The Possible Role of the Microbiota-Gut-Brain-Axis in Autism Spectrum Disorder
Abstract
:1. Introduction
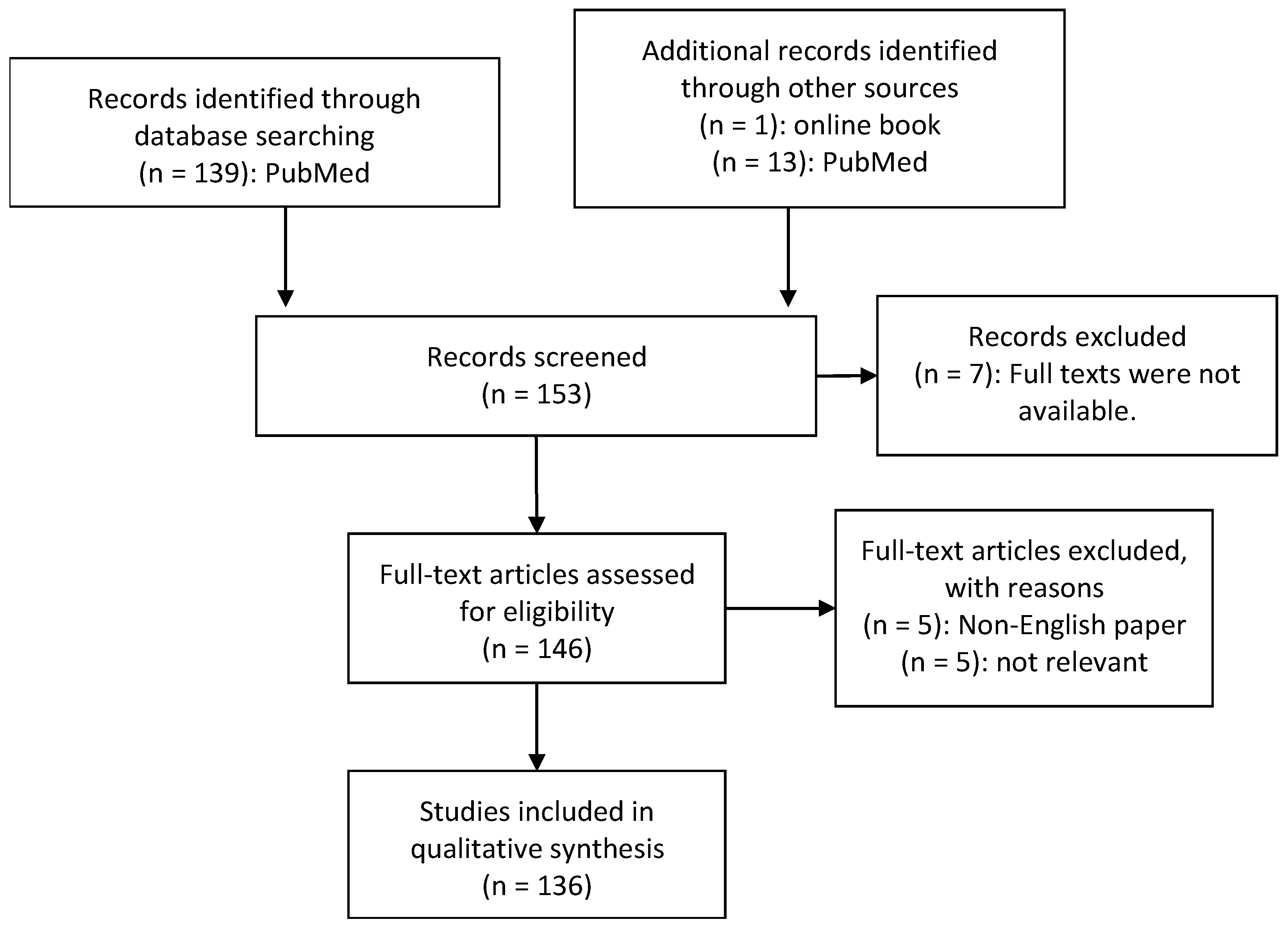
2. Material and Methods
- -
- Diagnosis of ASD by medical experts
- -
- Human studies of the microbiome or metabolome in autistic individuals
- -
- ASD individuals were compared to a matched control group
- -
- Collection and analysis of at least one of the following biomaterials: GI biopsies, faecal, urinary or blood samples
- -
- Analysis of bacterial genome by sequencing
- -
- Published in a peer-reviewed article
- -
- Availability of the full text publication
- -
- Availability of the paper in English
3. Comorbidities in Autism Spectrum Disorder
3.1. Definition
3.2. Gastrointestinal Symptoms
3.3. Increased Gut Permeability
3.4. Alterations in the Brain
3.5. Mitochondrial Dysfunction
4. Factors Influencing the Gut Microbiota
4.1. Definitions and Facts
4.2. Developmemt and Disruptions of Microbial Colonisation in Autism
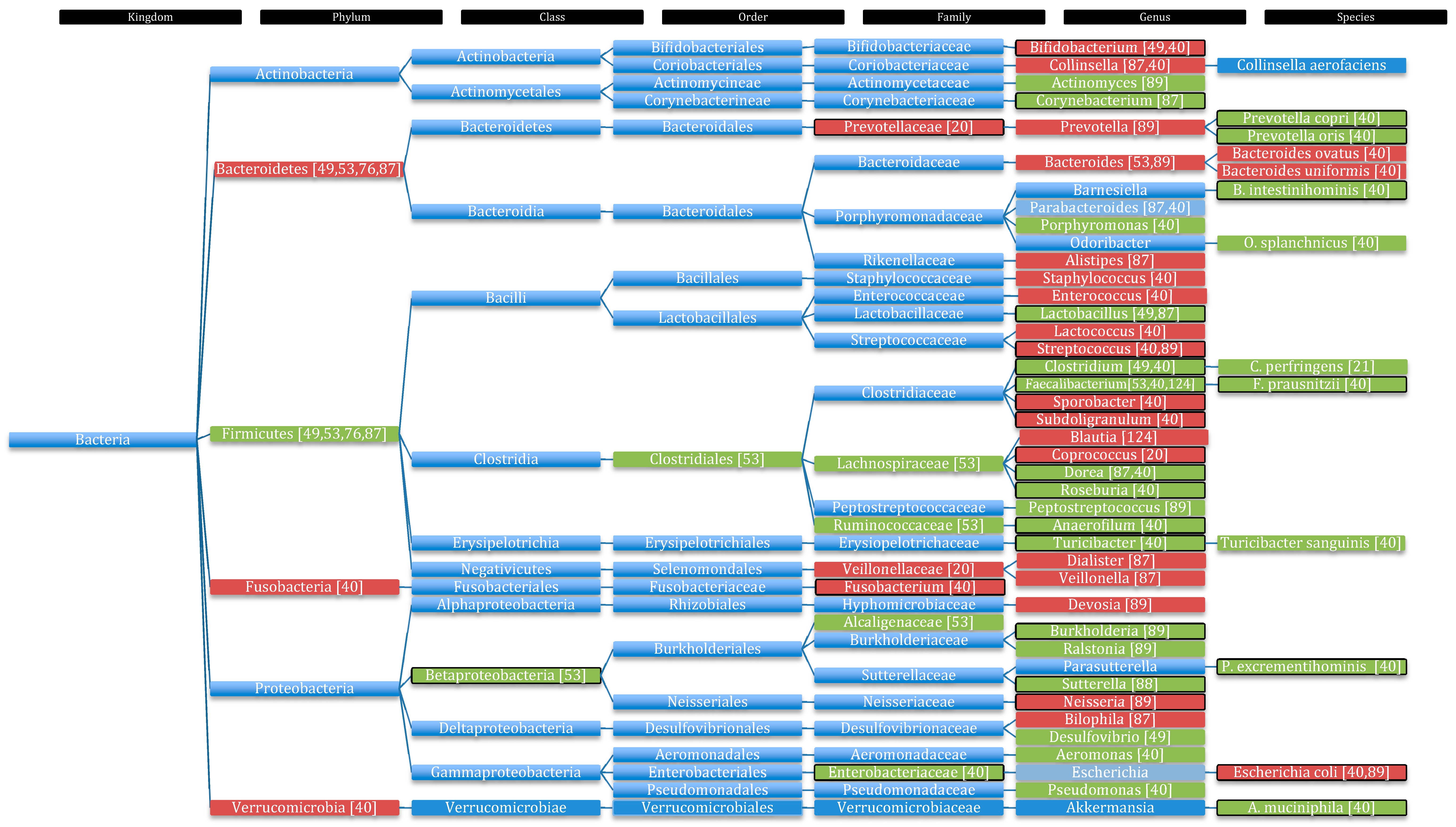
5. Alterations in Microbial Composition and Metabolic Profile in Autistic Individuals
5.1. Changes in Microbial Composition in Autistic Children
5.2. Altered Concentrations of Metabolites and Their Functional Consequences
5.2.1. Urinary Analysis
5.2.2. Blood Analysis
5.2.3. Short Chain Fatty Acids
5.3. Other Etiologies for ASD
6. Possible Therapeutic Measures Acting on the Microbiota
6.1. Prebiotics
6.2. Probiotics
6.3. Faecal Microbiota Transplantation
6.4. Other Treatment Strategies
7. Discussion and Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ASD | Autism Spectrum disorder |
| BBB | Blood-brain-barrier |
| BDNF | Brain derived neurotropic factor |
| CLDN | Claudine |
| DNA | Deoxyribonucleic acid |
| ETC | Electron transport chain |
| FMT | Faecal Microbiota transplantation |
| GABA | Gamma-amino-butyric acid |
| GI | Gastrointestinal |
| GLT2 | Glucose transporter 2 |
| HPA | Hypothalamus-pituitary-adrenal |
| MIA | Maternal immune activation |
| NADH | Nicotineamide-adenine-dinucleotide |
| NADPH | Nicotineamide-adenine-dinukleotide-phosphate |
| poly(I:C) | Polyinosinic:polycytidylic acid |
| ROS | Reactive oxygen species |
| SCFAs | Short chain fatty acids |
| SGLT | Sodium dependent glucose transporter 1 |
| spp | species |
| TPP | Thiamine-pyrophosphate |
References
- Santocchi, E.; Guiducci, L.; Fulceri, F.; Billeci, L.; Buzzigoli, E.; Apicella, F.; Calderoni, S.; Grossi, E.; Morales, M.A.; Muratori, F. Gut to brain interaction in Autism Spectrum Disorders: a randomized controlled trial on the role of probiotics on clinical, biochemical and neurophysiological parameters. BMC Psychiatry 2016, 16, 183. [Google Scholar] [CrossRef]
- Redinbo, M.R. The microbiota, chemical symbiosis, and human disease. J. Mol. Biol. 2014, 426, 3877–3891. [Google Scholar] [CrossRef]
- Viggiano, D.; Ianiro, G.; Vanella, G.; Bibbo, S.; Bruno, G.; Simeone, G.; Mele, G. Gut barrier in health and disease: focus on childhood. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 1077–1085. [Google Scholar] [PubMed]
- Rao, M.; Gershon, M.D. The bowel and beyond: the enteric nervous system in neurological disorders. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, E.Y.; McBride, S.W.; Hsien, S.; Sharon, G.; Hyde, E.R.; McCue, T.; Codelli, J.A.; Chow, J.; Reisman, S.E.; Petrosino, J.F.; et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 2013, 155, 1451–1463. [Google Scholar] [CrossRef] [PubMed]
- Dinan, T.G.; Cryan, J.F. The Microbiome-Gut-Brain Axis in Health and Disease. Gastroenterol. Clin. North. Am. 2017, 46, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Muszer, M.; Noszczynska, M.; Kasperkiewicz, K.; Skurnik, M. Human Microbiome: When a Friend Becomes an Enemy. Arch. Immunol Ther. Exp. (Warsz) 2015, 63, 287–298. [Google Scholar] [CrossRef] [PubMed]
- Petra, A.I.; Panagiotidou, S.; Hatziagelaki, E.; Stewart, J.M.; Conti, P.; Theoharides, T.C. Gut-Microbiota-Brain Axis and Its Effect on Neuropsychiatric Disorders With Suspected Immune Dysregulation. Clin. Ther. 2015, 37, 984–995. [Google Scholar] [CrossRef]
- Quigley, E.M. Basic Definitions and Concepts: Organization of the Gut Microbiome. Gastroenterol. Clin. North. Am. 2017, 46, 1–8. [Google Scholar] [CrossRef]
