Exploiting RAS Nucleotide Cycling as a Strategy for Drugging RAS-Driven Cancers
Abstract
1. Introduction
2. The RAS Gene Family
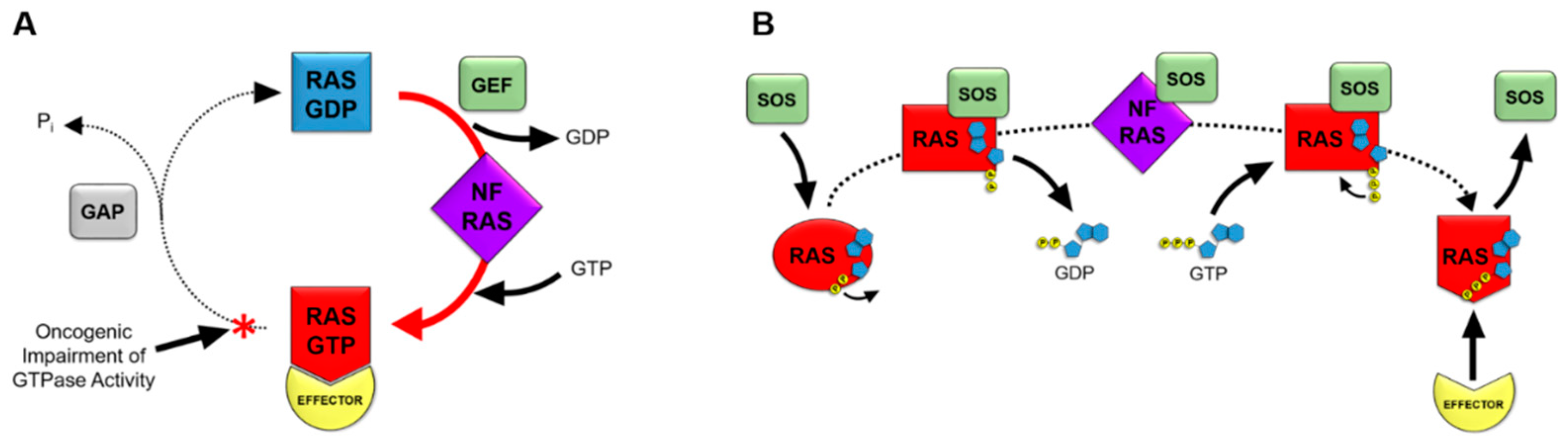
3. RAS Nucleotide Cycling
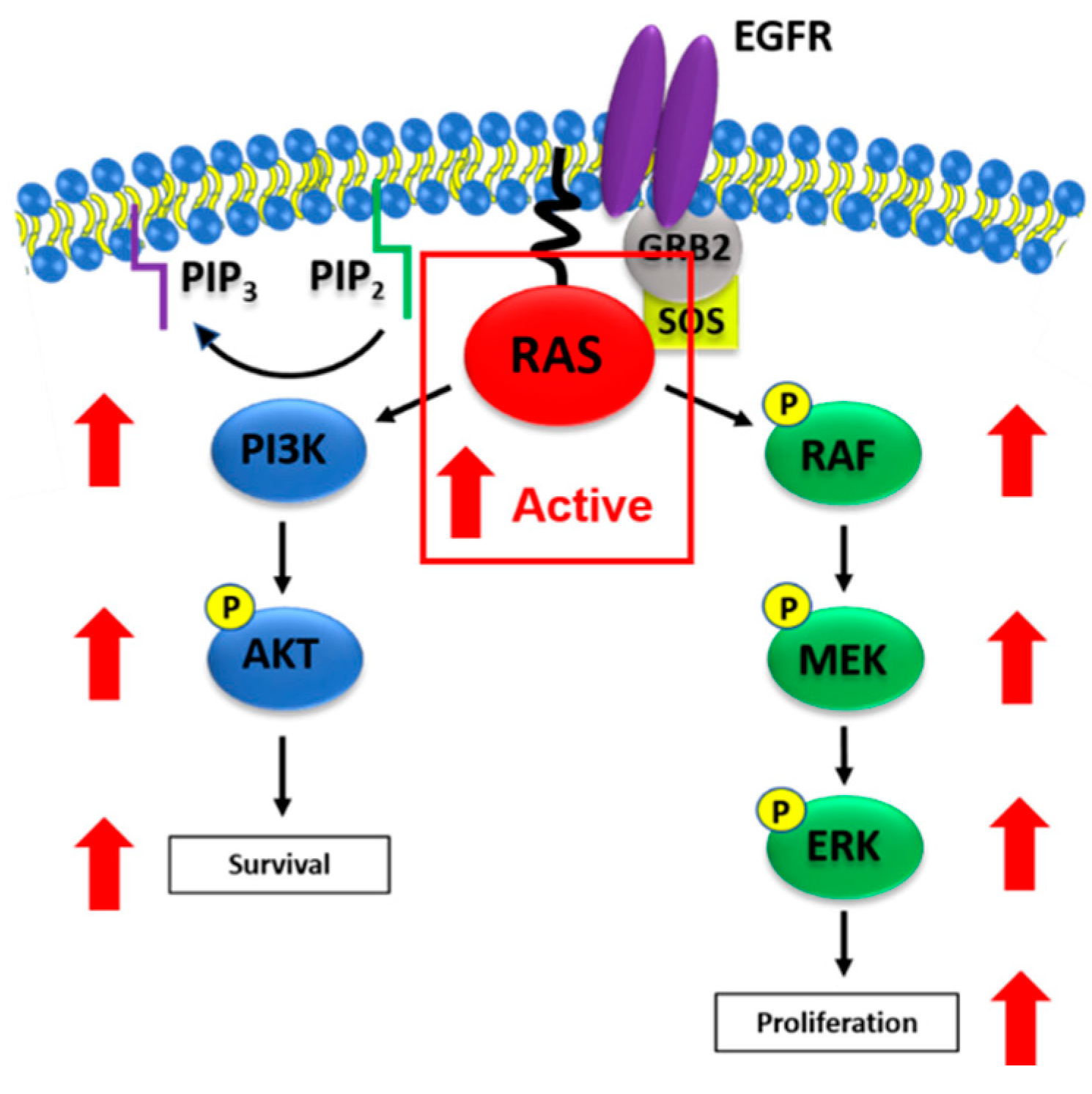
4. The RAS Signal Transduction Pathway
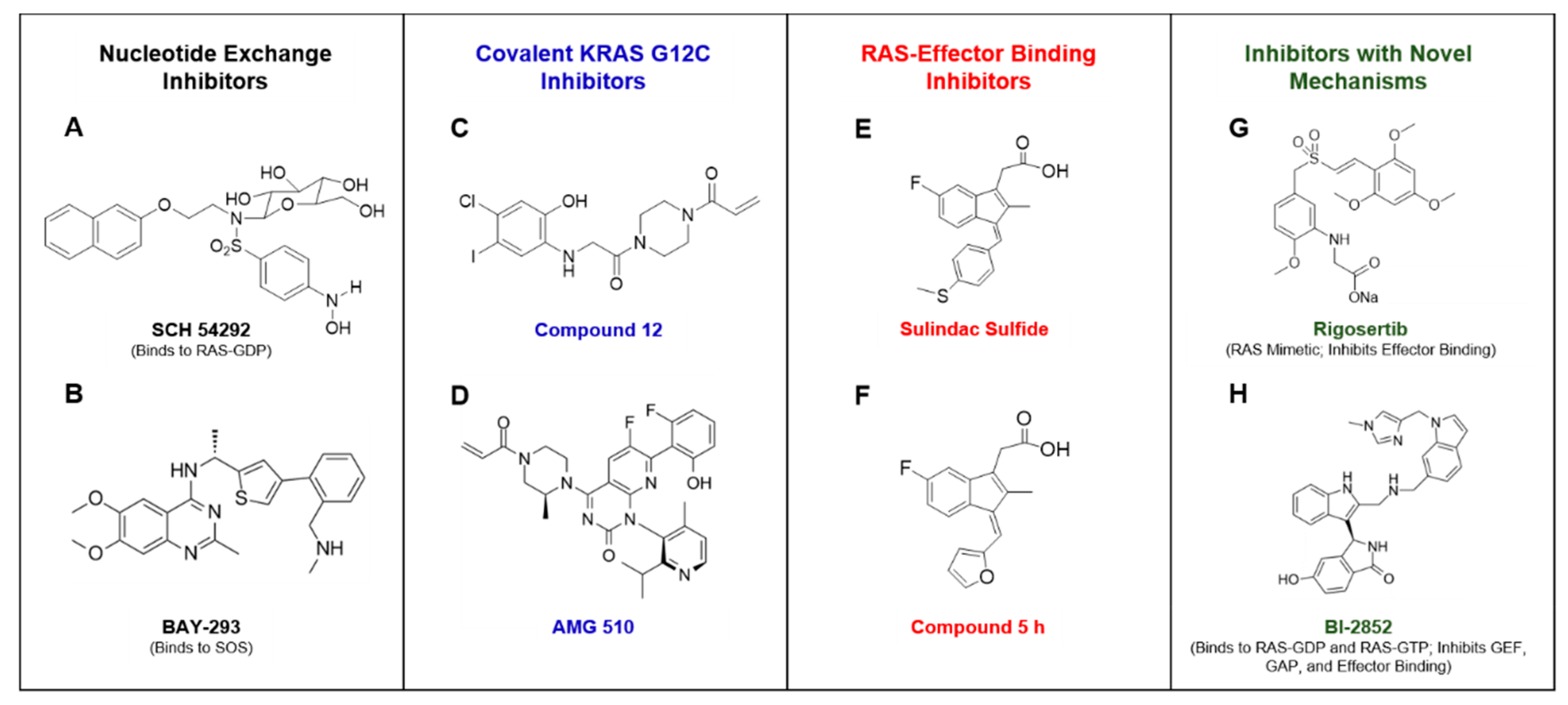
5. Approaches to Inhibit RAS
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Prior, I.A.; Lewis, P.D.; Mattos, C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012, 72, 2457–2467. [Google Scholar] [CrossRef] [PubMed]
- Stephen, A.G.; Esposito, D.; Bagni, R.K.; McCormick, F. Dragging Ras Back in the Ring. Cancer Cell 2014, 25, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Holderfield, M. Efforts to Develop KRAS Inhibitors. Cold Spring Harb. Perspect. Med. 2017, 8, a031864. [Google Scholar] [CrossRef] [PubMed]
- Downward, J. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer 2003, 3, 11–22. [Google Scholar] [CrossRef]
- Nash, G.M.; Gimbel, M.; Shia, J.; Nathanson, D.R.; Ndubuisi, M.I.; Zeng, Z.S.; Kemeny, N.; Patye, P.B. Kras Mutation Correlates with Accelerated Metastatic Progression in Patients with Colorectal Liver Metastases. Ann. Surg. Oncol. 2010, 17, 572–578. [Google Scholar] [CrossRef]
- Harvey, J.J. An Unidentified Virus which causes the Rapid Production of Tumours in Mice. Nature 1964, 204, 1104–1105. [Google Scholar] [CrossRef]
- Kirsten, W.H.; Mayer, L.A. Morphologic Responses to a Murine Erythroblastosis Virus. J. Natl. Cancer Inst. 1967, 39, 311–335. [Google Scholar]
- Cox, A.D.; Der, C.J. Ras History: The Saga Continues. Small GTPases 2010, 1, 2–27. [Google Scholar] [CrossRef]
- Cox, A.D.; Fesik, S.W.; Kimmelman, A.C.; Luo, J.; Der, C.J. Drugging the undruggable RAS: Mission Possible? Nat. Rev. Drug Discov. 2014, 13, 828–851. [Google Scholar] [CrossRef]
- Ostrem, J.M.; Peters, U.; Sos, M.L.; Wells, J.A.; Shokat, K.M. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 2013, 503, 548–551. [Google Scholar] [CrossRef]
