. The fast formation of new telomeres at the break points allows holocentric species a rapid karyotype evolution, including chromosome fissions and rearrangements [57,86] (Figure 6). A high karyotype variability between related holocentric species, e.g., within the genera Carex and Eleocharis, with chromosome numbers of 2n = 12–124 and 2n = 6–196, respectively, probably reflects this situation [87]. In addition, a chromosome number reduction to x = 3 from the possible basic number of x = 5 occurred in E. subarticulata [88]. An intraspecific chromosome number variability with 2n = 41–47 in E. kamtschatica [89] and 2n = 6–8 in E. maculosa [90] has been reported. On the other hand, a comparison of all known holocentric lineages with their closest related monocentric lineages revealed that the different speciation rates between mono- and holocentrics are not related to different centromere types [91].
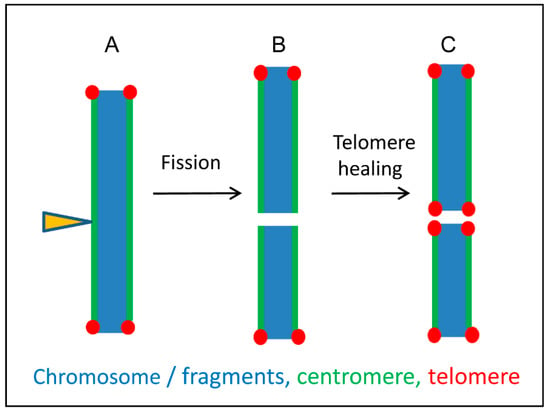
Figure 6.
A model illustrating possible karyotype alterations after fragmentation of holocentric chromosomes based on the interplay between holocentricity and telomere healing. (A) Irradiation of holocentric chromosomes (arrowhead) induces chromosome fragments. Centromere activity is present along all chromosome fragments. (B) Broken ends lack telomere repeats and become gradually healed (C) by de novo telomere syntheses. Based on Jankowska et al. [86].
Could it be that the development of holocentricity is advantageous to move large chromosomes during cell divisions? Obviously, this is not a reason because, similar to the large chromosomes of Luzula [41] and Rhynchospora [47,51], the much smaller chromosomes of Cuscuta [46], Drosera [92,93,94,95] and Chionographis [96,97] are holocentric, and the holokinetic chromosomes of the spider superfamily Dysderoidea are of different size [80]. On the other hand, the very large chromosomes of lilies, of Triticeae, of some legumes, and of conifers, for instance, are monocentric. Generally, holocentric species have smaller genomes compared to their monocentric relatives [87].
In contrast to possible evolutionary advantages, holocentric chromosomes might represent a challenge during nuclear division, because due to the extended centromere architecture, a merotelic attachment of spindle fibers, which may lead to chromosome mis-segregation, is more likely [57,60]. As a consequence, holocentric organisms had to adapt their meiotic processes [98].
Holocentric animals adapted the meiotic chromosome segregation by remodeling their chromosomes into functionally monocentric ones, as in the worm Caenorhabditis elegans, by adopting a telokinetic behavior as in Heteroptera bugs, and by “inverted meiosis” as in the citrus mealybug Planococcus citri [49]. The holokinetic plant species L. elegans [49,50], R. pubera and R. tenuis [55,99] display an inverted order of meiotic chromatid segregation. Sister chromatids separate already during meiosis I, while the homologues segregate in meiosis II [50].
6. Conclusions
Here, we show that centromere-specific antibodies combined with super-resolution microscopy are useful to identify the ultrastructure of the different centromere types and microtubule attachment sites. Recent analysis of species with nonclassical centromeres has demonstrated that in addition to monocentromeres, meta-polycentric chromosomes, cluster-, and line-like holocentromeres developed during plant evolution. The ultrastructural centromere investigations showing interspecific centromere variability, and in some species variability between mitotic and meiotic centromeres, suggest a development from monocentromeres towards holocentromeres. An opposite direction of evolution or independently from an original monocentromere organization is less likely. Whether meta-polycentromeres are an intermediate between mono- and holocentromeres is not confirmed, because until now, no lineage that contains species with both types of centromeres, meta-polycentric and holocentric, has been described. The centromere unit-based organization of holocentromeres is dynamic, thus allowing the formation of line- or cluster-like holocentromeres.
