High-Level Production of a Thermostable Mutant of Yarrowia lipolytica Lipase 2 in Pichia pastoris
Abstract
1. Introduction
2. Results
2.1. Lipase Activity Analysis for Transformants with Different Copies of the lip2 Gene
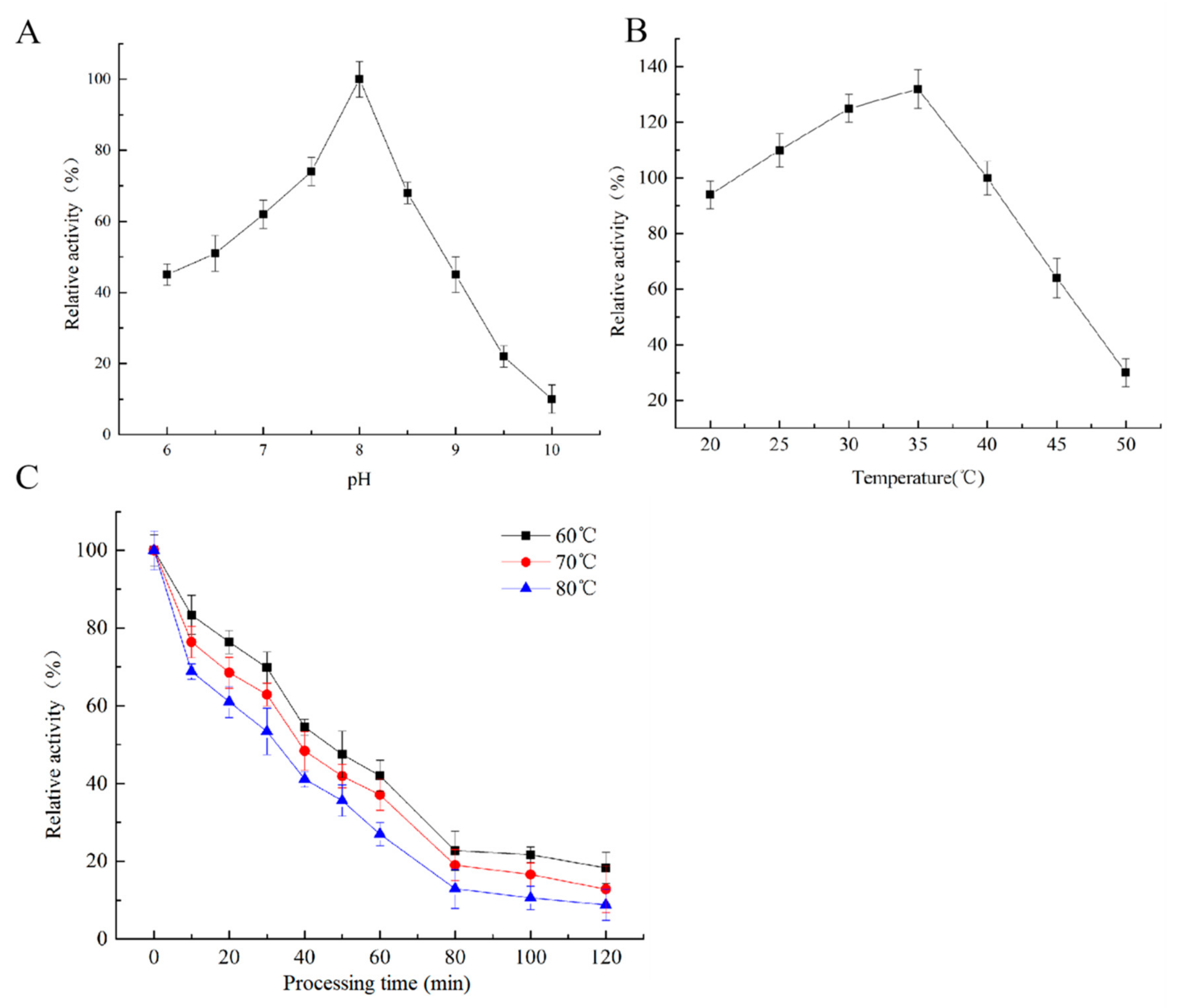
2.2. Characterization Analysis of the YlLip2 Mutant
2.3. Shaking Flask Culture Optimization for YlLip2 Mutant Production
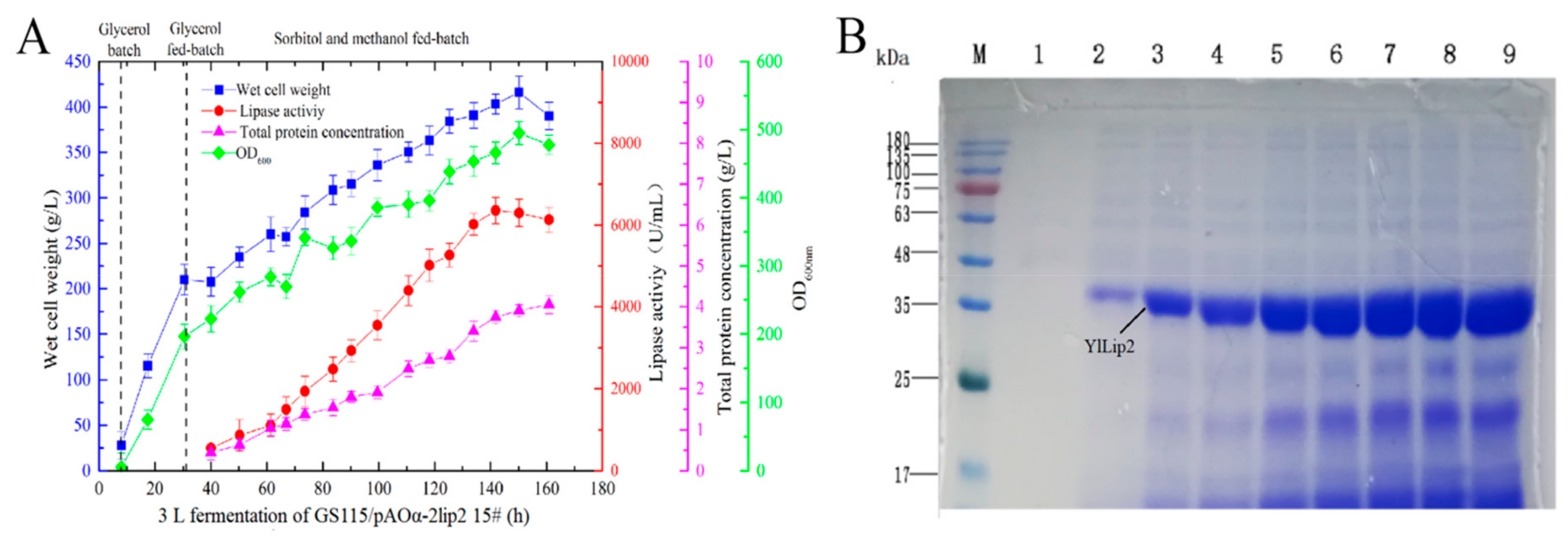
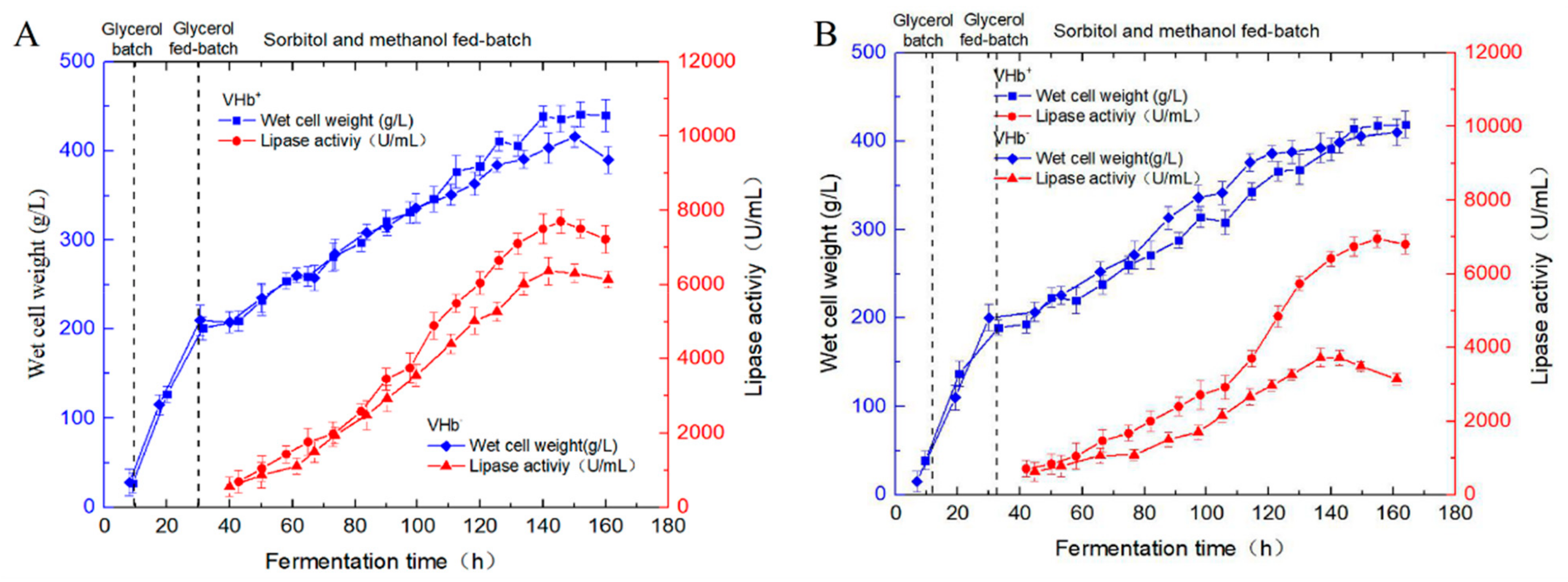
2.4. Fed-Batch Fermentation, SDS-PAGE, and Mass Spectrometry
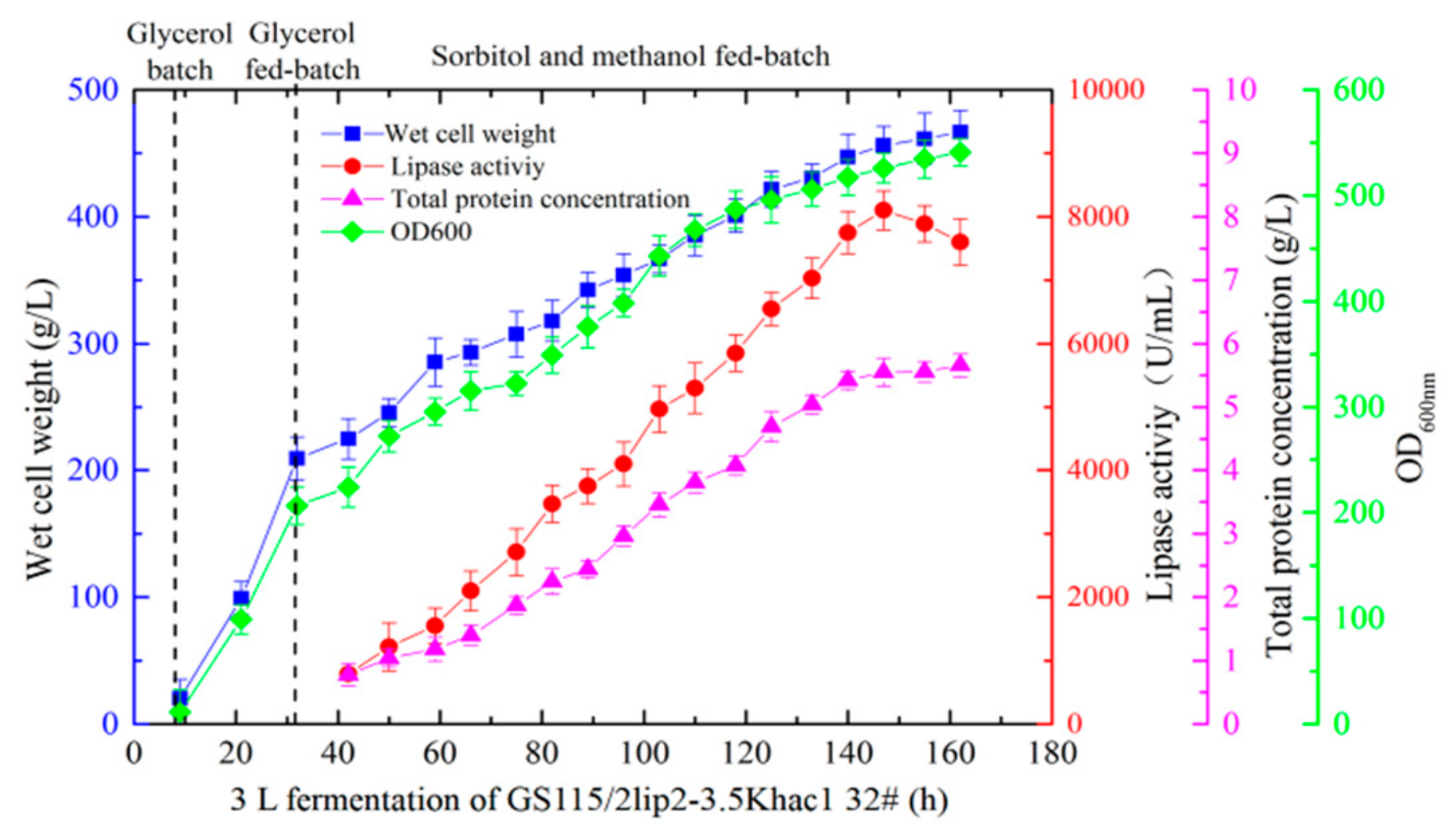
2.5. Effect of Coexpression Hac1p on YlLip2 Secretion
2.6. Influence of Coexpressing vgb on YlLip2 Production
2.7. Coexpression of Hac1p and VHb to Increase YlLip2 Expression Level
3. Discussion
4. Materials and Methods
4.1. Strains, Plasmids, and Media
4.2. Site-Directed Mutagenesis and Vector Construction
4.3. Transformation and Screening
4.4. Shaking Flask Cultures and Optimizing Culture Parameters
4.5. Lipase Activity and Total Protein Concentration
4.6. Biochemical Characterization Analysis
4.7. Determination of Gene Copy Numbers
4.8. Fed-Batch Fermentation
4.9. Biomass and VHb Activity Analysis
4.10. SDS-PAGE and Mass Spectrometry Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| DO | dissolved oxygen |
| ER | endoplasmic reticulum |
| GAP | glyceraldehyde-3-phosphate dehydrogenase gene |
| lip2 | Yarrowia lipolytica lipase 2 gene |
| OD600 | optical density at 600 nm |
| qPCR | quantitative PCR |
| rol | Rhizopus oryzae lipase gene |
| UPR | unfolded protein response |
| VHb | Vitreoscilla hemoglobin |
| vgb | Vitreoscilla hemoglobin gene |
| YNB | yeast nitrogen base with no amino acids |
| YlLip2 | Yarrowia lipolytica lipase 2 |
References