- Lichtman, J.S.; Sonnenburg, J.L.; Elias, J.E. Monitoring host responses to the gut microbiota. ISME J. 2015, 9, 1908–1915. [Google Scholar] [CrossRef] [PubMed]
- Latalova, K.; Hajda, M.; Prasko, J. Can gut microbes play a role in mental disorders and their treatment? Psychiatr. Danub. 2017, 29, 28–30. [Google Scholar] [CrossRef]
- Stilling, R.M.; Dinan, T.G.; Cryan, J.F. Microbial genes, brain & behaviour - epigenetic regulation of the gut-brain axis. Genes Brain Behav. 2014, 13, 69–86. [Google Scholar] [CrossRef]
- Marler, S.; Ferguson, B.J.; Lee, E.B.; Peters, B.; Williams, K.C.; McDonnell, E.; Macklin, E.A.; Levitt, P.; Margolis, K.G.; Beversdorf, D.Q.; et al. Association of Rigid-Compulsive Behavior with Functional Constipation in Autism Spectrum Disorder. J. Autism. Dev. Disord. 2017, 47, 1673–1681. [Google Scholar] [CrossRef]
- Fulceri, F.; Morelli, M.; Santocchi, E.; Cena, H.; Del Bianco, T.; Narzisi, A.; Calderoni, S.; Muratori, F. Gastrointestinal symptoms and behavioral problems in preschoolers with Autism Spectrum Disorder. Dig. Liver Dis. 2016, 48, 248–254. [Google Scholar] [CrossRef]
- Iovene, M.R.; Bombace, F.; Maresca, R.; Sapone, A.; Iardino, P.; Picardi, A.; Marotta, R.; Schiraldi, C.; Siniscalco, D.; Serra, N.; et al. Intestinal Dysbiosis and Yeast Isolation in Stool of Subjects with Autism Spectrum Disorders. Mycopathologia 2017, 182, 349–363. [Google Scholar] [CrossRef]
- Berding, K.; Donovan, S.M. Microbiome and nutrition in autism spectrum disorder: current knowledge and research needs. Nutr. Rev. 2016, 74, 723–736. [Google Scholar] [CrossRef] [PubMed]
- Buie, T. Potential Etiologic Factors of Microbiome Disruption in Autism. Clin. Ther. 2015, 37, 976–983. [Google Scholar] [CrossRef] [PubMed]
- Reddy, B.L.; Saier, M.H. Autism and our intestinal microbiota. J. Mol. Microbiol. Biotechnol. 2015, 25, 51–55. [Google Scholar] [CrossRef]
- Wakefield, A.J. The gut-brain axis in childhood developmental disorders. J. Pediatr Gastroenterol Nutr. 2002, 34 (Suppl. 1), S14–S17. [Google Scholar] [CrossRef]
- Kang, D.W.; Park, J.G.; Ilhan, Z.E.; Wallstrom, G.; Labaer, J.; Adams, J.B.; Krajmalnik-Brown, R. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS ONE 2013, 8, e68322. [Google Scholar] [CrossRef]
- Finegold, S.M.; Summanen, P.H.; Downes, J.; Corbett, K.; Komoriya, T. Detection of Clostridium perfringens toxin genes in the gut microbiota of autistic children. Anaerobe 2017, 45, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Zeidán Chuliá, F.; Moreira, J.C. Clostridium Bacteria and its Impact in Autism Research: Thinking “Outside The Box” of Neuroscience. J. Commun. Disord. Deaf Stud. Hearing Aids 2013, 1, 101. [Google Scholar] [CrossRef]
- Alam, R.; Abdolmaleky, H.M.; Zhou, J.R. Microbiome, inflammation, epigenetic alterations, and mental diseases. Am. J. Med. Genet. B Neuropsychiatr Genet. 2017, 174, 651–660. [Google Scholar] [CrossRef]
- Dinan, T.G.; Cryan, J.F. Gut instincts: microbiota as a key regulator of brain development, ageing and neurodegeneration. J. Physiol. 2017, 595, 489–503. [Google Scholar] [CrossRef]
- Fung, T.C.; Olson, C.A.; Hsiao, E.Y. Interactions between the microbiota, immune and nervous systems in health and disease. Nat. Neurosci. 2017, 20, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Diaz Heijtz, R. Fetal, neonatal, and infant microbiome: Perturbations and subsequent effects on brain development and behavior. Semin. Fetal. Neonatal. Med. 2016, 21, 410–417. [Google Scholar] [CrossRef]
- Madore, C.; Leyrolle, Q.; Lacabanne, C.; Benmamar-Badel, A.; Joffre, C.; Nadjar, A.; Laye, S. Neuroinflammation in Autism: Plausible Role of Maternal Inflammation, Dietary Omega 3, and Microbiota. Neural Plast. 2016, 2016, 3597209. [Google Scholar] [CrossRef] [PubMed]
- Principi, N.; Esposito, S. Gut microbiota and central nervous system development. J. Infect. 2016, 73, 536–546. [Google Scholar] [CrossRef]
- Martin, C.R.; Mayer, E.A. Gut-Brain Axis and Behavior. Nestle Nutr. Inst. Workshop Ser. 2017, 88, 45–53. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Li, S.; Gan, R.Y.; Zhou, T.; Xu, D.P.; Li, H.B. Impacts of gut bacteria on human health and diseases. Int. J. Mol. Sci. 2015, 16, 7493–7519. [Google Scholar] [CrossRef]
- Schumann, C.M.; Amaral, D.G. Stereological analysis of amygdala neuron number in autism. J. Neurosci. 2006, 26, 7674–7679. [Google Scholar] [CrossRef] [PubMed]