- Buhrman, G.; O′connor, C.; Zerbe, B.; Kearney, B.M.; Napoleon, R.; Kovrigina, E.A.; Vajda, S.; Kozakov, D.; Kovrigin, E.L.; Mattos, C. Analysis of Binding Site Hot Spots on the Surface of Ras GTPase. J. Mol. Biol. 2011, 413, 773–789. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, G.A.; Der, C.J.; Rossman, K.L. RAS isoforms and mutations in cancer at a glance. J. Cell Sci. 2016, 129, 1287–1292. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.A.; Mattos, C. The Ras–Membrane Interface: Isoform-Specific Differences in the Catalytic Domain. Mol. Cancer Res. 2015, 13, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Casey, P.J.; Solski, P.A.; Der, C.J.; Buss, J.E. p21ras is modified by a farnesyl isoprenoid. Proc. Natl. Acad. Sci. USA 1989, 86, 8323–8327. [Google Scholar] [CrossRef] [PubMed]
- Farnsworth, C.C.; Gelb, M.H.; Glomset, J.A. Identification of Geranylgeranyl-Modified Proteins in HeLa Cells. Science 1990, 247, 320–322. [Google Scholar] [CrossRef]
- Gutierrez, L.; Magee, A.I.; Marshall, C.J.; Hancock, J.F. Post-translational processing of p21ras is two-step and involves carboxyl-methylation and carboxy-terminal proteolysis. EMBO J. 1989, 8, 1093–1098. [Google Scholar] [CrossRef]
- Wong, K.A.; Russo, A.; Wang, X.; Chen, Y.J.; Lavie, A.; O’Bryan, J.P. A New Dimension to Ras Function: A Novel Role for Nucleotide-Free Ras in Class II Phosphatidylinositol 3-Kinase Beta (PI3KC2β) Regulation. PLoS ONE 2012, 7, e45360. [Google Scholar] [CrossRef]
- John, J.; Sohmen, R.; Feuerstein, J.; Linke, R.; Wittinghofer, A.; Goody, R.S. Kinetics of interaction of nucleotides with nucleotide-free H-ras p21. Biochemistry 1990, 29, 6058–6065. [Google Scholar] [CrossRef]
- Mori, K.; Hata, M.; Neya, S.; Hoshino, T. Common Semiopen Conformations of Mg2+-Free Ras, Rho, Rab, Arf, and Ran Proteins Combined with GDP and Their Similarity with GEF-Bound Forms. J. Am. Chem. Soc. 2005, 127, 15127–15137. [Google Scholar] [CrossRef]
- Boriack-Sjodin, P.A.; Margarit, S.M.; Bar-Sagi, D.; Kuriyan, J. The structural basis of the activation of Ras by Sos. Nature 1998, 394, 337–343. [Google Scholar] [CrossRef]
- Ye, M.; Shima, F.; Muraoka, S.; Liao, J.; Okamoto, H.; Yamamoto, M.; Tamura, A.; Yagi, N.; Ueki, T.; Kataoka, T. Crystal Structure of M-Ras Reveals a GTP-Bound “Off” State Conformation of Ras Family Small GTPases. J. Biol. Chem. 2005, 280, 31267–31275. [Google Scholar] [CrossRef] [PubMed]