Based on the distribution of spindle attachment sites at CENH3/CENP-A-containing or CENH3/CENP-A-free regions, a classification of different centromere types and subtypes has been suggested. No correlation exists between holocentricity and chromosome size. Holocentricity might be advantageous due to a higher tolerance of chromosome fragmentations.
Alternative mechanisms have developed to segregate holocentric chromosomes, showing that proper chromosome segregation during mitosis and meiosis may be performed not only by distinct monocentromeres. Elongated centromeres and the different types of holocentromeres also fulfill this essential task successfully.
Author Contributions
V.S. wrote the manuscript with input from all other authors. All authors have read and agreed to the published version of the manuscript.
Funding
This work has been supported by the Deutsche Forschungsgemeinschaft (Schu 762/11-1; HO 1779/32-1), DAAD/CAPES (57517412; 88881.144086/2017-01) and the Czech Science Foundation (17-09750S).
Acknowledgments
We thank Catrin Kaydamov for help to arrange the reference list.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Musacchio, A.; Desai, A. A molecular view of kinetochore assembly and function. Biology (Basel) 2017, 6, 5. [Google Scholar] [CrossRef] [PubMed]
- Henikoff, S.; Ahmad, K.; Malik, H.S. The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 2001, 293, 1098–1102. [Google Scholar] [CrossRef] [PubMed]
- Cleveland, D.W.; Mao, Y.; Sullivan, K.F. Centromeres and kinetochores: From epigenetics to mitotic checkpoint signaling. Cell 2003, 112, 407–421. [Google Scholar] [CrossRef]
- Plohl, M.; Meštrović, N.; Mravinac, B. Centromere identity from the DNA point of view. Chromosoma 2014, 123, 313–325. [Google Scholar] [CrossRef]
- Hoang, P.T.N.; Schubert, V.; Meister, A.; Fuchs, J.; Schubert, I. Variation in genome size, cell and nucleus volume, chromosome number and rDNA loci among duckweeds. Sci. Rep. 2019, 9, 3234. [Google Scholar] [CrossRef]
- Tran, T.D.; Cao, H.X.; Jovtchev, G.; Novák, P.; Vu, G.T.; Macas, J.; Schubert, I.; Fuchs, J. Chromatin organization and cytological features of carnivorous Genlisea species with large genome size differences. Front. Plant Sci. 2015, 6, 613. [Google Scholar] [CrossRef]
- Vu, G.T.H.; Schmutzer, T.; Bull, F.; Cao, H.X.; Fuchs, J.; Tran, D.T.; Jovtchev, G.; Pistrick, K.; Stein, N.; Pecinka, A.; et al. Comparative genome analysis reveals divergent genome size evolution in a carnivorous plant genus. Plant Genome 2015, 8, 1–14. [Google Scholar] [CrossRef]
- Fonsêca, A.; Ferreira, J.; dos Santos, T.R.; Mosiolek, M.; Bellucci, E.; Kami, J.; Gepts, P.; Geffroy, V.; Schweizer, D.; dos Santos, K.G.; et al. Cytogenetic map of common bean (Phaseolus vulgaris L.). Chromosome Res. 2010, 18, 487–502. [Google Scholar] [CrossRef]
- Da Costa Silva, S.; Marques, A.; dos Santos Soares Filho, W.; Mirkov, T.E.; Andrea Pedrosa-Harand, A.; Guerra, M. The cytogenetic map of the Poncirus trifoliata (L.) Raf.—a nomenclature system for chromosomes of all citric species. Trop. Plant Biol. 2011, 4, 99–105. [Google Scholar] [CrossRef]
- Heckmann, S.; Houben, A. Holokinetic centromeres. In Plant Centromere Biology; Jiang, J., Birchler, J.A., Eds.; Wiley-Blackwell: Ames, IA, USA, 2013; Volume 1, pp. 83–94. [Google Scholar]