- De Theije, C.G.; Bavelaar, B.M.; Lopes da Silva, S.; Korte, S.M.; Olivier, B.; Garssen, J.; Kraneveld, A.D. Food allergy and food-based therapies in neurodevelopmental disorders. Pediatr. Allergy Immunol. 2014, 25, 218–226. [Google Scholar] [CrossRef]
- Mayer, E.A.; Tillisch, K.; Gupta, A. Gut/brain axis and the microbiota. J. Clin. Invest. 2015, 125, 926–938. [Google Scholar] [CrossRef]
- Vuong, H.E.; Yano, J.M.; Fung, T.C.; Hsiao, E.Y. The Microbiome and Host Behavior. Annu. Rev. Neurosci. 2017, 40, 21–49. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.L.; Vieira-Silva, S.; Liston, A.; Raes, J. How informative is the mouse for human gut microbiota research? Dis. Model. Mech. 2015, 8, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Wu, Y.; Wu, B.L. Genetic architecture, epigenetic influence and environment exposure in the pathogenesis of Autism. Sci. China Life Sci. 2015, 58, 958–967. [Google Scholar] [CrossRef]
- Bik, E.M. The Hoops, Hopes, and Hypes of Human Microbiome Research. Yale J. Biol. Med. 2016, 89, 363–373. [Google Scholar]
- Li, Q.; Zhou, J.M. The microbiota-gut-brain axis and its potential therapeutic role in autism spectrum disorder. Neuroscience 2016, 324, 131–139. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef]
- De Angelis, M.; Piccolo, M.; Vannini, L.; Siragusa, S.; De Giacomo, A.; Serrazzanetti, D.I.; Cristofori, F.; Guerzoni, M.E.; Gobbetti, M.; Francavilla, R. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS ONE 2013, 8, e76993. [Google Scholar] [CrossRef]
- Mezzelani, A.; Raggi, M.E.; Marabotti, A.; Milanesi, L. Ochratoxin A as possible factor trigging autism and its male prevalence via epigenetic mechanism. Nutr. Neurosci. 2016, 19, 43–46. [Google Scholar] [CrossRef] [PubMed]
- Kushak, R.I.; Winter, H.S. Intestinal microbiota, metabolome and gender dimorphism in autism spectrum disorders. Res. Autism Spectr. Disord. 2018, 49, 65–74. [Google Scholar] [CrossRef]
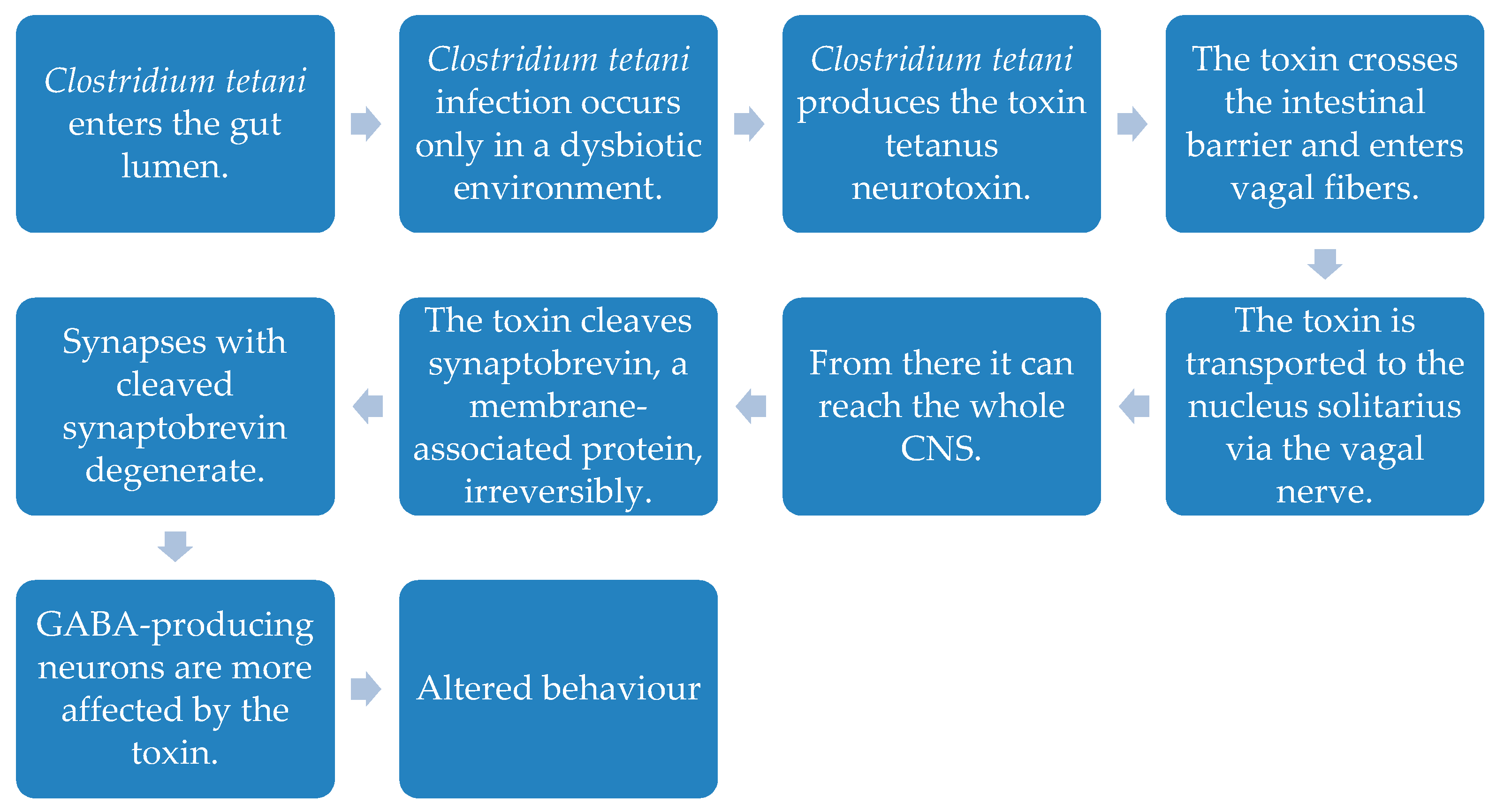
- Bolte, E.R. Autism and Clostridium tetani. Med. Hypotheses 1998, 51, 133–144. [Google Scholar] [CrossRef]
- Baio, J.; Wiggins, L.; Christensen, D.L.; Maenner, M.J.; Daniels, J.; Warren, Z.; Kurzius-Spencer, M.; Zahorodny, W.; Robinson Rosenberg, C.; White, T.; et al. Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years - Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2014. MMWR Surveill Summ. 2018, 67, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Association, A.P. Diagnostic and Statistical Manual of Mental Disorders (DSM-5®); American Psychiatric Publishing: Washington, DC, USA, 2013. [Google Scholar]
- Navarro, F.; Liu, Y.; Rhoads, J.M. Can probiotics benefit children with autism spectrum disorders? World J. Gastroenterol. 2016, 22, 10093–10102. [Google Scholar] [CrossRef]
- Manchia, M.; Fanos, V. Targeting aggression in severe mental illness: The predictive role of genetic, epigenetic, and metabolomic markers. Prog. Neuropsychopharmacol. Biol. Psychiatry 2017, 77, 32–41. [Google Scholar] [CrossRef]
- Mayer, E.A.; Padua, D.; Tillisch, K. Altered brain-gut axis in autism: comorbidity or causative mechanisms? Bioessays 2014, 36, 933–939. [Google Scholar] [CrossRef]
- Tomova, A.; Husarova, V.; Lakatosova, S.; Bakos, J.; Vlkova, B.; Babinska, K.; Ostatnikova, D. Gastrointestinal microbiota in children with autism in Slovakia. Physiol. Behav. 2015, 138, 179–187. [Google Scholar] [CrossRef]
- Ding, H.T.; Taur, Y.; Walkup, J.T. Gut Microbiota and Autism: Key Concepts and Findings. J. Autism Dev. Disord. 2017, 47, 480–489. [Google Scholar] [CrossRef]
- Hornig, M. The role of microbes and autoimmunity in the pathogenesis of neuropsychiatric illness. Curr. Opin. Rheumatol. 2013, 25, 488–795. [Google Scholar] [CrossRef]
- Naviaux, R.K. Metabolic features of the cell danger response. Mitochondrion 2014, 16, 7–17. [Google Scholar] [CrossRef]
- Williams, B.L.; Hornig, M.; Buie, T.; Bauman, M.L.; Cho Paik, M.; Wick, I.; Bennett, A.; Jabado, O.; Hirschberg, D.L.; Lipkin, W.I. Impaired carbohydrate digestion and transport and mucosal dysbiosis in the intestines of children with autism and gastrointestinal disturbances. PLoS ONE 2011, 6, e24585. [Google Scholar] [CrossRef]
- Van De Sande, M.M.; van Buul, V.J.; Brouns, F.J. Autism and nutrition: The role of the gut-brain axis. Nutr. Res. Rev. 2014, 27, 199–214. [Google Scholar] [CrossRef]