- Liu, T.; Zhao, Y.; Wang, X.; Li, X.; Yan, Y. A novel oriented immobilized lipase on magnetic nanoparticles in reverse micelles system and its application in the enrichment of polyunsaturated fatty acids. Bioresour. Technol. 2013, 132, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Reis, P.; Holmberg, K.; Watzke, H.; Leser, M.E.; Miller, R. Lipases at interfaces: A review. Adv. Colloid Interfac. 2009, 147–148, 237–250. [Google Scholar] [CrossRef] [PubMed]
- Borrelli, G.; Trono, D. Recombinant lipases and phospholipases and their use as biocatalysts for industrial applications. Int. J. Mol. Sci. 2015, 16, 20774–20840. [Google Scholar] [CrossRef] [PubMed]
- Vescovi, V.; Giordano, R.; Mendes, A.; Tardioli, P. Immobilized lipases on functionalized silica particles as potential biocatalysts for the synthesis of fructose oleate in an organic solvent/water system. Molecules 2017, 22, 212. [Google Scholar] [CrossRef]
- Pignede, G.; Wang, H.; Fudalej, F.; Gaillardin, C.; Seman, M.; Nicaud, J.M. Characterization of an extracellular lipase encoded by LIP2 in Yarrowia lipolytica. J. Bacteriol. 2000, 182, 2802–2810. [Google Scholar] [CrossRef]
- Barrera-Rivera, K.; Martínez-Richa, A. Yarrowia lipolytica extracellular lipase Lip2 as biocatalyst for the ring-opening polymerization of ε-caprolactone. Molecules 2017, 22, 1917. [Google Scholar] [CrossRef]
- Fickers, P.; Marty, A.; Nicaud, J.M. The lipases from Yarrowia lipolytica: genetics, production, regulation, biochemical characterization and biotechnological applications. Biotechnol. Adv. 2011, 29, 632–644. [Google Scholar] [CrossRef]
- Yu, M.; Lange, S.; Richter, S.; Tan, T.; Schmid, R.D. High-level expression of extracellular lipase Lip2 from Yarrowia lipolytica in Pichia pastoris and its purification and characterization. Protein Expr. Purif. 2007, 53, 255–263. [Google Scholar] [CrossRef]
- Yu, M.; Wen, S.; Tan, T. Enhancing production of Yarrowia lipolytica lipase Lip2 in Pichia pastoris. Eng. Life Sci. 2010, 10, 458–464. [Google Scholar] [CrossRef]