- Neumann, P.; Navrátilová, A.; Schroeder-Reiter, E.; Koblížková, A.; Steinbauerová, V.; Chocholová, E.; Novák, P.; Wanner, G.; Macas, J. Stretching the rules: Monocentric chromosomes with multiple centromere domains. PLoS Genet. 2012, 8, e1002777. [Google Scholar] [CrossRef]
- Neumann, P.; Pavlíková, Z.; Koblížková, A.; Fuková, I.; Jedličková, V.; Novák, P.; Macas, J. Centromeres off the hook: Massive changes in centromere size and structure following duplication of CenH3 gene in Fabeae species. Mol. Biol. Evol. 2015, 32, 1862–1879. [Google Scholar] [CrossRef] [PubMed]
- Neumann, P.; Schubert, V.; Fuková, I.; Manning, J.E.; Houben, A.; Macas, J. Epigenetic histone marks of extended meta-polycentric centromeres of Lathyrus and Pisum chromosomes. Front. Plant Sci. 2016, 7, 234. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.C.; Lee, C.C.; Kao, C.Y.; Chang, N.C.; Lin, C.C.; Shoemaker, D.; Wang, J. Evolution of long centromeres in fire ants. BMC Evol. Biol. 2016, 16, 189. [Google Scholar] [CrossRef] [PubMed]
- Brinkley, B.R.; Valdivia, M.M.; Tousson, A.; Brenner, S.L. Compound kinetochores of the Indian muntjac. Evolution by linear fusion of unit kinetochores. Chromosoma 1984, 91, 1–11. [Google Scholar] [CrossRef]
- Metcalfe, C.J.; Bulazel, K.V.; Ferreri, G.C.; Schroeder-Reiter, E.; Wanner, G.; Rens, W.; Obergfell, C.; Eldridge, M.D.; O’Neill, R.J. Genomic instability within centromeres of interspecific marsupial hybrids. Genetics 2007, 177, 2507–2517. [Google Scholar] [CrossRef]
- Paweletz, N.; Vig, B.K.; Finze, E.M. Evolution of compound centromeres. A new phenomenon. Cancer Genet. Cytogenet. 1989, 42, 75–86. [Google Scholar] [CrossRef]
- Clarke, L.; Carbon, J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature 1980, 287, 504–509. [Google Scholar] [CrossRef]
- Earnshaw, W.C.; Allshire, R.C.; Black, B.E.; Bloom, K.; Brinkley, B.R.; Brown, W.; Cheeseman, I.M.; Choo, K.H.; Copenhaver, G.P.; Deluca, J.G.; et al. Esperanto for histones: CENP-A, not CenH3, is the centromeric histone H3 variant. Chromosome Res. 2013, 21, 101–106. [Google Scholar] [CrossRef]
- Drinnenberg, I.A.; deYoung, D.; Henikoff, S.; Malik, H.S. Recurrent loss of CenH3 is associated with independent transitions to holocentricity in insects. eLife 2014, 3, e03676. [Google Scholar] [CrossRef]
- Akiyoshi, B.; Gull, K. Discovery of unconventional kinetochores in kinetoplastids. Cell 2014, 156, 1247–1258. [Google Scholar] [CrossRef]
- Akiyoshi, B. Evolution: A mosaic-type centromere in an early-diverging fungus. Curr. Biol. 2019, 29, R1184–R1186. [Google Scholar] [CrossRef]
- Navarro-Mendoza, M.I.; Pérez-Arques, C.; Panchal, S.; Nicolás, F.E.; Mondo, S.J.; Ganguly, P.; Pangilinan, J.; Grigoriev, I.V.; Heitman, J.; Sanyal, K.; et al. Early diverging fungus Mucor circinelloides lacks centromeric histone CENP-A and displays a mosaic of point and regional centromeres. Curr. Biol. 2019, 29, 3791–3802. [Google Scholar] [CrossRef] [PubMed]
- Van Hooff, J.J.; Tromer, E.; van Wijk, L.M.; Snel, B.; Kops, G.J. Evolutionary dynamics of the kinetochore network in eukaryotes as revealed by comparative genomics. EMBO Rep. 2017, 18, 1559–1571. [Google Scholar] [CrossRef] [PubMed]