- Fiorentino, M.; Sapone, A.; Senger, S.; Camhi, S.S.; Kadzielski, S.M.; Buie, T.M.; Kelly, D.L.; Cascella, N.; Fasano, A. Blood-brain barrier and intestinal epithelial barrier alterations in autism spectrum disorders. Mol. Autism 2016, 7, 49. [Google Scholar] [CrossRef]
- Fond, G.; Boukouaci, W.; Chevalier, G.; Regnault, A.; Eberl, G.; Hamdani, N.; Dickerson, F.; Macgregor, A.; Boyer, L.; Dargel, A.; et al. The “psychomicrobiotic”: Targeting microbiota in major psychiatric disorders: A systematic review. Pathol. Biol. (Paris) 2015, 63, 35–42. [Google Scholar] [CrossRef]
- Vuong, H.E.; Hsiao, E.Y. Emerging Roles for the Gut Microbiome in Autism Spectrum Disorder. Biol. Psychiatry 2017, 81, 411–423. [Google Scholar] [CrossRef]
- Wakefield, A.J.; Puleston, J.M.; Montgomery, S.M.; Anthony, A.; O’Leary, J.J.; Murch, S.H. Review article: the concept of entero-colonic encephalopathy, autism and opioid receptor ligands. Aliment. Pharmacol. Ther. 2002, 16, 663–674. [Google Scholar] [CrossRef]
- Wang, Y.; Kasper, L.H. The role of microbiome in central nervous system disorders. Brain Behav. Immun. 2014, 38, 1–12. [Google Scholar] [CrossRef]
- Kaelberer, M.M.; Buchanan, K.L.; Klein, M.E.; Barth, B.B.; Montoya, M.M.; Shen, X.; Bohórquez, D.V. A gut-brain neural circuit for nutrient sensory transduction. Science 2018, 361, eaat5236. [Google Scholar] [CrossRef]
- Han, W.; Tellez, L.A.; Perkins, M.H.; Perez, I.O.; Qu, T.; Ferreira, J.; Ferreira, T.L.; Quinn, D.; Liu, Z.-W.; Gao, X.-B.; et al. A Neural Circuit for Gut-Induced Reward. Cell 2018, 175, 665–678. [Google Scholar] [CrossRef]
- Rose, S.; Bennuri, S.C.; Murray, K.F.; Buie, T.; Winter, H.; Frye, R.E. Mitochondrial dysfunction in the gastrointestinal mucosa of children with autism: A blinded case-control study. PLoS ONE 2017, 12, e0186377. [Google Scholar] [CrossRef]
- Douglas-Escobar, M.; Elliott, E.; Neu, J. Effect of intestinal microbial ecology on the developing brain. JAMA Pediatr. 2013, 167, 374–379. [Google Scholar] [CrossRef] [PubMed]
- Mulle, J.G.; Sharp, W.G.; Cubells, J.F. The gut microbiome: a new frontier in autism research. Curr. Psychiatry Rep. 2013, 15, 337. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, C.S. Microbiome Disturbances and Autism Spectrum Disorders. Drug Metab. Dispos. 2015, 43, 1557–1571. [Google Scholar] [CrossRef] [PubMed]
- Rescigno, M. Intestinal microbiota and its effects on the immune system. Cell Microbiol 2014, 16, 1004–1013. [Google Scholar] [CrossRef]
- Rook, G.A.; Raison, C.L.; Lowry, C.A. Microbiota, immunoregulatory old friends and psychiatric disorders. Adv. Exp. Med. Biol. 2014, 817, 319–356. [Google Scholar] [CrossRef]
- Sharon, G.; Sampson, T.R.; Geschwind, D.H.; Mazmanian, S.K. The Central Nervous System and the Gut Microbiome. Cell 2016, 167, 915–932. [Google Scholar] [CrossRef]
- Groer, M.W.; Gregory, K.E.; Louis-Jacques, A.; Thibeau, S.; Walker, W.A. The very low birth weight infant microbiome and childhood health. Birth Defects Res. C Embryo Today 2015, 105, 252–264. [Google Scholar] [CrossRef]
- SM, O.M.; Stilling, R.M.; Dinan, T.G.; Cryan, J.F. The microbiome and childhood diseases: Focus on brain-gut axis. Birth Defects Res. C Embryo Today 2015, 105, 296–313. [Google Scholar] [CrossRef]
- Borre, Y.E.; O’Keeffe, G.W.; Clarke, G.; Stanton, C.; Dinan, T.G.; Cryan, J.F. Microbiota and neurodevelopmental windows: implications for brain disorders. Trends Mol. Med. 2014, 20, 509–518. [Google Scholar] [CrossRef]
- Dinan, T.G.; Stilling, R.M.; Stanton, C.; Cryan, J.F. Collective unconscious: how gut microbes shape human behavior. J. Psychiatr. Res. 2015, 63, 1–9. [Google Scholar] [CrossRef]
- Borre, Y.E.; Moloney, R.D.; Clarke, G.; Dinan, T.G.; Cryan, J.F. The impact of microbiota on brain and behavior: mechanisms & therapeutic potential. Adv. Exp. Med. Biol. 2014, 817, 373–403. [Google Scholar] [CrossRef]
- Rook, G.A.; Lowry, C.A.; Raison, C.L. Hygiene and other early childhood influences on the subsequent function of the immune system. Brain Res. 2015, 1617, 47–62. [Google Scholar] [CrossRef]
- Arora, S.K.; Dewan, P.; Gupta, P. Microbiome: Paediatricians’ perspective. Indian J. Med. Res. 2015, 142, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Dinan, T.G.; Cryan, J.F. The impact of gut microbiota on brain and behaviour: implications for psychiatry. Curr. Opin. Clin. Nutr. Metab. Care 2015, 18, 552–558. [Google Scholar] [CrossRef] [PubMed]