- Li, L.; Zhang, S.; Wu, W.; Guan, W.; Deng, Z.; Qiao, H. Enhancing thermostability of Yarrowia lipolytica lipase 2 through engineering multiple disulfide bonds and mitigating reduced lipase production associated with disulfide bonds. Enzyme Microb. Technol. 2019, 126, 41–49. [Google Scholar] [CrossRef]
- Wen, S.; Tan, T.; Zhao, H. Improving the thermostability of lipase Lip2 from Yarrowia lipolytica. J. Biotechnol. 2013, 164, 248–253. [Google Scholar] [CrossRef]
- Bordes, F.; Tarquis, L.; Nicaud, J.; Marty, A. Isolation of a thermostable variant of Lip2 lipase from Yarrowia lipolytica by directed evolution and deeper insight into the denaturation mechanisms involved. J. Biotechnol. 2011, 156, 117–124. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, Z. Engineering strategies for enhanced production of protein and bio-products in Pichia pastoris: A review. Biotechnol. Adv. 2018, 36, 182–195. [Google Scholar] [CrossRef]
- Macauley-Patrick, S.; Fazenda, M.L.; McNeil, B.; Harvey, L.M. Heterologous protein production using the Pichia pastoris expression system. Yeast 2005, 22, 249–270. [Google Scholar] [CrossRef]
- Ahmad, M.; Hirz, M.; Pichler, H.; Schwab, H. Protein expression in Pichia pastoris: recent achievements and perspectives for heterologous protein production. Appl. Microbiol. Biot. 2014, 98, 5301–5317. [Google Scholar] [CrossRef]
- Zhang, A.; Luo, J.; Zhang, T.; Pan, Y.; Tan, Y.; Fu, C.; Tu, F. Recent advances on the GAP promoter derived expression system of Pichia pastoris. Mol. Biol. Rep. 2009, 36, 1611–1619. [Google Scholar] [CrossRef]
- Cereghino, J.L.; Cregg, J.M. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol. Rev. 2000, 24, 45–66. [Google Scholar] [CrossRef]