- Schermelleh, L.; Heintzmann, R.; Leonhardt, H. A guide to super-resolution fluorescence microscopy. J. Cell Biol. 2010, 190, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, U.; Reilly, D.T.; Schroeder, C.M. Zooming in on biological processes with fluorescence nanoscopy. Curr. Opin. Biotechnol. 2013, 24, 646–653. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.R.; Ross, S.T.; Davidson, M.W. Structured illumination microscopy for superresolution. ChemPhysChem 2014, 15, 566–576. [Google Scholar] [CrossRef]
- Komis, G.; Šamajová, O.; Ovečka, M.; Šamaj, J. Super-resolution microscopy in plant cell imaging. Trends Plant Sci. 2015, 20, 834–843. [Google Scholar] [CrossRef]
- Nienhaus, K.; Nienhaus, G.U. Where do we stand with super-resolution optical microscopy? J. Mol. Biol. 2016, 428, 308–322. [Google Scholar] [CrossRef]
- Baroux, C.; Schubert, V. Technical Review: Microscopy and image processing tools to analyse plant chromatin—Practical considerations. In Plant Chromatin Dynamics: Methods and Protocols; Bemer, M., Baroux, C., Eds.; Humana Press: New York, NY, USA, 2018; Volume 1675, pp. 537–589. [Google Scholar]
- Schermelleh, L.; Ferrand, A.; Huser, T.; Eggeling, C.; Sauer, M.; Biehlmaier, O.; Drummen, G.P.C. Super-resolution microscopy demystified. Nat. Cell Biol. 2019, 21, 72–84. [Google Scholar] [CrossRef]
- Rouquette, J.; Cremer, C.; Cremer, T.; Fakan, S. Functional nuclear architecture studied by microscopy: Present and future. In International Review of Cell and Molecular Biology; Jeon, K.W., Ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2010; Volume 282, pp. 1–90. [Google Scholar]
- Han, R.; Li, Z.; Fan, Y.; Jiang, Y. Recent advances in super-resolution fluorescence imaging and its applications in biology. J. Genet. Genomics 2013, 40, 583–595. [Google Scholar] [CrossRef]
- Fornasiero, E.F.; Opazo, F. Super-resolution imaging for cell biologists: Concepts, applications, current challenges and developments. Bioessays 2015, 37, 436–451. [Google Scholar] [CrossRef] [PubMed]
- Kraus, F.; Miron, E.; Demmerle, J.; Chitiashvili, T.; Budco, A.; Alle, Q.; Matsuda, A.; Leonhardt, H.; Schermelleh, L.; Markaki, Y. Quantitative 3D structured illumination microscopy of nuclear structures. Nat. Protoc. 2017, 12, 1011–1028. [Google Scholar] [CrossRef] [PubMed]
- Schubert, V. Super-resolution microscopy – applications in plant cell research. Front. Plant Sci. 2017, 8, 531. [Google Scholar] [CrossRef] [PubMed]
- Demidov, D.; Schubert, V.; Kumke, K.; Weiss, O.; Karimi-Ashtiyani, R.; Buttlar, J.; Heckmann, S.; Wanner, G.; Dong, Q.; Han, F.; et al. Anti-phosphorylated histone H2AThr120: A universal microscopic marker for centromeric chromatin of mono- and holocentric plant species. Cytogenet. Genome Res. 2014, 143, 150–156. [Google Scholar] [CrossRef]
- Houben, A.; Wako, T.; Furushima-Shimogawara, R.; Presting, G.; Künzel, G.; Schubert, I.; Fukui, K. The cell cycle dependent phosphorylation of histone H3 is correlated with the condensation of plant mitotic chromosomes. Plant J. 1999, 18, 675–679. [Google Scholar] [CrossRef]
- Gernand, D.; Demidov, D.; Houben, A. The temporal and spatial pattern of histone H3 phosphorylation at serine 28 and serine 10 is similar in plants but differs between mono- and polycentric chromosomes. Cytogenet. Genome Res. 2003, 101, 172–176. [Google Scholar] [CrossRef]