- Mohajeri, M.H.; Brummer, R.J.M.; Rastall, R.A.; Weersma, R.K.; Harmsen, H.J.M.; Faas, M.; Eggersdorfer, M. The role of the microbiome for human health: From basic science to clinical applications. Eur. J. Nutr. 2018, 57 (Suppl. 1), 1–14. [Google Scholar] [CrossRef]
- Ghaisas, S.; Maher, J.; Kanthasamy, A. Gut microbiome in health and disease: Linking the microbiome-gut-brain axis and environmental factors in the pathogenesis of systemic and neurodegenerative diseases. Pharmacol. Ther. 2016, 158, 52–62. [Google Scholar] [CrossRef]
- Kang, D.W.; Adams, J.B.; Gregory, A.C.; Borody, T.; Chittick, L.; Fasano, A.; Khoruts, A.; Geis, E.; Maldonado, J.; McDonough-Means, S.; et al. Microbiota Transfer Therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: an open-label study. Microbiome 2017, 5, 10. [Google Scholar] [CrossRef]
- Curran, E.A.; Dalman, C.; Kearney, P.M.; Kenny, L.C.; Cryan, J.F.; Dinan, T.G.; Khashan, A.S. Association Between Obstetric Mode of Delivery and Autism Spectrum Disorder: A Population-Based Sibling Design Study. JAMA Psychiatry 2015, 72, 935–942. [Google Scholar] [CrossRef]
- Curran, E.A.; Cryan, J.F.; Kenny, L.C.; Dinan, T.G.; Kearney, P.M.; Khashan, A.S. Obstetrical Mode of Delivery and Childhood Behavior and Psychological Development in a British Cohort. J. Autism Dev. Disord. 2016, 46, 603–614. [Google Scholar] [CrossRef]
- Knight, R.; Callewaert, C.; Marotz, C.; Hyde, E.R.; Debelius, J.W.; McDonald, D.; Sogin, M.L. The Microbiome and Human Biology. Annu. Rev. Genomics Hum. Genet. 2017, 18, 65–86. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.; Stombaugh, J.; Lozupone, C.; Turnbaugh, P.J.; Gordon, J.I.; Knight, R. The mind-body-microbial continuum. Dialogues Clin. Neurosci. 2011, 13, 55–62. [Google Scholar]
- Labouesse, M.A.; Langhans, W.; Meyer, U. Long-term pathological consequences of prenatal infection: beyond brain disorders. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2015, 309, R1–R12. [Google Scholar] [CrossRef] [PubMed]
- Ibi, D.; Yamada, K. Therapeutic Targets for Neurodevelopmental Disorders Emerging from Animal Models with Perinatal Immune Activation. Int. J. Mol. Sci. 2015, 16, 28218–28229. [Google Scholar] [CrossRef] [PubMed]
- Lazaro, C.P.; Ponde, M.P.; Rodrigues, L.E. Opioid peptides and gastrointestinal symptoms in autism spectrum disorders. Rev. Bras. Psiquiatr. 2016, 38, 243–246. [Google Scholar] [CrossRef] [PubMed]
- Strati, F.; Cavalieri, D.; Albanese, D.; De Felice, C.; Donati, C.; Hayek, J.; Jousson, O.; Leoncini, S.; Renzi, D.; Calabro, A.; et al. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome 2017, 5, 24. [Google Scholar] [CrossRef]
- Williams, B.L.; Hornig, M.; Parekh, T.; Lipkin, W.I. Application of novel PCR-based methods for detection, quantitation, and phylogenetic characterization of Sutterella species in intestinal biopsy samples from children with autism and gastrointestinal disturbances. mBio 2012, 3, e00261-11. [Google Scholar] [CrossRef]
- Kushak, R.I.; Winter, H.S.; Buie, T.M.; Cox, S.B.; Phillips, C.D.; Ward, N.L. Analysis of the Duodenal Microbiome in Autistic Individuals: Association With Carbohydrate Digestion. J. Pediatr. Gastroenterol. Nutr. 2017, 64, e110–e116. [Google Scholar] [CrossRef]
- Mayer, E.A.; Knight, R.; Mazmanian, S.K.; Cryan, J.F.; Tillisch, K. Gut microbes and the brain: paradigm shift in neuroscience. J. Neurosci. 2014, 34, 15490–15496. [Google Scholar] [CrossRef]
- Gondalia, S.V.; Palombo, E.A.; Knowles, S.R.; Cox, S.B.; Meyer, D.; Austin, D.W. Molecular characterisation of gastrointestinal microbiota of children with autism (with and without gastrointestinal dysfunction) and their neurotypical siblings. Autism Res. 2012, 5, 419–427. [Google Scholar] [CrossRef]
- Son, J.S.; Zheng, L.J.; Rowehl, L.M.; Tian, X.; Zhang, Y.; Zhu, W.; Litcher-Kelly, L.; Gadow, K.D.; Gathungu, G.; Robertson, C.E.; et al. Comparison of Fecal Microbiota in Children with Autism Spectrum Disorders and Neurotypical Siblings in the Simons Simplex Collection. PLoS ONE 2015, 10, e0137725. [Google Scholar] [CrossRef]