- Valencia, A.; Chardin, P.; Wittinghofer, A.; Sander, C. The ras protein family: Evolutionary tree and role of conserved amino acids. Biochemistry 1991, 30, 4637–4648. [Google Scholar] [CrossRef] [PubMed]
- Geyer, M.; Schweins, T.; Herrmann, C.; Prisner, T.; Wittinghofer, A.; Kalbitzer, H.R. Conformational Transitions in p21Ras and in Its Complexes with the Effector Protein Raf-RBD and the GTPase Activating Protein GAP. Biochemistry 1996, 35, 10308–10320. [Google Scholar] [CrossRef]
- Spoerner, M.; Herrmann, C.; Vetter, I.R.; Kalbitzer, H.R.; Wittinghofer, A. Dynamic properties of the Ras switch I region and its importance for binding to effectors. Proc. Natl. Acad. Sci. USA 2001, 98, 4944–4949. [Google Scholar] [CrossRef]
- Shima, F.; Ijiri, Y.; Muraoka, S.; Liao, J.; Ye, M.; Araki, M.; Matsumoto, K.; Yamamoto, N.; Sugimoto, T.; Yoshikawa, Y.; et al. Structural Basis for Conformational Dynamics of GTP-bound Ras Protein. J. Biol. Chem. 2010, 285, 22696–22705. [Google Scholar] [CrossRef]
- Matsumoto, S.; Miyano, N.; Baba, S.; Liao, J.; Kawamura, T.; Tsuda, C.; Takeda, A.; Yamamoto, M.; Kumasaka, T.; Kataoka, T.; et al. Molecular Mechanism for Conformational Dynamics of Ras·GTP Elucidated from In-Situ Structural Transition in Crystal. Sci. Rep. 2016, 6, 25931. [Google Scholar] [CrossRef]
- Cherfils, J.; Zeghouf, M. Regulation of Small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 2013, 93, 269–309. [Google Scholar] [CrossRef]
- Ahmadian, M.R.; Stege, P.; Scheffzek, K.; Wittinghofer, A. Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat. Genet. 1997, 4, 686–689. [Google Scholar] [CrossRef]
- Heesen, H.T.; Gerwert, K.; Schlitter, J. Role of the arginine finger in Ras·RasGAP revealed by QM/MM calculations. FEBS Lett. 2007, 581, 5677–5684. [Google Scholar] [CrossRef]
- Scheffzek, K. The Ras-RasGAP Complex: Structural Basis for GTPase Activation and Its Loss in Oncogenic Ras Mutants. Science 1997, 277, 333–338. [Google Scholar] [CrossRef]
- Kötting, C.; Kallenbach, A.; Suveyzdis, Y.; Wittinghofer, A.; Gerwert, K. The GAP arginine finger movement into the catalytic site of Ras increases the activation entropy. Proc. Natl. Acad. Sci. USA 2008, 105, 6260–6265. [Google Scholar] [CrossRef] [PubMed]