- Weisshart, K.; Fuchs, J.; Schubert, V. Structured Illumination Microscopy (SIM) and Photoactivated Localization Microscopy (PALM) to analyze the abundance and distribution of RNA polymerase II molecules on flow-sorted Arabidopsis nuclei. Bio-Protocol 2016, 6, e1725. [Google Scholar] [CrossRef]
- Wanner, G.; Schroeder-Reiter, E.; Ma, W.; Houben, A.; Schubert, V. The ultrastructure of mono- and holocentric plant centromeres: An immunological investigation by structured illumination microscopy and scanning electron microscopy. Chromosoma 2015, 124, 503–517. [Google Scholar] [CrossRef]
- Furuyama, S.; Biggins, S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc. Natl. Acad. Sci. USA 2007, 104, 14706–14711. [Google Scholar] [CrossRef]
- Pluta, A.F.; Mackay, A.M.; Ainsztein, A.M.; Goldberg, I.G.; Earnshaw, W.C. The centromere: Hub of chromosomal activities. Science 1995, 270, 1591–1594. [Google Scholar] [CrossRef]
- Ishii, T.; Karimi-Ashtiyani, R.; Banaei-Moghaddam, A.M.; Schubert, V.; Fuchs, J.; Houben, A. The differential loading of two barley CENH3 variants into distinct centromeric substructures is cell type- and development-specific. Chromosome Res. 2015, 23, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Banaei-Moghaddam, A.M.; Schubert, V.; Kumke, K.; Weibeta, O.; Klemme, S.; Nagaki, K.; Macas, J.; González-Sánchez, M.; Heredia, V.; Gómez-Revilla, D.; et al. Nondisjunction in favor of a chromosome: The mechanism of rye B chromosome drive during pollen mitosis. Plant Cell 2012, 24, 4124–4134. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, L.; Neumann, P.; Jang, T.-S.; Klemme, S.; Schubert, V.; Koblížková, A.; Houben, A.; Macas, J. Mitotic spindle attachment to the holocentric chromosomes of Cuscuta europaea does not correlate with the distribution of CENH3 chromatin. Front. Plant Sci. 2020, 10, 1799. [Google Scholar] [CrossRef] [PubMed]
- Marques, A.; Schubert, V.; Houben, A.; Pedrosa-Harand, A. Restructuring of holocentric centromeres during meiosis in the plant Rhynchospora pubera. Genetics 2016, 204, 555–568. [Google Scholar] [CrossRef]
- Heckmann, S.; Schroeder-Reiter, E.; Kumke, K.; Ma, L.; Nagaki, K.; Murata, M.; Wanner, G.; Houben, A. Holocentric chromosomes of Luzula elegans are characterized by a longitudinal centromere groove, chromosome bending, and a terminal nucleolus organizer region. Cytogenet. Genome Res. 2011, 134, 220–228. [Google Scholar] [CrossRef]
- Heckmann, S.; Jankowska, M.; Schubert, V.; Kumke, K.; Ma, W.; Houben, A. Alternative meiotic chromatid segregation in the holocentric plant Luzula elegans. Nat. Commun. 2014, 5, 4979. [Google Scholar] [CrossRef]
- Heckmann, S.; Schubert, V.; Houben, A. Holocentric plant meiosis: First sisters, then homologues. Cell Cycle 2014, 13, 3623–3624. [Google Scholar] [CrossRef][Green Version]
- Marques, A.; Ribeiro, T.; Neumann, P.; Macas, J.; Novák, P.; Schubert, V.; Pellino, M.; Fuchs, J.; Ma, W.; Kuhlmann, M.; et al. Holocentromeres in Rhynchospora are associated with genome-wide centromere-specific repeat arrays interspersed among euchromatin. Proc. Natl. Acad. Sci. USA 2015, 112, 13633–13638. [Google Scholar] [CrossRef]