- Sreekrishna, K.; Brankamp, R.G.; Kropp, K.E.; Blankenship, D.T.; Tsay, J.T.; Smith, P.L.; Wierschke, J.D.; Subramaniam, A.; Birkenberger, L.A. Strategies for optimal synthesis and secretion of heterologous proteins in the methylotrophic yeast Pichia pastoris. Gene 1997, 190, 55–62. [Google Scholar] [CrossRef]
- Hohenblum, H.; Gasser, B.; Maurer, M.; Borth, N.; Mattanovich, D. Effects of gene dosage, promoters, and substrates on unfolded protein stress of recombinant Pichia pastoris. Biotechnol. Bioeng. 2004, 85, 367–375. [Google Scholar] [CrossRef]
- Jiao, L.; Zhou, Q.; Su, Z.; Xu, L.; Yan, Y. High-level extracellular production of Rhizopus oryzae lipase in Pichia pastoris via a strategy combining optimization of gene-copy number with co-expression of ERAD-related proteins. Protein Expr. Purif. 2018, 147, 1–12. [Google Scholar] [CrossRef]
- Jiao, L.; Zhou, Q.; Su, Z.; Yan, Y. Efficient heterologous production of Rhizopus oryzae lipase via optimization of multiple expression-related helper proteins. Int. J. Mol. Sci. 2018, 19, 3372. [Google Scholar] [CrossRef] [PubMed]
- Ron, D.; Walter, P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007, 8, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Elvira, M.; Torres-Quiroz, F.; Escamilla-Ayala, A.; Domínguez-Martin, E.; Escalante, R.; Kawasaki, L.; Ongay-Larios, L.; Coria, R. The unfolded protein response pathway in the yeast Kluyveromyces lactis. A comparative view among yeast species. Cells 2018, 7, 106. [Google Scholar] [CrossRef] [PubMed]
- Guerfal, M.; Ryckaert, S.; Jacobs, P.P.; Ameloot, P.; Van Craenenbroeck, K.; Derycke, R.; Callewaert, N. The HAC1 gene from Pichia pastoris: Characterization and effect of its overexpression on the production of secreted, surface displayed and membrane proteins. Microb. Cell Fact. 2010, 9. [Google Scholar] [CrossRef]
- Oh, M.H.; Cheon, S.A.; Kang, H.A.; Kim, J.Y. Functional characterization of the unconventional splicing of Yarrowia lipolytica HAC1 mRNA induced by unfolded protein response. Yeast 2010, 27, 443–452. [Google Scholar] [CrossRef]