- Louis, P. Does the human gut microbiota contribute to the etiology of autism spectrum disorders? Dig. Dis. Sci. 2012, 57, 1987–1989. [Google Scholar] [CrossRef]
- Argou-Cardozo, I.; Zeidan-Chulia, F. Clostridium Bacteria and Autism Spectrum Conditions: A Systematic Review and Hypothetical Contribution of Environmental Glyphosate Levels. Med. Sci. 2018, 6, 29. [Google Scholar] [CrossRef]
- Kantarcioglu, A.S.; Kiraz, N.; Aydin, A. Microbiota-Gut-Brain Axis: Yeast Species Isolated from Stool Samples of Children with Suspected or Diagnosed Autism Spectrum Disorders and In Vitro Susceptibility Against Nystatin and Fluconazole. Mycopathologia 2016, 181, 1–7. [Google Scholar] [CrossRef]
- Weston, B.; Fogal, B.; Cook, D.; Dhurjati, P. An agent-based modeling framework for evaluating hypotheses on risks for developing autism: effects of the gut microbial environment. Med. Hypotheses 2015, 84, 395–401. [Google Scholar] [CrossRef]
- Heberling, C.; Dhurjati, P. Novel systems modeling methodology in comparative microbial metabolomics: identifying key enzymes and metabolites implicated in autism spectrum disorders. Int. J. Mol. Sci. 2015, 16, 8949–8967. [Google Scholar] [CrossRef]
- Grimaldi, R.; Cela, D.; Swann, J.R.; Vulevic, J.; Gibson, G.R.; Tzortzis, G.; Costabile, A. In vitro fermentation of B-GOS: impact on faecal bacterial populations and metabolic activity in autistic and non-autistic children. FEMS Microbiol. Ecol. 2017, 93. [Google Scholar] [CrossRef]
- Xiong, X.; Liu, D.; Wang, Y.; Zeng, T.; Peng, Y. Urinary 3-(3-Hydroxyphenyl)-3-hydroxypropionic Acid, 3-Hydroxyphenylacetic Acid, and 3-Hydroxyhippuric Acid Are Elevated in Children with Autism Spectrum Disorders. Biomed. Res. Int. 2016, 2016, 9485412. [Google Scholar] [CrossRef]
- Clayton, T.A. Metabolic differences underlying two distinct rat urinary phenotypes, a suggested role for gut microbial metabolism of phenylalanine and a possible connection to autism. FEBS Lett. 2012, 586, 956–961. [Google Scholar] [CrossRef]
- Gevi, F.; Zolla, L.; Gabriele, S.; Persico, A.M. Urinary metabolomics of young Italian autistic children supports abnormal tryptophan and purine metabolism. Mol. Autism 2016, 7, 47. [Google Scholar] [CrossRef]
- Yang, Y.; Tian, J.; Yang, B. Targeting gut microbiome: A novel and potential therapy for autism. Life Sci. 2018, 194, 111–119. [Google Scholar] [CrossRef]
- Vasquez, A. Biological plausibility of the gut-brain axis in autism. Ann. N. Y. Acad. Sci. 2017, 1408, 5–6. [Google Scholar] [CrossRef] [PubMed]
- Yap, I.K.; Angley, M.; Veselkov, K.A.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. Urinary metabolic phenotyping differentiates children with autism from their unaffected siblings and age-matched controls. J. Proteome Res. 2010, 9, 2996–3004. [Google Scholar] [CrossRef] [PubMed]
- Lees, H.J.; Swann, J.R.; Wilson, I.D.; Nicholson, J.K.; Holmes, E. Hippurate: the natural history of a mammalian-microbial cometabolite. J. Proteome Res. 2013, 12, 1527–1546. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, M.; Francavilla, R.; Piccolo, M.; De Giacomo, A.; Gobbetti, M. Autism spectrum disorders and intestinal microbiota. Gut Microbes 2015, 6, 207–213. [Google Scholar] [CrossRef]
- Wynendaele, E.; Verbeke, F.; Stalmans, S.; Gevaert, B.; Janssens, Y.; Van De Wiele, C.; Peremans, K.; Burvenich, C.; De Spiegeleer, B. Quorum Sensing Peptides Selectively Penetrate the Blood-Brain Barrier. PLoS ONE 2015, 10, e0142071. [Google Scholar] [CrossRef] [PubMed]
- Marler, S.; Ferguson, B.J.; Lee, E.B.; Peters, B.; Williams, K.C.; McDonnell, E.; Macklin, E.A.; Levitt, P.; Gillespie, C.H.; Anderson, G.M.; et al. Brief Report: Whole Blood Serotonin Levels and Gastrointestinal Symptoms in Autism Spectrum Disorder. J. Autism Dev. Disord. 2016, 46, 1124–1130. [Google Scholar] [CrossRef]
- Reardon, S. Gut-brain link grabs neuroscientists. Nature 2014, 515, 175–177. [Google Scholar] [CrossRef]
- Jory, J. Abnormal fatty acids in Canadian children with autism. Nutrition 2016, 32, 474–477. [Google Scholar] [CrossRef]
- Weiser, M.J.; Butt, C.M.; Mohajeri, M.H. Docosahexaenoic Acid and Cognition throughout the Lifespan. Nutrients 2016, 8, 99. [Google Scholar] [CrossRef]
- Anwar, A.; Marini, M.; Abruzzo, P.M.; Bolotta, A.; Ghezzo, A.; Visconti, P.; Thornalley, P.J.; Rabbani, N. Quantitation of plasma thiamine, related metabolites and plasma protein oxidative damage markers in children with autism spectrum disorder and healthy controls. Free Radic. Res. 2016, 50, S85–S90. [Google Scholar] [CrossRef]