- Lawrimore, J.; Bloom, K. The regulation of chromosome segregation via centromere loops. Crit. Rev. Biochem. Mol. Biol. 2019, 54, 352–370. [Google Scholar] [CrossRef]
- Nagaki, K.; Kashihara, K.; Murata, M. Visualization of diffuse centromeres with centromere-specific histone H3 in the holocentric plant Luzula nivea. Plant Cell 2005, 17, 1886–1893. [Google Scholar] [CrossRef]
- Ribeiro, T.; Buddenhagen, C.E.; Thomas, W.W.; Souza, G.; Pedrosa-Harand, A. Are holocentrics doomed to change? Limited chromosome number variation in Rhynchospora Vahl (Cyperaceae). Protoplasma 2018, 255, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Cabral, G.; Marques, A.; Schubert, V.; Pedrosa-Harand, A.; Schlögelhofer, P. Chiasmatic and achiasmatic inverted meiosis of plants with holocentric chromosomes. Nat. Commun. 2014, 5, 5070. [Google Scholar] [CrossRef] [PubMed]
- Guerra, M.; Cabral, G.; Cuacos, M.; González-García, M.; González-Sánchez, M.; Vega, J.; Puertas, M.J. Neocentrics and holokinetics (holocentrics): Chromosomes out of the centromeric rules. Cytogenet. Genome Res. 2010, 129, 82–96. [Google Scholar] [CrossRef] [PubMed]
- Cuacos, M.; Franklin, F.C.H.; Heckmann, S. Atypical centromeres in plants-what they can tell us. Front. Plant Sci. 2015, 6, 913. [Google Scholar] [CrossRef] [PubMed]
- Rocha, D.M.; Marques, A.; Andrade, C.G.; Guyot, R.; Chaluvadi, S.R.; Pedrosa-Harand, A.; Houben, A.; Bennetzen, J.L.; Vanzela, A.L. Developmental programmed cell death during asymmetric microsporogenesis in holocentric species of Rhynchospora (Cyperaceae). J. Exp. Bot. 2016, 67, 5391–5401. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Schubert, V.; Martis, M.M.; Hause, G.; Liu, Z.; Shen, Y.; Conrad, U.; Shi, W.; Scholz, U.; Taudien, S.; et al. The distribution of α-kleisin during meiosis in the holocentromeric plant Luzula elegans. Chromosome Res. 2016, 24, 393–405. [Google Scholar] [CrossRef]
- Melters, D.P.; Paliulis, L.V.; Korf, I.F.; Chan, S.W. Holocentric chromosomes: Convergent evolution, meiotic adaptations, and genomic analysis. Chromosome Res. 2012, 20, 579–593. [Google Scholar] [CrossRef]
- Dernburg, A.F. Here, there, and everywhere: Kinetochore function on holocentric chromosomes. J. Cell Biol. 2001, 153, F33–F38. [Google Scholar] [CrossRef]
- Guerra, M.; Ribeiro, T.; Felix, L.P. Monocentric chromosomes in Juncus (Juncaceae) and implications for the chromosome evolution of the family. Bot. J. Linn. Soc. 2019, 191, 475–483. [Google Scholar] [CrossRef]
- Pimpinelli, S.; Goday, C. Unusual kinetochores and chromatin diminution in Parascaris. Trends Genet. 1989, 5, 310–315. [Google Scholar] [CrossRef]
- Wrensch, D.I.; Kethley, J.B.; Noton, R.A. Cytogenetics of holokinetic chromosomes and and inverted meiosis: Keys to evolutionary succes of mites, with generalization on eukaryotes. In Mites: Ecological and Evolutionary Analyses of Life History Patterns; Houck, M.A., Ed.; Springer: Boston, MA, USA, 1994; pp. 282–343. [Google Scholar]
- Mola, L.M.; Papeschi, A.G. Holokinetic chromosomes at a glance. J. Basic Appl. Genet. 2006, 17, 17–33. [Google Scholar]
- Lima de Faria, A. Genetics, origin and evolution of kinetochores. Hereditas 1949, 35, 422–444. [Google Scholar] [CrossRef]