- Graf, A.; Gasser, B.; Dragosits, M.; Sauer, M.; Leparc, G.G.; Tuchler, T.; Kreil, D.P.; Mattanovich, D. Novel insights into the unfolded protein response using Pichia pastoris specific DNA microarrays. BMC Genom. 2008, 9, 390. [Google Scholar] [CrossRef]
- Gasser, B.; Maurer, M.; Gach, J.; Kunert, R.; Mattanovich, D. Engineering of Pichia pastoris for improved production of antibody fragments. Biotechnol. Bioeng. 2006, 94, 353–361. [Google Scholar] [CrossRef]
- D’Anjou, M.C.; Daugulis, A.J. A rational approach to improving productivity in recombinant Pichia pastoris fermentation. Biotechnol. Bioeng. 2001, 72, 1–11. [Google Scholar] [CrossRef]
- Zhang, W.; Smith, L.A.; Plantz, B.A.; Schlegel, V.L.; Meagher, M.M. Design of methanol feed control in Pichia pastoris fermentations based upon a growth model. Biotechnol. Prog. 2002, 18, 1392–1399. [Google Scholar] [CrossRef]
- Jahic, M.; Veide, A.; Charoenrat, T.; Teeri, T.; Enfors, S.O. Process technology for production and recovery of heterologous proteins with Pichia pastoris. Biotechnol. Prog. 2006, 22, 1465–1473. [Google Scholar] [CrossRef]
- Wei, X.X.; Chen, G.Q. Applications of the VHb gene vgb for improved microbial fermentation processes. Method. Enzymol. 2008, 436, 273–287. [Google Scholar] [CrossRef]
- Wang, X.; Sun, Y.; Shen, X.; Ke, F.; Zhao, H.; Liu, Y.; Xu, L.; Yan, Y. Intracellular expression of Vitreoscilla hemoglobin improves production of Yarrowia lipolytica lipase LIP2 in a recombinant Pichia pastoris. Enzyme Microb. Tech. 2012, 50, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Jiao, L.; Zhou, Q.; Liu, W.; Yan, Y. New insight into the method of posttransformational vector amplification (PTVA) in Pichia pastoris. J. Microbiol. Meth. 2018, 148, 151–154. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Shen, X.; Zhao, H.; Sun, Y.; Liu, T.; Liu, Y.; Xu, L.; Yan, Y. Combined strategies for the improvement of heterologous expression of a His-tagged Yarrowia lipolytica lipase Lip2 in Pichia pastoris. Afr. J. Biotechnol. 2011, 10, 18503–18512. [Google Scholar]