- Hunter, J.C.; Manandhar, A.; Carrasco, M.A.; Gurbani, D.; Gondi, S.; Westover, K.D. Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations. Mol. Cancer Res. 2015, 13, 1325–1335. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, C.; Martin, G.A.; Wittinghofer, A. Quantitative Analysis of the Complex between p21 and the Ras-binding Domain of the Human Raf-1 Protein Kinase. J. Biol. Chem. 1995, 270, 2901–2905. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Viciana, P.; Warne, P.H.; Dhand, R.; Vanhaesebroeck, B.; Gout, I.; Fry, M.J.; Waterfield, M.D.; Downward, J. Phosphatidyl-3-OH Kinase as a Direct Target of Ras. Nature 1994, 370, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, M.C.; Er, E.E.; Blenis, J. The Ras-ERK and PI3K-mTOR pathways: Cross-talk and compensation. Trends Biochem. Sci. 2011, 36, 320–328. [Google Scholar] [CrossRef]
- Misale, S.; Fatherree, J.P.; Cortez, E.; Li, C.; Bilton, S.J.; Timonina, D.; Myers, D.T.; Lee, D.; Gomez-Caraballo, M.; Greenberg, M.; et al. KRAS G12C NSCLC Models Are Sensitive to Direct Targeting of KRAS in Combination with PI3K Inhibition. Clin. Cancer Res. 2018, 25, 796–807. [Google Scholar] [CrossRef]
- Young, A.; Lou, D.; McCormick, F. Oncogenic and Wild-type Ras Play Divergent Roles in the Regulation of Mitogen-Activated Protein Kinase Signaling. Cancer Discov. 2013, 3, 112–123. [Google Scholar] [CrossRef]
- Baines, A.T.; Xu, D.; Der, C.J. Inhibition of Ras for cancer treatment: The search continues. Future Med. Chem. 2011, 3, 1787–1808. [Google Scholar] [CrossRef]
- Lobell, R.B.; Liu, N.; Buser, C.A.; Davide, J.P.; DePuy, E.; Hamilton, K.; Koblan, K.S.; Lee, Y.; Mosser, S.; Motzel, S.L.; et al. Preclinical and clinical pharmacodynamic assessment of L-778,123, a dual inhibitor of farnesyl:protein transferase and geranylgeranyl:protein transferase type-I. Mol. Cancer Ther. 2002, 1, 747–758. [Google Scholar]
- Desolms, S.J.; Ciccarone, T.M.; MacTough, S.C.; Shaw, A.W.; Buser, C.A.; Ellis-Hutchings, M.; Fernandes, C.; Hamilton, K.A.; Huber, H.E.; Kohl, N.E.; et al. Dual Protein Farnesyltransferase−Geranylgeranyltransferase-I Inhibitors as Potential Cancer Chemotherapeutic Agents. J. Med. Chem. 2003, 46, 2973–2984. [Google Scholar] [CrossRef]