- Mangiola, F.; Ianiro, G.; Franceschi, F.; Fagiuoli, S.; Gasbarrini, G.; Gasbarrini, A. Gut microbiota in autism and mood disorders. World J. Gastroenterol 2016, 22, 361–368. [Google Scholar] [CrossRef]
- Pifer, R.; Sperandio, V. The Interplay between the Microbiota and Enterohemorrhagic Escherichia coli. Microbiol. Spectr. 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- Aw, W.; Fukuda, S. Toward the comprehensive understanding of the gut ecosystem via metabolomics-based integrated omics approach. Semin. Immunopathol. 2015, 37, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Lei, E.; Vacy, K.; Boon, W.C. Fatty acids and their therapeutic potential in neurological disorders. Neurochem. Int. 2016, 95, 75–84. [Google Scholar] [CrossRef]
- Liu, X.; Cao, S.; Zhang, X. Modulation of Gut Microbiota-Brain Axis by Probiotics, Prebiotics, and Diet. J. Agric. Food Chem. 2015, 63, 7885–7895. [Google Scholar] [CrossRef]
- Borghi, E.; Borgo, F.; Severgnini, M.; Savini, M.N.; Casiraghi, M.C.; Vignoli, A. Rett Syndrome: A Focus on Gut Microbiota. Int. J. Mol. Sci. 2017, 18, 344. [Google Scholar] [CrossRef]
- Frye, R.E.; Rose, S.; Chacko, J.; Wynne, R.; Bennuri, S.C.; Slattery, J.C.; Tippett, M.; Delhey, L.; Melnyk, S.; Kahler, S.G.; et al. Modulation of mitochondrial function by the microbiome metabolite propionic acid in autism and control cell lines. Transl. Psychiatry 2016, 6, e927. [Google Scholar] [CrossRef]
- Slattery, J.; MacFabe, D.F.; Kahler, S.G.; Frye, R.E. Enteric Ecosystem Disruption in Autism Spectrum Disorder: Can the Microbiota and Macrobiota be Restored? Curr. Pharm. Des. 2016, 22, 6107–6121. [Google Scholar] [CrossRef]
- Mohajeri, M.H.; La Fata, G.; Steinert, R.E.; Weber, P. Relationship between the gut microbiome and brain function. Nutr. Rev. 2018, 76, 481–496. [Google Scholar] [CrossRef]
- Rinaldi, A. Piecing together a different picture: A host of new studies on autism have begun decoding the longstanding puzzle of its causes. EMBO Rep. 2016, 17, 1690–1695. [Google Scholar] [CrossRef] [PubMed]
- Beales, D.L. Biome depletion in conjunction with evolutionary mismatches could play a role in the etiology of neurofibromatosis 1. Med. Hypotheses 2015, 84, 305–314. [Google Scholar] [CrossRef]
- Inoue, R.; Sakaue, Y.; Sawai, C.; Sawai, T.; Ozeki, M.; Romero-Perez, G.A.; Tsukahara, T. A preliminary investigation on the relationship between gut microbiota and gene expressions in peripheral mononuclear cells of infants with autism spectrum disorders. Biosci. Biotechnol. Biochem. 2016, 80, 2450–2458. [Google Scholar] [CrossRef]
- Diaz-Gerevini, G.T.; Repossi, G.; Dain, A.; Tarres, M.C.; Das, U.N.; Eynard, A.R. Beneficial action of resveratrol: How and why? Nutrition 2016, 32, 174–178. [Google Scholar] [CrossRef] [PubMed]
- La Fata, G.; Rastall, R.A.; Lacroix, C.; Harmsen, H.J.M.; Mohajeri, M.H.; Weber, P.; Steinert, R.E. Recent Development of Prebiotic Research-Statement from an Expert Workshop. Nutrients 2017, 9, 1376. [Google Scholar] [CrossRef] [PubMed]
- Sherwin, E.; Sandhu, K.V.; Dinan, T.G.; Cryan, J.F. May the Force Be With You: The Light and Dark Sides of the Microbiota-Gut-Brain Axis in Neuropsychiatry. CNS Drugs 2016, 30, 1019–1041. [Google Scholar] [CrossRef] [PubMed]
- Umbrello, G.; Esposito, S. Microbiota and neurologic diseases: potential effects of probiotics. J. Transl. Med. 2016, 14, 298. [Google Scholar] [CrossRef]
- Aroniadis, O.C.; Brandt, L.J. Fecal microbiota transplantation: past, present and future. Curr. Opin. Gastroenterol. 2013, 29, 79–84. [Google Scholar] [CrossRef]
- Petrof, E.O.; Claud, E.C.; Gloor, G.B.; Allen-Vercoe, E. Microbial ecosystems therapeutics: a new paradigm in medicine? Benef. Microbes 2013, 4, 53–65. [Google Scholar] [CrossRef]
- Ianiro, G.; Bibbo, S.; Gasbarrini, A.; Cammarota, G. Therapeutic modulation of gut microbiota: Current clinical applications and future perspectives. Curr. Drug Targets 2014, 15, 762–770. [Google Scholar] [CrossRef]
- Skosnik, P.D.; Cortes-Briones, J.A. Targeting the ecology within: The role of the gut-brain axis and human microbiota in drug addiction. Med. Hypotheses 2016, 93, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Whiteley, P. Nutritional management of (some) autism: A case for gluten- and casein-free diets? Proc. Nutr. Soc. 2015, 74, 202–207. [Google Scholar] [CrossRef] [PubMed]