- Lu, L.; Zhao, M.; Liang, S.; Zhao, L.; Li, D.; Zhang, B. Production and synthetic dyes decolourization capacity of a recombinant laccase from Pichia pastoris. J. Appl. Microbiol. 2009, 107, 1149–1156. [Google Scholar] [CrossRef]
- Sarramegna, V.; Demange, P.; Milon, A.; Talmont, F. Optimizing functional versus total expression of the human μ-opioid receptor in Pichia pastoris. Protein Expr. Purif. 2002, 24, 212–220. [Google Scholar] [CrossRef]
- Yurugi-Kobayashi, T.; Asada, H.; Shiroishi, M.; Shimamura, T.; Funamoto, S.; Katsuta, N.; Ito, K.; Sugawara, T.; Tokuda, N.; Tsujimoto, H.; et al. Comparison of functional non-glycosylated GPCRs expression in Pichia pastoris. Biochem. Biophys. Res. Commun. 2009, 380, 271–276. [Google Scholar] [CrossRef]
- Noseda, D.; Recúpero, M.; Blasco, M.; Ortiz, G.; Galvagno, M.A. Cloning, expression and optimized production in a bioreactor of bovine chymosin B in Pichia (Komagataella) pastoris under AOX1 promoter. Protein Expr. Purif. 2013, 92, 235–244. [Google Scholar] [CrossRef]
- Çalık, P.; Bayraktar, E.; İnankur, B.; Soyaslan, E.Ş.; Şahin, M.; Taşpınar, H.; Açık, E.; Yılmaz, R.; Özdamar, T. Influence of pH on recombinant human growth hormone production by Pichia pastoris. J. Chem. Technol. Biotechnol. 2010, 85, 1628–1635. [Google Scholar] [CrossRef]
- Katakura, Y.; Zhang, W.; Zhuang, G.; Omasa, T.; Kishimoto, M.; Goto, Y.; Suga, K. Effect of methanol concentration on the production of human β2-glycoprotein I domain V by a recombinant Pichia pastoris: A simple system for the control of methanol concentration using a semiconductor gas sensor. J. Ferment. Bioeng. 1998, 86, 482–487. [Google Scholar] [CrossRef]