- Raponi, M.; Winkler, H.; Dracopoli, N.C. KRAS mutations predict response to EGFR inhibitors. Curr. Opin. Pharmacol. 2008, 8, 413–418. [Google Scholar] [CrossRef] [PubMed]
- Knickelbein, K.; Zhang, L. Mutant KRAS as a critical determinant of the therapeutic response of colorectal cancer. Genes Dis. 2015, 2, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Gysin, S.; Salt, M.; Young, A.; McCormick, F. Therapeutic Strategies for Targeting Ras Proteins. Genes Cancer 2011, 2, 359–372. [Google Scholar] [CrossRef] [PubMed]
- Taveras, A.; Remiszewski, S.; Doll, R.; Cesarz, D.; Huang, E.; Kirschmeier, P.; Pramanik, B.; Snow, M.; Wang, Y.S.; Del Rosario, J.; et al. Ras oncoprotein inhibitors: The discovery of potent, ras nucleotide exchange inhibitors and the structural determination of a drug-protein complex. Bioorg. Med. Chem. 1997, 5, 125–133. [Google Scholar] [CrossRef]
- Maurer, T.; Garrenton, L.S.; Oh, A.; Pitts, K.; Anderson, D.J.; Skelton, N.J.; Fauber, B.P.; Pan, B.; Malek, S.; Stokoe, D.; et al. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc. Natl. Acad. Sci. USA 2012, 109, 5299–5304. [Google Scholar] [CrossRef]
- Sun, Q.; Burke, J.P.; Phan, J.; Burns, M.C.; Olejniczak, E.T.; Waterson, A.G.; Lee, T.; Rossanese, O.W.; Fesik, S.W. Discovery of Small Molecules that Bind to K-Ras and Inhibit Sos-Mediated Activation. Angew. Chem. 2012, 124, 6244–6247. [Google Scholar] [CrossRef]
- Winter, J.J.G.; Anderson, M.; Blades, K.; Brassington, C.; Breeze, A.L.; Chresta, C.; Embrey, K.; Fairley, G.; Faulder, P.; Finlay, M.R.V.; et al. Small Molecule Binding Sites on the Ras:SOS Complex Can Be Exploited for Inhibition of Ras Activation. J. Med. Chem. 2015, 58, 2265–2274. [Google Scholar] [CrossRef]
- Hillig, R.C.; Sautier, B.; Schroeder, J.; Moosmayer, D.; Hilpmann, A.; Stegmann, C.M.; Werbeck, N.D.; Briem, H.; Boemer, U.; Weiske, J.; et al. Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS–SOS1 interaction. Proc. Natl. Acad. Sci. USA 2019, 116, 2551–2560. [Google Scholar] [CrossRef]
- Ruess, D.A.; Heynen, G.J.; Ciecielski, K.J.; Ai, J.; Berninger, A.; Kabacaoglu, D.; Görgülü, K.; Dantes, Z.; Wörmann, S.M.; Diakopoulos, K.N.; et al. Po-201 mutant kras-driven cancers depend on ptpn11/shp2 phosphatase. Nat. Med. 2018, 24, 954–960. [Google Scholar] [CrossRef]