- Alfawaz, H.A.; Bhat, R.S.; Al-Ayadhi, L.; El-Ansary, A.K. Protective and restorative potency of Vitamin D on persistent biochemical autistic features induced in propionic acid-intoxicated rat pups. BMC Complement. Altern. Med. 2014, 14, 416. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C. Mental health: thinking from the gut. Nature 2015, 518, S12–S15. [Google Scholar] [CrossRef] [PubMed]
- Higashida, H.; Furuhara, K.; Yamauchi, A.M.; Deguchi, K.; Harashima, A.; Munesue, S.; Lopatina, O.; Gerasimenko, M.; Salmina, A.B.; Zhang, J.S.; et al. Intestinal transepithelial permeability of oxytocin into the blood is dependent on the receptor for advanced glycation end products in mice. Sci. Rep. 2017, 7, 7883. [Google Scholar] [CrossRef] [PubMed]

| Elevated Urinary Metabolites in ASD Patients | Decreased Urinary Metabolites in ASD Patients | ||
|---|---|---|---|
| N-methyl-2-pyridone-5-carboxamide | [104] | Glutamate | [104] |
| N-methyl nicotinic acid | [104] | Hippurate | [104] |
| N-methyl nicotinamide | [104] | Phenylacetylglutamine | [104] |
| N-acetyl glycoprotein fragments | [105] | ||
| Succinate | [105] | Melatonin | [101] |
| Acetate | [105] | Kynurenic acid | [101] |
| Taurine | [105] | ||
| Dimethylamine | [105] | ||
| 3-(3-hydroxyphenyl)-3-hydroxypropionic acid | [99] | ||
| 3-hydroxyphenylacetic acid | [99] | ||
| 3-hydroxyhippuric acid | [99] | ||
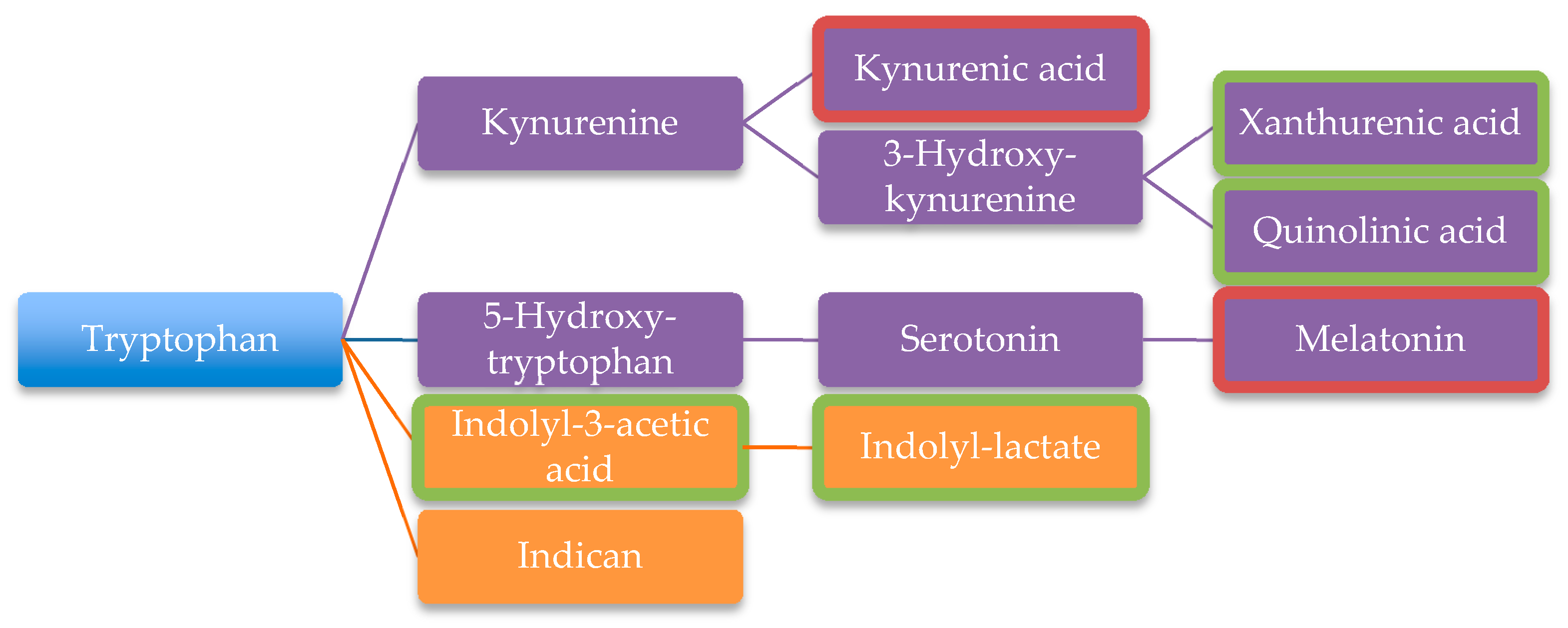
| Tryptophan degradation products | [106] | ||
| Xanthurenic acid | [101] | ||
| Quinolinic acid | [101] | ||
| Indolyl-3-acetic | [101] | ||
| Indolyl-lactate | [101] | ||
| P-cresol | [101,102] | ||
| TNF-α | [49] | ||
| Free amino acids | [40] | ||
| Sulphate, sulphite, thiosulphate | [104] |
| Elevated Metabolites in the Blood of ASD Patients | Decreased Metabolites in the Blood of ASD Patients | ||
|---|---|---|---|
| Serotonin (Candida, Streptococcus, Escherichia and Enterococcus spp) | [108] | Methionine | [104] |
| GABA (Lactobacillus and Bifidobacterium spp) | [98] | S-adenosylmethionine | [104] |
| p-cresol (Clostridia spp) | [104] | Homocysteine | [104] |
| Lipopolysaccharides (gram negative bacteria) | [56,113] | Glutathione | [104] |
| Docosahexaenoic acid | [110] | ||
| Eicosapentaenoic acid | [110] | ||
| Arachidonic acid | [110] | ||
| Thiamine-pyrophosphate | [112] |
| Probiotic Used | Effects | Studies |
|---|---|---|
| Children Dophilus (Bifidobacterium, Lactobacillus, Streptococcus spp) | Amelioration of Firmicutes to Bacteroidetes ratio | Humans [49] |
| Lactobacillus rhamnosus, Bifidobacterium infantis, Bifidobacterium longus, Lactobacillus helveticus, Lactobacillus reuteri and Lactobacillus paracasei | Improvement of GI problems | Humans [106] |
| Lactobacillus reuteri | - Reversing inflammation caused through lipopolysaccharides - Stimulating the production of Oxytocin | Rats [46] |
| Bifidobacterium spp | Elevation of arachidonic and docosahexaenoic acid concentration in the brain | Mice [117] |
| Lactobacillus rhamnosus | - Reduction of corticosterone levels - Alterations of GABA levels | Mice [117] |
| Bacteroides fragilis | - Improvement of autistic features - Correction of tight junction proteins expression in the colon | Mice [5] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Srikantha, P.; Mohajeri, M.H. The Possible Role of the Microbiota-Gut-Brain-Axis in Autism Spectrum Disorder. Int. J. Mol. Sci. 2019, 20, 2115. https://doi.org/10.3390/ijms20092115
Srikantha P, Mohajeri MH. The Possible Role of the Microbiota-Gut-Brain-Axis in Autism Spectrum Disorder. International Journal of Molecular Sciences. 2019; 20(9):2115. https://doi.org/10.3390/ijms20092115
Chicago/Turabian StyleSrikantha, Piranavie, and M. Hasan Mohajeri. 2019. "The Possible Role of the Microbiota-Gut-Brain-Axis in Autism Spectrum Disorder" International Journal of Molecular Sciences 20, no. 9: 2115. https://doi.org/10.3390/ijms20092115
APA StyleSrikantha, P., & Mohajeri, M. H. (2019). The Possible Role of the Microbiota-Gut-Brain-Axis in Autism Spectrum Disorder. International Journal of Molecular Sciences, 20(9), 2115. https://doi.org/10.3390/ijms20092115