- Cereghino, G.; Cereghino, J.; Ilgen, C.; Cregg, J. Production of recombinant proteins in fermenter cultures of the yeast Pichia pastoris. Curr. Opin. Biotechnol. 2002, 13, 329–332. [Google Scholar] [CrossRef]
- Zhang, L.; Li, Y.; Wang, Z.; Xia, Y.; Chen, W.; Tang, K. Recent developments and future prospects of Vitreoscilla hemoglobin application in metabolic engineering. Biotechnol. Adv. 2007, 25, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Dikshit, K.L.; Webster, D.A. Cloning, characterization and expression of the bacterial globin gene from Vitreoscilla in Escherichia coli. Gene 1988, 70, 377–386. [Google Scholar] [CrossRef]
- Wu, J.; Wang, S.; Fu, W. Lower temperature cultures enlarge the effects of Vitreoscilla hemoglobin expression on recombinant Pichia pastoris. Int. J. Mol. Sci. 2012, 13, 13212–13226. [Google Scholar] [CrossRef]
- Zhou, Q.; Jiao, L.; Qiao, Y.; Wang, Y.; Xu, L.; Yan, J.; Yan, Y. Overexpression of GRAS Rhizomucor miehei lipase in Yarrowia lipolytica via optimizing promoter, gene dosage and fermentation parameters. J. Biotechnol. 2019, 306, 16–23. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Abad, S.; Kitz, K.; Hormann, A.; Schreiner, U.; Hartner, F.S.; Glieder, A. Real-time PCR-based determination of gene copy numbers in Pichia pastoris. Biotechnol. J. 2010, 5, 413–420. [Google Scholar] [CrossRef]
- Sinha, J.; Plantz, B.A.; Inan, M.; Meagher, M.M. Causes of proteolytic degradation of secreted recombinant proteins produced in methylotrophic yeast Pichia pastoris: Case study with recombinant ovine interferon-tau. Biotechnol. Bioeng. 2005, 89, 102–112. [Google Scholar] [CrossRef]
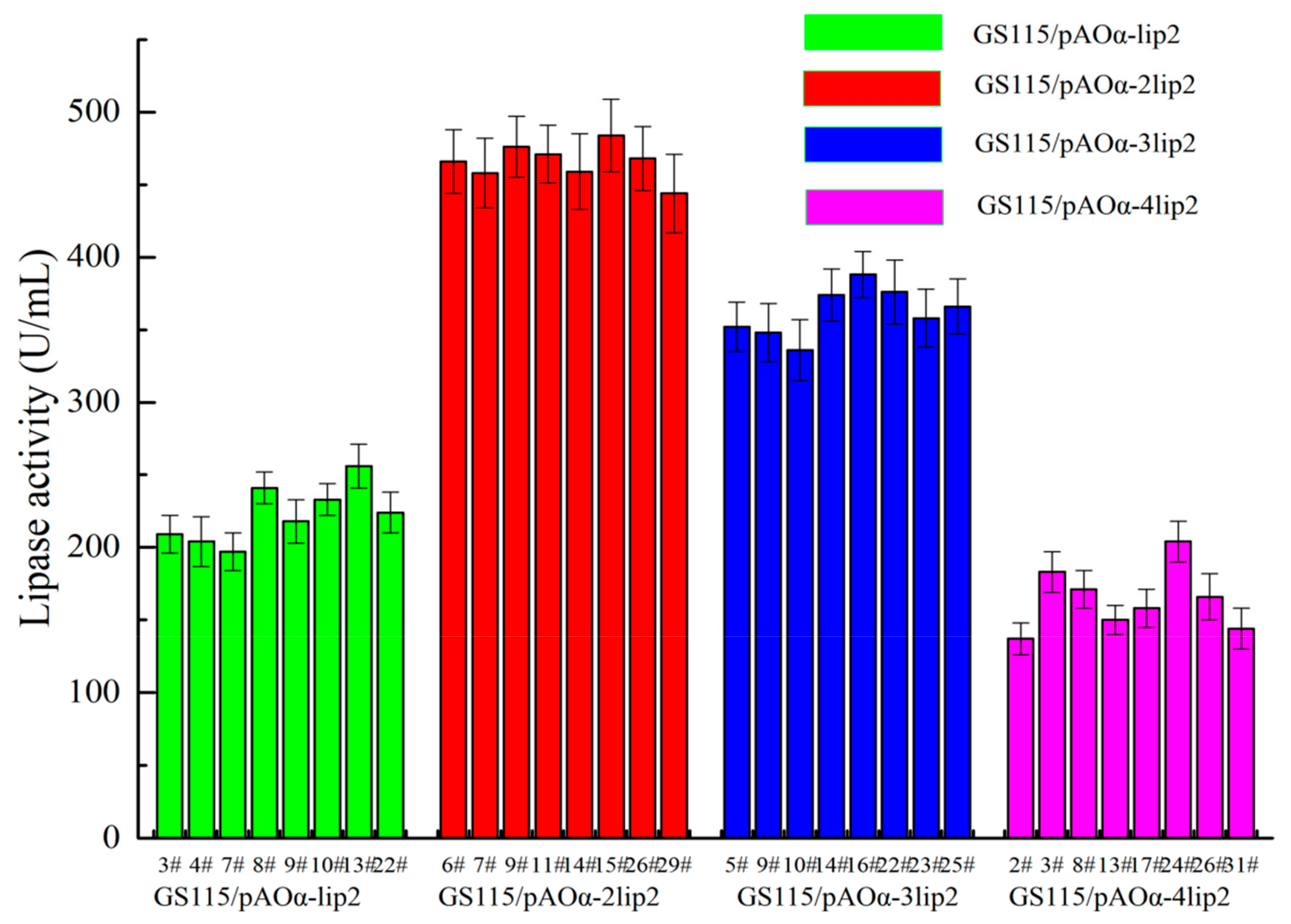

| Recombinant Strains | Genes Copy Number | ||
|---|---|---|---|
| lip2 | hac1 | vgb | |
| GS115 | - | 1 | - |
| GS115/pAOα-lip2 #13 | 1 | - | - |