- Lu, H.; Liu, C.; Velazquez, R.; Wang, H.; Dunkl, L.M.; Kazic-Legueux, M.; Haberkorn, A.; Billy, E.; Manchado, E.; Brachmann, S.M.; et al. SHP2 Inhibition Overcomes RTK-Mediated Pathway Re-Activation in KRAS Mutant Tumors Treated with MEK Inhibitors. Mol. Cancer Ther. 2019, 18, 1323–1334. [Google Scholar] [CrossRef]
- Hong, L.; Guo, Y.; Basuray, S.; Agola, J.O.; Romero, E.; Simpson, D.S.; Schroeder, C.E.; Simons, P.; Waller, A.; Garcia, M.; et al. A Pan-GTPase Inhibitor as a Molecular Probe. PLoS ONE 2015, 10, e0134317. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.; Kamada, Y.; Sameshima, T.; Yaguchi, M.; Niida, A.; Sasaki, S.; Miwa, M.; Ohkubo, S.; Sakamoto, J.I.; Kamaura, M.; et al. K-Ras(G12D)-selective inhibitory peptides generated by random peptide T7 phage display technology. Biochem. Biophys. Res. Commun. 2017, 484, 605–611. [Google Scholar] [CrossRef] [PubMed]
- Lito, P.; Solomon, M.; Hansen, R.; Li, L.S.; Rosen, N. Abstract LB-071: Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science 2016, 351, 604–608. [Google Scholar] [CrossRef] [PubMed]
- Patricelli, M.P.; Janes, M.R.; Li, L.S.; Hansen, R.; Peters, U.; Kessler, L.V.; Chen, Y.; Kucharski, J.M.; Feng, J.; Ely, T.; et al. Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State. Cancer Discov. 2016, 6, 316–329. [Google Scholar] [CrossRef]
- Lim, S.M.; Westover, K.D.; Ficarro, S.B.; Harrison, R.A.; Choi, H.G.; Pacold, M.E.; Carrasco, M.; Hunter, J.; Kim, N.D.; Xie, T.; et al. Therapeutic Targeting of Oncogenic K-Ras by a Covalent Catalytic Site Inhibitor. Angew. Chem. Int. Ed. Engl. 2014, 53, 199–204. [Google Scholar] [CrossRef]
- Xiong, Y.; Lu, J.; Hunter, J.; Li, L.; Scott, D.; Choi, H.G.; Lim, S.M.; Manandhar, A.; Gondi, S.; Sim, T.; et al. Covalent Guanosine Mimetic Inhibitors of G12C KRAS. ACS Med. Chem. Lett. 2017, 8, 61–66. [Google Scholar] [CrossRef]
- Research AA for C. AMG 510 First to Inhibit “Undruggable” KRAS. 2019. Available online: https://cancerdiscovery.aacrjournals.org/content/early/2019/06/11/2159-8290.CD-NB2019-073 (accessed on 18 September 2019).