- Câmara, A. Posição actual do problemado centromero. Genética Ibérica 1953, 5, 1–33. [Google Scholar]
- Castro, D. The structure of the centromere and its functioning. Genética Ibérica 1950, 2, 201–209. [Google Scholar]
- Moore, G.; Aragón-Alcaide, L.; Roberts, M.; Reader, S.; Miller, T.; Foote, T. Are rice chromosomes components of a holocentric chromosome ancestor? Plant Mol. Biol. 1997, 35, 17–23. [Google Scholar] [CrossRef]
- Schrader, F. The role of the kinetochore in the chromosomal evolution of the Heteroptera and Homoptera. Evolution 1947, 1, 134–142. [Google Scholar] [CrossRef]
- Sybenga, J. Specialization in the behavior of chromosomes on the meiotic spindle. Genetica 1981, 57, 143–151. [Google Scholar] [CrossRef]
- Sybenga, J. Cytogenetics in Plant Breeding, 1st ed.; Springer: Berlin/Heidelberg, Germany, 1992; p. 469. [Google Scholar]
- Vaarama, A. Cytological observation on Pleurozium schreberi, with special reference to centromere evolution. Ann. Bot. Soc. Zool. Bot. Fenn. Vanamo 1954, 28, 1–59. [Google Scholar]
- Swanson, C.P. Cytology and Cytogenetics, 1st ed.; Prentice-Hall, Inc.: Englewood Cliffs, NJ, USA, 1957; p. 596. [Google Scholar]
- Greilhuber, J. Chromosomes of the monocotyledons (general aspects). In Monocotyledons: Systematics and Evolution; Rudall, P.J., Cribb, P.J., Cutler, D.F., Humphries, C.J., Eds.; Royal Botanic Gardens: Kew, UK, 1995; Volume 2, pp. 379–414. [Google Scholar]
- Villasante, A.; Abad, J.P.; Méndez-Lago, M. Centromeres were derived from telomeres during the evolution of the eukaryotic chromosome. Proc. Natl. Acad. Sci. USA 2007, 104, 10542–10547. [Google Scholar] [CrossRef]
- Villasante, A.; Méndez-Lago, M.; Abad, J.P.; Montejo de Garcíni, E. The birth of the centromere. Cell Cycle 2007, 6, 2872–2876. [Google Scholar] [CrossRef][Green Version]
- Malik, H.S.; Henikoff, S. Conflict begets complexity: The evolution of centromeres. Curr. Opin. Genet. Dev. 2002, 12, 711–718. [Google Scholar] [CrossRef]
- Zedek, F.; Bureš, P. Holocentric chromosomes: From tolerance to fragmentation to colonization of the land. Ann. Bot. 2018, 121, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Král, J.; Forman, M.; Korinkova, T.; Lerma, A.C.R.; Haddad, C.R.; Musilova, J.; Rezac, M.; Herrera, I.M.A.; Thakur, S.; Dippenaar-Schoeman, A.S.; et al. Insights into the karyotype and genome evolution of haplogyne spiders indicate a polyploid origin of lineage with holokinetic chromosomes. Sci. Rep. 2019, 9, 3001. [Google Scholar] [CrossRef] [PubMed]
- Stimpson, K.M.; Matheny, J.E.; Sullivan, B.A. Dicentric chromosomes: Unique models to study centromere function and inactivation. Chromosome Res. 2012, 20, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Drinnenberg, I.A.; Akiyoshi, B. Evolutionary lessons from species with unique kinetochores. Prog. Mol. Subcell. Biol. 2017, 56, 111–138. [Google Scholar] [PubMed]
- Zedek, F.; Bureš, P. Pest arthropods with holocentric chromosomes are more resistant to sterilizing ionizing radiation. Radiat. Res. 2019, 191, 255–261. [Google Scholar] [CrossRef]
- Hughes-Schrader, S.; Ris, H. The diffuse spindle attachment of coccids, verified by the mitotic behavior of induced chromosome fragments. J. Exp. Zool. 1941, 87, 429–456. [Google Scholar] [CrossRef]