| GS115/pAOα-2lip2 #15 | 2 | - | - |
| GS115/pAOα-3lip2 #16 | 3 | - | - |
| GS115/pAOα-4lip2 #24 | 4 | - | - |
| GS115/2lip2-3.5Khac1 #32 | 2 | 2 | - |
| GS115/2lip2-ZAvgb #13 | 2 | - | 1 |
| GS115/2lip2-hac1-vgb #21 | 2 | 2 | 1 |
| Host Strains | YlLip2 Mutants | Half-Life Time | Lipase Activity | Reference |
|---|---|---|---|---|
| P. pastoris X-33 | S214 + 69 + 196 + 197 + 210 | 24.76 min at 70 °C | 86 U/mL in shaking flask | [10] |
| P. pastoris X-33 | S214 + 69 + 196 + 197 + 210 + 216 | 101.93 min at 70 °C | 66 U/mL in shaking flask | [10] |
| Y. lipolytica JMY1212 | Cys 244Ala | 36 min at 75 °C | 2.02 U/mL in shaking flask | [12] |
| P. pastoris GS115 | Cys 244Ala | over 30 min at 80 °C | 2630 U/mL in shaking flask | This study |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Q.; Su, Z.; Jiao, L.; Wang, Y.; Yang, K.; Li, W.; Yan, Y. High-Level Production of a Thermostable Mutant of Yarrowia lipolytica Lipase 2 in Pichia pastoris. Int. J. Mol. Sci. 2020, 21, 279. https://doi.org/10.3390/ijms21010279
Zhou Q, Su Z, Jiao L, Wang Y, Yang K, Li W, Yan Y. High-Level Production of a Thermostable Mutant of Yarrowia lipolytica Lipase 2 in Pichia pastoris. International Journal of Molecular Sciences. 2020; 21(1):279. https://doi.org/10.3390/ijms21010279
Chicago/Turabian StyleZhou, Qinghua, Zhixin Su, Liangcheng Jiao, Yao Wang, Kaixin Yang, Wenjuan Li, and Yunjun Yan. 2020. "High-Level Production of a Thermostable Mutant of Yarrowia lipolytica Lipase 2 in Pichia pastoris" International Journal of Molecular Sciences 21, no. 1: 279. https://doi.org/10.3390/ijms21010279
APA StyleZhou, Q., Su, Z., Jiao, L., Wang, Y., Yang, K., Li, W., & Yan, Y. (2020). High-Level Production of a Thermostable Mutant of Yarrowia lipolytica Lipase 2 in Pichia pastoris. International Journal of Molecular Sciences, 21(1), 279. https://doi.org/10.3390/ijms21010279