- O’Bryan, J.P. Pharmacological Targeting of RAS: Recent Success with Direct Inhibitors. Pharmacol. Res. 2019, 139, 503–511. [Google Scholar] [CrossRef]
- Schmitz, A.L.; Schrage, R.; Gaffal, E.; Charpentier, T.H.; Wiest, J.; Hiltensperger, G.; Morschel, J.; Hennen, S.; Häußler, D.; Horn, V.; et al. A cell-permeable inhibitor to trap Gαq proteins in the empty pocket conformation. Chem. Biol. 2014, 21, 890–902. [Google Scholar] [CrossRef]
- Müller, M.P.; Jeganathan, S.; Heidrich, A.; Campos, J.; Goody, R.S. Nucleotide based covalent inhibitors of KRas can only be efficient in vivo if they bind reversibly with GTP-like affinity. Sci. Rep. 2017, 7, 3687. [Google Scholar] [CrossRef]
- Keeton, A.B.; Salter, E.A.; Piazza, G.A. The RAS-Effector Interaction as a Drug Target. Cancer Res. 2017, 77, 221–226. [Google Scholar] [CrossRef]
- Kato-Stankiewicz, J.; Hakimi, I.; Zhi, G.; Zhang, J.; Serebriiskii, I.; Guo, L.; Edamatsu, H.; Koide, H.; Menon, S.; Eckl, R.; et al. Inhibitors of Ras/Raf-1 interaction identified by two-hybrid screening revert Ras-dependent transformation phenotypes in human cancer cells. Proc. Natl. Acad. Sci. USA 2002, 99, 14398–14403. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Rabbitts, T.H. Interfering with RAS–effector protein interactions prevent RAS-dependent tumour initiation and causes stop–start control of cancer growth. Oncogene 2010, 29, 6064–6070. [Google Scholar] [CrossRef] [PubMed]
- Trinh, T.B.; Upadhyaya, P.; Qian, Z.; Pei, D. Discovery of a Direct Ras Inhibitor by Screening a Combinatorial Library of Cell-Permeable Bicyclic Peptides. ACS Comb. Sci. 2016, 18, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Welsch, M.E.; Kaplan, A.; Chambers, J.M.; Stokes, M.E.; Bos, P.H.; Zask, A.; Zhang, Y.; Sanchez-Martin, M.; Badgley, M.A.; Huang, C.S.; et al. Multivalent Small-Molecule Pan-RAS Inhibitors. Cell 2017, 168, 878–889. [Google Scholar] [CrossRef] [PubMed]
- Athuluri-Divakar, S.K.; Carpio, R.V.D.; Dutta, K.; Baker, S.J.; Cosenza, S.C.; Basu, I.; Gupta, Y.K.; Reddy, M.R.; Ueno, L.; Hart, J.R.; et al. A Small Molecule RAS-Mimetic Disrupts RAS Association with Effector Proteins to Block Signaling. Cell 2016, 165, 643–655. [Google Scholar] [CrossRef] [PubMed]
- McGee, J.H.; Shim, S.Y.; Lee, S.J.; Swanson, P.K.; Jiang, S.Y.; Durney, M.A.; Verdine, G.L. Exceptionally High-Affinity Ras Binders That Remodel Its Effector Domain. J. Biol. Chem. 2018, 293, 3265–3280. [Google Scholar] [CrossRef]
- Cruz-Migoni, A.; Canning, P.; Quevedo, C.E.; Bataille, C.J.R.; Bery, N.; Miller, A.; Russell, A.J.; Phillips, S.E.V.; Carr, S.B.; Rabbitts, T.H. Structure-based development of new RAS-effector inhibitors from a combination of active and inactive RAS-binding compounds. Proc. Natl. Acad. Sci. USA 2019, 116, 2545–2550. [Google Scholar] [CrossRef]
- Herrmann, C.; Block, C.; Geisen, C.; Haas, K.; Weber, C.; Winde, G.; Möröy, T.; Müller, O. Sulindac sulfide inhibits Ras signaling. Oncogene 1998, 17, 1769–1776. [Google Scholar] [CrossRef]
- Piazza, G.A.; Rahm, A.L.; Krutzsch, M.; Sperl, G.; Paranka, N.S.; Gross, P.H.; Brendel, K.; Burt, R.W.; Alberts, D.S.; Pamukcu, R. Antineoplastic drugs sulindac sulfide and sulfone inhibit cell growth by inducing apoptosis. Cancer Res. 1995, 55, 3110–3116. [Google Scholar]