- Schrader, F. Notes an the mitotic behavior of long chromosomes. Cytologia 1935, 6, 422–430. [Google Scholar] [CrossRef]
- Jankowska, M.; Fuchs, J.; Klocke, E.; Fojtová, M.; Polanská, P.; Fajkus, J.; Schubert, V.; Houben, A. Holokinetic centromeres and efficient telomere healing enable rapid karyotype evolution. Chromosoma 2015, 124, 519–528. [Google Scholar] [CrossRef]
- Bureš, P.; Zedek, F.; Marková, M. Holocentric chromosomes. In Plant Genome Diversity, Physical Structure, Behaviour and Evolution of Plant Genomes; Greilhuber, J., Dolezel, J., Wendel, J.F., Eds.; Springer: Vienna, Austria, 2013; Volume 2, pp. 187–208. [Google Scholar]
- Da Silva, C.R.M.; González-Elizondo, M.S.; Vanzela, A.L.L. Reduction of chromosome number in Eleocharis subarticulata (Cyperaceae) by multiple translocations. Bot. J. Linn. Soc. 2005, 149, 457–464. [Google Scholar] [CrossRef]
- Yano, O.; Hoshino, A.T. Cytological studies of aneuploidy in Eleocharis kamtschatica (Cyperaceae). Cytologia 2006, 71, 141–147. [Google Scholar] [CrossRef][Green Version]
- Da Silva, C.R.; González-Elizondo, M.S.; Laforga Vanzela, A.L. Chromosome reduction in Eleocharis maculosa (Cyperaceae). Cytogenet. Genome Res. 2008, 122, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Márquez-Corro, J.I.; Martín-Bravo, S.; Spalink, D.; Luceño, M.; Escudero, M. Inferring hypothesis-based transitions in clade-specific models of chromosome number evolution in sedges (Cyperaceae). Mol. Phylogenet. Evol. 2019, 135, 203–209. [Google Scholar] [CrossRef]
- Kolodin, P.; Cempírková, H.; Bureš, P.; Horová, L.; Veleba, A.; Francová, J.; Adamec, L.; Zedek, F. Holocentric chromosomes may be an apomorphy of Droseraceae. Plant Syst. Evol. 2018, 304, 1289–1296. [Google Scholar] [CrossRef]
- Sheikh, S.A.; Kondo, K. Differential staining with orcein, Giemsa, CMA, and DAPI for comparative chromosome study of 12 species of Australian Drosera (Droseraceae). Am. J. Bot. 1995, 82, 1278–1286. [Google Scholar] [CrossRef]
- Sheikh, S.A.; Kondo, K.; Hoshi, Y. Study of diffused centromeric nature of Drosera chromosomes. Cytologia 1995, 60, 43–47. [Google Scholar] [CrossRef]
- Veleba, A.; Šmarda, P.; Zedek, F.; Horová, L.; Šmerda, J.; Bureš, P. Evolution of genome size and genomic GC content in carnivorous holokinetics (Droseraceae). Ann. Bot. 2017, 119, 409–416. [Google Scholar] [CrossRef]
- Tanaka, N.; Tanaka, N. Chromosome Studies in Chionographis (Liliaceae) I. On the holokinetic nature of chromosomes in Chionographis japonica Maxim. Cytologia 1977, 42, 753–763. [Google Scholar] [CrossRef]
- Tanaka, N.; Tanaka, N. Chromosome studies in Chionographis (Liliaceae). II. Morphological characteristics of the somatic chromosomes of four Japanese members. Cytologia 1979, 44, 935–949. [Google Scholar] [CrossRef]
- Marques, A.; Pedrosa-Harand, A. Holocentromere identity: From the typical mitotic linear structure to the great plasticity of meiotic holocentromeres. Chromosoma 2016, 125, 669–681. [Google Scholar] [CrossRef]
- Marques, A.; Schubert, V.; Houben, A.; Pedrosa-Harand, A. Loss of the line-like holocentromere structure during inverted meiosis in a holocentric plant. Cytogenet. Genome Res. 2016, 148, 128. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).