- Piazza, G.A.; Rahm, A.K.; Finn, T.S.; Fryer, B.H.; Li, H.; Stoumen, A.L.; Pamukcu, R.; Ahnen, D.J. Apoptosis primarily accounts for the growth-inhibitory properties of sulindac metabolites and involves a mechanism that is independent of cyclooxygenase inhibition, cell cycle arrest, and p53 induction. Cancer Res. 1997, 57, 2452–2459. [Google Scholar]
- Lawson, K.R.; Ignatenko, N.A.; Piazza, G.A.; Cui, H.; Gerner, E.W. Influence of K-ras activation on the survival responses of Caco-2 cells to the chemopreventive agents sulindac and difluoromethylornithine. Cancer Epidemiol. Biomark. Prev. 2000, 9, 1155–1162. [Google Scholar]
- Piazza, G.A.; Keeton, A.B.; Tinsley, H.N.; Gary, B.D.; Whitt, J.D.; Mathew, B.; Thaiparambil, J.; Coward, L.; Gorman, G.; Li, Y.; et al. A novel sulindac derivative that does not inhibit cyclooxygenases but potently inhibits colon tumor cell growth and induces apoptosis with antitumor activity. Cancer Prev. Res. 2009, 2, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Thompson, H.J.; Jiang, C.; Lu, J.; Mehta, R.G.; Piazza, G.A.; Paranka, N.S.; Pamukcu, R.; Ahnen, D.J. Sulfone metabolite of sulindac inhibits mammary carcinogenesis. Cancer Res. 1997, 57, 267–271. [Google Scholar] [PubMed]
- Karaguni, I.M.; Glüsenkamp, K.H.; Langerak, A.; Geisen, C.; Ullrich, V.; Winde, G.; Möröy, T.; Müller, O. New indene-derivatives with anti-proliferative properties. Bioorg. Med. Chem. Lett. 2002, 12, 709–713. [Google Scholar] [CrossRef]
- Waldmann, H.; Karaguni, I.M.; Carpintero, M.; Gourzoulidou, E.; Herrmann, C.; Brockmann, C.; Oschkinat, H.; Müller, O. Sulindac-Derived Ras Pathway Inhibitors Target the Ras–Raf Interaction and Downstream Effectors in the Ras Pathway. Angew. Chem. Int. Ed. 2004, 43, 454–458. [Google Scholar] [CrossRef]
- Kessler, D.; Gmachl, M.; Mantoulidis, A.; Martin, L.J.; Zoephel, A.; Mayer, M.; Gollner, A.; Covini, D.; Fischer, S.; Gerstberger, T.; et al. Drugging an undruggable pocket on KRAS. Proc. Natl. Acad. Sci. USA 2019, 116, 15823–15829. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mattox, T.E.; Chen, X.; Maxuitenko, Y.Y.; Keeton, A.B.; Piazza, G.A. Exploiting RAS Nucleotide Cycling as a Strategy for Drugging RAS-Driven Cancers. Int. J. Mol. Sci. 2020, 21, 141. https://doi.org/10.3390/ijms21010141
Mattox TE, Chen X, Maxuitenko YY, Keeton AB, Piazza GA. Exploiting RAS Nucleotide Cycling as a Strategy for Drugging RAS-Driven Cancers. International Journal of Molecular Sciences. 2020; 21(1):141. https://doi.org/10.3390/ijms21010141
Chicago/Turabian StyleMattox, Tyler E., Xi Chen, Yulia Y. Maxuitenko, Adam B. Keeton, and Gary A. Piazza. 2020. "Exploiting RAS Nucleotide Cycling as a Strategy for Drugging RAS-Driven Cancers" International Journal of Molecular Sciences 21, no. 1: 141. https://doi.org/10.3390/ijms21010141
APA StyleMattox, T. E., Chen, X., Maxuitenko, Y. Y., Keeton, A. B., & Piazza, G. A. (2020). Exploiting RAS Nucleotide Cycling as a Strategy for Drugging RAS-Driven Cancers. International Journal of Molecular Sciences, 21(1), 141. https://doi.org/10.3390/ijms21010141




